Abstract
Previously, the OEP16.1 channel pore in the outer envelope membrane of mature pea (Pisum sativum) chloroplasts in vitro has been characterized to be selective for amino acids. Isolation of OEP16.2, a second OEP16 isoform from pea, in the current study allowed membrane localization and gene expression of OEP16 to be followed throughout seed development and germination of Arabidopsis thaliana and P. sativum. Thereby it can be shown on the transcript and protein level that the isoforms OEP16.1 and OEP16.2 in both plant species are alternating: whereas OEP16.1 is prominent in early embryo development and first leaves of the growing plantlet, OEP16.2 dominates in late seed development stages, which are associated with dormancy and desiccation, as well as early germination events. Further, OEP16.2 expression in seeds is under control of the phytohormone abscisic acid (ABA), leading to an ABA-hypersensitive phenotype of germinating oep16 knockout mutants. In consequence, the loss of OEP16 causes metabolic imbalance, in particular that of amino acids during seed development and early germination. It is thus concluded that in vivo OEP16 most probably functions in shuttling amino acids across the outer envelope of seed plastids.
Keywords: Abscisic acid, amino acid transport, Arabidopsis, outer envelope protein, pea, plastid, seed development, seed metabolite content
Introduction
Seed development and germination of seed plants in general can be subdivided into a series of interlocking processes (for overviews, see Finkelstein et al., 2008; Holdsworth et al., 2008a; Angelovici et al., 2010). Immediately after fertilization, morphogenesis determines embryo development and leads to the establishment of all embryonic structures until the heart stage. Seed maturation then begins with a transition from maternal—represented by the seed coat compartment—to filial control (Weber et al., 2005), during which embryos and endosperm grow by cell division. First in seed maturation, a period of reserve substance accumulation includes metabolic reorganization and synthesis of storage compounds such as starch, storage proteins, and oil. Later on, a major loss of water leads to desiccation-tolerant, mature dry seeds. In parallel, initiation and establishment of dormancy accompany these two seed maturation phases. Thus, acquirement of desiccation-tolerant seeds is highly associated with dormancy, allowing the dry embryo to maintain viability in the dormant stage as well as to switch to germination under the respective favourable conditions (see below). Seed dormancy is maximal in harvest-ripe seeds, but afterwards declines during dry storage in the after-ripening stage. Mature dry seeds contain a large number of mRNA species, which are mainly associated with metabolism, protein synthesis, and degradation, and thus provide important transcripts for early stages of germination (Nakabayashi et al., 2005). Expression of these genes in ripening seeds is mostly induced by the phytohormone abscisic acid (ABA) as indicated by an over-representation of cis-regulatory ABA-responsive elements (ABREs) in their promoter regions. Dormancy in Arabidopsis embryos in general is non-deep and is imposed by endosperm and testa tissue. However, for subsequent germination, dormancy has to be relieved by a concerted sequence of processes, which are after-ripening (i.e. dry storage of seeds), followed by cold (or nitrate) treatment, and finally light exposure (for a review, see Holdsworth et al., 2008b). In contrast to ABA, which acts as a positive regulator of dormancy, gibberellins release dormancy and promote progression of germination. In Arabidopsis, the endosperm tissue in mature, dry seeds, in addition to controlling seed dormancy has been proposed also to be an important regulator of the germination potential (for a review, see Holdsworth et al., 2008a). Water uptake during germination finally can be described by several steps such as imbibition, reactivation of metabolic processes, radicle emergence through the endosperm and testa layers, as well as radicle elongation.
