Abstract
The cytosolic mevalonate (MVA) pathway in Hevea brasiliensis latex is the conventionally accepted pathway which provides isopentenyl diphosphate (IPP) for cis-polyisoprene (rubber) biosynthesis. However, the plastidic 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway may be an alternative source of IPP since its more recent discovery in plants. Quantitative RT-PCR (qRT-PCR) expression profiles of genes from both pathways in latex showed that subcellular compartmentalization of IPP for cis-polyisoprene synthesis is related to the degree of plastidic carotenoid synthesis. From this, the occurrence of two schemes of IPP partitioning and utilization within one species is proposed whereby the supply of IPP for cis-polyisoprene from the MEP pathway is related to carotenoid production in latex. Subsequently, a set of latex unique gene transcripts was sequenced and assembled and they were then mapped to IPP-requiring pathways. Up to eight such pathways, including cis-polyisoprene biosynthesis, were identified. Our findings on pre- and post-IPP metabolic routes form an important aspect of a pathway knowledge-driven approach to enhancing cis-polyisoprene biosynthesis in transgenic rubber trees.
Keywords: cis-polyisoprene, gene expression, Hevea brasiliensis, isopentenyl diphosphate, isoprenoids, latex pathways, rubber biosynthesis, transcriptome
Introduction
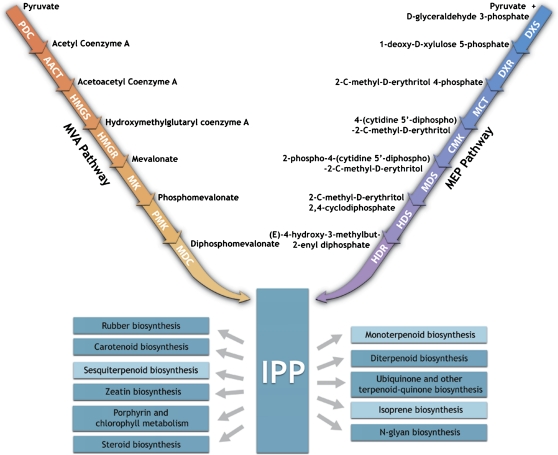
Natural rubber is found in the latex of the Hevea brasiliensis tree and consists of high molecular weight cis-polyisoprene produced from the isoprenoid pathway (Chappell, 1995) (Fig. 1). From a cell biology point of view, latex is essentially the cytoplasmic content of laticifers or latex vessels (Moir, 1959; Gomez and Moir, 1979; d’Auzac and Jacob, 1989; de Faÿ and Jacob, 1989). As cytoplasm, latex contains organelles typical of eukaryotic cells but of particular interest to rubber biosynthesis are the latex-specific organelles i.e. rubber particles and Frey-Wyssling particles. IPP incorporation into cis-polyisoprene occurs on the surface of rubber particles in the cytosol (Archer et al., 1963; Archer and Audley, 1987; Kekwick, 1989) while the Frey-Wyssling particle is a specialized plastid which contains carotenoids thus giving a yellowish colour to the latex of some rubber tree clones (Dickenson, 1969; Gomez and Moir, 1979; d’Auzac and Jacob, 1989).
Fig. 1.
General pathway of isoprenoid biosynthesis. IPP is a common intermediate of numerous isoprenoids and may be generated by the cytosolic MVA or the plastidic MEP pathway. Pathways which subsequently utilize IPP for synthesis of different classes of isoprenoids are shown. Isoprenoid end-products, including rubber (cis-polyisoprene), which are found in rubber tree latex are indicated within darker blue boxes. Among these, rubber, steroid, and N-glycan biosynthesis are cytosolic; zeatin biosynthesis partially cytosolic and plastidic; and the rest plastidic. PDC, pyruvate dehydrogenase complex; AACT, acetyl coenzyme A acetyltransferase; HMGS, hydroxymethyglutaryl coenzyme A synthase; HMGR, hydroxymethyglutaryl coenzyme A reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MDC, diphosphomevalonate decarboxylase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase.
Evidence for the MVA pathway route to rubber was first established based on experiments involving incubation of latex with 14C-labelled intermediates (Kekwick, 1989). More recently, plants were found to contain both cytosolic MVA and plastidic MEP pathways and that metabolic crossover of pathway intermediates such as IPP occur between subcellular compartments (Eisenreich et al., 2001; Bick and Lange, 2003; Rodríguez-Concepción, 2006, 2010; Kirby and Keasling, 2009). In the rubber tree, the existence of the MEP pathway was supported by the identification of 1-deoxy-D-xylulose 5-phosphate synthase (DXS) sequences from latex transcriptome sequencing (Ko et al., 2003; Chow et al., 2007). This led to the idea that the MEP pathway synthesizes IPP for carotenoids in Frey-Wyssling particles but could, in addition, provide IPP for cis-polyisoprene synthesis. Evidence supporting this is important as a basis for choosing the appropriate IPP-generating pathway to manipulate in the production of transgenic rubber trees with higher rubber yield. Therefore, the metabolic routes to the synthesis of IPP, the common intermediate of isoprenoid biosynthesis, was examined first using qRT-PCR analysis of MVA and MEP pathway gene expression in rubber tree latex that had been treated with ethylene and in two rubber tree clones (or varieties) that were comparable in age and growth conditions but differed in latex carotenoid content. Secondly, conventional and next generation sequencing were used to determine the extent of metabolic outcomes of IPP in latex. Our results provide an overview of the synthesis and utilization of IPP in latex isoprenoid biosynthesis.
