Abstract
Aminoacyl-tRNA synthetases (aaRSs) ensure faithful translation of mRNA into protein by coupling an amino acid to a set of tRNAs with conserved anticodon sequences. Here, we show that in mitochondria of Saccharomyces cerevisiae, a single aaRS (MST1) recognizes and aminoacylates two natural tRNAs that contain anticodon loops of different size and sequence. Besides a regular 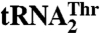 with a threonine (Thr) anticodon, MST1 also recognizes an unusual
with a threonine (Thr) anticodon, MST1 also recognizes an unusual 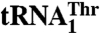 , which contains an enlarged anticodon loop and an anticodon triplet that reassigns the CUN codons from leucine to threonine. Our data show that MST1 recognizes the anticodon loop in both tRNAs, but employs distinct recognition mechanisms. The size but not the sequence of the anticodon loop is critical for
, which contains an enlarged anticodon loop and an anticodon triplet that reassigns the CUN codons from leucine to threonine. Our data show that MST1 recognizes the anticodon loop in both tRNAs, but employs distinct recognition mechanisms. The size but not the sequence of the anticodon loop is critical for 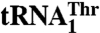 recognition, whereas the anticodon sequence is essential for aminoacylation of
recognition, whereas the anticodon sequence is essential for aminoacylation of 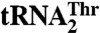 . The crystal structure of MST1 reveals that, while lacking the N-terminal editing domain, the enzyme closely resembles the bacterial threonyl-tRNA synthetase (ThrRS). A detailed structural comparison with Escherichia coli ThrRS, which is unable to aminoacylate
. The crystal structure of MST1 reveals that, while lacking the N-terminal editing domain, the enzyme closely resembles the bacterial threonyl-tRNA synthetase (ThrRS). A detailed structural comparison with Escherichia coli ThrRS, which is unable to aminoacylate 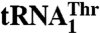 , reveals differences in the anticodon-binding domain that probably allow recognition of the distinct anticodon loops. Finally, our mutational and modeling analyses identify the structural elements in MST1 (e.g., helix α11) that define tRNA selectivity. Thus, MTS1 exemplifies that a single aaRS can recognize completely divergent anticodon loops of natural isoacceptor tRNAs and that in doing so it facilitates the reassignment of the genetic code in yeast mitochondria.
, reveals differences in the anticodon-binding domain that probably allow recognition of the distinct anticodon loops. Finally, our mutational and modeling analyses identify the structural elements in MST1 (e.g., helix α11) that define tRNA selectivity. Thus, MTS1 exemplifies that a single aaRS can recognize completely divergent anticodon loops of natural isoacceptor tRNAs and that in doing so it facilitates the reassignment of the genetic code in yeast mitochondria.
Keywords: protein synthesis, anticodon recognition
Aminoacyl-tRNA synthetases (aaRSs) provide the ribosome with aminoacyl-tRNA substrates for protein synthesis (1, 2). The ability of aaRSs to precisely match a particular anticodon sequence in tRNA with the cognate amino acid is critical to maintain fidelity of mRNA translation. The coupling of amino acids to tRNA is a two-step process. In the first step, an aaRS activates the amino acid with ATP, and in the second, it promotes the transfer of the aminoacyl group from the aminoacyl-AMP conjugate to the 3′-end of tRNA. To maintain fidelity during protein synthesis, an aaRS thus needs to select the correct amino acid and tRNA substrates from a large pool of structurally similar molecules in the cell. The active-site groove of the aaRS plays a role of the first selection sieve that prevents activation of most noncognate amino acids (3, 4), and a cis-editing site (5) or free standing editing domain (6–8) further proofreads misactivated amino acids. In contrast, the selection of tRNA substrates depends on the ability of aaRSs to establish interactions with a unique set of identity elements present in a given tRNA (9, 10). The major identity elements reside in the anticodon loop and the amino acid acceptor stem of tRNA (9). Except for tRNAAla (11) and tRNASer (12), mutations in the anticodon loop result in significant loss of aminoacylation efficiency (9), suggesting that the isoacceptor tRNAs must carry similar anticodon loops or else they would not be recognized by a given aaRS.
In the mitochondria of certain yeast species, including Saccharomyces cerevisiae, the CUN (N denotes A, U, G, and C) codons have been reassigned from leucine (Leu) to threonine (Thr). This is due to the presence of an unusual 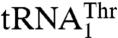 with an enlarged 8-nucleotide anticodon loop and a UAG anticodon (13) that typically reads the Leu codons. In addition to
with an enlarged 8-nucleotide anticodon loop and a UAG anticodon (13) that typically reads the Leu codons. In addition to 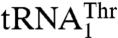 , yeast mitochondria also contain a canonical
, yeast mitochondria also contain a canonical 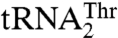 with a UGU anticodon that reads the standard Thr codons (ACN). It has been shown that the anticodon sequence of
with a UGU anticodon that reads the standard Thr codons (ACN). It has been shown that the anticodon sequence of 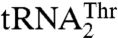 is essential for recognition by threonyl-tRNA synthetase (ThrRS). In particular, both bacterial and yeast cytosolic ThrRSs recognize G35 and U36 of the anticodon loop (14, 15). Furthermore, the crystal structure of the Escherichia coli ThrRS-tRNAThr complex reveals that G35 and U36 form specific interactions with residues of the anticodon-binding domain of ThrRS (16). Taken together, these findings argue that an unusual S. cerevisiae mitochondrial
is essential for recognition by threonyl-tRNA synthetase (ThrRS). In particular, both bacterial and yeast cytosolic ThrRSs recognize G35 and U36 of the anticodon loop (14, 15). Furthermore, the crystal structure of the Escherichia coli ThrRS-tRNAThr complex reveals that G35 and U36 form specific interactions with residues of the anticodon-binding domain of ThrRS (16). Taken together, these findings argue that an unusual S. cerevisiae mitochondrial 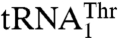 might be recognized by a distinct ThrRS. However, we have recently shown that, in spite of the dramatic differences in their anticodon loop sequences, both
might be recognized by a distinct ThrRS. However, we have recently shown that, in spite of the dramatic differences in their anticodon loop sequences, both 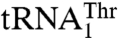 and
and 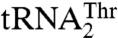 are aminoacylated with similar efficiencies by a single S. cerevisiae mitochondrial ThrRS (MST1) (17). This raised an immediate question as to how MST1 is able to recognize the isoacceptor tRNAs that carry different size anticodon loops. Here, we provide structural and functional evidence that MST1 recognizes anticodon loops as the major identity elements in both
are aminoacylated with similar efficiencies by a single S. cerevisiae mitochondrial ThrRS (MST1) (17). This raised an immediate question as to how MST1 is able to recognize the isoacceptor tRNAs that carry different size anticodon loops. Here, we provide structural and functional evidence that MST1 recognizes anticodon loops as the major identity elements in both 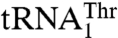 and
and 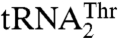 . The plasticity in substrate recognition allows MST1 to threonylate the unusual
. The plasticity in substrate recognition allows MST1 to threonylate the unusual 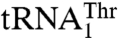 with an enlarged anticodon loop and a reassigned anticodon while maintaining activity for the canonical
with an enlarged anticodon loop and a reassigned anticodon while maintaining activity for the canonical 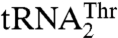 . This explains how a single aaRS is capable of recognizing completely different anticodon loops present in natural isoacceptor tRNAs.
