Abstract
The chloroplast ATP synthase catalyzes the light-driven synthesis of ATP and acts as a key feedback regulatory component of photosynthesis. Arabidopsis possesses two homologues of the regulatory γ subunit of the ATP synthase, encoded by the ATPC1 and ATPC2 genes. Using a series of mutants, we show that both these subunits can support photosynthetic ATP synthesis in vivo with similar specific activities, but that in wild-type plants, only γ1 is involved in ATP synthesis in photosynthesis. The γ1-containing ATP synthase shows classical light-induced redox regulation, whereas the mutant expressing only γ2-ATP synthase (gamma exchange-revised ATP synthase, gamera) shows equally high ATP synthase activity in the light and dark. In situ redox titrations demonstrate that the regulatory thiol groups on γ2-ATP synthase remain reduced under physiological conditions but can be oxidized by the strong oxidant diamide, implying that the redox potential for the thiol/disulphide transition in γ2 is substantially higher than that for γ1. This regulatory difference may be attributed to alterations in the residues near the redox-active thiols. We propose that γ2-ATP synthase functions to catalyze ATP hydrolysis-driven proton translocation in nonphotosynthetic plastids, maintaining a sufficient transthylakoid proton gradient to drive protein translocation or other processes. Consistent with this interpretation, ATPC2 is predominantly expressed in the root, whereas modifying its expression results in alteration of root hair development. Phylogenetic analysis suggests that γ2 originated from ancient gene duplication, resulting in divergent evolution of functionally distinct ATP synthase complexes in dicots and mosses.
Keywords: plant development, bioenergetics
Light-driven linear electron flow (LEF) in photosynthesis stores energy in reduced NADPH. It is also coupled to the translocation of protons into the thylakoid lumen, generating an electrochemical gradient of protons, the proton motive force (pmf), across the thylakoid. The pmf in turn drives the synthesis of ATP from ATP and Pi via the CFO-CF1 ATP synthase (ATP synthase), storing energy in the form of the photophosphorylation potential. The plastid ATP synthase complex consists of nine different subunits. Four of these subunits (I, II, III14, and IV, also called b, b’, c, and a) form the integral membrane CFO subcomplex and the remaining five subunits make up the extrinsic CF1 subcomplex (α3, β3, γ, δ, and ε) that contains the catalytic sites of ATP synthase (1, 2). The CFO subcomplex drives the synthesis of ATP at the CF1 subcomplex via the rotational-catalysis/binding-change mechanism (3–5).
The ATP synthase is regulated at several levels (6). It has been known for some time that the ATP synthase requires a substantial pmf to become activated (7–9). This pmf-activation is thought to involve interaction between the FO and F1 subcomplexes because detachment of FO activates ATP hydrolysis (10). A second level of regulation occurs in plants and green algae via thioredoxin-mediated thiol modulation of the γ subunit, altering the extent of pmf required to activate the complex (4). The structural basis of the regulation is assigned to a sequence of nine amino acid residues containing two Cys residues (Cys199–Cys205, in Arabidopsis thaliana numbering) able to form an intrapeptide disulfide bond upon oxidation (11). In the light, electron flow from photosystem I (PSI) reduces thioredoxin-f via ferredoxin:thioredoxin oxidoreductase (12), which in turn reduces the γ subunit, forming an ATP synthase that is activated with a relatively low pmf of about 50 mV (13). After dark adaptation for tens of minutes, the γ subunit cysteine residues become oxidized, imposing a higher pmf requirement for activation, of about 100 mV, thus resulting in ATP synthase inactivation, preventing rapid hydrolysis of ATP. Redox-regulation via thioredoxin is highly sensitive to light, and acts as a “switch,” rapidly transitioning between inhibition in the dark and full activity at even very low light intensities (14).
The ATP synthase is also fine-tuned in the light, during steady-state photosynthesis in response to changes in metabolism and/or environmental conditions, e.g., endogenous CO2 levels or drought stress (15–17). The resulting changes in proton efflux adjust the steady-state pmf and subsequently affect the regulation of light capture and electron transfer. The mechanism of this fine tuning is not yet clear, though a number of mechanisms have been proposed, including drawing down of inorganic phosphate and phosphorylation (18).
Arabidopsis thaliana possesses two genes that appear to code ATP synthase γ subunits, ATPC1 and ATPC2 (At4G04640 and At1G15700), located on chromosomes 4 and 1, respectively (19, 20), and showing 73% sequence similarity (19). Both Arabidopsis γ homologues contain the domain responsible for the redox regulation of the enzyme (19). The current study aims to answer the intriguing questions: In which tissues are γ1 and γ2 localized? Can both ATPC homologues function in ATP synthesis? Are their regulatory and functional properties different? What functions might they serve in the plant?
Results and Discussion
As described in detail below, this study used a series of Arabidopsis plant lines, wild type (Columbia), two mutant lines lacking ATPC2, atpc2 [transfer-DNA (T-DNA) inserted in chl1_5402949], atpc2_2 (T-DNA inserted in chl 1_5404314), a mutant lacking ATPC1 (dpa1) (21) complemented with 35S∷ATPC1, and a mutant line lacking ATPC1but expressing ATPC2 behind a 35S promoter gamma exchange-revised ATP synthase (gamera).
The γ2 Subunit Is Chloroplast Localized and Expressed at Very Low Levels.