Materials and methods
Plant material and experimental trees
Latex samples were harvested from 17-year-old rubber trees under bi-weekly tapping in the Rubber Research Institute of Malaysia Research Station. Because these trees were not newly tapped for this work, the effect of ethylene treatment or tree clone variation on latex gene expression by qRT-PCR were not due to immediate wounding. Ethylene treatment was administered by applying ethephon (2-chloroethane phosphonic acid; 2.5% v/v) above the length of the tapping cut on the tree trunk. For analysis of the effect of ethylene, latex was collected from a group of five trees (tree clone RRIM 600) at these intervals: 1, 3, and 8 d after ethephon application. Concurrently, latex was collected from a corresponding group of five control (untreated) trees in the same plot on the same days to avoid the effects of day-to-day variation. For analysis of the tree clone effect, latex was collected on the same day from three trees each of clones RRIM 600 and PB 235. The same latex samples were used for total solids determination in latex. For mRNAseq data generation, latex was collected from three trees of clone RRIM 600.
RNA isolation and quality assessment
All latex samples for RNA isolation were collected directly into denaturing buffer and stored at –80 °C if not processed immediately. Total RNA isolation was performed as previously described by Kush et al. (1990). RNA integrity was assessed using the Agilent 2100 bioanalyser (Agilent Technologies, USA) based on the RNA Integrity Number (RIN). RIN values of total RNAs for qRT-PCR analysis were between 6–7.5 and for mRNAseq data generation was 8.
Latex ultracentrifugation and determination of total solids content
Latex samples of clones RRIM 600 and PB 235 were collected into chilled conical flasks on ice. The separation of latex into rubber and non-rubber zones (Moir, 1959) was performed by centrifuging 30 ml latex at 53 000 g for 1 h at 4 °C. As a means of estimating latex rubber content, the determination of total solids content from clones RRIM 600 and PB 235 was done by drying 20 ml of latex from each clone (in triplicate) in a Petri dish at 50 °C. Latex samples were dried to a thin film and the weight recorded for 48 h or until the weight became constant. Latex was collected from both clones on two different occasions for total solids content determination.
qRT-PCR analysis
Specific primers for genes encoding enzymes of the isoprenoid biosynthesis pathway were designed using the Beacon Designer version 4.0 software (Premier Biosoft International, USA) (see Supplementary Table S1a at JXB online). Determination of primer efficiency, SYBR Green quantification of transcript level by qRT-PCR (including ‘no template’ and ‘no reverse transcriptase’ control reactions) and melt curve analysis of amplification products were performed using the Rotor-Gene 3000 Real Time Thermal Cycler (Corbett Research, Australia) as previously described by Chow et al. (2007). All qRT-PCR reactions were performed in triplicate. The transcript level of each candidate gene was expressed as the Pfaffl ratio where CP was the cycle threshold generated by qRT-PCR and the 18S ribosomal RNA gene (18S rRNA) was the normalizer and calibrator (Pfaffl, 2001). The 18S rRNA gene expression level was 10 000 unless otherwise stated. The suitability of the 18S rRNA gene as a housekeeping gene was determined by the two-tailed Student’s t test for CP values obtained from RNA samples being compared (see Supplementary Table S1 and Fig. S1 at JXB online). For the analysis of the effect of ethylene, fold change in gene expression on three different days resulting from ethylene treatment was expressed as the average transcript level (Pfaffl ratio) of each gene in ethylene-treated latex over the average transcript level (Pfaffl ratio) in control latex (see Supplementary Table S1b, c at JXB online).
Statistical analysis of qRT-PCR expression data
CP values were presented as means ±standard deviation. Statistical significance between transcript levels (Pfaffl ratios) of candidate genes in samples compared was assessed using Student’s t test with P values indicated.
