Abstract
The unprecedented genetic diversity found at vertebrate MHC (major histocompatibility complex) loci influences susceptibility to most infectious and autoimmune diseases. The evolutionary explanation for how these polymorphisms are maintained has been controversial. One leading explanation, antagonistic coevolution (also known as the Red Queen), postulates a never-ending molecular arms race where pathogens evolve to evade immune recognition by common MHC alleles, which in turn provides a selective advantage to hosts carrying rare MHC alleles. This cyclical process leads to negative frequency-dependent selection and promotes MHC diversity if two conditions are met: (i) pathogen adaptation must produce trade-offs that result in pathogen fitness being higher in familiar (i.e., host MHC genotype adapted to) vs. unfamiliar host MHC genotypes; and (ii) this adaptation must produce correlated patterns of virulence (i.e., disease severity). Here we test these fundamental assumptions using an experimental evolutionary approach (serial passage). We demonstrate rapid adaptation and virulence evolution of a mouse-specific retrovirus to its mammalian host across multiple MHC genotypes. Critically, this adaptive response results in trade-offs (i.e., antagonistic pleiotropy) between host MHC genotypes; both viral fitness and virulence is substantially higher in familiar versus unfamiliar MHC genotypes. These data are unique in experimentally confirming the requisite conditions of the antagonistic coevolution model of MHC evolution and providing quantification of fitness effects for pathogen and host. These data help explain the unprecedented diversity of MHC genes, including how disease-causing alleles are maintained.
Keywords: host-pathogen, antibiotic resistance, endangered species, pathogen escape of adaptive immunity
Classical MHC genes are characterized by unprecedented levels of allelic diversity. To use an extreme example, over 2,300 alleles have been identified for the human HLA-B gene (1). How this diversity is maintained has been an evolutionary puzzle for decades, but it is now generally assumed that some process of balancing selection is operating (2). Various mechanisms have been put forth to explain MHC evolution with antagonistic coevolution (leading to rare allele advantage), heterozygote advantage, and mating preferences being the leading candidates (3–5). The relative importance of these nonmutually exclusive mechanisms has been controversial and difficult to assess (6–9), although all likely contribute to maintaining MHC diversity. Here, we test critical assumptions of the antagonistic coevolution model.
J. B. S. Haldane was the first researcher to frame antagonistic coevolution between hosts and their pathogens as the diversifying selection responsible for the unprecedented serological diversity caused by MHC polymorphisms (10). Antagonistic coevolution proposes a never-ending molecular arms race whereby pathogen populations evolve to escape immune recognition by common MHC alleles circulating in host populations, which in turn provides a selective advantage to hosts carrying rare MHC alleles (9, 11). This cyclical process leads to negative frequency-dependent selection (12) and can promote MHC diversity if two requisite conditions are met: (i) adaptations that benefit pathogen fitness in one host MHC genotype must be costly to pathogen fitness when infecting hosts carrying an unfamiliar MHC genotype (i.e., antagonistic pleiotropy), and (ii) these patterns of adaptation must produce correlated patterns of virulence (i.e., disease severity). These fitness trade-offs, which arise as a consequence of pathogen adaptation to specific MHC genotypes, provides a selective advantage to hosts carrying unfamiliar (i.e., rare) MHC genotypes through increased resistance to infectious disease. As MHC allelic diversity increases over time in a population, so too does the chance that some of these alleles will confer susceptibility to autoimmune disease (2). Furthermore, because the same MHC allele can confer resistance against one infectious agent and susceptibility to another (13), it is likely that all MHC alleles in a host population confer susceptibility to at least some pathogens. Thus, antagonistic coevolution can help explain MHC evolution, but can also explain why MHC diversity is associated with susceptibility to most infectious and autoimmune diseases. Finally, evidence in support of antagonistic coevolution also offers partial solutions to problems associated with endangered species, antibiotic resistance, emerging infectious diseases, and the evolution of sex (14, 15).
MHC genes are the most polymorphic loci in vertebrates (2), and encode proteins that function to bind and present peptide fragments generated by the breakdown of self and foreign (e.g., viral) proteins to circulating T lymphocytes. Foreign-peptide/MHC complexes that activate T lymphocytes trigger the onset of an adaptive immune response (16). Genetic diversity found within the MHC controls a significant portion of the variation in susceptibility to infectious disease (17), and the importance of classic MHC genes as prime targets of pathogen adaptation is evidenced by examples of MHC escape mutants in a variety of viral pathogens (18–20).
Direct experimental support for antagonistic coevolution as a mechanism promoting MHC diversity is limited. Recent work from invertebrates, which do not have adaptive immune systems or MHC molecules, has shown that antagonistic coevolution can accelerate the rate of molecular evolution in pathogen populations (21) and that it is capable of maintaining genetic diversity in host populations (as measured by microsatellite variability) (22). Explicit work on vertebrate MHC diversity and patterns of pathogen adaptation has shown that MHC genes control the trajectory of pathogen adaptation (23) and that viruses carrying an MHC-escape epitope specific for one host MHC are lost when exposed to another disparate host MHC; this implies a pathogen fitness cost associated with adaptation to a specific host MHC (24). These observed patterns are all consistent with predicted outcomes of antagonistic coevolution. However, no study has experimentally tested both requisite conditions in a single system and in a quantitative fashion (i.e., across multiple pathogen and host genotypes). This test is required to evaluate if antagonistic coevolution can produce the type and intensity of selection necessary to explain MHC evolution.