The primary sequences of γ1 (coded by ATPC1) and γ2 (coded by ATPC2) contain apparent N-terminal transit peptides of 50 and 60 amino acids, respectively, suggesting that γ2, like γ1, is targeted to the chloroplast. The subcellular localization of γ2 in chloroplast was confirmed by fluorescence microscopy on transiently transformed Arabidopsis protoplasts expressing γ2-GFP-fusion protein (Fig. S1).
The ATPC1 gene was found to be highly expressed in photosynthetic tissues, and disrupting this gene abolished photosynthetic growth implying that it is essential for photosynthesis (21, 22). In contrast, ATPC2 was expressed at low levels in photosynthetic tissues, presumably insufficient to complement the ATPC1 knockout (21). Previous mass spectroscopy-based proteome analysis (23) was able to detect the γ2 subunit in wild-type chloroplasts, but we were unable to detect it using immunoblot analysis (Fig. S2), likely indicating very low expression. Knocking out this gene did not markedly affect growth (Fig. S2A) or photosynthesis (Fig. 1), implying that it plays little or no role in photosynthesis.
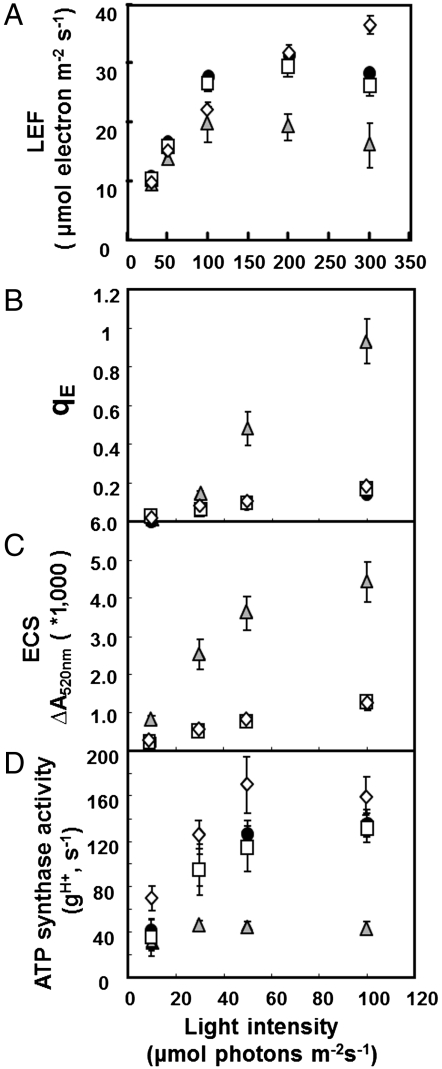
Fig. 1.
Wild-type Columbia (black circles), atpc2 (open squares), gamera (gray triangles), and dpa1 is complemented with 35S∷ATPC1 (open diamond) were compared for differences in actinic light intensity-dependence of LEF (A), energy-dependent exciton quenching (qE) (B), light-induced pmf (estimated by analysis of the electrochromic shift (ECSt) (C), and proton conductivity across the thylakoid membrane (gH+) (D), based on ECS decay kinetics. Data is for attached leaves (n = 3–4).
γ2 Can Form an Active ATP Synthase, Suppressing the Loss of γ1.
To determine whether γ2 could, if expressed, support ATP synthase activity, we generated plants containing exclusively γ2—rather than γ1-ATP synthase by expressing ATPC2 under control of the constitutive 35S promoter in the ATPC1 null mutant background (see Supporting Information) (21). The resulting mutant was termed gamera. Using Western blot analysis with antibodies specific to γ1 and γ2 (Fig. S2 B and C), we confirmed that gamera lacked detectable levels of the γ1 subunit, but gained γ2 subunit. On the other hand, leaves from wild type and the atpc2 mutant contained γ1, but no detectable γ2. Both γ1 in wild type and γ2 in gamera were found exclusively in the insoluble (membrane) fractions, indicating that these were likely bound into the ATP synthase complexes. All independent gamera transformants showed a lower than wild type accumulation of ATP synthase protein subunits, despite high levels of ATPC2 mRNA expression, suggesting that the γ2-ATP synthase may be less stable than γ1-ATP synthase (Fig. S2 B and C).
The gamera mutant was able to grow photoautrophically (Fig. S2A), indicating that higher (than wild type) expression levels of γ2 were able to functionally replace γ1 in the ATP synthase. However, the growth rates were somewhat slower in gamera than in wild type, resulting in rosette diameter 30% smaller in 4 wk old plants (Fig. S2A).
Photosynthetic Properties of Gamera.
Fig. 1A shows the dependence of LEF, measured by saturation-pulse chlorophyll fluorescence yield analysis (24) on photosynthetically active radiation (PAR) for wild type, atpc2 and gamera. The initial slope of LEF against PAR was only slightly (approximately 10%) reduced in gamera compared to wild type or atpc2, indicating that the maximal quantum efficiency of photosynthesis was similar for all three genotypes. This result is also consistent with our observation that atpc2 and gamera had minimal effects on maximal photosystem II (PSII) quantum efficiency estimated by saturating-pulse fluorescence yield changes in dark-adapted leaves (0.83 ± 0.0019, 0.83 ± 0.0012, and 0.80 ± 0.0013 for wild type, atpc2, and gamera, respectively). However, LEF saturated at about 100 μmol photons m-2 s-1 in gamera compared to about 200 μmol photons m-2 s-1 for wild type and atpc2, whereas gamera reached a maximum LEF of approximately 33% (approximately 20 μmol electrons m-2 s-1) compared to wild type and atpc2, indicating that gamera had a rate-limitation at steps beyond PSII, most likely at the ATP synthase.