. This explains how a single aaRS is capable of recognizing completely different anticodon loops present in natural isoacceptor tRNAs.
Results
MST1 Recognizes Distinct Anticodon Loops of Two tRNAThr Species.
We have previously shown that S. cerevisiae MST1 recognizes both 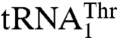 and
and 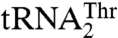 as substrates, whereas neither Candida albicans MST1 nor E. coli ThrRS is able to aminoacylate the unusual
as substrates, whereas neither Candida albicans MST1 nor E. coli ThrRS is able to aminoacylate the unusual 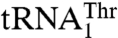 (17). This suggests that MST1 has coevolved with
(17). This suggests that MST1 has coevolved with 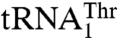 to complete the CUN codon reassignment in yeast mitochondria. Given the diverse sequences in the anticodon loops of
to complete the CUN codon reassignment in yeast mitochondria. Given the diverse sequences in the anticodon loops of 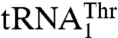 and
and 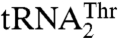 , we hypothesized that MST1 might not interact with the anticodon loop of these tRNAs in a manner resembling AlaRS and SerRS (11, 12). Surprisingly, mutational and biochemical results show that the anticodon loops of both tRNAThr species are recognized by S. cerevisiae MST1. For
, we hypothesized that MST1 might not interact with the anticodon loop of these tRNAs in a manner resembling AlaRS and SerRS (11, 12). Surprisingly, mutational and biochemical results show that the anticodon loops of both tRNAThr species are recognized by S. cerevisiae MST1. For 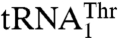 , deleting U32 in the anticodon loop decreased aminoacylation efficiency (kcat/Km) 13-fold, mutations G37A and U38A reduced the threonylation efficiency 4- and 15-fold respectively, whereas mutations in positions 35 and 36 did not have any effect on aminoacylation (Table 1). In addition, changing the
, deleting U32 in the anticodon loop decreased aminoacylation efficiency (kcat/Km) 13-fold, mutations G37A and U38A reduced the threonylation efficiency 4- and 15-fold respectively, whereas mutations in positions 35 and 36 did not have any effect on aminoacylation (Table 1). In addition, changing the 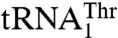 anticodon from UAG to UGU did not affect either the kcat or Km value. In contrast, replacing the UGU anticodon of
anticodon from UAG to UGU did not affect either the kcat or Km value. In contrast, replacing the UGU anticodon of 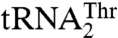 with UAG decreased aminoacylation efficiency 180-fold, whereas additionally mutating positions 37 and 38 did not affect the aminoacylation activity further. Furthermore, transplanting the entire anticodon loop of
with UAG decreased aminoacylation efficiency 180-fold, whereas additionally mutating positions 37 and 38 did not affect the aminoacylation activity further. Furthermore, transplanting the entire anticodon loop of 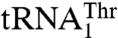 to
to 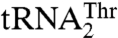 rescued the aminoacylation efficiency 17-fold compared to the G35A/U36G mutant. Our results suggest that MST1 recognizes the size of the enlarged anticodon loop as well as U38 in
rescued the aminoacylation efficiency 17-fold compared to the G35A/U36G mutant. Our results suggest that MST1 recognizes the size of the enlarged anticodon loop as well as U38 in 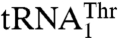 and the G35/U36 sequence of the anticodon loop in
and the G35/U36 sequence of the anticodon loop in 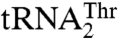 . Thus, S. cerevisiae MST1 represents an aaRS capable of recognizing completely different anticodon loops present in two natural isoacceptor tRNA substrates.
. Thus, S. cerevisiae MST1 represents an aaRS capable of recognizing completely different anticodon loops present in two natural isoacceptor tRNA substrates.
Table 1.
Threonylation activities of tRNAThr variants by MST1
| Anticodon loop | kcat, min-1 | Km, μM | kcat/Km, min-1 μM-1 | Relative kcat/Km* | |
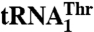 |
|||||
| WT† | UUUUAGGU | 2.8 ± 0.4‡ | 0.29 ± 0.09 | 10.3 ± 2.3 | 100 |
| ΔU32 | UUUAGGU | 0.85 ± 0.15 | 1.2 ± 0.3 | 0.79 ± 0.31 | 7.7 |
| A35G/G36U | UUUUGUGU | 2.7 ± 1.0 | 0.33 ± 0.22 | 9.0 ± 2.2 | 87 |
| G37A | UUUUAGAU | 2.9 ± 0.6 | 1.2 ± 0.4 | 2.4 ± 0.3 | 23 |
| U38A | UUUUAGGA | 1.3 ± 0.3 | 1.8 ± 0.5 | 0.69 ± 0.04 | 6.7 |
| A73G | UUUUAGGU | 0.37 ± 0.12 | 0.50 ± 0.34 | 0.89 ± 0.32 | 8.6 |
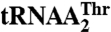 |
|||||
| WT† | UUUGUAA | 2.3 ± 0.1 | 0.44 ± 0.04 | 5.4 ± 0.6 | 52 |
| G35A/U36G | UUUAGAA | 0.076 ± 0.009 | 3.1 ± 1.9 | 0.030 ± 0.012 | 0.29 |
| G35A/U36G/A37G | UUUAGGA | 0.1 ± 0.05 | 0.94 ± 0.09 | 0.11 ± 0.05 | 1.1 |
| G35A/U36G/A37G/A38U | UUUAGGU | 0.063 ± 0.006 | 2.3 ± 0.6 | 0.028 ± 0.006 | 0.27 |
| insU32/G35A/U36G | UUUUAGAA | 0.058 ± 0.01 | 1.2 ± 0.1 | 0.05 ± 0.01 | 0.49 |
| insU32/G35A/U36G/A37G/A38U | UUUUAGGU | 0.78 ± 0.03 | 1.6 ± 0.1 | 0.5 ± 0.03 | 4.9 |
*The kcat/Km values are relative to WT 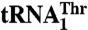 .
.
†The results for WT tRNAs were taken from ref. 17.
‡The results are the average of at least three repeats with standard deviations indicated.
MST1 Is a Structural Homologue of the Bacterial but not the Archaeal ThrRS.