Here, we conducted serial passage of Friend virus complex (25), a mouse-specific pathogen, through a series of genetically identical individuals from each of three MHC-congenic inbred mouse strains (BALB/cdd, BALB/ckk, and BALB/cbb). Serial passage experimentation involves repeated transmission of a pathogen through a series of hosts and is a proven technique for observing pathogen adaptation in experimental time (26, 27). Although transmission factors can influence pathogen evolution, serial passage relaxes this selective constraint on pathogen adaptation, allowing one to focus on the importance of genetic interactions between host and pathogen in determining patterns of pathogen adaptation. The incorporation of congenic mice into our experimental design allowed us to explicitly analyze how MHC genotype influences evolved patterns of pathogen fitness and virulence. Results from our experiments provide direct experimental support for antagonistic coevolution with pathogens as a mechanism capable of favoring MHC polymorphism in vertebrate populations.
Results
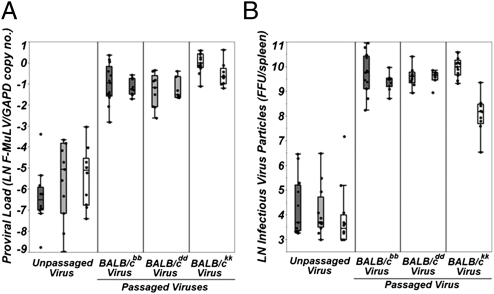
To quantify both the presence and magnitude of adaptation in response to serial passage, we compared pathogen fitness measures (proviral load and infectious particle counts) between groups of animals (n = 6–10 individuals per group) from the same host MHC genotype infected with either unpassaged virus or their respective postpassage stocks (two replicate stocks per genotype) for each of the three host MHC genotypes (six total postpassage stocks) (Fig. S1). Proviral loads from animals infected with postpassage viruses were significantly higher than those from animals infected with unpassaged virus (ANOVA, F1,93 = 403.77, P < 0.0001), representing an ∼54-fold mean increase in pathogen fitness (Fig. 1A). Similarly, infectious virus particle counts demonstrated a highly significant 109-fold increase in mean pathogen fitness after serial passage (Fig. 1B) (ANOVA, F1,95 = 737.38, P < 0.0001). When each replicate passage line is analyzed independently, all comparisons between animals from each genotype infected with unpassaged or postpassage viruses are highly significant for both measures of pathogen fitness (Table S1). Therefore, pathogen adaptation during serial passage of Friend virus complex produced pathogen stocks with substantially enhanced fitness.
Fig. 1.
Pathogens rapidly increase fitness during serial passage. Serial passage of virus through BALB/cbb (dark gray boxplots), BALB/cdd (light gray boxplots), and BALB/ckk (white boxplots) host genotypes produces virus stocks with significantly higher fitness. Data above are shown on a natural log scale. Both proviral load measures (A) and infectious particle counts (B) are significantly higher in animals infected with passaged vs. unpassaged viruses across all three MHC-congenic mouse strains.
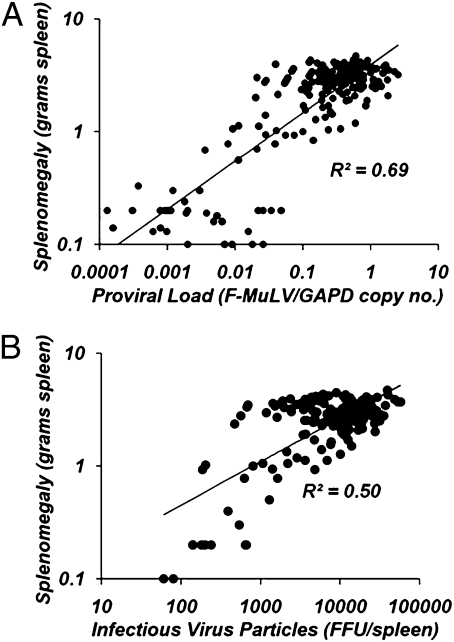
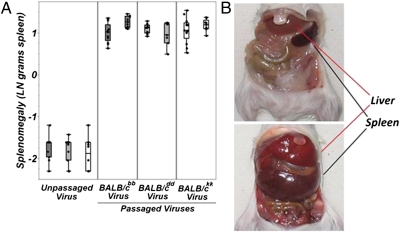
Virulence associated with the acute phase of infection with Friend virus complex is gross splenomegaly and hepatomegaly (spleen and liver enlargement, respectively) because of the rapid induction of clonal proliferation of virally infected cells within these organs (28, 29). We focused on splenomegaly as the virulence response variable to compare against measures of pathogen fitness because the spleen is the primary site of virus replication (28). First, we observed strong positive correlations between our measures of pathogen fitness and splenomegaly [splenomegaly × provirus: R2 = 0.69, ANOVA, F1,200 = 445.42, P < 0.0001 (Fig. 2A); splenomegaly × infectious virus particles: R2 = 0.50, ANOVA, F1,204 = 257.75, P < 0.0001 (Fig. 2B)]. Second, comparison of spleen weights among the same groups of animals represented in Fig. 1 revealed that infection with postpassage stocks resulted in a highly significant 18-fold increase in mean virulence associated with infection by postpassage virus stocks (ANOVA, F1,93 = 2126.98, P < 0.0001) (Fig. 3). Again, when replicate passage lines are analyzed independently, all comparisons between animals from each genotype infected with unpassaged or passaged viruses are highly significant (Table S2). Combined, these results indicate that pathogen fitness and virulence are positively correlated and that the observed increase in pathogen fitness of our postpassage stocks results in a dramatically more virulent disease.