We confirmed a lower in vivo ATP synthase activity for gamera by probing proton flux and light-induced pmf by measuring the decay of the electrochromic shift (ECS) (Fig. 1) as described previously (15, 16, 25). The extent of the rapid decay of ECS upon switching off actinic light, termed ECSt, which reflects the light-driven thylakoid pmf was three- to fourfold larger in gamera than in wild type or atpc2 (Fig. 1C) suggesting that gamera generated a larger light-induced pmf than wild type or atpc2, consistent with a slow rate of proton efflux through the ATP synthase. Supporting this conclusion, gamera produced a substantially higher extent of energy-dependent exciton quenching (qE) than wild type or atpc2 (Fig. 1B), implying a more acidic lumen pH as a result of increased pmf.
The increased pmf in gamera could be attributed to a decrease in efflux of protons from the lumen. Fig. 1D plots the relative conductivity of the thylakoid membrane to protons (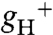 ), which predominantly reflects ATP synthase activity and can be estimated by the decay kinetics of the ECS (15, 24). The
), which predominantly reflects ATP synthase activity and can be estimated by the decay kinetics of the ECS (15, 24). The 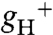 in atpc2 was similar to that in wild type, but remained consistently lower in gamera, indicating slower proton efflux. Western blot analysis showed that ATP synthase β-subunit (Fig. S3) decreased substantially in gamera compared to wild type and atpc2, implying that the total ATP synthase content, rather than decreased specific activity accounted for the lower
in atpc2 was similar to that in wild type, but remained consistently lower in gamera, indicating slower proton efflux. Western blot analysis showed that ATP synthase β-subunit (Fig. S3) decreased substantially in gamera compared to wild type and atpc2, implying that the total ATP synthase content, rather than decreased specific activity accounted for the lower 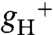 in gamera.
in gamera.
Analysis of atpc1-complemented with ATPC1 (35S∷ATPC1 in dpa1) showed no differences from wild type in growth, photosynthetic performance, or in vivo ATP synthase activity (Fig. 1) indicating that the lowering of ATP synthase content or activity in gamera was due to substitution of ATPC1 by ATPC2, rather than secondary effects of altering ATPC promoter regions.
γ2-ATP Synthase Is Equally Active in Light- and Dark-Adapted Leaves.
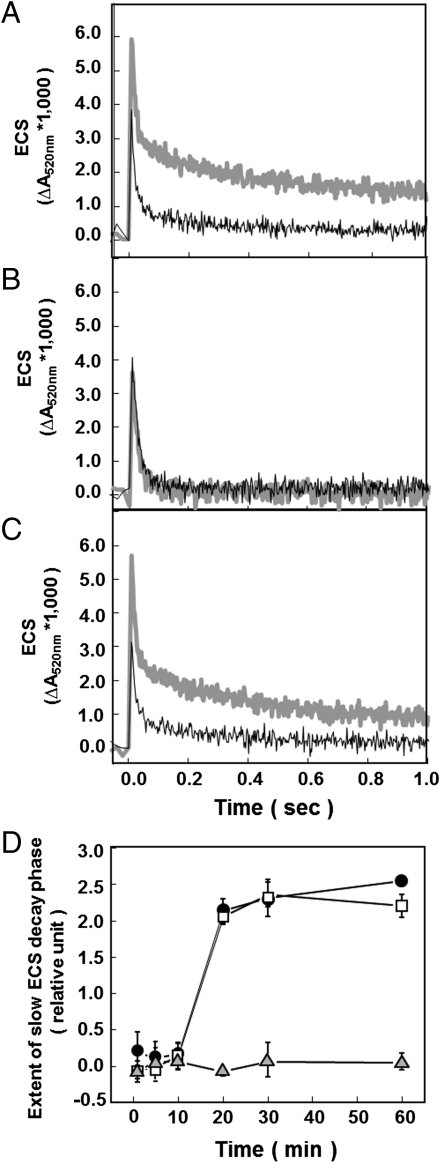
We analyzed ATP synthase activity in attached leaves by exciting PSI and PSII with short (100 μs), nonsaturating LED pulses to rapidly generate pmf, and probing the resulting proton flux through the ATP complex via the decay of the ECS absorbance change at 520 nm (13, 14) (Fig. 2 A–C). A few seconds after preillumination, ATP synthase activity of wild type remained active even at low pmf, as indicated by rapid nearly monophasic flash-induced ECS decay-with a fast phase of approximately 25 ms constituting approximately 80% of the decay, and a slower phase with lifetime longer than 250 ms, showing that the γ subunit was predominantly reduced. Upon extensive (> 20 min). dark adaptation, the extent of the slow ECS decay phase markedly increased, reflecting oxidation of the γ subunit, which imposes an increased pmf requirement for activation of the ATP synthase. Owing to the slower decay of ECS in dark-adapted leaves, the contribution of the cytochrome b6f complex turnover to the electric field can be seen as a rise in ECS on the approximately 10 ms time scale. In light-adapted material, this phase is masked by the rapid decay of ECS. The extent of slow ECS decay phase showed a sigmoidal dependence on dark adaptation (Fig. 2D), with a pronounced lag phase from 0 to 10 min after preillumination, followed by a approximately 10 min half time decrease in activity. The sigmoidal oxidation kinetics were previously interpreted as reflecting the equilibration of γ subunit thiols redox state with that of a larger pool of redox carriers (14). Almost identical results were obtained for atpc2, indicating that γ2 is not required for regulation of ATP synthase. In contrast with wild type and atpc2, the ECS decay kinetics in gamera remained constant with a nearly monophasic decay showing halftime of about 21 ms, even after extensive (> 60 min) dark-adaptation (Fig. 2B), indicating that γ2-ATP synthase is not down-regulated by thiol oxidation in the dark.