The switch from reserve accumulation to desiccation stages in seed development is accompanied by pronounced changes in transcripts and metabolites (reviewed in Angelovici et al., 2010). In consequence, not only the reserve accumulation phase but also the desiccation period during seed maturation is a prerequisite for mobilization of stored metabolites throughout germination of seeds. Highly active metabolic processes during desiccation and dormancy are thus related to those dominating seed germination, thereby bridging seed maturation with germination. Modulation of metabolic processes in turn involves biosynthesis and intracellular redistribution of solutes and nutrients. During seed development many of the required metabolites, however, are synthesized and/or metabolized in plastids. The plastid organelle family of higher plants, which evolved after endosymbiosis of a progenitor of today’s cyanobacteria with an ancient mitochondria-containing eukaryotic cell (Gould et al., 2008), conducts vital biosynthetic functions throughout plant growth and development (for a review, see Tetlow et al., 2005). In contrast to photosynthetically active chloroplasts, storage plastids are heterotrophic organelles that convert photosynthates derived from source tissue into storage compounds that can be mobilized during plant development (for an overview, see Lopez-Juez and Pyke, 2005). Amyloplasts (starch) in the endosperm of seeds, in cotyledons, tubers, or fruits, elaioplasts (oil) in seeds of oilseed plants, and chromoplasts (carotenoids) in flowers and fruits represent the major storage-type plastids. Besides photosynthesis, biosynthetic capacities of plastids include primary assimilation of nitrogen and sulphur, and synthesis of amino acids, fatty acids, tetrapyrroles, nucleic acids, hormones, and many other compounds of secondary metabolism. Subsequently, these plastid-derived metabolic intermediates are delivered to other cell compartments and provided for the synthesis of complex compounds such as polypeptides or storage sugars, or may even serve as signalling molecules. The importance of plastids, in particular during embryo and seed development, is underlined by the fact that the loss of function of plastid-localized proteins very often results in embryo lethality (Bryant et al., 2011, and references therein). Besides protein import and translation, functions of embryo-essential plastid proteins are linked to synthesis of indispensable metabolites such as amino acids, vitamins, nucleotides, and fatty acids. Since plastids, like their Gram-negative ancestors, are delimited by two membranes, solute and metabolite transport across both of these envelopes represents a central bottleneck within the intracellular network during seed development and germination (for overviews on plastid solute transport, see Philippar and Soll, 2007; Linka and Weber, 2010).
In the plastid outer envelope membrane, members of the OEP16 solute channels (for outer envelope protein of 16 kDa) are found to be specifically expressed in embryo and seed development (for an overview, see Pudelski et al., 2010). Originally, OEP16—designated as the OEP16.1 isoform in the following—was isolated as a 16 kDa protein from the outer envelope membrane of pea (Pisum sativum) chloroplasts (Pohlmeyer et al., 1997). The corresponding recombinant protein Ps-OEP16.1 formed a slightly cation-selective, high conductance channel when reconstituted into black lipid bilayers in vitro. Further, Ps-OEP16.1 in liposome swelling assays displayed a strikingly high selectivity for amino acids and amines. The predicted OEP16 pore has a diameter of ∼1 nm, but was impermeable to 3-phosphoglycerate and uncharged sugars such as fructose, glucose, sucrose, and sorbitol (Pohlmeyer et al., 1997). Therefore, Ps-OEP16.1 represents the plastid OEP channel with the highest selectivity and shows functional characteristics similar to those of specific metabolite channels in the outer membrane of Gram-negative bacteria (for an overview, see Duy et al., 2007). OEP16 consists of four membrane-spanning α-helices and most probably is built by OEP16 homo-oligomers (Pohlmeyer et al., 1997; Steinkamp et al., 2000; Linke et al., 2004). The α-helical structure of OEP16 is in contrast to other known OEPs, which form a β-barrel pore in the plastid outer envelope membrane like porin-like channels from the outer membrane of Gram-negative bacteria (compare Duy et al., 2007). During evolution, presumably before seed plants and bryophytes split, gene duplication in an ancestor of all land plants resulted in OEP16.2, a second isoform (Drea et al., 2006). Interestingly, all OEP16.2 proteins from angiosperms thereby gained an additional exon in the loop sequence connecting the first and the second membrane-spanning helices. This peptide loop—called the S-domain by Drea and co-workers—is the potential selectivity filter of the channel (Steinkamp et al., 2000; Linke et al., 2004), and might thus indicate that OEP16.2 has a slightly different substrate specificity from that of OEP16.1. In addition, the S-domain is specific for seed plants because it is absent in the OEP16.2 orthologues from mosses and ferns. Therefore, it is speculated that with the S-domain in OEP16.2, a seed-specific function was introduced for the OEP16 proteins (Drea et al., 2006).
Apart from the in vitro biochemical and electrophysiological characterization of Ps-OEP16.1, studies on the in vivo function in recent years have focused on OEP16 proteins and mutants in the model plant Arabidopsis thaliana (Philippar et al., 2007; Pollmann et al., 2007; Pudelski et al., 2009; Samol et al., 2011a, b). Here the isoforms At-OEP16.1 (At2g29800), At-OEP16.2 (At4g16160), and At-OEP16.4 (At3g62880) comprise the OEP16 subfamily, which groups into the PRAT (pre-protein and amino acid transporter) superfamily (Murcha et al., 2007; Pudelski et al., 2010). Common for these PRAT proteins is a four α-helical secondary structure, the presence of a TIM17 domain, and the absence of a classical chloroplast transit peptide. According to various transcriptomic analyses and its presence determined by several proteomic approaches in isolated Arabidopsis chloroplasts, cauliflower proplastids, and purified chloroplast envelope membranes from Arabidopsis, pea, and maize, the abundance and importance of the OEP16.1 isoform in leaf tissue is evident (Froehlich et al., 2003; Ferro et al., 2003; Kleffmann et al., 2004; Zybailov et al., 2008; Bräutigam et al., 2008a, b; Bräutigam and Weber, 2009). RNA of OEP16.2 instead shows strong and exclusive localization in seed, embryo, cotyledon, and pollen, while OEP16.4 transcripts are present ubiquitously at a lower level (for an overview, see Pudelski et al., 2010). The presence of OEP16.2 in seed development was confirmed by in situ hybridization and promoter–GUS (β-glucuronidase) analysis when Drea et al. (2006) detected the highest activity of the Arabidopsis OEP16.2 promoter, which contains four ABREs, in embryos of mature, dry, and germinating seeds of transformed tobacco plants.
