Abstract
The structural eye lens protein γD-crystallin is a major component of cataracts, but its conformation when aggregated is unknown. Using expressed protein ligation, we uniformly 13C labeled one of the two Greek key domains so that they are individually resolved in two-dimensional (2D) IR spectra for structural and kinetic analysis. Upon acid-induced amyloid fibril formation, the 2D IR spectra reveal that the C-terminal domain forms amyloid β-sheets, whereas the N-terminal domain becomes extremely disordered but lies in close proximity to the β-sheets. Two-dimensional IR kinetics experiments show that fibril nucleation and extension occur exclusively in the C-terminal domain. These results are unexpected because the N-terminal domain is less stable in the monomer form. Isotope dilution experiments reveal that each C-terminal domain contributes two or fewer adjacent β-strands to each β-sheet. From these observations, we propose an initial structural model for γD-crystallin amyloid fibrils. Because only 1 μg of protein is required for a 2D IR spectrum, even poorly expressing proteins can be studied under many conditions using this approach. Thus, we believe that 2D IR and protein ligation will be useful for structural and kinetic studies of many protein systems for which IR spectroscopy can be straightforwardly applied, such as membrane and amyloidogenic proteins.
Cataracts are a protein misfolding disease caused by the aggregation of lens crystallin proteins into insoluble deposits that blur vision (1, 2). Because these proteins are not regenerated, damage from UV radiation, oxidative stress, and other chemical modifications accumulates with time (1, 2). As a result, over 50% of the population over 55 develops age-related cataracts (2). Additionally, numerous mutations that destabilize crystallin protein folds are linked to inherited and juvenile-onset cataracts (1). Although the causative factors associated with this disease are known, the structures of the aggregates and the mechanisms by which they form are unknown.
Like other protein aggregation diseases such as type II diabetes mellitus and Alzheimer’s disease, the molecular structures of proteins in cataracts are difficult to determine. Atomic-level structures have been obtained for some amyloid aggregates of peptides using NMR spectroscopy (3, 4) and X-ray crystallography (5). However, the most widely used techniques for studying aggregate structures and aggregation mechanisms are circular dichroism spectroscopy, fluorescence spectroscopy, and transmission electron microscopy, which provide little detailed structural information. Two-dimensional (2D) IR spectroscopy is emerging as an important tool for studying protein aggregates such as amyloid fibrils (6–8) because it provides bond-by-bond structural resolution on kinetically evolving samples (6, 8–10). Two-dimensional IR spectroscopy probes secondary structure through cross peak couplings and solvent exposure through 2D lineshapes. Its bond-specific structural resolution comes from isotope labeling. Mechanistic information is obtained from rapid-scan methods that track the kinetics of aggregate formation (6). Spectra can be calculated from molecular dynamics simulations allowing structural models to be tested (11–15). Isotope-edited 2D IR spectroscopy using 13C = 18O labeled backbone amides has been used to study the fibril structure and mechanism of fibril formation for amyloid beta (Aβ) (8) and the human islet amyloid polypeptide (hIAPP) (6, 7), as well as many membrane-bound (13, 16) and soluble polypeptides (9, 10).
Thus far, isotope-edited infrared spectroscopy has been limited to peptides that can be made by solid phase synthesis, which is how the 13C = 18O labels are incorporated (17). However, solid phase synthesis is limited to peptides of about 45–70 residues. Even if longer sequences could be synthesized, a single 13C = 18O label would eventually be obscured by Glu, Asp, and Arg side chains, as well as natural abundance 13C = 16O bonds, all of which absorb in the same frequency range (18). Nonnatural amino acids and deuterium labels are well-resolved in protein IR spectra, but only report on local environment not structure (19–21). In this article, we describe an alternative approach in which we isotope label an entire section of a protein rather than a single C═O bond. To do so, we express labeled and unlabeled segments of the protein separately, and then reconstitute the full-length protein using expressed protein ligation (22), which is a variant of native chemical ligation (23). We show that the unlabeled and 13C labeled regions are spectroscopically distinguishable, enabling the acquisition of structural and kinetic information on large proteins. We apply our technique to study amyloid fibril formation in the 173 amino acid eye lens protein γD-crystallin and learn that only one of its two structurally homologous domains forms the β-sheet core of the fibrils.