Sequence generation, analysis, and annotation
Sanger expressed sequence tag (EST) sequences were generated from the latex cDNA library reported previously and assembled into unique transcripts (UTs) as described by Chow et al. (2007). A total of 10 760 088 next generation (NG) reads (35 nt) were generated using a 150 bp fragment library constructed from latex total RNA via the Illumina Fast Track mRNAseq sequencing service (San Diego, USA). NG reads were assembled with Velvet 0.7.63 (Zerbino and Birney, 2008) using a hash length range of 15 to 31 in order to determine the optimal hash value for assembly. NG contigs (≥100 nt) were co-assembled with Sanger UTs (≥50 nt) using Phrap (www.phrap.org) based on the same parameters as those used for Sanger EST assembly. Sanger UTs, NG contigs, and hybrid contigs were classified into arbitrary nucleotide size classes in a composite histogram for the evaluation of size improvement after co-assembly.
BlastN (Altschul et al., 1990) analysis was used for mapping NG contigs (≥100 nt) to Sanger UTs where only hits with 100% matches to Sanger UTs were accepted. Rubber sequences containing complete protein coding regions were manually identified and compiled from the GenBank non-redundant database (www.ncbi.nlm.nih.gov). Megablast (Altschul et al., 1990) analysis was used to map these complete protein coding regions to Sanger UTs, NG contigs, and hybrid contigs. Functional annotation of the hybrid contigs was carried out using the BLAST2GO pipeline (blast2go.org) (Conesa et al., 2005). The hybrid contigs were also subjected to automated and manual curation for known pathways based on the KEGG Pathway database (Kanehisa and Goto, 2000).
Data availability
DNA sequence data have been deposited in the following National Centre for Biotechnology Information (NCBI) databases: latex ESTs in dbEST (accession nos. HS990039–HS999999, JG000001–JG014330), NG reads in the Sequence Read Archive (SRA) (accession no. SRA028457).
Results and discussion
Induction of MVA and MEP pathway gene expression due to latex ethylene stimulation
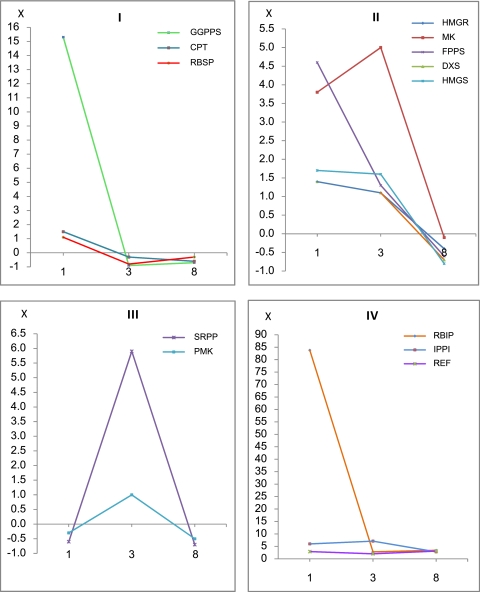
To address the question of whether IPP from the MEP pathway is used for cis-polyisoprene synthesis, a panel of candidate Hevea genes representing pre-IPP (MVA and MEP pathways) and post-IPP steps was selected for qRT-PCR expression profiling (see Supplementary Table S1a at JXB online). Within this panel, genes for post-IPP rubber biosynthesis steps were cis-prenyltransferase (CPT) (Asawatreratanakul et al., 2003), a rubber elongation factor (REF) (Dennis and Light, 1989), a small rubber particle protein (SRPP) (Oh et al., 1999), a rubber biosynthesis stimulator protein (RBSP) (Yusof et al., 2000), and a patatin-like inhibitor protein of rubber biosynthesis (RBIP) (Yusof et al., 1998), all of which have been demonstrated to affect IPP incorporation in vitro into rubber. The hypothesis was that the MEP pathway should not be affected under conditions where rubber biosynthesis was increased if IPP from this pathway was not utilized for cis-polyisoprene formation. Rubber trees (tree clone RRIM 600) were first treated with ethephon as the application of such ethylene-releasing chemicals to the tapping cut leads to prolonged latex flow and, thus, increased rubber production (Abraham et al., 1971). The effect of ethylene was confirmed by noting that the volume of latex collected from treated trees (after final drip) was higher than from control trees (data not shown). Subsequently, gene expression was compared between ethylene-treated and control trees at three time points after treatment (days 1, 3, and 8) based on the Hevea 18S rRNA gene as the housekeeping gene. The qRT-PCR results showed that all gene transcripts were induced by ethylene and that there were four trends of gene expression based on transcript level fold change over three sampling days (Fig. 2; see Supplementary Table S1b, c at JXB online). In particular, the induction of CPT, REF, SRPP, RBSP, and RBIP which are involved in post-IPP steps indicated that ethylene stimulation led to an increase in cis-polyisoprene synthesis. However, since both MVA and MEP pathway candidate genes were induced, the source of IPP for cis-polyisoprene synthesis was not distinguishable. Therefore, it was concluded that, while this experiment showed that ethylene increased rubber biosynthesis, it did not rule out the MEP pathway as an additional source of IPP.