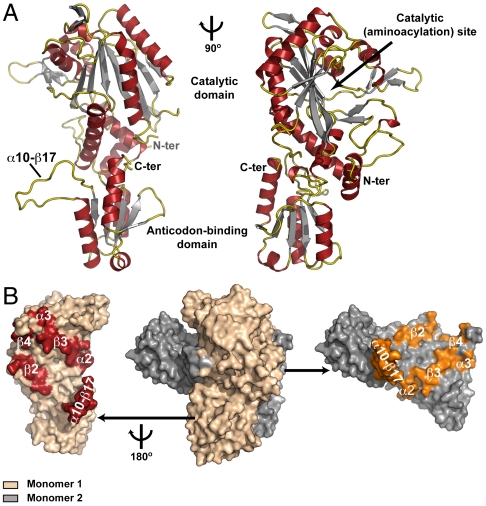
To facilitate structural studies on the mechanism(s) by which MST1 recognizes distinct anticodon loops, we have determined a crystal structure of apo MST1. The tetragonal crystals, which contained a monomer of apo MST1 in the asymmetric unit, diffracted X-rays to 2.1-Å resolution. The crystal structure revealed that MST1, although lacking the entire editing domain in its N terminus, adopts a class II aaRS fold (Fig. 1A, Fig. S1, and Table S1). At its N terminus, MST1 contains a large aminoacylation domain (residues 1–339) that presumably binds the amino acid substrate, ATP, and the acceptor arm of tRNAThr (Fig. 1A). The aminoacylation domain contains a zinc (Zn2+) ion, which is coordinated with the side chains of Cys133, His184, and His319 (Fig. 2A). The presence of a tightly bound Zn2+ suggests the location of the active-site groove, which catalyzes activation of threonine and the subsequent transfer of the threonyl group onto tRNAThr (Fig. 1A, Right). A smaller C-terminal domain of MST1 (residues 340–462) serves as the anticodon-binding domain that recognizes certain features in the anticodon loops in tRNAThr isoacceptors (Fig. 1A).
Fig. 1.
MST1 adopts a class II aaRS fold with a dimer as a biological unit. (A) Ribbon diagram of the crystal structure of apo MST1 determined at 2.1-Å resolution. The structure is presented from two angles with the arrow pointing into the active site. Secondary structure elements are colored as follows: α helices are red, β strands are gray, and loops are gold. (B) Surface representation of the MST1 homodimer (center) with the secondary structure elements from both domains contributing to the dimer interface highlighted (Left and Right). The beige monomer is rotated 180° clockwise around the vertical axis and translated away from the gray monomer thus fully exposing the dimer interface. The interacting elements are in red and orange, respectively.
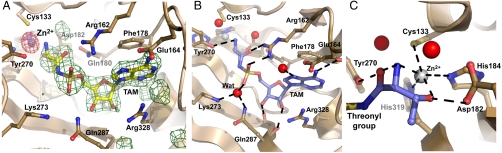
Fig. 2.
The catalytic site in the MST1-threonyl adenylate complex is poised for promoting the aminoacyl transfer. (A) Threonyl sulfamoyl adenylate (TAM) and Zn2+ bind to the active site of MST1. The experimental Fo - Fc electron density map (green mash) contoured at 3σ and calculated to 2.0 Å showing the positive peak for TAM. The positive peak (8σ) in the anomalous difference map (red mash) confirms the presence of Zn2+ in the active site of MST1. TAM (gold balls-and-sticks) and Zn2+ (gray sphere) derived from the final round of structure refinement are superimposed over the corresponding experimental maps. MST1 residues are beige. (B) TAM (blue ball-and-sticks) extensively interacts with Zn2+ (gray), water molecules (red spheres), and the active-site residues of MST1 (beige). The putative hydrolytic water is labeled as Wat. (C) The threonyl group interacts with Zn2+, which provides basis for the amino acid selectivity. Both α-amino and γ-hydroxyl of threonine interact with Zn2+. Tyr270, Asp182, and surrounding water molecules (red spheres) further stabilize the binding of the threonyl moiety. The main hydrogen bonds are shown with dashed lines.
Although a monomer in the asymmetric unit, MST1 is a stable homodimer in solution like all other class II aaRSs (Fig. 1B). The crystal packing analysis of all MST1 crystal forms also suggested that the enzyme is indeed in dimeric form. In the majority of known ThrRSs the homodimers are formed through head-to-head interactions between the aminoacylation domains. In the case of MST1, however, both the aminoacylation and anticodon-binding domains significantly contribute to the stability of the homodimer (Fig. 1B). In particular, a long loop connecting helix α10 with the strand β17 (α10–β17 loop) in the anticodon-binding domain of one MST1 monomer interacts extensively with the residues in helix α2 and strand β3 of the aminoacylation domain of the other MST1 monomer (Figs. 1 A and B). This peculiar cross-subunit interaction stabilizes the MST1 homodimer and perhaps contributes to its compactness. Indeed, our analysis revealed that MST1 buries a significantly larger surface area on homodimer formation (approximately 10,000 Å2) than the E. coli (5,340 Å2), Staphylococcus aureus (6,560 Å2), and Aeropyrum pernix ThrRS (7,720 Å2). While it remains to be seen if this additional interaction has any physiological significance, it is important to mention that α10–β17 loop interacts with elements of the aminoacylation domain that support the floor of the active-site crevice.
The fact that MST1 lacks the editing domain prompted us to speculate about the evolutionary origins of the enzyme. Several crystal structures of the prokaryotic orthologues are known, but only the ThrRS from an archaeon A. pernix lacks the editing domain. This led us to speculate that MST1 might be a structural descendant of the archaeal and not of the bacterial enzyme. However, the superpositioning of the aminoacylation and anticodon-binding domains of MST1 onto the corresponding domains in A. pernix [Protein Data Bank (PDB) ID code 3A32] resulted with an rmsd value of 2.3 Å (Fig. S2C), whereas the same calculation using the E. coli (PDB ID code 1QF6) and S. aureus ThrRS (PDB ID code 1NYR) yielded much smaller rmsd values of 1.23 and 1.05 Å, respectively (Figs. S2 A and B). Thus, our findings suggest that MST1 most likely evolved from the full-length bacterial ThrRS and not from the archaeal enzyme that lacks the cis-editing domain. The loss of the editing domain could be compensated by an unknown in trans editing factor, by higher amino acid specificity of the catalytic domain, or by a better tolerance of the Thr → Ser substitutions by the mitochondrial proteins.
Binding of Threonyl Adenylate Stabilizes the “Open” Conformation of MST1 and Induces Conformational Rearrangements in the Active Site that Are Important for Aminoacyl Transfer.