γD-crystallin is a member of the γ-crystallin family of monomeric, soluble structural eye lens proteins which constitute about 20% of the total lens proteins and are a major component of insoluble cataract deposits (24). Native γD-crystallin consists of two Greek key domains that are each 80–90 residues long, connected by a short loop (Fig. 1) (25). Numerous modes of aggregation have been proposed for the two-domain structure, including amorphous precipitation, domain-swap oligomerization, and amyloid fibril formation (1, 26, 27). In vitro, fibrillar deposits of γ-crystallins are created by UV damage, low pH, and chemical and thermal denaturation, although it is unclear to what extent each form (or others) contribute to in vivo cataracts (1, 27–29). We utilize acid initiated aggregation because it reproducibly generates fibrillar aggregates of γ-crystallins and because low pH conditions encountered during lens development may be relevant to juvenile cataract formation (27). γD-crystallin, and each of its isolated domains, forms amyloid fibrils in vitro at pH 3 and 37 °C (27). Although there is no domain-specific structural information in the fibril state, fibrils formed from the isolated N-terminal domain (NTD) more closely resemble fibrils from full-length γD-crystallin than do fibrils from the C-terminal domain (CTD), as characterized by FTIR spectroscopy and transmission electron microscopy (TEM) imaging (27). Moreover, the NTD is less stable (30, 31) and has partially unfolded states at pH 7. Although the relationship between domain stability, partially unfolded states, and aggregation propensity is unclear (32), these results have led to the suggestion that the NTD is most likely to form the β-sheets of the amyloid fibril core (33). We show that it is actually the CTD of full-length human γD-crystallin that forms the β-sheets of the amyloid fibril core in low pH aggregates.
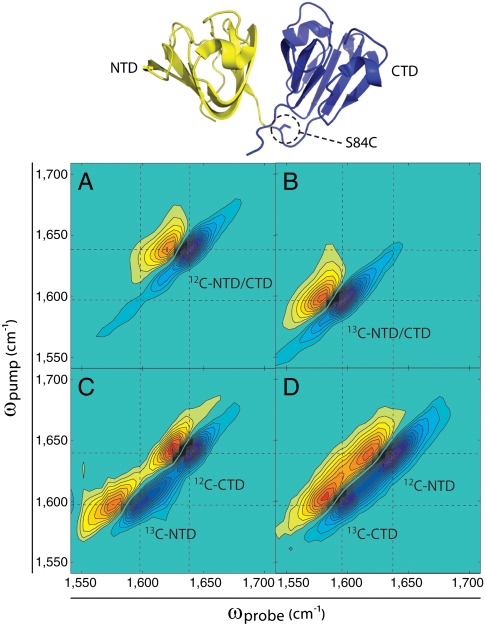
Fig. 1.
Two-dimensional IR spectroscopy of native human γD-crystallin. (Top) Crystal structure of human γD-crystallin indicating the division of domains in the segmental labeling scheme. (Bottom, A–D) Two-dimensional IR spectra of full-length γD-crystallin at pD = 7. Frequencies and FWHM node slopes are summarized in Table S1.
Results
Human γD-crystallin and its individual domains were expressed in Escherichia coli grown in either unlabeled or 13C-enriched media. Segmentally labeled samples were prepared from separately expressed NTD (residues 1–83) and CTD (84–173) using expressed protein ligation to reconstitute the full-length protein (Fig. S1A). The resulting protein contains a residual N-terminal Met and a conservative S84C substitution at the point of ligation. No change in fold stability relative to the wild type was observed in chemical denaturation experiments (Fig. S1B) and the mutations are not expected to alter the protein’s aggregation properties under mildly acidic conditions.
Two-dimensional IR spectra of natively folded γD-crystallin at pD = 7 with four different isotope labeling schemes are shown in Fig. 1 A–D. The spectra span the region of the amide I band, which is largely caused by backbone carbonyl stretch motions that correlate to protein secondary structures. In the spectrum of unlabeled γD-crystallin (Fig. 1A), the most intense features are a pair of positive and negative peaks at ωpump = 1,637 cm-1. The peaks occur in pairs because the 2D IR pulse sequence probes both the fundamental transition (ν = 0 → 1) and its sequence band (ν = 1 → 2), which give rise to the negative (green-blue) and positive (yellow-orange) peaks, respectively. We assign these features to the antiparallel β-sheets of the Greek keys, because they make up the bulk of the overall protein fold and the assignment is consistent with the observed frequency. The amide I intensity extends to 1,675 cm-1 because the eigenstates of β-sheets cover a wide frequency range, and because other secondary structures contribute to the high frequency side of the spectrum (15, 18). Intensity below 1,610 cm-1 is largely due to side chain absorbances and the presence of 13C at its natural abundance of 1.1% (18). More detailed descriptions of the structural origins of the spectra could be obtained by calculating 2D IR spectra from molecular dynamics simulations of the Protein Data Bank structure (11–15), but because the folds of the Greek keys are so similar, one domain cannot be distinguished from the other.