Fig. 2.
Redox state of the ATP synthase probed by the decay of flash-induced ECS signals. Flash-induced relaxation kinetics of ECS were measured in wild type (A), gamera (B), and atpc2 (C) at 1 (thin black line) and 60 min (thick gray line) dark adaptation following after 2 min preillumination at 100 μmol photons m-2 s-1 red actinic light. The extent of slow ECS decay phase of ECS kinetics after brief subsaturating actinic illumination in wild type (black circles), atpc2 (open squares), and gamera (gray triangles) (D). Data is for attached leaves with n = 3.
Equilibrium Redox Titrations of γ1 and γ2-ATP Synthase.
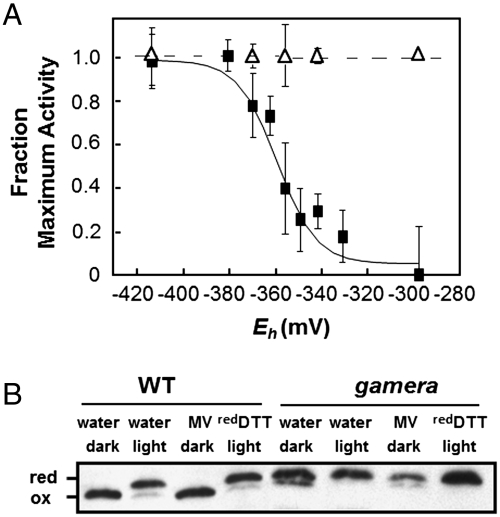
In situ equilibrium redox titrations (22), were used to assess the redox properties of γ1- and γ2-ATP synthase in wild type and gamera, respectively (Fig. 3A). In wild type, the ECS decay was rapid at low redox potential and slow at high potential, where γ1 was expected to be reduced and oxidized, respectively. Assuming a stromal pH of 7.5, we estimate a redox midpoint potential for the γ1 subunit of approximately -356 mV (versus standard hydrogen electrode) with an n = 2 redox behavior for the thiol/disulfide interconversion, similar to that reported previously (22). By contrast, ECS decay in gamera remained constant with changing the redox potential (Fig. 3A) over the entire range accessible by varying reduced and oxidized DTT (approximately -300 to -420 mV versus standard hydrogen electrode). The maximal activity of the ATP synthase under reducing conditions, as judged by the ECS decay, was about twofold higher in the wild type than in gamera, presumably reflecting the overall differences in ATP synthase content (Figs. S2 B and C).
Fig. 3.
Equilibrium redox titration (22) of thiol/disulfide regulatory groups in the γ subunit of the chloroplast ATP synthase in wild type (WT) (closed squares) and gamera (opened triangles) (A). In situ fractional ATP synthase activity was estimated by probing ECS decay kinetics (ΔA520) as described in the text. (n = 4–6). Detection of reduced (red) and oxidized (ox) γ subunit using gel shift assay, described in text (B). The redox states of γ1 and γ2 subunit thiols in wild type and gamera were visualized by AMS-labeling followed by SDS/PAGE and immnoblotting after treatment of leaf discs with water as control, MV, and reduced DTT under light (approximately 50 μmol photons m-2 s-1) or dark as described in Materials and Methods.
To determine whether the lack of redox regulation of γ2-ATP synthase was caused by a shift in the redox potential of the thiol/disulfide transition or by a change in the effects of such transition on the activity of the enzyme, we assayed the redox state of the γ1 and γ2 subunits directly, based on modification of free sulfhydryl groups with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonate (AMS) followed by differential separation by sodium dodecyl sulfate polyacrylamide gel electrophoresis (26) (Fig. 3B). Wild-type leaves infiltrated with water showed classical light-induced changes in redox state of γ1, with the higher molecular weight reduced form, in light-adapted leaves. In contrast, we saw no light-induced changes in the apparent molecular weight of γ2 in gamera leaves infiltrated with water. To ensure maximal physiological differences in redox states, we repeated these experiments using two conditions designed to reflect the extremes of in vivo redox states: 1) oxidizing conditions in dark-adapted leaves in the presence of methyl viologen (MV) to prevent accumulation of electrons on thioredoxin (13); 2) reducing conditions in leaves illuminated with 300 μmol photons m-2 s-1 actinic light and infiltrated with DTT (Fig. 3B). As with water-infiltrated leaves, γ1 showed a higher molecular weight under reducing and not under oxidizing conditions, clearly indicating the expected redox transition that altered AMS binding. However, in gamera, the molecular weight of γ2 remained higher, even under these more extreme redox conditions, implying that its redox potential was shifted towards higher potentials with respect to γ1. To determine whether γ2 is oxidizable under more oxidizing conditions, we infiltrated leaves with the strong thiol oxidant, diamide, and observed both oxidation of thiols (Fig. S3A) and a decrease in ATP synthase activity in vivo (Fig. S3B), indicating that γ2 is oxidizable but at a more oxidizing redox potential, probably out of the range set by the thioredoxin system in vivo.