Fig. 2.
Four trends of fold change in transcript level (I–IV) of rubber biosynthesis pathway genes after ethylene treatment as shown by qRT-PCR expression analysis. The x-axis shows three sampling days (days 1, 3, and 8 after ethylene treatment) and the y-axis shows gene expression fold change compared with untreated trees. Fold change in transcription resulting from ethylene treatment was expressed as the average transcript level (Pfaffl ratio) of each gene in ethylene-treated latex over the average transcript level (Pfaffl ratio) in control latex. A positive fold change indicated the induction of transcription by ethylene while a negative fold change indicated repression. (see Supplementary Table S1b, c at JXB online). REF, rubber elongation factor; SRPP, small rubber particle protein; RBSP, rubber biosynthesis stimulator protein; RBIP, rubber biosynthesis inhibitor protein; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; IPPI, isopentenyl diphosphate isomerase; CPT, cis-prenyltransferase; HMGS, hydroxymethyglutaryl coenzyme A synthase; HMGR, hydroxymethyglutaryl coenzyme A reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase.
Expression of MVA- and MEP-specific genes in relation to carotenoid production in latex
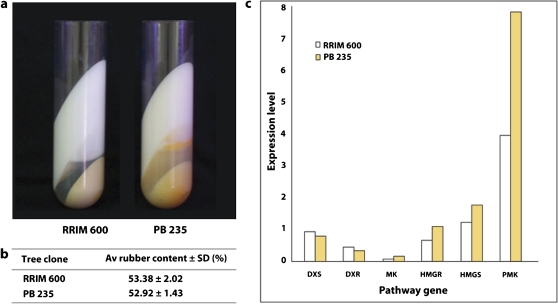
It was next investigated whether the MEP pathway contributes IPP to rubber biosynthesis by utilizing latex carotenoid content as a marker for pathway activity. Latex from PB 235, is visibly more yellow than that from RRIM 600 due to a higher amount of carotenoid-containing Frey-Wyssling particles. In ultracentrifuged latex, the presence of Frey-Wyssling particles is indicated by the yellow-orange pigment where they occur in the separated latex zones (Fig. 3) (Moir, 1959). RRIM 600 and PB 235 trees that were of the same age and grown under the same field management conditions were found for this experiment. This was an important condition to ensure that qRT-PCR expression profiles from two different tree clones were comparable. The latex rubber content in these two clones was determined first and it was found that both contained the same amount of rubber based on total solids content (Fig. 3). Subsequently, these two clones were chosen for comparative qRT-PCR expression of gene transcripts specific to the MVA and MEP pathways. Since clone PB 235 produced more carotenoids, expression of the MEP pathway enzymes, DXS and DXR, in PB 235 latex would be expected to be higher due to greater metabolic flux in this IPP-generating route. Instead, expression of MEP pathway gene transcript levels in PB 235 was lower than in RRIM 600 while, conversely, MVA pathway gene transcript levels in PB 235 were higher than in RRIM 600 (Fig. 4; see Supplementary Fig. S1 at JXB online). Therefore, to explain how the same amount of rubber is made by both clones in spite of these expression trends, it is plausible that, in RRIM 600 latex where less carotenoids are produced (and where MEP pathway gene transcription was noted to be higher), plastidic IPP is transported to the cytosol to compensate for the lower amount of IPP produced via the MVA pathway.
Fig. 3.
Comparative expression of MVA and MEP pathway-specific genes in rubber tree clones varying in latex carotenoid content. (a) Ultracentrifugation of RRIM 600 and PB 235 latex produced separated zones viewed from the side of the centrifuge tubes. PB 235 latex contains more carotenoids as shown by the more intense yellow-orange zones. (b) Latex from clones RRIM 600 and PB 235 contain the same amount of rubber based on total solids content. (c) Expression trends of gene transcripts specific to the MVA (HMGR, HMGS, PMK, MK) and MEP (DXS, DXR) pathways in tree clones RRIM 600 and PB 235. Both clones showed the same expression trends using latex RNAs from two biological replicates (see Supplementary Fig. S1 at JXB online). Av, average; SD, standard deviation; HMGS, hydroxymethyglutaryl coenzyme A synthase; HMGR, hydroxymethyglutaryl coenzyme A reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase.
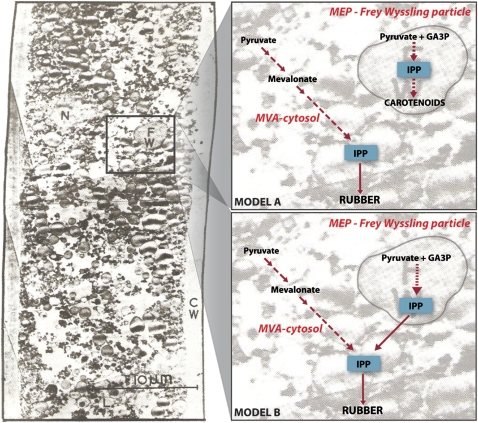
Fig. 4.