Although OEP16.1 and OEP16.2 genes have been sequenced in various plant species (Drea et al., 2006), an OEP16.2 isoform of pea has not been discovered so far. Further, data on the expression specificity of OEP16.2 are based on transcript analysis only. To analyse and compare membrane localization and orientation of the OEP16.1 and OEP16.2 isoforms, OEP16.2 was therefore isolated from P. sativum. This enabled the expression pattern and hormonal regulation of both OEP16 isoforms to be followed at the transcript and protein level in two plant species (Arabidopsis and pea). Especially during seed development and germination, OEP16.1 and OEP16.2 showed an alternating and complementing expression pattern. Because OEP16.2 proved to be specifically induced by the phytohormone ABA in late seed maturation and early germination, the focus was on these tissues for analysis of OEP16 loss-of-function mutants in Arabidopsis. Thereby, it can be shown that the plastid outer envelope proteins OEP16, and in particular the isoform OEP16.2, affect metabolic fluxes during ABA-controlled seed development and germination.
Materials and methods
Plant material and growth conditions
Experiments on A. thaliana were performed with ecotype Col-0 (Lehle Seeds, Round Rock, TX, USA) and oep16 mutant plants. The oep16 triple mutant oep16.1/2/4 was generated by crossing the double mutant oep16.1-1/oep16.4-2 with oep16.2-1, both representing knockout lines for OEP16.1, OEP16.4, and OEP16.2, respectively (Philippar et al., 2007). In the subsequent F3 generation, the following mutant lines were selected by PCR genotyping: (i) oep16.1/2/4, homozygous for all three OEP16 alleles; (ii) OEP16.1/2/4-WT, corresponding to the respective wild-type genotypes for OEP16.1, OEP16.2, and OEP16.4; and with (iii) oep16.1/2; (iv) oep16.1/4; and (v) oep16.2/4 all possible combinations of double mutants, containing the wild-type form of the appropriate third OEP16 isoform. The original single OEP16 T-DNA insertion lines oep16.1-1 (SALK_024018), oep16.2-1 (SAIL #1377_1225_B03), and oep16.4-2 (SALK_109275) as well as PCR genotyping conditions are described in Philippar et al. (2007). Except for oep16.1/4, additional T-DNA insertions identified in some progeny of oep16.1-1 (for details, see Pudelski et al., 2009) were absent in oep16 double and triple mutant lines. Loss of OEP16 transcripts and proteins was concomitantly controlled by RT-PCR and immunoblots (see Supplementary Fig. S1 available at JXB online).
Before sowing, Arabidopsis seeds were surface-sterilized with 5% hypochloride and ethanol. To synchronize germination, all seeds were kept at 4° C for 2–3 d. Seedlings were grown on plates [0.3% Gelrite medium (Roth, Karlsruhe, Germany); 1% d-sucrose; 0.5× MS salts, pH 5.7], and mature plants were gown on soil (Stender substrate A210, Stender AG, Schermbeck, Germany). Plant growth occurred in growth chambers with a 16 h light (21° C; photon flux density of 100 μmol m−2 s−1) and 8 h dark (16° C) cycle. ABA germination assays were performed essentially as described previously (Drechsel et al., 2010). For germination on nylon filters, the filters were placed into Petri dishes and soaked in liquid medium.
Wild-type pea plants (P. sativum L., cv. ‘Arvica’, Prague, Czech Republic) were grown on sand in the greenhouse under a 16 h light (220 μmol m−2 s−1)/8 h dark regime at 21° C. The pea mutants snf34 (SnRK1 antisense, named Vic-34 in Radchuk et al., 2006) and anti-ABA as well as the corresponding wild-type lines are described in Radchuk et al. (2006) and Radchuk et al. (2010a), respectively.