Fig. 2.
Pathogen fitness is positively correlated with virulence. Regression analyses between measures of pathogen fitness and virulence were performed. Both proviral load (A) and infectious particle counts (B) demonstrate positive and highly significant correlations with our measure of virulence (splenomegaly).
Fig. 3.
Pathogen adaptation results in a significantly more virulent infection. (A) Measures of virulence were compared between the same groups of animals used in the experiments summarized in Fig. S1. Based on an analysis of pooled data, infection with postpassage vs. unpassaged virus stocks was associated with a highly significant increase in spleen weight. Data shown on natural log scale. (B) Typical size difference between the spleens and livers of BALB/c animals infected with either unpassaged (Upper) or postpassage (Lower) virus stocks.
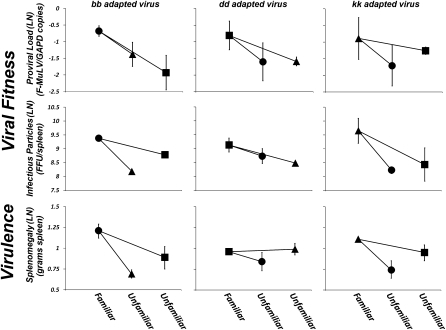
To determine whether pathogen adaptation results in fitness trade-offs between MHC genotypes, groups of animals (n = 6–10 individuals per group) from each host MHC genotype were independently infected with all six postpassage stocks (three postpassage stocks per replicate experiment × two replicates). The antagonistic coevolution hypothesis requires trade-offs in pathogen fitness that result in passaged pathogens being significantly more fit when infecting hosts carrying a familiar versus an unfamiliar MHC genotype. A least-squares multiple regression analysis was used to construct a model that tested for a significant host genotype by virus genotype (genotype*genotype) interaction effect on patterns of pathogen adaptation (see Tables S3 and S4 for details of model construction). Based on this analysis, infectious virus particles and proviral loads demonstrated highly significant genotype*genotype interaction effects (infectious particle counts: ANOVA, F1,10 = 6.35, P < 0.0001; proviral load: ANOVA, F1,10 = 4.57, P < 0.0001), and indicate that adapted pathogens are significantly more fit when infecting their familiar host genotype versus an unfamiliar host genotype in all 12 comparisons (Fig. 4 and Table S4). Importantly, there were no underlying susceptibility differences between these MHC genotypes (Table 1), indicating that observed differences in adapted pathogen fitness between familiar and unfamiliar hosts is a consequence of the adaptive response by the pathogen rather than preexisting variation in susceptibility among host genotypes.
Fig. 4.
Pathogens adapt to specific MHC genotypes, and this results in lower fitness and virulence when infecting unfamiliar MHC genotypes. Pathogen adaptation to specific MHC genotypes is predicted to result in strong host genotype by pathogen genotype interactions. To test this prediction six groups of animals (6–10 individuals per group) from each of our three host genotypes were independently infected with all six postpassage stocks (two passage lines were conducted in each of the three host genotypes) and interaction effects were tested using a least-square general linearized model. Symbols reflect host genotypes (●, BALB/cbb hosts; ■, BALB/cdd hosts; ▲, BALB/ckk hosts) and represent the least-square mean of two replicate tests (error bars represent the range of means for each test). Data are shown on a natural log scale. Comparisons of patterns of pathogen adaptation and virulence between virus stocks (denoted bb passaged virus, dd passaged virus, and kk passaged virus) across the three host genotypes shows that pathogen fitness and virulence is host genotype-specific. Viral fitness (12 of 12 comparisons) and disease virulence (5 of 6 comparisons) are higher when a passaged virus stock is exposed to a familiar (i.e., its’ host genotype-of-passage) versus an unfamiliar (a genotype it hasn't been passaged through) MHC genotype. On average (based on least-squares means shown above), viral fitness is 114% (x = −0.79 F-MuLV/GAPDH ratio versus x = −1.56 F-MuLV/GAPDH ratio) and 149% (x = 9.41 FFU/spleen vs. x = 8.50 FFU/spleen) higher in familiar vs. unfamiliar host genotypes based on proviral load and infectious particles, respectively. Virulence is 28% (x = 1.10 g spleen vs. x = 0.86 g spleen) higher in familiar vs. unfamiliar host genotypes.