Spatial models of subcellular trafficking of IPP for cis-polyisoprene biosynthesis in latex. The electron micrograph on the left panel shows a section of a laticifer (latex vessel) and its subcellular structures (courtesy of Rubber Research Institute of Malaysia; CW, cell wall; FW, Frey-Wyssling; L, lutoid; N, nucleus). On the right are extrapolated panels showing two schemes of subcellular trafficking of IPP. Model A: In clones with a high carotenoid content, IPP utilization is partitioned whereby the MVA pathway supplies IPP for cis-polyisoprene synthesis in the cytosol while the MEP pathway supplies IPP for carotenoid biosynthesis in Frey-Wyssling particles. Clone PB 235 is an example. Model B: In clones which do not produce large amounts of carotenoids, IPP for cis-polyisoprene synthesis is supplied by both MVA and MEP pathways whereby IPP generated by the latter is transported from the Frey-Wyssling particle to the cytosol. Clone RRIM 600 is an example.
From the preceding observations, two models of IPP partitioning and utilization in latex are proposed that are dependent on the carotenoid content of different rubber tree clones (Fig. 4). In clones with a high carotenoid content (for example, PB 235), partitioning of IPP exists between carotenoid biosynthesis in Frey-Wyssling particles and cis-polyisoprene synthesis in the rubber particles of the cytosol. In clones which do not produce large amounts of carotenoids (for example, RRIM 600), the MEP pathway is an alternative provider of IPP for cis-polyisoprene synthesis in the rubber particles of the cytosol (Fig. 4). Here, cis-polyisoprene synthesis utilizes the IPP pool in the cytosol which is contributed by both the MVA and MEP pathways. In RRIM 600, this would also explain why MVA and MEP pathway genes were induced by ethylene (Fig. 2; see Supplementary Table S1b, c at JXB online), given that IPP from both pathways is channelled to make rubber. Previously, feeding of RRIM 600 seedlings with 13C-labelled 1-deoxy-D-xylulose triacetate (an intermediate derivative of the MEP pathway) implied that the MEP pathway contributed IPP for carotenoid biosynthesis only but not for rubber biosynthesis (Sando et al., 2008). It is therefore postulated further that subcellular partitioning of IPP is dependent on developmental stage. It is possible that, in a clone like RRIM 600, the plant initially generates IPP via independent cytosolic and plastidic routes in a young seedling but switches to the utilization of both routes in order to meet increased rubber biosynthesis demand in a mature rubber tree. The relative contribution of the MVA and MEP pathways in making different isoprenoid products and cross-talk between these pathways have been reported in plants (Kasahara et al., 2002, 2004; Laule et al., 2003; Dudareva et al., 2005; Towler et al., 2007; Skopurinska-Tudek et al., 2008). However, our observations point to the occurrence of two schemes of IPP partitioning and utilization within one species, either of which may operate according to changing physiological requirements in latex. Therefore, this suggests yet another level of control of isoprenoid biosynthesis in plants.
Mapping of post-IPP branch pathways by latex transcriptome analysis
From our expression profiling results, the choice of IPP generation pathway to enhance or repress should be based on the nature of IPP partitioning and utilization in the rubber tree clone to be genetically transformed. However, even if an increased level of cytosolic IPP was achieved, competition from other isoprenoid products in latex may divert IPP away from the cis-polyisoprene branch. Therefore, transcriptome sequences were generated to survey post-IPP products in latex. High-throughput sequencing of up to 10 000 Sanger ESTs from clone RRIM 600 was previously adopted for gene discovery in latex (Chow et al., 2007; Mat-Isa et al., 2007, 2009). Since a prerequisite for substantial coverage of pathways is a transcriptome resource with deep gene coverage, the Sanger EST collection was enlarged to 35 000 sequences and, in addition, NG data based on Illumina mRNAseq were generated. Subsequently, co-assembly of Sanger and NG sequences resulted in 23 257 hybrid contigs (Table 1). The rationale of hybrid assembly was to combine the benefits of Sanger (longer transcript assemblies) and NG (deeper sequence coverage) sequencing methods so that a significantly more comprehensive latex transcriptome could be obtained. At various stages of developing this resource, validation measures were introduced to ensure that the resulting hybrid contigs had been assembled correctly: optimization of hash length during NG contig assembly (see Supplementary Fig. S2 at JXB online), mapping of Sanger UTs to NG contigs (see Supplementary Fig. S2 at JXB online), analysis of hybrid contig size distribution (see Supplementary Fig. S3 at JXB online) and mapping of rubber complete protein coding regions to hybrid contigs (see Supplementary Tables S2 and S3 at JXB online). As a result, the 23 257 hybrid contigs obtained represent a deep latex transcriptome resource. Hybrid contigs and their functional annotations are shown in Supplementary Tables S4 and S5, respectively, at JXB online.