The active site in the aminoacylation domain of all aaRSs promotes formation of the aminoacyl-AMP conjugate and the subsequent transfer of the amino acid onto the 3′ hydroxyl (or 2′ hydroxyl) group of A76 on the substrate tRNA. To provide further evidence that MST1 is a genuine ThrRS and to probe its mechanism of amino acid selection, we determined the crystal structure of MST1 complexed with a nonhydrolyzable analog of threonyl-AMP, threonyl sulfamoyl adenylate (TAM) (Fig. 2, Fig. S3, and Table S1). The binary complex structure revealed that TAM binds in the presumed active-site crevice and that its binding promotes conformational rearrangements in the active site that are essential for the subsequent aminoacyl transfer reaction (Fig. 2 and Fig. S3). On a global level, helix α4, strands β5 and β6, and the loop β5–β6 rotate approximately 10° counterclockwise around the vertical axis when the structure is oriented as in Fig. S3A. These three elements form a lid atop the active site, and their concerted movement yields an “open” conformation of the enzyme that presumably allows TAM to bind the active site (Fig. S3A). Increased mobility in this part of the structure is reflected in a complete disorder of the β5–β6 loop on TAM binding (Fig. S3A). However, because the same conformational change was observed in the apo-MST1 structure derived from another crystal form (see SI Results and Discussion and Fig. S4), we propose that TAM binding does not promote but rather stabilizes the open conformation of MST1.
Additional conformational rearrangements occur in the active site where TAM interacts with the Zn2+ ion and a number of the amino acid side chains (Fig. 2 and Fig. S3B). The conserved residues Phe178 and Arg328 stack the adenine ring and place its amino group within a hydrogen bond distance from the carbonyl oxygen of Leu175 and the side-chain atoms of Glu164 (Fig. 2B). The placement of the adenine ring is further stabilized by hydrogen bonds between its N1 and N3 atoms and the amide nitrogen of Leu175 and Oγ of Ser 325, respectively (not shown). Further, the O2′ and O3′ hydroxyls in the ribose form hydrogen bonds with the carbonyl oxygens of Gln287 and Val288, respectively (Fig. 2B). These interactions stabilize the 3′ endo configuration of the ribose ring in TAM that is bound to MST1. Perhaps the most interesting observation is that our binary complex crystals contain the putative hydrolytic water in the active site (Fig. 2B). This water mediates interactions between the active-site residues and the sulfamoyl group, which is a mimic of the phosphoryl moiety. In particular, the catalytic Lys273 and the side chain of Gln287 orient a water molecule for attack onto the sulfur of the sulfamoyl group (Fig. 2B). A structural comparison with apo MST1 reveals that the Lys273 side chain rotates toward the hydrolytic water and sulfamoyl on TAM binding (Fig. S3B). Thus, in the crystal containing the binary complex we captured the conformation of the active site poised to promote the transfer of threonine from threonyl adenylate onto A76 of tRNAThr. The sulfamoyl group is further anchored in the active-site crevice through interactions between the nonbridging oxygen O1 and the guanidinium group of Arg162 (Fig. 2B). Finally, the threonyl moiety interacts with both the amino acid residues and Zn2+ (Fig. 2C). The guanidinium group of Arg162 forms a hydrogen bond with the carbonyl oxygen, Tyr270 and Zn2+ interact with the α-amino group, whereas Asp182 and Zn2+ coordinate the Oγ of the threonyl moiety (Fig. 2C). The subtle movements of Gln180, Asp182, and Tyr270 further facilitate the appropriate positioning of threonyl adenylate in the active site (Fig. S3C). In conclusion, our results show unambiguously that MST1 is a genuine ThrRS. The enzyme binds with high affinity the analog of the threonyl-AMP conjugate through interactions with a tightly bound Zn2+ ion. The conformation of the active site of MST1 in our crystal of the binary complex is poised for catalysis of the aminoacyl transfer.
Modeling of the MST1-tRNAThr Complex and Mutational Study Suggest Residues Important for tRNA Selectivity.
Perhaps the most interesting feature of MST1 is its ability to recognize completely different anticodon loops, which, in turn, encode the same amino acid. The anticodon loops in 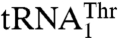 and
and 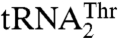 are not only different in their length but also in their sequence (Fig. 3A). Because we were unable to obtain a crystal structure of MST1 in complex with either of the natural tRNAThr species, we modeled the binary complex between MST1 and a regular
are not only different in their length but also in their sequence (Fig. 3A). Because we were unable to obtain a crystal structure of MST1 in complex with either of the natural tRNAThr species, we modeled the binary complex between MST1 and a regular 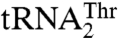 using the crystal structure of the E. coli binary ThrRS-tRNAThr complex as a model. Our modeling results reveal that the anticodon-binding domain of MST1 adopts a more open conformation when compared with the E. coli enzyme (Fig. 3B). In particular, strands β19 and β20 as well as the connecting loops adopt an orientation that could create more space for the binding of the enlarged anticodon loop of
using the crystal structure of the E. coli binary ThrRS-tRNAThr complex as a model. Our modeling results reveal that the anticodon-binding domain of MST1 adopts a more open conformation when compared with the E. coli enzyme (Fig. 3B). In particular, strands β19 and β20 as well as the connecting loops adopt an orientation that could create more space for the binding of the enlarged anticodon loop of 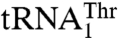 (Fig. 3B). This suggested that the solvent-exposed residues in this part of the anticodon-binding domain of MST1 might be responsible for tRNA discrimination.
(Fig. 3B). This suggested that the solvent-exposed residues in this part of the anticodon-binding domain of MST1 might be responsible for tRNA discrimination.
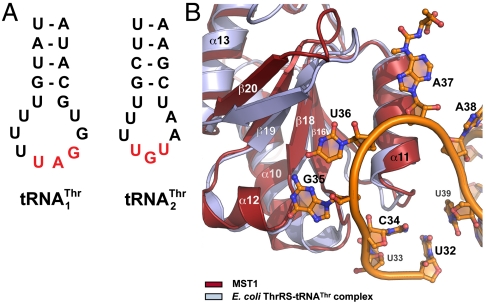
Fig. 3.
Structural modeling suggests residues in MST1 important for tRNA selectivity. (A) The secondary structure diagrams of the anticodon stem-loops of the mitochondrial 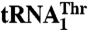 and
and 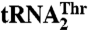 are shown. The main differences are in the anticodon sequence (red bold letters) and in the length of the anticodon loop. MST1 is capable of binding and charging both tRNAs regardless of the differences in sequence and structure. (B) Comparison of MST1 with E. coli ThrRS complexed with tRNAThr (PDB ID code 1QF6) suggests the anticodon-binding domain of MST1 adopts a more open conformation that might be compatible with different anticodon loops found in
are shown. The main differences are in the anticodon sequence (red bold letters) and in the length of the anticodon loop. MST1 is capable of binding and charging both tRNAs regardless of the differences in sequence and structure. (B) Comparison of MST1 with E. coli ThrRS complexed with tRNAThr (PDB ID code 1QF6) suggests the anticodon-binding domain of MST1 adopts a more open conformation that might be compatible with different anticodon loops found in 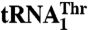 and
and 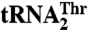 . The catalytic and anticodon domains of E.coli ThrRS (light blue) were superimposed onto the corresponding domains of MST1 (red), and the resulting rotation–translation matrix was applied onto tRNAThr (orange).