Table 1.
Susceptibility is equivalent among host genotypes
| Effect | Mean ± SE | Test |
| Infectious virus particles | ANOVA: F2,33 = 0.07, P = 0.93 | |
| BALB/cdd (n = 12) | 115.33 FFU/spleen ± 52.76 | |
| BALB/ckk (n = 12) | 149 FFU/spleen ± 105.42 | |
| BALB/cbb (n = 12) | 152.8 FFU/spleen ± 61.24 | |
| Proviral load | ANOVA: F2,30 = 0.94, P = 0.40 | |
| BALB/cdd (n = 11) | 0.01 F-MuLV/GAPDH copy no. ± 0.01 | |
| BALB/ckk (n = 10) | 0.01 F-MuLV/GAPDH copy no. ± 0.01 | |
| BALB/cbb (n = 12) | 0.004 F-MuLV/GAPDH copy no. ± 0.009 | |
| Splenomegaly | ANOVA: F2,33 = 0.05, P = 0.95 | |
| BALB/cdd (n = 12) | 0.17 g ± 0.02 | |
| BALB/ckk (n = 12) | 0.16 g ± 0.02 | |
| BALB/cbb (n = 12) | 0.18 g ± 0.02 |
Comparisons of unpassaged virus fitness and virulence measures between groups of infected animals from each of the three host genotypes. There are no significant differences in measures of pathogen fitness or virulence between these host MHC genotypes. Based on these results, we conclude that there are no underlying susceptibility differences between these MHC genotypes to infection with our unpassaged virus stock.
When comparing the severity of disease (as measured by splenomegaly) associated with infection by passaged pathogens, disease virulence was observed to be significantly higher in familiar versus unfamiliar host genotypes in five of six total comparisons (ANOVA, F1,10 = 4.01, P = 0.0001) (Fig. 4 and Table S4). Again, this result was not because of underlying differences in susceptibility among host genotypes (Table 1).
Discussion
Antagonistic coevolution predicts fitness trade-offs associated with pathogen adaptation. Adaptations that benefit pathogen fitness in one host MHC genotype must be costly to pathogen fitness when infecting hosts carrying other MHC genotypes (i.e., antagonistic pleiotropy). Without such a trade-off antagonistic coevolution would not drive negative frequency-dependent selection as there would be no selective advantage for rare host genotypes. Results from our experiments demonstrate that pathogen adaptation is host MHC genotype-specific. We conclude that pathogen adaptation to the familiar host MHC genotype produces trade-offs in pathogen fitness by reducing the reproductive output of adapted viruses when infecting hosts carrying unfamiliar MHC genotypes. Although previous work has shown that interactions between host and pathogen genotypes are important for determining patterns of pathogen fitness and virulence associated with infection (30), our dataset is unique in that it provides direct experimental support for fitness trade-offs associated with a pathogen's adaptation to specific host MHC genotypes, thus confirming the first major assumption of the antagonistic coevolution model of MHC evolution.
To maintain negative frequency-dependent selection, patterns of pathogen adaptation must produce correlated patterns of virulence. Otherwise, host individuals carrying rare MHC alleles gain no selective advantage. In our experiments, passaged pathogens were most fit and most virulent when infecting a familiar host MHC versus an unfamiliar host MHC. Based on these data, we conclude that pathogen adaptation to specific MHC genotypes is costly to host fitness because enhanced exploitation by adapted pathogens results in more virulent disease in hosts with familiar MHC genotypes. This finding confirms the second major assumption of the antagonistic coevolution model of MHC evolution and provides the necessary selective advantage to hosts carrying rare or otherwise unfamiliar MHC genotypes: enhanced resistance to infectious disease.
The data presented here support antagonistic coevolution as a viable mechanism explaining the evolution and maintenance of MHC polymorphism in vertebrate populations. Theoretical models have predicted that continuous cycles of adaptation and counter adaptation between hosts and their multiple pathogens can maintain large numbers of MHC alleles in host populations (9). As a result of these coevolutionary phenomena, associations between MHC allelic diversity and susceptibility to infectious and autoimmune diseases are predicted to be a natural consequence of antagonistic coevolution; that is, each MHC allele will typically have a subset of diseases to which it is resistant and a subset to which it is susceptible (2). Importantly, the evolution and maintenance of these susceptibility alleles may in turn influence MHC evolution. For example, if a trade-off existed between the benefits of possessing more MHC alleles and the fitness costs associated with their cumulative susceptibilities, one would predict the evolution of an optimal degree of individual MHC diversity, which has been observed in some systems (31). Finally, previous models have also predicted that when pathogens adapt to a given MHC allele it would likely be driven to extinction, but these models only work in the absence of trade-offs (i.e., antagonistic pleiotropy) (6). Thus, the data presented here places the antagonistic coevolution model of MHC evolution on solid empirical footing because one of its most important criticisms is removed.