Fig. 1B shows the 2D IR spectrum of uniformly 13C-labeled γD-crystallin. This spectrum is nearly identical to that of the unlabeled protein except that it is shifted down in frequency by 39 cm-1, consistent with the change in effective mass upon 13C substitution of single amide bond. Overlays of diagonal slices taken from the spectra illustrate this similarity (Fig. S2A). Because 2D IR frequencies and lineshapes are strongly influenced by coupling between amide I modes within secondary structure elements, this similarity indicates that coupling in the unlabeled sample is preserved upon uniform 13C labeling. It also confirms that the amide I band is well-represented by local modes that are largely decoupled from other vibrational modes within the protein, thereby validating the exciton description that is usually used to model the amide I band in calculations (11–15). Most importantly for the purposes of this paper, the 2D IR spectra of 12C and 13C proteins are interchangeable via displacement along the diagonal, allowing quantitative comparisons between the spectra of labeled and unlabeled proteins.
The Greek key features in Fig. 1 A and B are elongated along the diagonal due to the inhomogeneous distribution of vibrational frequencies within the sample (18), which is related to the structural distribution of the protein. This distribution may be quantified by measuring the slope of the nodal line between the fundamental and overtone signals (34). Perfectly homogeneous lineshapes would result in vertical nodal lines, whereas perfectly inhomogeneous lineshapes would result in nodal line slopes of 1, parallel with the diagonal. Because most 2D IR lineshapes derived from condensed-phase samples have both homogeneous and inhomogeneous components the node slope is a convenient method for determining their relative contributions. The FWHM node slopes of the main amide I bands in unlabeled and labeled samples are 1.30 and 1.25, respectively (see Table S1 for a summary of all frequencies and node slopes). These values reflect the mixture of homogeneous and inhomogeneous contributions associated with the structural disorder of the natively folded protein and quantify the high degree of similarity in the unlabeled and labeled 2D IR lineshapes.
To resolve the individual domains of γD-crystallin, we use segmental uniform 13C labeling. Two-dimensional IR spectra of native N-terminally and C-terminally labeled full-length γD-crystallin are shown in Fig. 1 C and D. In each of these spectra, two pairs of peaks are observed along the diagonal: one at ωpump ≈ 1,638 cm-1 arising from the unlabeled portion of the protein and a second at ωpump ≈ 1,596 cm-1 arising from the labeled portion. The frequency separation is not exactly 39 cm-1, because the NTD features are more elongated than those of the CTD. Node slopes indicate that the NTD has a larger amount of structural disorder than the CTD (Table S1). This result is consistent with previous fluorescence studies in which the NTD was shown to be less kinetically and thermodynamically stable than the CTD under these conditions (30, 31).
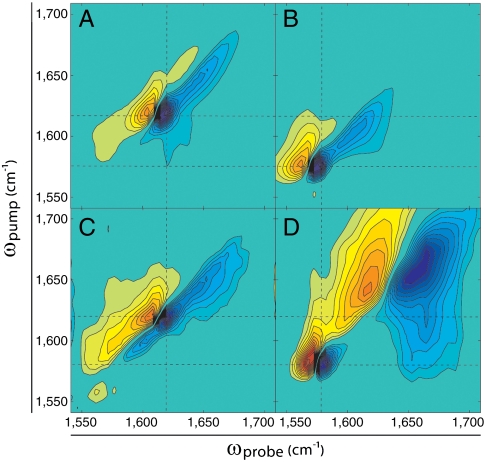
Incubation of γD-crystallin at pD = 3 and 37 °C causes the formation of amyloid fibrils (27). Two-dimensional IR spectra of fibrillar samples are shown in Fig. 2, with the same four isotope labeling schemes as Fig. 1 A–D. The spectra of the fibrils are markedly different from those of the corresponding natively folded proteins, and are dominated by two distinct peak pairs along the diagonal with very different lineshapes. In the spectrum of unlabeled γD-crystallin aggregates (Fig. 2A), a narrow peak pair appears at 1,617 cm-1 and an extremely elongated pair at 1,645 cm-1 that spans from 1,630 to 1,685 cm-1. The 1,617 cm-1 peak pair is a clear signature of amyloid fibrils as recognized by the frequency, anharmonic shift, and narrow linewidth associated with an extended β-sheet architecture (7, 8, 15). A similar peak pair appears in fibril spectra of Aβ (8) and hIAPP (6, 7). The elongated peak pair at 1,645 cm-1, on the other hand, is due to disordered structures that result in an extremely inhomogeneous lineshape and a blue shift relative to the Greek key features of the native protein. The fibril spectrum of labeled γD-crystallin, shown in Fig. 2B, is very similar to that of the unlabeled fibrils, but is once again shifted down in frequency so that the amyloid peak pair appears at 1,575 cm-1 and the disordered peak appears at 1,605 cm-1. Diagonal slices through Fig. 2 A and B are overlaid for comparison in Fig. S2B. What is striking about the data is the difference between the aggregated (Fig. 2) and natively folded (Fig. 1) spectra. The node slopes of the unlabeled aggregate sample are 1.61 and 1.11 for the 1,617 cm-1 and 1,645 cm-1 peaks, respectively (Table S1). Clearly, the amyloidogenic pathway of γD-crystallin aggregation involves a massive structural rearrangement in which a large portion of the protein forms highly ordered extended β-sheets whereas another region becomes extremely disordered with no recognizable secondary structure.