. The catalytic and anticodon domains of E.coli ThrRS (light blue) were superimposed onto the corresponding domains of MST1 (red), and the resulting rotation–translation matrix was applied onto tRNAThr (orange).
To further understand the recognition of 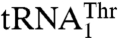 and
and 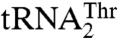 , we mutated 15 solvent-exposed residues in the anticodon-binding domain of MST1 that might be in close proximity of the anticodon loop (Fig. 4). The N432A, R434A, and R439A variants all showed weaker binding for
, we mutated 15 solvent-exposed residues in the anticodon-binding domain of MST1 that might be in close proximity of the anticodon loop (Fig. 4). The N432A, R434A, and R439A variants all showed weaker binding for 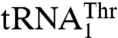 , as indicated by the increased Km values (Table 2). However, mutants N432A and R439A, but not R434A, displayed increased Km for
, as indicated by the increased Km values (Table 2). However, mutants N432A and R439A, but not R434A, displayed increased Km for 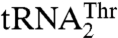 as well, which suggests that Arg434 is specific for
as well, which suggests that Arg434 is specific for 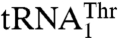 recognition. In contrast, mutants T357A, Q362A, and Y405A displayed increased Km values for
recognition. In contrast, mutants T357A, Q362A, and Y405A displayed increased Km values for 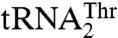 and not for
and not for 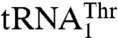 , suggesting their possible role in recognizing
, suggesting their possible role in recognizing 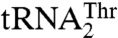 but not
but not 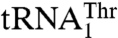 . Our mutational and biochemical analysis confirmed that MST1 employs different mechanisms to bind the anticodon loops of
. Our mutational and biochemical analysis confirmed that MST1 employs different mechanisms to bind the anticodon loops of 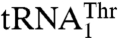 and
and 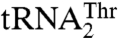 .
.
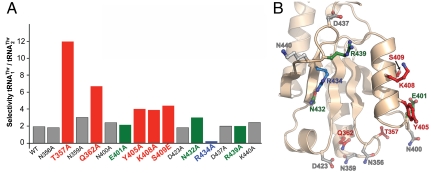
Fig. 4.
Mutational studies suggest that residues in helix α11 and loop β16–α10 contribute to 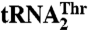 recognition, whereas Arg434 is critical for
recognition, whereas Arg434 is critical for 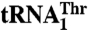 binding. (A) The biochemical analysis of MST1 mutants and their ability to bind and aminoacylate
binding. (A) The biochemical analysis of MST1 mutants and their ability to bind and aminoacylate 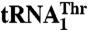 and
and 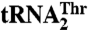 is shown. The blue bar designates Arg434, which is critical for binding
is shown. The blue bar designates Arg434, which is critical for binding 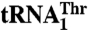 but not for
but not for 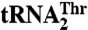 . Red bars designate residues in MST1 that contribute to
. Red bars designate residues in MST1 that contribute to 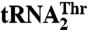 recognition. Green bars highlight the side chains that contribute to binding both mitochondrial tRNAThr species. Finally, gray bars designate the amino acid side chains that are not involved in tRNAThr recognition. (B) The side chains of the amino acids used in the mutational and biochemical studies are mapped onto the structure of the anticodon-binding domain of MST1 (beige). The mutated side chains are colored as in A.
recognition. Green bars highlight the side chains that contribute to binding both mitochondrial tRNAThr species. Finally, gray bars designate the amino acid side chains that are not involved in tRNAThr recognition. (B) The side chains of the amino acids used in the mutational and biochemical studies are mapped onto the structure of the anticodon-binding domain of MST1 (beige). The mutated side chains are colored as in A.
Table 2.
Threonylation of tRNAThr by MST1 variants
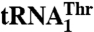 |
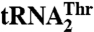 |
Selectivity* | |||||
| kcat, min-1 | Km, μM | kcat/Km, min-1 μM-1 | kcat, min-1 | Km, μM | kcat/Km, min-1 μM-1 | ||
| WT† | 2.8 ± 0.4‡ | 0.29 ± 0.09 | 10.3 ± 2.3 | 2.3 ± 0.1 | 0.44 ± 0.04 | 5.4 ± 0.6 | 1.9 |
| N356A | 3.6 ± 0.7 | 0.31 ± 0.05 | 11.6 ± 0.3 | 3.2 ± 1.0 | 0.51 ± 0.15 | 6.5 ± 2.0 | 1.8 |
| T357A | 4.8 ± 0.7 | 0.47 ± 0.11 | 10.2 ± 0.9 | 0.6 ± 0.1 | 0.83 ± 0.31 | 0.8 ± 0.4 | 12.0 |
| N359A | 5.2 ± 0.6 | 0.30 ± 0.08 | 18.3 ± 5.0 | 3.5 ± 0.2 | 0.59 ± 0.07 | 6.0 ± 0.6 | 3.0 |
| Q362A | 7.0 ± 0.2 | 0.46 ± 0.16 | 16.5 ± 6.1 | 2.7 ± 0.4 | 1.23 ± 0.42 | 2.5 ± 1.1 | 6.7 |
| N400A | 3.0 ± 0.6 | 0.24 ± 0.05 | 12.6 ± 4.4 | 2.6 ± 0.5 | 0.64 ± 0.41 | 5.2 ± 2.5 | 2.4 |
| E401A | 4.4 ± 0.5 | 0.13 ± 0.04 | 34.9 ± 5.3 | 4.0 ± 0.2 | 0.27 ± 0.13 | 16.6 ± 7.0 | 2.1 |
| Y405A | 4.9 ± 1.2 | 0.30 ± 0.11 | 17.0 ± 2.8 | 3.6 ± 0.7 | 0.94 ± 0.42 | 4.2 ± 1.3 | 4.0 |
| K408A | 5.6 ± 0.8 | 0.49 ± 0.15 | 11.7 ± 1.8 | 1.8 ± 0.5 | 0.69 ± 0.24 | 3.0 ± 1.5 | 3.9 |
| S409E | 3.0 ± 0.4 | 0.28 ± 0.04 | 10.8 ± 0.02 | 1.0 ± 0.3 | 0.49 ± 0.19 | 2.5 ± 1.8 | 4.4 |
| D423A | 4.0 ± 0.5 | 0.20 ± 0.02 | 20.2 ± 2.0 | 3.3 ± 0.3 | 0.31 ± 0.06 | 11.1 ± 3.1 | 1.8 |
| N432A | 3.0 ± 0.8 | 0.59 ± 0.22 | 5.2 ± 0.7 | 2.4 ± 0.4 | 1.40 ± 0.32 | 1.7 ± 0.2 | 3.0 |
| R434A | 2.0 ± 0.3 | 0.83 ± 0.29 | 2.7 ± 1.1 | 6.2 ± 0.3 | 0.44 ± 0.13 | 14.8 ± 4.8 | 0.2 |
| D437A | 3.2 ± 0.1 | 0.24 ± 0.13 | 15.8 ± 6.8 | 2.7 ± 0.3 | 0.37 ± 0.13 | 7.9 ± 3.3 | 2.0 |
| R439A | 7.1 ± 0.8 | 0.95 ± 0.40 | 9.0 ± 5.4 | 6.0 ± 1.0 | 1.39 ± 0.37 | 4.5 ± 1.4 | 2.0 |
| K440A | 6.7 ± 1.6 | 0.48 ± 0.19 | 16.2 ± 7.6 | 5.3 ± 2.2 | 0.78 ± 0.06 | 6.8 ± 3.1 | 2.4 |
*Selectivity was calculated as the ratio of kcat/Km values for 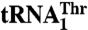 over
over 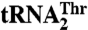 .