Antagonistic coevolution is not mutually exclusive of other mechanisms that can contribute to MHC diversity, such as heterozygote advantage and mating preferences (8). A future challenge will be to design experiments that measure the relative contribution of each of these mechanisms to the unprecedented genetic diversity of MHC genes. This challenge will be particularly difficult when attempting to discriminate between heterozygote advantage and antagonistic coevolution because both mechanisms accumulate their fitness consequences over the lifetime of infections experienced by a host.
There are two caveats that must be made anytime one uses MHC-congenic inbred strains (or any congenic strains). First, the construction of MHC-congenic strains causes 4–20 mb of the DNA regions flanking the MHC to also come from the donor strain. Second, there is evidence that some mutations occurring in inbred lines become fixed slowly over time and so this causes congenic strains to also differ at these new fixed mutations (32). Thus, it cannot be known with 100% certainty that all of the pathogen adaptation effects reported are a result exclusively of the classic MHC antigen-presenting genes, as is often assumed. However, given the central role MHC genes play in immune-mediated selection on pathogens (18), the fact that each of these host genotypes possess completely different sets of MHC genes, and the consistency in our results, it seems likely that MHC genes are playing the major role in determining these observed patterns.
The results presented here have implications beyond MHC evolution, because they suggest that a host population with low MHC genetic diversity would select for more virulent pathogens. If this suggestion is true, many livestock breeds and endangered species that exhibit reduced genetic diversity may be particularly sensitive to the consequences of rapid pathogen adaptation. Additionally, the prophylactic use of antibiotics on domesticated livestock places a selective pressure on pathogen populations to evolve antibiotic resistance, and as a consequence has been implicated in the emergence of antibiotic-resistant strains of human pathogens (33). Reintroduction of critical genetic diversity into livestock species through selective breeding could have the dual benefit of reducing the impact of pathogens on productivity as well as reducing our use of antibiotics. Similarly, 89% of attempts to reintroduce captive-bred endangered species fail (34). One contributing factor could be the evolution of more virulent diseases in founding populations with low diversity. Preservation of host genetic diversity critical to host immune defenses could help ameliorate this grim statistic, which has particular importance in an era where a quarter of all vertebrate species are predicted to be endangered by 2050 (34). Finally, emerging infectious diseases represent an increasing threat to animal as well as human health. This study suggests that it is important to consider what role genetic variation in host populations plays in limiting the evolution and spread of more virulent pathogens.
Materials and Methods
Host.
MHC-congenic BALB/c mice (BALB/cdd, BALB/ckk, and BALB/cbb; d, k, and b designate distinct MHC haplotypes) were purchased from Jackson Laboratories and bred under specific pathogen-free conditions at the University of Utah. Use of three MHC-congenic strains of BALB/c mice allowed us to experimentally isolate the influence of genetic variation within the MHC region on patterns of pathogen adaptation. All experimental animals were females between 2 and 6 mo of age. All animal use was in compliance with federal regulations and the guidelines set forth by the University of Utah's Institutional Animal Care and Use Committee (08-10017).
Pathogen.
A National Institutes of Health 3T3 cell line (3-6a) containing a biological clone of the Friend virus complex was kindly provided by Sandra Ruscetti (National Institute of Allergy and Infectious Diseases, Bethesda, MD). This biological clone was grown in tissue culture to produce a stock of unpassaged virus. This stock also served as the pathogen stock used to infect animals to obtain baseline fitness and virulence estimates during the test phase of this experiment. This cell line expresses the integrated genomes of two viruses that act synergistically to produce the disease associated with Friend virus complex infection; the replication-competent Friend murine leukemia virus (F-MuLV) and the defective spleen focus forming virus (SFFV). F-MuLV is a replication competent virus and SFFV must coinfect the same cell as F-MuLV in order for its genome to be packaged into functional viral particles using F-MuLV structural proteins. As SFFV replication is dependent upon F-MuLV, we rationalized that it was sufficient to limit our analyses to measures of F-MuLV fitness. However, to test whether measures of F-MuLV fitness adequately reflect responses in the SFFV population we compared F-MuLV with SFFV proviral loads from the same individuals in a random subset of our test animals and found a highly significant and positive association between F-MuLV and SFFV fitness (Fig. S2) (R2 = 0.72, F1,64 = 169.96, P < 0.0001). Therefore, we conclude that measures of adaptive responses based on F-MuLV fitness values are also representative for SFFV.
Infections and Passages.