Evidence That γ2-ATP Synthases Have a Moonlighting Function in Root Morphogenesis.
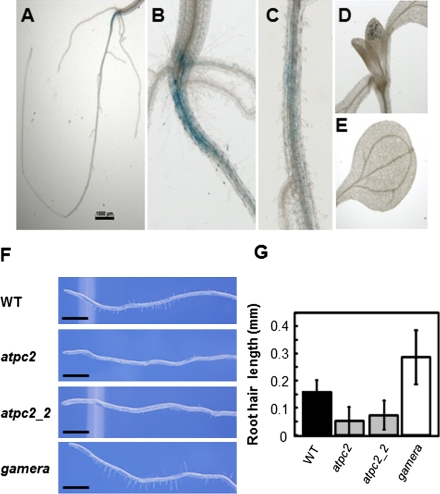
The gene expression profiles of ATPC1 and ATPC2 in different tissues were investigated using the publicly available software Genevestigator (27) (Fig. S4). In leaves, ATPC1 expression was about 100-fold higher than that of ATPC2 consistent with the failure to observe accumulation of γ2 protein in wild-type leaves using γ2-specific antibodies (Fig. S2B). However, in radicle or mature root tissue, ATPC2 expression was approximately equal to or above that of ATPC1, suggesting a function of γ2 in roots. To confirm the expression pattern of ATPC2, we constructed plants with β-glucouronidase (GUS) driven by ATPC1 (ATPC1-GUS) and ATPC2 promoters (Supporting Information). In contrast to ATPC1-GUS, which showed a strong staining over the whole seedling (Fig. S5), ATPC2-GUS expression was absent in the green tissues and appeared to be localized along the root and at the root-hypocotyl junction (Fig. 4 A–E), again suggesting a role of γ2 in root development.
Fig. 4.
Evidence for a function of ATPC2 in development. ATPC2∷GUS staining in 10 d old seedlings in root system (A), hypocotyl-root junction (B), primary root (C), young leaves (D), and cotyledons (E). Representative photographs of root hairs of wild type (WT), two separate atpc2 lines, and gamera (F). Scale bars 1.00 mm. Root hair lengths measured in wild type, two atpc2 lines, and gamera (G) (n = 32–274).
We therefore analyzed the root morphology in wild type, gamera, and atpc2 knock out lines. In 7 d old seedlings, two separate alleles of atpc2 had developmentally immature root hairs. The atpc2 mutant showed 60–80% fewer root hairs than wild type or gamera (Fig. 4F). The average length of root hair in atpC2 was about 50% that of wild type. Strikingly, gamera, with overexpressed ATPC2, showed dramatically (approximately 2.5 times) longer root hairs than wild type (Fig. 4 F and G).
Although the precise function of γ2-ATP synthase is currently unknown, our results indicate that it plays a role, either directly or indirectly, in development of nonphotosynthetic tissues, resulting in strong effects on root morphology.
Species Distribution, Phylogeny, and Structural Aspects of ATPC Genes.
Comparison of sequenced genomes showed that dicots contain two or more homological duplicates (or multiplicates) of the γ subunit (Fig. S6). The monocots Zea mayes and Oryza sativa, the green algae Ostreococcus lucimarinus and Chlamydomonas reinhardtii, or cyanobacteria (see also below) showed only a single copy, suggesting a possible role of the “secondary-duplicate” γ2-like subunit specifically in dicots. In all dicots, one homologue is clearly γ1-like, possessing a regulatory loop region with two cysteine residues. In many cases, though, one or more of the “γ2-like” subunit duplicates have clear differences in the redox regulatory loop, probably rendering it redox-insensitive. For examples, grape (Vitis vinifera), poplar (Populus trichocarpa), and lotus (Lotus japonicus, Nelumbo nucifera) are missing one of the two regulatory cysteine residues, whereas tobacco (Nicotiana tabacum), barrel clover (Medicago truncatula), felon herb (Artemisia indica), apple (Malus pumila var. domestica), and tomato (Solanum lycopersicum) are missing both. Soybean (Glycine max), castor bean (Ricinus communis), and selaginella (Selaginella tamariscina) γ-homologues retain both cysteine residues, but also contains the same threonine to glutamate substitution in the regulatory loop region as in Arabidopsis γ2 (Fig. S6).
We investigated the possible evolutionary origins of the diversity of primary and secondary γ subunit homologues. The occurrence of duplicate (or multiplicate) genes coding for the γ subunit of the plastidic ATP sythase was screened in available genomes of uni- and multicellular representatives of viridiplantae and of the evolutionary ancestors of their plastids, i.e., the cyanobacteria. Multiple copies of ATPC were only detected in multicellular species whereas both cyanobacteria and unicellular green algae contain only one ATPC gene strongly corroborating the hypothesis put forward above that the duplicate genes are expressed differentially to adapt different types of tissues to their specific requirements. Consequently, duplicate versions of the gene are found in many subgroups of multicellular virdiplantae down to the very early branching mosses. An evolutionary analysis shows a far-going congruence between the phylogeny of these γ subunit genes and that of their parent species (Fig. S7).