.
†The results for WT tRNAs were taken from ref. 17.
‡The results are the average of at least three repeats with standard deviations indicated.
Discussion
Distinct Mechanisms of Anticodon Recognition by MST1.
AaRSs have evolved characteristic patterns for recognition of their cognate tRNAs. Each aaRS typically identifies a unique set of nucleotides on tRNAs, most notably in the anticodon loop and acceptor stem. It was thus surprising that yeast arginyl-tRNA synthetase was shown to recognize different nucleotides in the anticodon loops of the naked tRNAArg and tRNAAsp transcripts (18). However, the native tRNAAsp, which harbors an m1G37 modification, does not serve as a substrate for ArgRS (19). A nucleotide insertion at the anticodon loop of tRNAGly leads to a frame-shift suppressor tRNA (20, 21), although its recognition by the cognate aaRS remains unclear. The first example of dual-mode recognition of biological tRNAs by a single aaRS came from the mammalian seryl-tRNA synthetase (SerRS) (22), which recognizes distinct T-loop sequences in two mitochondrial tRNASer species. Structural and mutational studies show that the distal α helix and C terminus, which are unique to mitochondrial SerRSs, are involved in binding both tRNASer isoacceptors (23). It has also been shown that different sets of SerRS residues are responsible for binding the two tRNASer species. Here, we present structural and functional evidence that yeast mitochondrial ThrRS recognizes its tRNA substrates in a different way: Unlike the mammalian mitochondrial SerRS that does not recognize the anticodon loop, MST1 specifically interacts with the divergent anticodon loops in 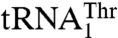 and
and 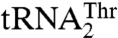 .
.
We have shown that 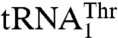 has recently evolved from mitochondrial tRNAHis and is distant from
has recently evolved from mitochondrial tRNAHis and is distant from 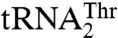 in the phylogeny (17). MST1 has also evolved since the split between Candida and Saccharomyces to recognize both mitochondrial tRNAThr species. The structure of S. cerevisiae MST1 shows that its anticodon-binding domain adopts a distinct conformation when compared with the corresponding domains in bacterial apo ThrRSs (Fig. 3B). In particular, strands β19 and β20 adopt a more open conformation, which would position them farther away from the anticodon loop. This structural rearrangement might be critical for accommodation of the larger anticodon loop of
in the phylogeny (17). MST1 has also evolved since the split between Candida and Saccharomyces to recognize both mitochondrial tRNAThr species. The structure of S. cerevisiae MST1 shows that its anticodon-binding domain adopts a distinct conformation when compared with the corresponding domains in bacterial apo ThrRSs (Fig. 3B). In particular, strands β19 and β20 adopt a more open conformation, which would position them farther away from the anticodon loop. This structural rearrangement might be critical for accommodation of the larger anticodon loop of 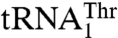 . In agreement with this proposal, our mutational studies show that Arg434 in β19 is critical for the binding of
. In agreement with this proposal, our mutational studies show that Arg434 in β19 is critical for the binding of 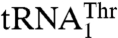 (Table 2). The equivalent residue of Arg434 in E. coli ThrRS is Arg609, which has been shown to interact with the base of U36 (16). However, Arg434 in MST1 does not contribute to the aminoacylation of the regular
(Table 2). The equivalent residue of Arg434 in E. coli ThrRS is Arg609, which has been shown to interact with the base of U36 (16). However, Arg434 in MST1 does not contribute to the aminoacylation of the regular 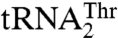 , suggesting that the context of the anticodon-binding domain in MST1 promoted the change-of-function of Arg434 to specifically recognize
, suggesting that the context of the anticodon-binding domain in MST1 promoted the change-of-function of Arg434 to specifically recognize 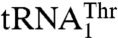 . This is further supported by the data that show that MST1 recognizes G37 and U38, but not A35 and U36 in
. This is further supported by the data that show that MST1 recognizes G37 and U38, but not A35 and U36 in 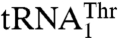 . Moreover, our data suggest that the enlarged size of the anticodon loop is important for threonylation of
. Moreover, our data suggest that the enlarged size of the anticodon loop is important for threonylation of 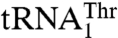 . Thus, presence of an additional nucleotide in
. Thus, presence of an additional nucleotide in 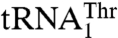 is likely to have an effect at the structural level, which is then sensed by Arg434 and in lesser part by Asn432 and Arg439.
is likely to have an effect at the structural level, which is then sensed by Arg434 and in lesser part by Asn432 and Arg439.