Fig. 2.
(A–D) Two-dimensional IR spectra of aggregated full-length γD-crystallin at pD = 3. Isotope labeling schemes are identical to those described in Fig. 1. Frequencies and FWHM node slopes are summarized in Table S1.
Segmental 13C labeling allows one to assign structures to the individual domains. The 2D IR spectra of N-terminally labeled and C-terminally labeled proteins in the fibril state, shown in Fig. 2 C and D respectively, still contain narrow amyloid and broad disordered peak pairs, but their frequencies depend on the labeling scheme. In the N-terminally labeled sample, the amyloid peak pair appears at ωpump = 1,617 cm-1, identical to that in the unlabeled sample (Fig. 2A). Thus, the unlabeled CTD contributes to the amyloid β-sheets. However, because the disordered peak reaches to 1,675 cm-1, a portion of the CTD must be disordered because that frequency is too high to be reached by isotope labeled residues. In addition, no narrow amyloid signal is visible at 1,575 cm-1, indicating that the NTD does not contribute to the β-sheets of the fibril core.
To better resolve other features of the NTD, we turn to the spectrum of the C-terminally labeled sample (Fig. 2D). The CTD amyloid peak pair appears at ωpump = 1,575 cm-1, and signal from the NTD stretches from 1,620 to 1,700 cm-1 indicating that it is extremely disordered. We do not resolve any features indicative of secondary structure in the NTD, and conclude that it is completely structurally disordered in the fibril. The frequency range spanned in Fig. 2D appears larger than in Fig. 2C, because better spectral isolation of the features results in a greater fraction of the contour levels outlining the random coil features. Additionally, cross peaks are evident between the CTD and NTD, which appear as a narrow ridge at ωprobe ≈ 1,570 cm-1, and a strong cross peak below the diagonal at ωpump ≈ 1,570 cm-1. The cross peaks indicate that the domains are closely associated in the fibril even though they have very different conformations. Together, these results indicate that amyloid β-sheet-forming regions of γD-crystallin are localized exclusively in the CTD, and that the NTD assumes a highly disordered structure that lies in close proximity to the β-sheets of the fibril core.
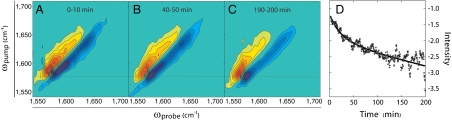
A major advantage of 2D IR spectroscopy over other structurally sensitive biophysical techniques is its ability to collect spectra on-the-fly, so that detailed information about protein structural kinetics can be obtained on timescales ranging from seconds to hours (6). Kinetics experiments provide information about aggregation mechanisms because they can identify transient structural intermediates that are not apparent in steady-state spectra. We monitored the kinetics of C-terminally labeled γD-crystallin fibril formation in situ over the course of 200 min. C-terminal labeling was chosen because the two domains are well-resolved in the fibril spectra. Fig. 3 A–C show early, middle, and late 2D IR spectra extracted from the kinetics data. At early times, the labeled β-sheet mode appears at about ωpump = 1,592 cm-1, and evolves toward the lower frequency until it reaches the amyloid frequency in the late spectrum. Because frequency provides a measure of the size and structural disorder of the β-sheets (7, 15), these spectra indicate that β-sheets increase in size and become more ordered as the fibrils form. The growth of negative fundamental intensity at 1,575 cm-1 as a function of time is plotted in Fig. 3D, with a double exponential fit that we assign to a fast fibril nucleation phase and a slow fibril growth phase, consistent with the observed frequency shifts. If a minor species is the cause of aggregation, then it must account for < 5% of the population, based on the resolution of the spectra. At 200 min, the fibril structure has not yet equilibrated to the 18-h spectrum in Fig. 2D, indicating that there are slow timescales to fibril structure formation, especially in the high frequency region associated with the unlabeled NTD. Slow fibril reorganization processes have been observed in other systems (35), so it is likely that structural rearrangements of the NTD occur on a timescale of hours. Based on these kinetics, we conclude that the CTD itself serves to nucleate aggregation and form the core of the fibrils accompanied by slower kinetic timescales associated with fibril annealing.
Fig. 3.
Aggregation kinetics of full-length γD-crystallin (12C-NTD/13C-CTD) at pD = 3 and 37 °C. (A) Early, (B) mid, and (C) late aggregate spectra averaged over 10 min intervals. (D) Growth of amyloid fibril signal at 1,575 cm-1 fit to a double exponential with time constants of 23 and 511 min. Dashed line marks the frequency that the CTD fibrillar β-sheet ultimately adopts (see Fig. 2D).