Table 1.
Statistics of sequence assemblies
| Number of sequences | Total length (nt) | |
| Sanger sequences for assembly | 34 152 | |
| Sanger UTs (≥50 nt) | 8 618 | 5 415 737 |
| Consensi | 3 999 | |
| Singletons | 4 619 | |
| NG contigs (≥100 nt) | 29 751 | 5 594 713 |
| Input sequence for hybrid assembly | ||
| (Sanger UTs + NG contigs) | 38 369 | |
| Hybrid contigs | 23 257 | 8 078 653 |
| Consensi | 5 924 | |
| Singletons | 17 333 |
Assembly of Sanger sequences and next generation (NG) reads resulted in 8618 unique transcripts (UTs) and 29 751 NG contigs, respectively. The rationale of hybrid assembly (23 257 contigs) was to combine the benefits of Sanger (longer transcript assemblies) and NG (deeper sequence coverage) sequencing methods so that a significantly more comprehensive latex transcriptome could be obtained.
Having annotated a transcriptome-wide set of latex genes, the pathways downstream of IPP formation were discovered by mapping the 23 257 hybrid contigs to biochemical pathways from all organisms in the KEGG Pathway database (Kanehisa and Goto, 2000). Hybrid contigs encoding enzymes of isoprenoid biosynthesis were mapped to the KEGG pathway named as ‘terpenoid backbone biosynthesis’. This pathway shows 11 classes of metabolic destinations of IPP, including cis-polyisoprene. Downstream of IPP, up to eight (of 11) branch pathways were found, including the branch leading to cis-polyisoprene, which showed hits to hybrid contigs (Fig. 1). Unlike cis-polyisoprene, investigations of non-rubber products are limited and had been done in isolation. Nonetheless, literature on sterols, tocotrienols, tocopherols, plastoquinone, carotenoids, and dolichols in latex were consistent with the non-rubber isoprenoids that were identified (Dunphy et al., 1965; Kekwick, 1989; Tateyama et al., 1999; Phatthiya et al., 2007). More importantly, additional classes of non-rubber isoprenoid end-products, not previously reported in literature, have been identified through this pathway mapping. Although there is no information on relative metabolic flux between post-IPP pathways in latex at this stage, some or all of these non-rubber pathways may be suitable targets for down-regulation so that post-IPP metabolic flux can be concentrated on cis-polyisoprene synthesis.
Harnessing latex isoprenoid pathway information for enhancing cis-polyisoprene synthesis
Knowledge of IPP generation and utilization in latex isoprenoid biosynthesis is a useful guide to the rational design of gene constructs for engineering transgenic trees with increased flux in the rubber branch. This is essential because evaluation of a transgene effect in latex can only be conducted meaningfully after transgenic rubber plants reach maturity and produce sufficient latex for analysis. New information is provided on gene expression trends which are potentially useful as indicators of isoprenoid pathway behaviour. Firstly, the question of whether the MEP pathway contributes IPP towards rubber was answered through insights gained into the dual schemes of IPP partitioning and utilization in latex. Therefore, a prerequisite for choosing to manipulate either the MVA or MEP pathway lies in the determination of the nature of IPP trafficking between the cytosol and the plastid in the rubber tree clone to be transformed. Secondly, the spectrum of non-rubber pathways that was found in latex serves as a roadmap for developing strategies of redirecting IPP from competing pathways to cis-polyisoprene biosynthesis. In this respect, the hybrid contig collection is a valuable and extensive source of gene transcripts for the design of relevant over-expression or anti-sense constructs for enhancing cis-polyisoprene biosynthesis in transgenic rubber trees. Nonetheless, given the complexity of isoprenoid biosynthesis, future elucidation of other aspects such as a clearer understanding of metabolic fluxes and other pathway regulatory features would serve to further fine-tune approaches in metabolic engineering of rubber biosynthesis in vivo.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. qRT-PCR analysis of MVA and MEP pathway-specific gene transcript levels in RRIM 600 and PB 235.
Supplementary Fig. S2. NG contig assembly and validation.
Supplementary Fig. S3. Contig size distribution across three assembly data sets: Sanger UTs, NG contigs, and hybrid contigs.
Supplementary Table S1. qRT-PCR analysis of transcript levels of rubber biosynthesis pathway genes in ethylene-treated trees.
Supplementary Table S2. Interrogation of Sanger UTs, NG contigs, and hybrid contigs with 195 annotated rubber cDNAs to obtain a measure of complete protein coding regions.
Supplementary Table S3. List of 195 rubber cDNAs with complete protein coding regions, functional description, and tissue of origin.
Supplementary Table S4. Hybrid contig sequences in fasta format.