In contrast, MST1 specifically recognizes G35 and U36 in the anticodon sequence of 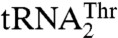 (Table 1). This clearly resembles the tRNA-recognition mechanism employed by the bacterial and yeast cytosolic ThrRSs. Because Arg434 does not partake in binding of the anticodon of
(Table 1). This clearly resembles the tRNA-recognition mechanism employed by the bacterial and yeast cytosolic ThrRSs. Because Arg434 does not partake in binding of the anticodon of 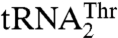 (Table 2), the question was raised as to what elements in MST1 might be responsible for the anticodon recognition. Our results suggest that residues in helix α11 (Tyr405 and in lesser part K408 and S409) and loop β16–α10 (Thr357 and Gln362) preferentially bind
(Table 2), the question was raised as to what elements in MST1 might be responsible for the anticodon recognition. Our results suggest that residues in helix α11 (Tyr405 and in lesser part K408 and S409) and loop β16–α10 (Thr357 and Gln362) preferentially bind 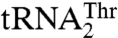 . The structural modeling suggests that Tyr405, and perhaps K408 and S409, might be important for binding of the 3′-end of the anticodon loop (A37 and A38), whereas Thr357 and Gln362 might interact with the anticodon (Fig. S5 B and C). The possible role in anticodon recognition of Thr357, which is conserved among mitochondrial ThrRSs that recognize both
. The structural modeling suggests that Tyr405, and perhaps K408 and S409, might be important for binding of the 3′-end of the anticodon loop (A37 and A38), whereas Thr357 and Gln362 might interact with the anticodon (Fig. S5 B and C). The possible role in anticodon recognition of Thr357, which is conserved among mitochondrial ThrRSs that recognize both 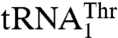 and
and 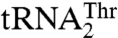 , but absent from bacterial ThrRSs or C. albicans MST1, is quite intriguing. The T357A mutant of MST1 shows a 7-fold reduced kcat/Km for
, but absent from bacterial ThrRSs or C. albicans MST1, is quite intriguing. The T357A mutant of MST1 shows a 7-fold reduced kcat/Km for 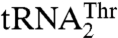 (Table 2). Because structural modeling suggests that Thr357 could interact with either U34 or G35 (Fig. S5C), it may well be that Thr357 may have evolved to compensate for the loss of interaction between Arg434 and the anticodon loop of
(Table 2). Because structural modeling suggests that Thr357 could interact with either U34 or G35 (Fig. S5C), it may well be that Thr357 may have evolved to compensate for the loss of interaction between Arg434 and the anticodon loop of 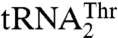 . Also, Thr357 and Gln362 are located in loop β16–α10, which precedes the long α10–β17 loop that is important for MST1 homodimerization (see MST1 Is a Structural Homologue of the Bacterial but not the Archaeal ThrRS and Fig. 1B). Thus, it is plausible that the interaction of Thr357 and Gln362 with the anticodon loop is relayed via α10–β17 loop to the active site of the other MST1 monomer. Consequently, the tRNA binding to one MST1 monomer could modulate the catalytic activity of the other monomer.
. Also, Thr357 and Gln362 are located in loop β16–α10, which precedes the long α10–β17 loop that is important for MST1 homodimerization (see MST1 Is a Structural Homologue of the Bacterial but not the Archaeal ThrRS and Fig. 1B). Thus, it is plausible that the interaction of Thr357 and Gln362 with the anticodon loop is relayed via α10–β17 loop to the active site of the other MST1 monomer. Consequently, the tRNA binding to one MST1 monomer could modulate the catalytic activity of the other monomer.
Although our data provide compelling evidence that different residues in MST1 contribute to recognition of different anticodon loops in two isoacceptor tRNAs, the detailed recognition mechanism remains elusive. Based on our findings, there could be two possible explanations as to how MST1 recognizes distinct anticodon loops in 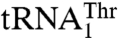 and
and 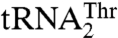 . In one, the same site in the anticodon-binding domain utilizes different residues that recognize specific features in one but not the other tRNA. Alternatively, the anticodon-binding domain could harbor two distinct sites each responsible for binding one anticodon loop. It is important to note that these sites need not be in close proximity, in which this situation would be somewhat analogous to aaRSs that carry out the editing reaction, and where the 3′-end of the tRNA translocates from one pocket to the other. Because our results cannot distinguish between these two possibilities, further structural studies of the binary MST1-tRNAThr complexes are warranted. In conclusion, our work shows that minor evolutionary changes in tRNA and aaRS genes could establish novel interactions that facilitate the reassignment of the genetic code.
. In one, the same site in the anticodon-binding domain utilizes different residues that recognize specific features in one but not the other tRNA. Alternatively, the anticodon-binding domain could harbor two distinct sites each responsible for binding one anticodon loop. It is important to note that these sites need not be in close proximity, in which this situation would be somewhat analogous to aaRSs that carry out the editing reaction, and where the 3′-end of the tRNA translocates from one pocket to the other. Because our results cannot distinguish between these two possibilities, further structural studies of the binary MST1-tRNAThr complexes are warranted. In conclusion, our work shows that minor evolutionary changes in tRNA and aaRS genes could establish novel interactions that facilitate the reassignment of the genetic code.
The Mechanism of Amino Acid Selection and Editing by MST1.
The editing function of aaRSs ensures that only a cognate amino acid is attached to a given tRNA and is thus essential for faithful translation of the genetic information. While different isoacceptor tRNAs harbor distinct structural elements, similar amino acids often lack any recognizable motifs that could be used in the selection process. For instance, ThrRS must be able to specifically select threonine from a pool of amino acids that includes valine and serine. Typically, the bacterial and archaeal ThrRSs employ cis-editing domains that hydrolyze misacylated seryl-tRNAThr but not threonyl-tRNAThr (24–26). In some archaeal species, the catalytic and editing domains are expressed as separate proteins that act in concert to prevent formation of the mischarged tRNAThr (27). In Mycoplasma, ThrRSs harbor dysfunctional editing domains with unknown physiological function (28). On the other hand, in striking contrast to the cytosolic and mitochondrial eukaryotic ThrRSs, yeast MST1 lacks the entire cis-editing domain (Fig. S2 A and B). This raises a question about a possible mechanism by which MST1 prevents the misacylation of tRNAThr and the subsequent misincorporation in the nascent protein of serine instead of threonine.
Perhaps the simplest explanation is that the catalytic site in MST1 efficiently selects for threonine over serine or valine. The discrimination against valine relies on the presence of Zn2+ in the active site. Our structure shows that Zn2+ coordinates both the α-amino and γ-hydroxyl groups the threonyl moiety. Because valine contains methyl in place of the side-chain hydroxyl, steric and electrostatic repulsions would prevent binding of valine in the active-site groove as previously suggested (29). However, this mechanism does not explain the basis for the rejection of serine. In fact, there is nothing in the aminoacylation site of MST1 that would prevent formation of seryl-AMP.
An intriguing question is why the editing site of MST1 is dispensable in yeast mitochondria. Mitochondrial phenylalanyl-tRNA synthetases (PheRSs) also lack a cis-editing domain (30), but their aminoacylation active sites are more selective toward amino acids than that of the cytosolic PheRSs. The increased selectivity for amino acid substrates of the mitochondrial PheRSs reduces the overall rate of misacylation to a tolerable level (31). On the other hand, the removal of the N-terminal domain in E. coli ThrRS activates pretransfer editing that hydrolyzes misactivated seryl-AMP (32). Thus, it could well be that the loss of the editing domain in MST1 either improves amino acid selectivity or accentuates the pretransfer editing function that acts on seryl-AMP conjugates. Alternatively, some unknown protein factor may act as an in trans editing enzyme.
Although it is obvious that a drastic mistranslation of mRNA could have detrimental effects on the overall health of the organism, there could be instances when mistranslation could be one of the adaptation mechanisms that allow the organism to survive. For instance, a recent study on a bacterial ThrRS has revealed that its editing function is inactivated under oxidative stress conditions (33). Also, it has been proposed that mistranslation could be well tolerated, if not desired, in bacteria and eukaryotes under certain stress conditions (34–36). In conclusion, while the physiological impact of ThrRS editing defects remains unclear, it is evident that future biochemical and structural studies on the mechanisms that govern the fidelity of ThrRS are warranted.