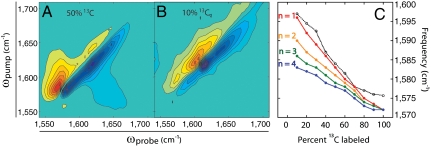
In amyloid fibrils formed by the approximately 40 residue peptides hIAPP (3) and Aβ (4), each polypeptide chain contributes one β-strand to each β-sheet. In longer amyloid-forming proteins, structures have been proposed in which each protein contributes several strands to the same β-sheet (4). Because the CTD of γD-crystallin is 90 residues long it could double back one or more times, contributing a strand each time. We have used isotope dilution to estimate the number of strands that each protein molecule contributes to the fibril β-sheets. It is well-established that the amide I frequency of β-sheets depends on the number of β-strands in the sheet (7, 15) because vibrational excitons become more delocalized as the size of the sheet is increased. Normally, the amide I vibrational modes delocalize across the β-sheets regardless of whether the strands come from the same protein or not. However, coupling is less effective between strands of different isotope content (7, 8) and delocalization is only significant across strands of the same isotopic composition. In a sample made from a large majority of 12C labeled protein and a small minority of 13C labeled protein, it is improbable for two 13C proteins to be sequentially stacked in the amyloid fibrils. As a result, the amide I vibrational modes will only be delocalized among 13C residues in the same protein and the amide I frequency of the 13C band reflects the number of β-strands that each individual protein contributes to the fibril. To implement this approach, we collected a series of 2D IR spectra for fibrils formed from varying ratios of 12C and uniformly labeled 13C proteins. The frequencies and cross peaks depend on the 13C/12C ratio, which reflects the delocalization of the amide I vibrational modes (see representative spectra in Fig. 4 A and B). For example, a large cross peak appears between the 12C and 13C amyloid peaks in the 50/50 mixture (Fig. 4A) because this ratio has the highest likelihood for adjacent labeled and unlabeled proteins. We focus on the frequency shift of the 13C labeled amyloid peaks. At 10% labeled protein incorporation, where labeled monomers are effectively isolated in the fibril, the labeled amyloid peak exists as a low frequency shoulder on the unlabeled amyloid peak (ωpump = 1,597 cm-1 in Fig. 4B). As the proportion of labeled protein increases, this peak grows in intensity and decreases in frequency until 100% of the proteins are labeled and the frequency matches that of the labeled spectrum (ωpump ≈ 1,575 cm-1 in Fig. 2B). The frequency trend is plotted in Fig. 4C, taken from diagonal slices of the spectra shown in Fig. S3.
Fig. 4.
Isotope dilution study of γD-crystallin amyloid fibrils. (A) Fifty percent labeled, (B) 10% labeled, (C) comparison of experimental labeled amyloid frequencies (dashed line) with simulated values (solid lines) for model fibrils with n consecutive strands per protein.
To interpret the experimental amide I frequencies, we modeled idealized β-sheets using an excitonic coupling Hamiltonian that is well-established for simulations of protein amide I vibrational modes (7, 15). Structural models and simulations are described in-depth in SI Materials and Methods. Briefly, parallel β-sheet structures containing 36 strands of 10 residues were generated, based on the average length of continuous strands in native γD-crystallin (7–8 residues) (25) and Aβ/hIAPP (10–12 residues) (3, 4, 11). Each strand was randomly assigned a frequency of either 1,604 or 1,644 cm-1, to represent either a labeled or unlabeled protein. Environmental disorder was simulated by randomly varying the local mode frequencies around the assigned values in a Gaussian distribution with an FWHM of 10 cm-1. The ratio of labeled to unlabeled strands was set to mimic the experimental fractions. The Hamiltonian was diagonalized and the infrared spectrum calculated from the eigenvectors and amide I transition dipoles from which the isotope labeled frequency was extracted. The simulation was repeated for multiple consecutive strands per protein by assigning frequencies to groups of strands rather than individually. Importantly, the structural models used in spectral simulations were identical aside from the position of 13C labels. The resulting simulated frequencies are plotted alongside the experimental data in Fig. 4C.
The modeling indicates that the β-sheet frequency at low ratios (10–30%) of labeled proteins depends sensitively on the number of consecutive labeled strands. The sensitivity is lost at higher ratios where the simulations all converge to the 100% labeled frequency. Low ratios are most sensitive because β-sheet frequencies scale with size, but ultimately reach an asymptote set by the coupling strength and the structural disorder of the protein and its environment (7, 15). A comparison of simulation to experiment reveals that the best agreement is for n = 1, indicating that each γD-crystallin CTD contributes one β-strand to each β-sheet of the amyloid fibril core. However, the data may also be consistent with n = 2, in which case each CTD contributes two strands, because our model does not include off-diagonal disorder in the Hamiltonian that arises from structural disorder in the fibril. More sophisticated calculations that include molecular dynamics simulations can incorporate the effects of disorder more realistically and therefore provide a better estimate of the frequencies of dilute labeled β-strands in a variety of configurations (11–15). Although the current level of analysis only provides an estimate of the number of consecutive strands, it allows us to rule out native-like structures (n = 4) and long β-solenoids and provides a guide for the design of more specific labeling schemes.