The spleen is a primary site of replication for this virus (28). Virus was serially passaged via intraperitoneal injection of spleen supernatants harvested from infected animals from the previous round of infection. Briefly, animals were killed and their spleens were removed and mechanically homogenized in an equivalent wt/vol ratio of 1× PBS (0.1 g = 100 μL diluents). Spleens were diluted in this manner as an approximate means of standardizing dose size between rounds of infection. During the final test phase however, pathogen stocks were titered using an infectious particle assay and dosage was standardized across experimental groups of animals with all test animals receiving approximately 1,000 focus forming units (FFU) (i.e., infectious particles). Spleen homogenates were transferred to 1.5-mL Eppendorf tubes and centrifuged for 5 min at 10,600 × g to fractionate homogenized tissue and supernatant. Spleen supernatant was decanted and stored at −70 °C until use. To initiate the next round of infection, animals were infected with 200 μL of thawed spleen supernatant. To avoid reductions in infectious titer because of repeated freeze-thaw cycles, spleen supernatants were only thawed once. The duration of a single passage round was 12 d with some animals being killed earlier if it was determined by visual inspection that they were not likely to survive to day 12 postinfection (all infections lasted between 10 and 12 d). Two animals were infected per round of infection and their spleen supernatants pooled to both increase the likelihood of successful passage and increase volumes of passage stocks. All passage lines had 10 rounds of serial passage through their respective hosts. Five of six passage lines were started from a stock that had been passaged in BALB/cdd animals for two rounds, which was done to overcome the following technical issue. The biological clone virus used to create this stock was derived from tissue culture and was therefore likely to be adapted to replicating within an in vitro environment. Therefore, selection pressures during the initial rounds of serial passage could be focused on readapting the virus to be an efficient “mouse-replicator” instead of an efficient replicator within specific mouse genotypes. This finding could have the effect of decreasing signals of genotype-specific adaptation at the test phase of our experiments. We rationalized that two initial rounds of passage through mice before divergence of passage lines would minimize this effect. We began one BALB/cdd passage line directly from unpassaged virus, which allowed us to test if two extra rounds of serial passage in BALB/cdd animals significantly impacted the magnitude of the adaptive response. There is no significant difference in proviral loads (ANOVA, F1,14 = 0.039, P = 0.85), infectious virus particle counts (ANOVA, F1,16 = 0.014, P = 0.0.91), or virulence (ANOVA, F1,14 = 1.55, P = 0.23) between groups of BALB/cdd animals infected with their 10- vs. 12-round postpassage stocks. Moreover, effects of virus genotype were statistically controlled for during the analysis of genotype-specific pathogen adaptation, which removes any potential biases in our data because of these additional two rounds of passage.
Virulence Measures.
Erythroblasts (stem-cell progenitors of erythrocytes) are the primary target of F-MuLV and SFFV. Under normal circumstances, these cells migrate from the bone marrow to the spleen, and secondarily the liver, where terminal differentiation into mature erythrocytes occurs in response to appropriate environmental signals. During infection the defective SFFV expresses a fusion envelope protein (gp55) that constitutively activates the host erythropoietin cell-surface receptor. The growth hormone erythropoietin is the natural ligand for this receptor, which under normal circumstances instructs the cell to clonally proliferate. Constitutive activation by the viral gp55 causes uncontrolled cellular proliferation of SFFV-infected cells (29), which leads to gross enlargement of both the spleen and liver (28). As the spleen is the primary site of viral replication, we measured spleen weight to estimate disease virulence.
Pathogen Fitness Measures.
To estimate pathogen fitness, a quantitative PCR (qPCR) assay was developed to measure the number of integrated retroviral genomes (proviruses) in DNA isolated from the spleens of infected individuals. Additionally, an indirect immunofluorescence assay was also used to measure the number of infectious virus particles in infected spleen supernatants.
Proviral load.
A qPCR assay was developed to measure the number of integrated retroviral genomes in the DNA of infected animals. Briefly, DNA was extracted from 100 μL of homogenized spleen supernatant thawed once using the DNeasy extraction kit (Qiagen). The presence of high quality DNA was confirmed by spectrophotometric analysis (Nanodrop Technologies). DNA extracts were stored at −4 °C in their elution buffer until use in reactions. Immediately before reaction setup, DNA concentrations of test animals were standardized by diluting all samples to ∼20 ng/uL in HPLC water. Reactions were optimized at 10 μL final reaction volumes. Assays were performed using the Roche Lightcycler 2.0 system in conjunction with the Lightcycler DNA SYBR Green Reaction Kit (Roche). All qPCR experiments were carried out in four sequential steps: a 10-min denaturation step, 45 PCR cycles (10 s at 95 °C, 3 s at 65 °C, 3 s at 72 °C), a melt analysis measuring fluorescence intensity every 0.1 °C increase from 65 °C to 95 °C, and a final cooling step down to 40 °C. Final primer concentrations used in reactions was 0.5 μM.