Experimental Procedures
Cloning, Mutagenesis and General Methods.
S. cerevisiae MST1 gene was cloned into pET28a expression vector (Novagen) with an N-terminal six-His tag. Expression of recombinant proteins was induced at 37 °C for 4 h with 0.5 mM isopropyl β-D-1-thiogalactopyranoside in Escherichia coli strain BL21-codon plus in Luria–Bertani media. His-tagged proteins were purified according to standard procedures. Mitochondrial tRNA genes were cloned into pUC18 vector (GenScript). Mutations in MST1 and tRNA genes were introduced using QuikChange Site-Directed Mutagenesis Kit (Stratagene).
In Vitro Assays.
In vitro tRNA transcripts were obtained using the T7 RNA polymerase runoff procedure as described (37). Aminoacylation experiments were performed as described (38) in the presence of 100 mM Na-HEPES pH 7.2, 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 25 μM [3H] Thr (100 μCi/mL), 0.2–9 μM tRNA transcripts, and 10–300 nM aaRSs.
Supplementary Material
Acknowledgments.
We thank Ilka Heinemann, Franz Lang, Patrick O’Donoghue, and Dan Su for inspiring discussions. J.L. was a Brown-Coxe Postdoctoral Fellow, and C.C. had support from the Beckman Scholars Program of the Arnold and Mabel Beckman Foundation. This work was supported by Grant GM022854 from the National Institute of General Medical Sciences (to D.S.) and by a grant from the American Cancer Society, Illinois Division, Inc. (to M.S.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences (Contract W-31-109-Eng-38).
Footnotes
The authors declare no conflict of interest.
Data deposition: Coordinates and structure factors for structures described have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3UGQ, apo-MST1 high-resolution crystal form; 3UGT, apo-MST1 low-resolution crystal form; 3UH0, MST1-TAM complex).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200109109/-/DCSupplemental.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008;9:242–253. doi: 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 3.Fersht AR, Kaethner MM. Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry. 1976;15:3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 4.Mascarenhas AP, An S, Rosen AE, Martinis SA, Musier-Forsyth K. Fidelity Mechanisms of the Aminoacyl-tRNA Synthetases. In: RajBhandary UL, Köhrer C, editors. Protein Engineering. New York: Springer-Verlag; 2008. pp. 153–200. [Google Scholar]
- 5.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 6.Ahel I, Korencic D, Ibba M, Söll D. Trans-editing of mischarged tRNAs. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An S, Musier-Forsyth K. Trans-editing of Cys-tRNAPro by Haemophilus influenzae YbaK protein. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 8.Chong YE, Yang XL, Schimmel P. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J Biol Chem. 2008;283:30073–30078. doi: 10.1074/jbc.M805943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guth EC, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25:531–542. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou YM, Schimmel P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry. 1989;28:6800–6804. doi: 10.1021/bi00443a003. [DOI] [PubMed] [Google Scholar]
- 12.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Tzagoloff A. Assembly of the mitochondrial membrane system: Sequences of yeast mitochondrial valine and an unusual threonine tRNA gene. Cell. 1979;18:47–53. doi: 10.1016/0092-8674(79)90352-0. [DOI] [PubMed] [Google Scholar]
- 14.Nameki N, et al. Similarities and differences in tRNA identity between Escherichia coli and Saccharomyces cerevisiae: Evolutionary conservation and divergence. Nucleic Acids Symp Ser. 1995;34:205–206. [PubMed] [Google Scholar]
- 15.Nameki N, Asahara H, Hasegawa T. Identity elements of Thermus thermophilus tRNAThr. FEBS Lett. 1996;396:201–207. doi: 10.1016/0014-5793(96)01094-0. [DOI] [PubMed] [Google Scholar]
- 16.Sankaranarayanan R, et al. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 17.Su D, et al. An unusual tRNAThr derived from tRNAHis reassigns in yeast mitochondria the CUN codons to threonine. Nucleic Acids Res. 2011;39:4866–4874. doi: 10.1093/nar/gkr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sissler M, Giegé R, Florentz C. Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J. 1996;15:5069–5076. [PMC free article] [PubMed] [Google Scholar]
- 19.Putz J, Florentz C, Benseler F, Giegé R. A single methyl group prevents the mischarging of a tRNA. Nat Struct Biol. 1994;1:580–582. doi: 10.1038/nsb0994-580. [DOI] [PubMed] [Google Scholar]
- 20.Murgola EJ, Prather NE, Mims BH, Pagel FT, Hijazi KA. Anticodon shift in tRNA: A novel mechanism in missense and nonsense suppression. Proc Natl Acad Sci USA. 1983;80:4936–4939. doi: 10.1073/pnas.80.16.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prather NE, Murgola EJ, Mims BH. Nucleotide insertion in the anticodon loop of a glycine transfer RNA causes missense suppression. Proc Natl Acad Sci USA. 1981;78:7408–7411. doi: 10.1073/pnas.78.12.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimada N, Suzuki T, Watanabe K. Dual mode recognition of two isoacceptor tRNAs by mammalian mitochondrial seryl-tRNA synthetase. J Biol Chem. 2001;276:46770–46778. doi: 10.1074/jbc.M105150200. [DOI] [PubMed] [Google Scholar]
- 23.Chimnaronk S, Gravers Jeppesen M, Suzuki T, Nyborg J, Watanabe K. Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J. 2005;24:3369–3379. doi: 10.1038/sj.emboj.7600811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dock-Bregeon AC, et al. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol Cell. 2004;16:375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Beebe K, Merriman E, Ribas De Pouplana L, Schimmel P. A domain for editing by an archaebacterial tRNA synthetase. Proc Natl Acad Sci USA. 2004;101:5958–5963. doi: 10.1073/pnas.0401530101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain T, et al. Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea. EMBO J. 2006;25:4152–4162. doi: 10.1038/sj.emboj.7601278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korencic D, et al. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc Natl Acad Sci USA. 2004;101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA. 2011;108:9378–83. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sankaranarayanan R, et al. Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase. Nat Struct Biol. 2000;7:461–465. doi: 10.1038/75856. [DOI] [PubMed] [Google Scholar]
- 30.Roy H, Ling J, Alfonzo J, Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds NM, et al. Cell-specific differences in the requirements for translation quality control. Proc Natl Acad Sci USA. 2010;107:4063–4068. doi: 10.1073/pnas.0909640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minajigi A, Francklyn CS. Aminoacyl transfer rate dictates choice of editing pathway in threonyl-tRNA synthetase. J Biol Chem. 2010;285:23810–23817. doi: 10.1074/jbc.M110.105320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruan B, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds NM, Lazazzera BA, Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 36.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy H, Ling J, Irnov M, Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.