Discussion and Conclusions
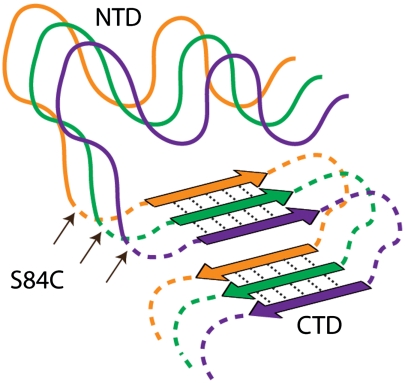
We have demonstrated that uniform, segmental 13C labeling of proteins, in combination with rapid-scan 2D IR spectroscopy, is a powerful tool for the analysis of protein structures and kinetics in samples that are difficult to study with traditional structural biology techniques. Using 2D IR spectroscopy of segmentally labeled human γD-crystallin, we showed that segmental labeling allows the spectral resolution of specific regions of sequence in large proteins. We found that 2D IR lineshapes are maintained upon efficient 13C labeling, and that small structural and dynamic differences in the domains of human γD-crystallin are reflected in their 2D IR node slopes. Through these methods, we evaluated the secondary structures of its individual domains in the fibrillar state, and learned that the in vitro acid-induced amyloid fibril formation process is dominated by contributions from the CTD at all stages of the mechanism. From isotope dilution experiments, we put a constraint on the number of β-strands that each γD-crystallin protein contributes to each β-sheet in the fibril state. One fibril model structure, which best accounts for our findings, is shown in Fig. 5. Here, much of the CTD forms extended parallel β-sheets in the fibril core, with each molecule contributing one β-strand to each β-sheet. A minimum of two stacked β-sheets is required to account for the approximately 10 Å reflection in X-ray diffraction data (27). Although the stacked β-sheets are modeled here as arising from a single column of protein molecules with two β-strands separated by loops, more complex fibril structures are also possible. For instance, more than two stacked β-sheets may exist in the fibril, and they may arise from multiple columns of protein molecules as has been observed in hIAPP (3) and Aβ (4) fibrils. Regions of the CTD and the entire NTD assume disordered states with no discernible secondary structure, but lie in close proximity to the β-sheets.
Fig. 5.
One possible structural model of three γD-crystallin monomers in an amyloid fibril, with n = 1. NTDs (solid lines) form random coil, whereas CTDs form a mixture of β-strands and coil-like loops (dashed lines). Additional models consistent with the current data are discussed in the text.
The results presented in this paper are unexpected in light of previous experiments that suggested that the NTD of γD-crystallin is more likely than the CTD to form the β-sheets of the fibril core. Aggregation experiments on truncated variants showed that both domains can form amyloid fibrils (27), and sequence analysis identified aggregation-prone regions in both domains (33). However, our results indicate that only the CTD forms β-sheets in the core of acid-induced fibrils. Thus, the covalent linkage between domains appears to play a critical role in determining the fibril structure. Differences in the aggregation behavior of the NTD may stem from steric constraints imposed by the attached CTD or specific interactions with the CTD that modulate the protein’s folding free energy landscape. The latter effect has been observed during refolding experiments performed at pH 7 (30), and our observation of closely associated β-sheets and coil in acid-induced fibrils suggests that it may also occur in misfolded states. Although our results clearly show that the CTD forms β-sheets in the fibril state and strongly suggest that it contains the fibril nucleation site, domain-level labeling does not resolve the specific structural transitions that result in aggregation. Recent reports have suggested that locally unfolded states may play a large role in aggregation (32), potentially resolving the discrepancy between previous truncation (27) and fold stability studies (30) and the results presented here. Because domain stabilities in lens crystallins are highly dependent on conditions (1), it is possible that a number of different aggregation-prone states may be accessed as conditions are varied, leading to different β-sheet architectures in the aggregates. Using the techniques described in this paper, these structural variations may be assessed across a wide range of physiologically relevant conditions in samples where all important structural elements of the protein are left intact.