F-MuLV primer sets (forward: GGGGACTCTGCATAGGAACA, reverse: GATTAAGCACGGTGGCAGAT) [designed from the sequenced FV57 F-MuLV genome (National Center for Biotechnology Information Accession no. X02794)] and SFFV primer sets (Kindly provided by Kim Hasenkrug, National Institute of Allergy and Infectious Diseases, Hamilton, MT) were used to measure proviral loads of the respective viruses. Both primer sets were confirmed to be specific for their respective viral genomes (Fig. S3) and were also confirmed to amplify infectious virus administered to animals and not related endogenous retroviral sequences that exist naturally in multiple copies within the genomes of mice (Fig. S4). Amplicon lengths for F-MuLV and SFFV PCR products are 159 and 109 bp, respectively.
Multiple controls were used to minimize assay variability. First, to control for concentration differences between samples in a given experiment, virus measures were normalized to the reference gene GAPDH (GAPD) [forward: CTGGAGAAACCTGCCAAGTA, reverse: TGTTGCTGTAGCCGTATTCA (RealTimePrimers)]. Thus, all proviral load measures reported are virus/GAPDH ratio values. The GAPDH amplicon is 223 bp in length. Second, a single DNA sample was designated as our “calibrator” and was included in every qPCR experiment. Each calculated virus/GAPDH ratio value was also normalized to the virus/GAPDH ratio of the calibrator sample. This control minimizes the effect of inconsistent PCR efficiencies between experiments. Finally, to control for differences in PCR efficiencies between our primer sets, we constructed standard curves for each. These curves are used to define the amplification efficiency of each primer set (F-MuLV primer set = 2.05, SFFV primer set = 1.98, GAPDH primer set = 1.86). Quantitative PCR measures are subsequently adjusted based on these values. Raw F-MuLV/GAPDH ratio data were adjusted by spleen weight before analysis (ratio × spleen weight = total provirus/spleen).
Infectious particle assay.
During tissue collection, small (<50 μL) aliquots of infected spleen supernatants were frozen at −70 °C for later use in infectious particle assays. Serial dilutions of spleen supernatants in 1XPBS (1/10, 1/30, 1/90) were used to inoculate wells of a TC24 plate containing 4 × 103 Mus dunni cells. Four days postinoculation, cell monolayers were fixed in 95% ethanol for 5 min and then washed twice with TNE buffer. Fixed cells were then overlaid with 150 μL of a 1/500 dilution of virus-specific antibody overnight (anti-RLV gp70) (provided by Sandra Ruscetti, National Institute of Allergy and Infectious Diseases, Bethesda, MD). After 24 h, cells were washed twice with TNE buffer and then overlaid with a horseradish peroxidase-conjugated secondary antibody for 3 h (Millipore; cat. #AP-186P). To provide a substrate for bound peroxidase to act on, cells were washed twice in TNE buffer and then treated with an AEC solution (Millipore). Infected cells appear pink and are readily quantified using a standard dissecting microscope. The lower limit of detection for this assay was 200 FFU/mL. As many infected animals had titers below this level (especially animals infected with unpassaged virus), we conservatively assigned these animals the highest possible titer (i.e., 200 FFU/mL). Raw infectious virus particle titers were adjusted by spleen weight before analysis (titer × spleen weight = total FFU/spleen).
Statistical Analyses.
All statistical analyses were carried out using the statistical package JMP Start Statistics version 9.0 (SAS), and data were graphically represented using Microsoft Excel and Microsoft Paint. Data were natural log-transformed when appropriate to meet assumptions of normality. A two-tailed one-way ANOVA was used as the statistical test for all pair-wise comparisons. To test for a significant main effect of “MHC familiarity” on patterns of pathogen adaptation and virulence, a linear model incorporating the main effects of host genotype and virus genotype, and the host genotype*virus genotype interaction effect was constructed. See Table S3 for a detailed description of the rationale used to identify the most appropriate linear model for our analyses. Statistics reported in the manuscript are based on analyses of pooled BALB/cbb, BALB/cdd, and BALB/ckk datasets, unless otherwise stated. Significant effects are conserved when replicate experiments are analyzed independently (see relevant SI Tables).
Supplementary Material
Acknowledgments
We thank Dr. Sandra Ruscetti and Dr. Kim Hasenkrug for their extremely useful insights on working with Friend virus complex; Drs. Ruscetti and Hasenkrug were also instrumental in the development of tissue culture methods and techniques for assaying viral fitness by providing cell lines, antibodies, and protocols. We also thank Dr. Fred Adler for his useful comments regarding the manuscript, experimental design, and statistical analyses; Linda Morrison for her assistance in the design and optimization of assays; and two anonymous reviewers for their helpful insights. This work was supported by National Science Foundation Grant DEB 0918969 (to W.K.P. and Fred Adler); National Science Foundation Doctoral Dissertation Improvement Grant DEB 0910052 (to J.K.); National Institutes of Health Training Grant T32AI055434 (to J.K.); and National Science Foundation Educational Outreach Grant DGE 08-41233 (to J.K. and J.S.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112633109/-/DCSupplemental.