The tree furthermore demonstrates that duplication of the ATPC gene had occurred frequently and independently during the evolution of multicellular plants. The two species of mosses, for example, as well as apple trees and glycines feature relatively recent duplication events within each of the mentioned lineages (Fig. S7). By contrast, an early duplication event apparently has given rise to two distinct ATPC genes in the ancestor of angiosperms with subsequent vertical inheritance of both copies towards most of the extant angiosperms. The resulting two clusters contain the above-analyzed ATPC1 and ATPC2 genes from Arabidopsis, respectively. The cluster encompassing the ATPC1 gene is substantially more compact (i.e., characterized by shorter branch lengths) than that containing the ATPC2 gene. This is in line with the ATPC1 gene operating under “standard” conditions in redox regulated ATP synthases, i.e., subjected to strong and unifunctional evolutionary constraints, whereas the secondary gene has evolved to fulfil different and possibly diverse functions in the various plants examined.
Conclusions: What Is the Function of γ2-ATP Synthase in Arabidopsis?
We found that the regulatory behavior of the chloroplast ATP synthase is markedly altered by substitution of γ2 by γ1. Wild type and atpc2 plants, which express only γ1-ATP synthase in leaves, show classical light-dark regulation of ATP synthase activity via the thioredoxin system (Fig. 2). In contrast, gamera, which expresses exclusively γ2-ATP synthase, shows light-dark-insensitive ATP synthase activity, due to a more positive redox potential for the regulatory thiol groups (Fig. 3A).
Because γ2-ATP synthase is equally active in the light and dark, we propose that it may function to catalyze low levels of ATP-driven proton translocation in nonphotosynthetic tissues or in the dark, (i.e., proton pumping into the lumen driven by ATP hydrolysis) to maintain sufficient transthylakoid proton gradient when γ1-ATP synthase is inactivated. The proton gradient across the thylakoid membrane is required for nonphotosynthesis processes, for example, ion transport or the twin-arginine translocation pathway for targeting plastid proteins into the thylakoid lumen or membrane (28–30) especially in nonphotosynthetic organelles such as etioplasts that develop without photosynthetic electron flow to reduce thioredoxin. It is also possible that γ2-ATP synthase allows for ATP synthesis in nonphotosynthetic plastids, via a chlororespiratory pathway (31). In accord with these views, we showed that γ2-ATP synthase is expressed in the root-hypocotyl junction (Fig. 4B), and altering its expression level had striking effects on root hair morphology (Fig. 4 F and G). This is consistent with recent proteomic analysis that shows that, unlike most photosynthetic proteins and complexes, the ATP synthase is already present in dark-adapted etioplasts (32). Although a full investigation of the function of γ2-ATP synthase in roots is beyond the scope of this paper, these observations support a role of γ2 not in photosynthesis but in development or maintenance/function of roots or other nonphotosynthetic tissues. Phylogenetic analyses (Fig. S7) likewise suggest the evolution of (possibly multiple) emergent “moonlight” functions for ATP synthase through divergent evolution of the γ subunits.
Materials and Methods
Plant Materials and Growth Conditions.
Wild-type Arabidopsis thaliana (ecotype Columbia) and ATPC2 T-DNA knockout lines, atpc2 (T-DNA is inserted in chl1_5402949) and gamera in which ATPC2 overexpressed in the ATPC1 T-DNA knock out line (dpa1) (21) (see SI Materials and Methods) were grown on soil under continuous light period at 20–30 μmol photons m-2 s-1 at 22 °C for 4 wks, as described previously (21).
In Vivo Spectroscopic Assays.
Maximal PSII quantum efficiency, LEF, and energy-dependent exciton quenching (qE) were estimated from saturation-pulse chlorophyll a fluorescence yield measurements using the instrumentation and methods described previously (33). The value of steady-state fluorescence yield with all PSII centers closed, FM’, was obtained after at least 15 min illumination, at which point the photosynthetic rates and fluorescence parameters were stable. The value of FM’’in fluorescence yield was estimated after 10 min of dark relaxation. Relative extents of steady-state, light-induced pmf (ECSt) and the conductivity of the ATP synthase to protons (gH+) were estimated from dark interval relaxation kinetics of absorbance changes associated with the ECS as described previously (15, 16). Flash-induced relaxation kinetics (FIRK) experiments were performed on infiltrated leaf discs described previously (17).
Equilibrium Redox Titrations.
Redox titrations were performed in situ in fully expanded detached leaves vacuum-infiltrated with varying ratios of oxidized and reduced 20 mM DTT solutions for 30 min incubation in the dark, as described previously (22). The activity of the ATP synthase was then monitored using the FIRK of the ECS signal as described in (13) using the instrumentation described in (17). The equilibrium redox potential (Eh) for DTT at pH 7.5 was calculated according to the Nernst equation, using Em,7.5 = -356 mV for DTT.
Determination of Redox States of γ Subunits.
The redox state of γ subunit of chloroplast ATP synthase was probed using the binding of AMS followed by nonreducing SDS/PAGE (26). To achieve oxidizing conditions, leaf 1 cm discs were infused with 0.1% Tween 20 and 20 mM Tricine containing 100 μM of MV followed by incubation for 30 min in darkness at room temperature. Reducing conditions were achieved by vacuum-infiltration of the leaf discs with 20 mM reduced DTT as a reducing agent (17) followed by illumination for 5 min by 100 μmol photons m-2 s-1.