Supplementary Table S5. BLAST2GO annotations of hybrid contigs.
Acknowledgments
The authors thank Paul Dear for critical reading of the manuscript and Mohd. Firdaus Raih for useful insights and opinions. Siti-Zakiah Zailani and V. Mony Rajan are thanked for their valuable assistance in the rubber laboratory and field experiments. Khairil-Anuar Zainal is gratefully acknowledged for producing the manuscript figures. This work was funded by the Malaysian Rubber Board and the Agro-Biotechnology Institute of the Malaysian Ministry of Science, Technology, and Innovation.
Glossary
Abbreviations
- EST
expressed sequence tag
- IPP
isopentenyl diphosphate
- MEP
2-C-methyl-D-erythritol 4-phosphate
- MVA
mevalonate
- NG
next generation
- UT
unique transcript
References
- Abraham PD, Blencowe JW, Chua SE, Gomez JB, Moir GFJ, Pakianathan SW, Sekhar BC, Southorn WA, Wycherley PR. Novel stimulants and procedures in the exploitation of Hevea. II. Pilot trial using (2-chloroethyl)-phosphonic acid (ethephon) and acetylene with various tapping systems. Journal of the Rubber Research Institute of Malaya. 1971;23:90–113. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Archer BL, Audley BG. New aspects of rubber biosynthesis. Botanical Journal of the Linnean Society. 1987;94:181–196. [Google Scholar]
- Archer BL, Audley BG, Cockbain EG, McSweeney GP. The biosynthesis of rubber. Incorporation of mevalonate and isopentenyl pyrophosphate into rubber by Hevea brasiliensis latex fractions. Biochemical Journal. 1963;89:565–574. doi: 10.1042/bj0890565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asawatreratanakul K, Zhang YW, Wititsuwannakul D, Wititsuwannakul R, Takahashi S, Rattanapittayaporn A, Koyama T. Molecular cloning, expression and characterisation of cDNA encoding cis-prenyltransferases from Hevea brasiliensis. A key factor participating in natural rubber biosynthesis. European Journal of Biochemistry. 2003;270:4671–4680. doi: 10.1046/j.1432-1033.2003.03863.x. [DOI] [PubMed] [Google Scholar]
- Bick JA, Lange BM. Metabolic cross-talk between cytosolic and plastidial pathways of isoprenoid biosynthesis: unidirectional transport of intermediates across the chloroplast envelope membrane. Archives of Biochemistry and Biophysics. 2003;415:146–154. doi: 10.1016/s0003-9861(03)00233-9. [DOI] [PubMed] [Google Scholar]
- Chappell J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annual Reviews of Plant Physiology. 1995;46:521–547. doi: 10.1104/pp.107.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow KS, Wan KL, Mat Isa MN, Bahari A, Tan SH, Harikrishna K, Yeang HY. Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. Journal of Experimental Botany. 2007;58:2429–2440. doi: 10.1093/jxb/erm093. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualisation and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- d’Auzac J, Jacob JL. The composition of latex from Hevea brasiliensis as a laticiferous cytoplasm. In: d’Auzac J, Jacob JL, Chrestin L, editors. Physiology of rubber tree latex. Boca Raton, FL: CRC Press; 1989. pp. 59–96. [Google Scholar]
- de Faÿ E, Jacob JL. Anatomical organisation of the laticiferous system in the bark. In: d’Auzac J, Jacob JL, Chrestin L, editors. Physiology of rubber tree latex. Boca Raton, FL: CRC Press; 1989. pp. 3–14. [Google Scholar]
- Dennis MS, Light DR. Rubber elongation factor from Hevea brasiliensis. Identification, characterisation and role in rubber biosynthesis. Journal of Biological Chemistry. 1989;264:18608–18617. [PubMed] [Google Scholar]
- Dickenson PB. Electron microscopical studies of latex vessel system of Hevea brasiliensis. Journal of the Rubber Research Institute of Malaya. 1969;21:543–559. [Google Scholar]
- Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proceedings of the National Academy of Sciences, USA. 2005;102:933–938. doi: 10.1073/pnas.0407360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy PJ, Whittle KJ, Pennock JF, Morton RA. Identification and estimation of tocotrienols in Hevea latex. Nature. 1965;207:521–522. [Google Scholar]
- Eisenreich W, Rohdich F, Bacher A. Deoxyxylulose phosphate pathway to terpenoids. Trends in Plant Science. 2001;6:78–84. doi: 10.1016/s1360-1385(00)01812-4. [DOI] [PubMed] [Google Scholar]
- Gomez JB, Moir GFJ. The ultracytology of latex vessels in Hevea brasiliensis. 1979 Monograph No. 4. Malaysian Rubber Research and Development Board, Kuala Lumpur. [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. Journal of Biological Chemistry. 2002;277:45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H. Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. Journal of Biological Chemistry. 2004;279:14049–14054. doi: 10.1074/jbc.M314195200. [DOI] [PubMed] [Google Scholar]