References
- 1.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2011;39(Database issue):D1171–D1176. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. The nature of selection on the major histocompatibility complex. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 3.Penn D, Potts W. The evolution of mating preferences and major histocompatibility genes. Am Nat. 1999;153(2):145–164. doi: 10.1086/303166. [DOI] [PubMed] [Google Scholar]
- 4.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annu Rev Ecol Evol Syst. 2006;37:159–186. [Google Scholar]
- 6.Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Boer RJ, Borghans JAM, van Boven M, Keşmir C, Weissing FJ. Heterozygote advantage fails to explain the high degree of polymorphism of the MHC. Immunogenetics. 2004;55:725–731. doi: 10.1007/s00251-003-0629-y. [DOI] [PubMed] [Google Scholar]
- 8.Piertney SB, Oliver MK. The evolutionary ecology of the major histocompatibility complex. Heredity (Edinb) 2006;96:7–21. doi: 10.1038/sj.hdy.6800724. [DOI] [PubMed] [Google Scholar]
- 9.Borghans JA, Beltman JB, De Boer RJ. MHC polymorphism under host-pathogen coevolution. Immunogenetics. 2004;55:732–739. doi: 10.1007/s00251-003-0630-5. [DOI] [PubMed] [Google Scholar]
- 10.Haldane JBS. Disease and evolution. Ric Sci. 1949;19(Suppl A):68–75. [Google Scholar]
- 11.Potts WK, Slev PR. Pathogen-based models favoring MHC genetic diversity. Immunol Rev. 1995;143:181–197. doi: 10.1111/j.1600-065x.1995.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 12.Ayala FJ, Campbell CA. Frequency-dependent selection. Annu Rev Ecol Syst. 1974;5:115–138. [Google Scholar]
- 13.McClelland EE, Penn DJ, Potts WK. Major histocompatibility complex heterozygote superiority during coinfection. Infect Immun. 2003;71:2079–2086. doi: 10.1128/IAI.71.4.2079-2086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebert D, Hamilton WD. Sex against virulence: The coevolution of parasitic diseases. Trends Ecol Evol. 1996;11:79–82. doi: 10.1016/0169-5347(96)81047-0. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton WD, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proc Natl Acad Sci USA. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy KP, Travers P, Janeway C, Walport M. Immunobiology. NY: Garland Science; 2008. [Google Scholar]
- 17.Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2:967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 18.McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 19.Furukawa Y, Kubota R, Tara M, Izumo S, Osame M. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood. 2001;97:987–993. doi: 10.1182/blood.v97.4.987. [DOI] [PubMed] [Google Scholar]
- 20.Erickson AL, et al. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 2001;15:883–895. doi: 10.1016/s1074-7613(01)00245-x. [DOI] [PubMed] [Google Scholar]
- 21.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berenos C, Wegner KM, Schmid-Hempel P. Antagonistic coevolution with parasites maintains host genetic diversity: An experimental test. Proc Biol Sci. 2011;278:218–224. doi: 10.1098/rspb.2010.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClelland EE, Adler FR, Granger DL, Potts WK. Major histocompatibility complex controls the trajectory but not host-specific adaptation during virulence evolution of the pathogenic fungus Cryptococcus neoformans. Proc Biol Sci. 2004;271:1557–1564. doi: 10.1098/rspb.2004.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedrich TC, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med. 2004;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 25.Friend C. Cell-free transmission in adult Swiss mice of a disease having the character of a leukemia. J Exp Med. 1957;105:307–318. doi: 10.1084/jem.105.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert D. Experimental evolution of parasites. Science. 1998;282:1432–1435. doi: 10.1126/science.282.5393.1432. [DOI] [PubMed] [Google Scholar]
- 27.Slev PR, Potts WK. Disease consequences of pathogen adaptation. Curr Opin Immunol. 2002;14:609–614. doi: 10.1016/s0952-7915(02)00381-3. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf D, Furth J, Buffett RF. Pathogenesis of mouse leukemia caused by Friend virus. Cancer Res. 1959;19:52–58. [PubMed] [Google Scholar]
- 29.Li JP, D'Andrea AD, Lodish HF, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 30.Grech K, Watt K, Read AF. Host-parasite interactions for virulence and resistance in a malaria model system. J Evol Biol. 2006;19:1620–1630. doi: 10.1111/j.1420-9101.2006.01116.x. [DOI] [PubMed] [Google Scholar]
- 31.Wegner KM, Kalbe M, Kurtz J, Reusch TB, Milinski M. Parasite selection for immunogenetic optimality. Science. 2003;301:1343. doi: 10.1126/science.1088293. [DOI] [PubMed] [Google Scholar]
- 32.McClelland EE, et al. Infection-dependent phenotypes in MHC-congenic mice are not due to MHC: Can we trust congenic animals? BMC Immunol. 2004;5:14. doi: 10.1186/1471-2172-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angulo FJ, Nargund VN, Chiller TC. Evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J Vet Med B Infect Dis Vet Public Health. 2004;51:374–379. doi: 10.1111/j.1439-0450.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 34.Frankham R, Ballou JD, Briscoe DA. Conservation Genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.