Strongly oxidizing conditions were obtained by similar infiltration with 0, 0.5, or 5 mM diamide. After treatments, the discs were frozen in liquid nitrogen and ground with mortar and pestle, then extracted in 50 mM Tris·HCl (pH 8.0), 2 mM MgCl2, and 10 mM NaCl. The extractions were fractionated to soluble and insoluble protein by centrifugation. Protein precipitates were washed with acetone and dissolved in freshly prepared solution containing 1% SDS, 50 mM Tris·HCl (pH 8.0), and 15 mM AMS, as described in (26). Proteins were separated by SDS/PAGE using running buffer lacking reducing agent, 1% SDS, 50 mM Tris·HCl (pH 8.0), and 10% glycerol. Relative contents of γ subunits in wild type and gamera were estimated by Western blotting, as described in (33), using specific antibodies raised against γ1 and γ2 (see Fig. S2B). The antibody raised against a γ2 peptide shows no cross-reactivity with γ1, whereas that raised against the γ1 peptide reacts with both γ1 and γ2.
GUS Assay.
Seedlings were washed in water after soaking in 3.7% formaldehyde for 10 min. The seedlings were then incubated in 1 mM 5-bromo-4-chloro-3-indolyl β-D-glucuronide cyclohexylammonium salt, 100 mM potasium phosphate buffer (pH 7.0), 1 mM EDTA, 1 mM potassium hexacyanoferrate, 1 mM potassium hexacyanoferrate, and 0.3% Triton X-100 overnight at 37 °C. For visualization of blue staining the chlorophyll was removed by incubation in 70% ethanol (34).
Observation of Root Phenotype.
Wild type, two homozygous ATPC2 T-DNA knockout lines, atpc2 (T-DNA inserted in chl1_5402949), atpc2_2 (T-DNA inserted in chl 1_5404314), and gamera were surface sterilized and germinated on Murashige and Skoog plates without sucrose in vertical transparent plates as in (21) . Plants were examined using light microscopy (Leica M165FC) after 5-d of growth. Tissue lengths were measured from recorded images using Image J (http://rsbweb.nih.gov/ij/). The lengths of root hairs were measured on the lower 5 mm of roots on four to five plants of each plant type (n = 32–274).
Supplementary Material
ACKNOWLEDGEMENTS.
We are grateful to Dr. Donald Ort (University of Illinois) and Dr. John Froelich (Michigan State University) for stimulating discussions, Dr. Gregory Howe (Michigan State University) for assistance with root microscopy, and Dr. Alice Barkan (University of Oregon) for the CF1-β antibodies. The work was supported by National Research Initiative (2008-35318-04665 supporting A.K. and A.D.) from the US Department of Agriculture National Institute of Food and Agriculture,and the US Department of Energy, Office of Science, Basic Energy Sciences Program (DE-FG02-04ERl5559 supporting .KK.) to D.M.K., and by the Deutsche Forschungsgemeinschaft (DFG), Sonderforschungsbereich Transregio 1 (SFB TR1) to J.M.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115728109/-/DCSupplemental.
References
- 1.Richter ML. Gamma-epsilon interactions regulate the chloroplast ATP synthase. Photosynth Res. 2004;79:319–329. doi: 10.1023/B:PRES.0000017157.08098.36. [DOI] [PubMed] [Google Scholar]
- 2.Groth G, Strotmann H. New results about structure, function and regulation of the chloroplast ATP synthase (CFoCF1) Physiol Plant. 1999;106:142–148. [Google Scholar]
- 3.Capaldi RA, Aggeler R. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem Sci. 2002;27:154–160. doi: 10.1016/s0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- 4.Junesch U, Graber P. The rate of ATP-synthesis as a function of ΔpH and Dy catalyzed by the active, reduced H+-ATPase from chloroplasts. FEBS Lett. 1991;294:275–278. doi: 10.1016/0014-5793(91)81447-g. [DOI] [PubMed] [Google Scholar]
- 5.Seelert H, Dencher NA, Muller DJ. Fourteen protomers compose the oligomer III of the proton-rotor in spinach chloroplast ATP synthase. J Mol Biol. 2003;333:337–344. doi: 10.1016/j.jmb.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 6.Ort DR, Oxborought K. In situ regulation of chloroplast coupling factor activity. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:269–291. [Google Scholar]
- 7.Walker JE. The regulation of catalysis in ATP synthase. Curr Opin Struct Biol. 1994;4:912–918. doi: 10.1016/0959-440x(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 8.Ort DR, Grandoni P, Ortiz-Lopez A, Hangarter RP. Control of photophosphorylation by regulation of the coupling factor. In: Zelitch I, editor. Perspectives in Biochemical and Genetic Regulation Photosynthesis. New York: Wiley–Liss; 1990. pp. 159–173. [Google Scholar]
- 9.Mills JD, Mitchell P. Modulation of coupling factor ATPase activity in intact chloroplasts: Reversal of thiol modulation in the dark. Biochim Biophys Acta. 1982;679:75–83. [Google Scholar]
- 10.Richter ML, Samra HS, He F, Giessel AJ, Kuczera KK. Coupling proton movement to ATP synthesis in the chloroplast ATP synthase. J Bioenerg Biomembr. 2005;37:467–473. doi: 10.1007/s10863-005-9493-9. [DOI] [PubMed] [Google Scholar]