- Kekwick RGO. The formation of isoprenoids in Hevea latex. In: d’Auzac J, Jacob JL, Chrestin L, editors. Physiology of rubber tree latex. Boca Raton, FL: CRC Press; 1989. pp. 145–164. [Google Scholar]
- Ko JH, Chow KS, Han KH. Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubber tree) Plant Molecular Biology. 2003;53:479–492. doi: 10.1023/B:PLAN.0000019119.66643.5d. [DOI] [PubMed] [Google Scholar]
- Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annual Review of Plant Biology. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- Kush A, Goyvaerts E, Chye ML, Chua NH. Laticifer-specific gene expressions in Hevea brasiliensis (rubber tree) Proceedings of the National Academy of Sciences, USA. 1990;87:1787–1790. doi: 10.1073/pnas.87.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule O, Fűrholz A, Chang HS, Zhu T, Wang X, Heifetz PB, Gruissem W, Lange M. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2003;100:6866–6871. doi: 10.1073/pnas.1031755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mat-Isa MN, Chow KS, Mohamad AFH, Shahrum MY, Hoh CC, Mohd-Amin MR, Zainal KA, Yeang HY, Wan KL. NRESTdb: access to the transcriptome of natural rubber latex. Journal of Rubber Research. 2009;12:229–238. [Google Scholar]
- Mat-Isa MN, Mohamad AFH, Shahrum MY, Hoh CC, Mohd-Amin MR, Zainal KA, Yeang HY, Wan KL, Chow KS. 2007. NRESTdb: a Natural Rubber Expressed Sequence Tag Database ( http://genome.ukm.my/nrestdb/). Molecular Biology Database Collection entry no. 921. Nucleic Acids Research 35.
- Moir GFJ. Ultracentrifugation and staining of Hevea latex. Nature. 1959;184:1626–1628. [Google Scholar]
- Oh SK, Kang H, Shin DH, Yang J, Chow KS, Yeang HY, Wagner B, Breiteneder H, Han KY. Isolation, characterisation and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. Journal of Biological Chemistry. 1999;274:17132–17138. doi: 10.1074/jbc.274.24.17132. [DOI] [PubMed] [Google Scholar]
- Phatthiya A, Takahashi S, Chareonthiphakorn N, Koyama T, Wititsuwannakul D, Wititsuwannakul R. Cloning and expression of the gene encoding solanesyl diphosphate synthase from Hevea brasiliensis. Plant Science. 2007;172:824–831. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M. Early steps in isoprenoid biosynthesis: multilevel regulation of the supply of common precursors in plant cells. Phytochemistry Reviews. 2006;5:1–15. [Google Scholar]
- Rodríguez-Concepción M. Supply of precursors for carotenoid biosynthesis in plants. Archives of Biochemistry and Biophysics. 2010;504:118–122. doi: 10.1016/j.abb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Sando T, Takeno S, Watanabe N, Okumoto H, Kuzuyama T, Yamashita A, Hattori M, Ogasawara N, Fukusaki E, Kobayashi A. Cloning and characterisation of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway genes of a natural-rubber producing plant Hevea brasiliensis. Bioscience, Biotechnology and Biochemistry. 2008;72:2903–2917. doi: 10.1271/bbb.80387. [DOI] [PubMed] [Google Scholar]
- Skopurinska-Tudek K, Poznanski J, Wojcik J, et al. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of dolichols in plants. Journal of Biological Chemistry. 2008;283:21024–21035. doi: 10.1074/jbc.M706069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateyama S, Wititsuwannakul R, Wititsuwannakul D, Sagami H, Ogura K. Dolichols of rubber plant, ginkgo and pine. Phytochemistry. 1999;51:11–15. [Google Scholar]
- Towler MJ, Weathers PJ. Evidence of artemisinin production from IPP stemming from both the mevalonate and the nonmevalonate pathways. Plant Cell Reports. 2007;26:2129–2136. doi: 10.1007/s00299-007-0420-x. [DOI] [PubMed] [Google Scholar]
- Yusof F, Chow KS, Ward MA, Walker JM. A stimulator protein of rubber biosynthesis from Hevea brasiliensis latex. Journal of Rubber Research. 2000;3:232–249. [Google Scholar]
- Yusof F, Ward MA, Walker JM. Purification and characterisation of an inhibitor of rubber biosynthesis from C-serum of Hevea brasiliensis latex. Journal of Rubber Research. 1998;1:95–110. [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequence data have been deposited in the following National Centre for Biotechnology Information (NCBI) databases: latex ESTs in dbEST (accession nos. HS990039–HS999999, JG000001–JG014330), NG reads in the Sequence Read Archive (SRA) (accession no. SRA028457).