The experiments described in this paper provide structural information about the individual domains of γD-crystallin because we placed the ligation site between them. Thus, our structural model does not contain information about the lengths of individual β-strands, the total number of β-sheets, or the topology of the loops and sheets in the fibril structure. However, it will be possible to obtain higher-resolution data on subdomain structures by choosing other locations for ligation. Labeled segments as small as 10% of the total sequence can be resolved in 2D IR spectra, as was demonstrated in isotope dilution experiments. Even in cases where ligation is inefficient, the methods described here are practical because of the small sample requirements for 2D IR spectroscopy. The experiments presented in this paper were performed at protein concentrations of 6 to 60 μM with sample volumes of < 2 μL, meaning that samples require less than 1 μg of protein. These concentrations and volumes are much smaller than are typically used in NMR experiments (4), and so a single synthesis provides enough sample to measure 2D IR spectra under many different conditions. We expect these methods to be particularly useful in identifying a variety of amyloid, native-like, and loop-sheet insertion aggregates (36) based on frequency shifts induced by vibrational coupling between labeled β-strands. One will also be able to identify α-helices and other secondary structural features using segmental labeling. Thus, we expect 2D IR spectroscopy with uniform segmental labeling to be useful in the study of difficult systems such as protein aggregates and membrane proteins.
Materials and Methods
Full-length γD-crystallin and each of its isolated N-terminal (residues 1–83) and C-terminal (84–173) domains were expressed in E. coli. Expressed protein ligation generated full-length proteins from N- and C-terminal domains. Samples were exchanged into deuterated buffer (20 mM sodium phosphate, 100 mM NaCl, pD = 7) at a protein concentration of 60 μM for analysis. Aggregation was initiated by shifting the pD to 3 using DCl followed by incubation at 37 °C for 18 h. Two-dimensional IR spectra were collected as described previously (17). Node slopes in 2D IR spectra were determined from linear fits to the points of zero intensity between fundamental and overtone peaks, across the diagonal FWHM of the fundamental peaks. Aggregation kinetics were monitored in situ using a water-jacketed sample cell for temperature control. Two-dimensional IR spectra for the isotope dilution study were obtained from mixtures of labeled and unlabeled γD-crystallin prepared prior to the initiation of aggregation. Experimental frequencies were determined by interpolation of ωpump maxima from diagonal slices (Fig. S3). Simulations of one-dimensional IR spectra for idealized β-sheets were performed according to methods described previously (7). The incorporation of labeled strands was simulated by shifting the local mode frequencies of randomly selected groups of 1, 2, 3, and 4 strands by 40 cm-1 in proportions reflecting the experimental ratios. Additional details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments.
Support for this research was provided by the National Science Foundation through a Collaborative Research in Chemistry (CRC) Grant 0832584.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117704109/-/DCSupplemental.
References
- 1.Wang Y, King JA. Protein Misfolding Diseases. New York: Wiley; 2010. Cataract as a protein-aggregation disease; pp. 487–515. [Google Scholar]
- 2.Bloemendal H, et al. Aging and vision: Structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Luca S, Yau W-M, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tycko R. Molecular structure of amyloid fibrils: Insights from solid-state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 5.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim S-H, et al. Two-dimensional IR spectroscopy and isotope labeling defines the pathway of amyloid formation with residue-specific resolution. Proc Natl Acad Sci USA. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strasfeld DB, Ling YL, Gupta R, Raleigh DP, Zanni MT. Strategies for extracting structural information from 2D IR spectroscopy of amyloid: Application to islet amyloid polypeptide. J Phys Chem B. 2009;113:15679–15691. doi: 10.1021/jp9072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Two-dimensional infrared spectra of isotopically diluted amyloid fibrils from Aβ40. Proc Natl Acad Sci USA. 2008;105:7720–7725. doi: 10.1073/pnas.0802993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith AW, Tokmakoff A. Probing local structural events in β-hairpin unfolding with transient nonlinear infrared spectroscopy. Angew Chem Int Ed. 2007;46:7984–7987. doi: 10.1002/anie.200701172. [DOI] [PubMed] [Google Scholar]
- 10.Kolano C, Helbing J, Kozinski M, Sander W, Hamm P. Watching hydrogen-bond dynamics in a β-turn by transient two-dimensional infrared spectroscopy. Nature. 2006;444:469–472. doi: 10.1038/nature05352. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, et al. 2D IR spectroscopy of human amylin fibrils reflects stable β-sheet structure. J Am Chem Soc. 2011;133:16062–16071. doi: 10.1021/ja204035k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang W, Abramavicius D, Voronine DV, Mukamel S. Simulation of two-dimensional infrared spectroscopy of amyloid fibrils. Proc Natl Acad Sci USA. 2007;104:14233–14236. doi: 10.1073/pnas.0700392104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woys AM, et al. 2D IR line shapes probe ovispirin peptide conformation and depth in lipid bilayers. J Am Chem Soc. 2010;132:2832–2838. doi: 10.1021/ja9101776. [DOI] [PubMed] [Google Scholar]