- 11.Nalin CM, McCarty RE. Role of a disulfide bond in the gamma subunit in activation of the ATPase of chloroplast coupling factor 1. J Biol Chem. 1984;259:7275–7280. [PubMed] [Google Scholar]
- 12.Schwarz O, Schurmann P, Strotmann H. Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J Biol Chem. 1997;272:16924–16927. doi: 10.1074/jbc.272.27.16924. [DOI] [PubMed] [Google Scholar]
- 13.Kramer DM, Crofts AR. Activation of the chloroplast ATPase measured by the electrochromic change in leaves of intact plants. Biochim Biophys Acta. 1989;976:28–41. [Google Scholar]
- 14.Kramer DM, et al. Regulation of coupling factor in field-grown sunflower: A Redox model relating coupling factor activity to the activities of other thioredoxin-dependent chloroplast enzymes. Photosynth Res. 1990;26:213–222. doi: 10.1007/BF00033134. [DOI] [PubMed] [Google Scholar]
- 15.Kanazawa A, Kramer DM. In vivo modulation of nonphotochemical exciton quenching (NPQ) by regulation of the chloroplast ATP synthase. Proc Natl Acad Sci USA. 2002;99:12789–12794. doi: 10.1073/pnas.182427499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz JA, et al. Plasticity in light reactions of photosynthesis for energy production and photoprotection. J Exp Bot. 2004;56:395–406. doi: 10.1093/jxb/eri022. [DOI] [PubMed] [Google Scholar]
- 17.Kohzuma K, et al. The long-term responses of the photosynthetic proton circuit to drought. Plant Cell Environ. 2009;32:209–219. doi: 10.1111/j.1365-3040.2008.01912.x. [DOI] [PubMed] [Google Scholar]
- 18.Takizawa K, Cruz JA, Kramer DM. Depletion of stromal inorganic phosphate induces high “energy-dependent” antenna exciton quenching (qE) by decreasing proton conductivity at CFO-CF1 ATP synthase. Plant Cell Environ. 2008;31:235–243. doi: 10.1111/j.1365-3040.2007.01753.x. [DOI] [PubMed] [Google Scholar]
- 19.Inohara N, et al. Two genes, atpC1 and atpC2, for the gamma subunit of Arabidopsis thaliana chloroplast ATP synthase. J Biol Chem. 1991;266:7333–7338. [PubMed] [Google Scholar]
- 20.Legen J, Misera S, Herrmann RG, Meurer J. Map positions of 69 Arabidopsis thaliana genes of all known nuclear encoded constituent polypeptides and various regulatory factors of the photosynthetic membrane: a case study. DNA Res. 2001;8:53–60. doi: 10.1093/dnares/8.2.53. [DOI] [PubMed] [Google Scholar]
- 21.Bosco CD, et al. Inactivation of the chloroplast ATP synthase gamma subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. J Biol Chem. 2004;279:1060–1069. doi: 10.1074/jbc.M308435200. [DOI] [PubMed] [Google Scholar]
- 22.Wu G, Ortiz-Flores G, Ortiz-Lopez A, Ort DR. A point mutation in atpC1 raises the redox potential of the Arabidopsis chloroplast ATP synthase gamma-subunit regulatory disulfide above the range of thioredoxin modulation. J Biol Chem. 2007;282:36782–36789. doi: 10.1074/jbc.M707007200. [DOI] [PubMed] [Google Scholar]
- 23.Kleffmann T, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 24.Baker NR, Harbinson J, Kramer DM. Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant, Cell Environ. 2007;30:1107–1125. doi: 10.1111/j.1365-3040.2007.01680.x. [DOI] [PubMed] [Google Scholar]
- 25.Avenson TJ, Cruz JA, Kanazawa A, Kramer DM. Regulating the proton budget of higher plant photosynthesis. Proc Natl Acad Sci USA. 2005;102:9709–9713. doi: 10.1073/pnas.0503952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konno H, Suzuki T, Bald D, Yoshida M, Hisabori T. Significance of the epsilon subunit in the thiol modulation of chloroplast ATP synthase. Biochem Biophys Res Commun. 2004;318:17–24. doi: 10.1016/j.bbrc.2004.03.179. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller M, Klosgen RB. The Tat pathway in bacteria and chloroplasts (review) Mol Membr Biol. 2005;22:113–121. doi: 10.1080/09687860500041809. [DOI] [PubMed] [Google Scholar]
- 29.Sargent F. The twin-arginine transport system: Moving folded proteins across membranes. Biochem Soc Trans. 2007;35:835–847. doi: 10.1042/BST0350835. [DOI] [PubMed] [Google Scholar]
- 30.Robinson C, Bolhuis A. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta. 2004;1694:135–147. doi: 10.1016/j.bbamcr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Hausler RE, et al. Chlororespiration and grana hyperstacking: How an Arabidopsis double mutant can survive despite defects in starch biosynthesis and daily carbon export from chloroplasts. Plant Physiol. 2009;149:515–533. doi: 10.1104/pp.108.128124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ploscher M, Reisinger V, Eichacker LA. Proteomic comparison of etioplast and chloroplast protein complexes. J Proteomics. 2011;74:1256–1265. doi: 10.1016/j.jprot.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 33.Livingston AK, Cruz JA, Kohzuma K, Dhingra A, Kramer DM. An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell. 2010;22:221–233. doi: 10.1105/tpc.109.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ditengou FA, et al. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:18818–18823. doi: 10.1073/pnas.0807814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.