- 14.Dijkstra AG, Jansen TlC, Knoester J. Modeling the vibrational dynamics and nonlinear infrared spectra of coupled amide I and II modes in peptides. J Phys Chem B. 2011;115:5392–5401. doi: 10.1021/jp109431a. [DOI] [PubMed] [Google Scholar]
- 15.Hahn S, Kim S-S, Lee C, Cho M. Characteristic two-dimensional IR spectroscopic features of antiparallel and parallel β-sheet polypeptides: Simulation studies. J Chem Phys. 2005;123:084905. doi: 10.1063/1.1997151. [DOI] [PubMed] [Google Scholar]
- 16.Manor J, et al. Gating mechanism of the influenza A M2 channel revealed by 1D and 2D IR spectroscopies. Structure. 2009;17:247–254. doi: 10.1016/j.str.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middleton CT, Woys AM, Mukherjee SS, Zanni MT. Residue-specific structural kinetics of proteins through the union of isotope labeling, mid-IR pulse shaping, and coherent 2D IR spectroscopy. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamm P, Zanni M. Concepts and Methods of 2D Infrared Spectroscopy. New York: Cambridge University Press; 2011. [Google Scholar]
- 19.Naraharisetty SRG, et al. C−D modes of deuterated side chain of leucine as structural reporters via dual-frequency two-dimensional infrared spectroscopy. J Phys Chem B. 2009;113:4940–4946. doi: 10.1021/jp8112446. [DOI] [PubMed] [Google Scholar]
- 20.Thielges MC, et al. Two-dimensional IR spectroscopy of protein dynamics using two vibrational labels: A site-specific genetically encoded unnatural amino acid and an active site ligand. J Phys Chem B. 2011;115:11294–11304. doi: 10.1021/jp206986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Getahun Z, et al. Using nitrile-derivatized amino acids as infrared probes of local environment. J Am Chem Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 22.Flavell RR, Muir TW. Expressed protein ligation (EPL) in the study of signal transduction, ion conduction, and chromatin biology. Acc Chem Res. 2009;42:107–116. doi: 10.1021/ar800129c. [DOI] [PubMed] [Google Scholar]
- 23.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 24.Su S, et al. Proteomic analysis of human age-related nuclear cataracts and normal lens nuclei. Invest Ophthalmol Visual Sci. 2011;52:4182–4191. doi: 10.1167/iovs.10-7094. [DOI] [PubMed] [Google Scholar]
- 25.Basak A, et al. High-resolution X-ray crystal structures of human γD-crystallin (1.25 Å) and the R58H mutant (1.15 Å) associated with aculeiform cataract. J Mol Biol. 2003;328:1137–1147. doi: 10.1016/s0022-2836(03)00375-9. [DOI] [PubMed] [Google Scholar]
- 26.Das P, King JA, Zhou R. Aggregation of γ-crystallins associated with human cataracts via domain swapping at the C-terminal β-strands. Proc Natl Acad Sci USA. 2011;108:10514–10519. doi: 10.1073/pnas.1019152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papanikolopoulou K, et al. Formation of amyloid fibrils in vitro by human γD-crystallin and its isolated domains. Mol Vision. 2008;14:81–89. [PMC free article] [PubMed] [Google Scholar]
- 28.Fatima U, Sharma S, Guptasarma P. Structures of differently aggregated and precipitated forms of γB crystallin: An FTIR spectroscopic and EM study. Protein Pept Lett. 2010;17:1155–1162. doi: 10.2174/092986610791760414. [DOI] [PubMed] [Google Scholar]
- 29.Ecroyd H, Carver JA. Crystallin proteins and amyloid fibrils. Cell Mol Life Sci. 2009;66:62–81. doi: 10.1007/s00018-008-8327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flaugh SL, Kosinski-Collins MS, King J. Interdomain side-chain interactions in human γD crystallin influencing folding and stability. Protein Sci. 2005;14:2030–2043. doi: 10.1110/ps.051460505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosinski-Collins MS, Flaugh SL, King J. Probing folding and fluorescence quenching in human γD crystallin Greek key domains using triple tryptophan mutant proteins. Protein Sci. 2004;13:2223–2235. doi: 10.1110/ps.04627004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brubaker WD, et al. Separating instability from aggregation propensity in γS-crystallin variants. Biophys J. 2011;100:498–506. doi: 10.1016/j.bpj.2010.12.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahin E, et al. Computational design and biophysical characterization of aggregation-resistant point mutations for γD crystallin illustrate a balance of conformational stability and intrinsic aggregation propensity. Biochemistry. 2011;50:628–639. doi: 10.1021/bi100978r. [DOI] [PubMed] [Google Scholar]
- 34.Kwak K, Rosenfeld DE, Fayer MD. Taking apart the two-dimensional infrared vibrational echo spectra: More information and elimination of distortions. J Chem Phys. 2008;128:204505. doi: 10.1063/1.2927906. [DOI] [PubMed] [Google Scholar]
- 35.Petty SA, Decatur SM. Intersheet rearrangement of polypeptides during nucleation of β-sheet aggregates. Proc Natl Acad Sci USA. 2005;102:14272–14277. doi: 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanism of Z-α1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.