Isolation of Ps-OEP16.2 cDNA
Degenerated oligonucleotide primers, directed toward conserved regions of known plant OEP16.2 isoforms (Drea et al., 2006) and designed according to the sequence of OEP16.2 from Medicago truncatula and the P. sativum codon usage, were used to amplify a corresponding region from reverse-transcribed pea seedling RNA. Using the SMART RACE cDNA Amplification Kit (Clontech) in combination with gene-pecific primers, overlapping N- and C-terminal fragments of Ps-OEP16.2 were subsequently isolated. The corresponding full-length cDNA was generated in a single PCR step by using primers flanking the 5' and 3' ends of the predicted open reading frame. For primer sequences, see Supplementary Table S1 at JXB online.
Protein extraction
Seed material from pea or Arabidopsis was homogenized in liquid nitrogen and resuspended in H2O. For protein isolation, 200 μl of this suspension were extracted by adding 800 μl of methanol (99.9%), 200 μl of chloroform (99%), and 600 μl of H2O. Solutions were mixed by inversion of the reaction tube until phase separation (∼10 min). After centrifugation for 10 min, 16 500 g at 4° C, the aqueous top phase was removed and discarded. A 600 μl aliquot of methanol was added and thoroughly mixed to precipitate proteins from the interphase. Proteins were collected by centrifugation for 5 min, 16 500 g at 4° C. The pellet was dried completely at 37° C and proteins were resuspended in 100 μl of extraction buffer [50 mM TRIS-HCl, pH 8; 50 mM EDTA; 2% LiDS; 10 mM dithiothreitol (DTT); 0.1 mM phenylmethylsulphonyl fluoride (PMSF)].
For protein extraction from leaf and seedlings, tissue was ground in liquid nitrogen, mixed with an equal volume of extraction buffer (50 mM TRIS-HCl, pH 8; 50 mM EDTA; 2% LiDS; 10 mM DTT; 0.1 mM PMSF) and incubated for 30 min on ice. Afterwards, insoluble cell debris was separated by centrifugation for 15 min, 16 000 g at 4° C.
Preparation and thermolysin treatment of plastid envelope membrane vesicles
Separated outer and inner envelope fractions from pea chloroplasts were isolated as described in Waegemann et al. (1992) from leaf tissue of 10-day-old pea plantlets. To detect OEP16.2 in outer envelope membranes, very young, 7-day-old, leaves were used.
For treatment with the protease thermolysin, outer envelope vesicles were precipitated at 100000g for 15 min at 4° C and resuspended in buffer ‘pwII’ (330mM sorbitol; 50mM HEPES, pH 7.6; 0.5mM CaCl2,). After determination of the total protein content, 2μg of thermolysin dissolved in pwII buffer were added per 1μg of envelope protein, and proteolysis was allowed to proceed for 0, 15, 30, and 45min on ice in the dark. Outer envelope membranes that were solubilized by the addition of 1% Triton X-100 prior to thermolysin treatment served as a control. All reactions were stopped by adding EDTA at a final concentration of 5 mM, separated by SDS–PAGE, and subjected to immunoblot analysis.
Immunoblot analysis
To generate antibodies against Ps-OEP16.2, the corresponding full-length cDNA was subcloned via EcoRI/XhoI into pET21a (Novagen) plasmid vector, resulting in a C-terminal fusion with a polyhistidine (His6) tag. The generated plasmid was used for overexpression of recombinant Ps-OEP16.2 after transforming Escherichia coli BL(DE3) cells (Novagen). Rapidly growing cells with a density of OD600=0.6 were induced with 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) for 3 h at 37° C. Afterwards, pelleted cells were resuspended in lysis solution [50 mM TRIS-HCl, pH 8.0, 25% sucrose (w/v), 1 mM EDTA, 100 mg ml−1 DNase] and sonicated three times for 30 s. Inclusion bodies were collected by centrifugation at 4° C, 20 000 g for 30 min and solubilized in buffer A (20 mM NaPP, pH 8.0, 100 mM NaCl, 2 mM β-mercaptoethanol, 8M urea) by sonification. Insoluble material was pelleted by centrifugation at 4° C, 20 000 g for 10 min. The supernatant, which contained the majority of the recombinant Ps-OEP16.2, was loaded onto an NiNTA affinity column (Qiagen), washed with buffer A with ascending imidazole concentrations (0, 20, 50 mM), and eluted with 100 mM imidazole. The resulting purified, recombinant Ps-OEP16.2 protein was used to generate antiserum in rabbit (Pineda Antibody Service, Berlin, Germany).
All other antisera were raised in rabbit against heterologously expressed proteins as described previously for Arabidopsis At-OEP16.1, At-OEP16.2 (Philippar et al., 2007), At-OEP37 (Goetze et al., 2006), and At-PIC1 (Duy et al., 2007), as well as pea Ps-OEP16.1 (Pohlmeyer et al., 1997) and Ps-VDAC (Clausen et al., 2004), respectively.
Appropriate amounts of organellar or total cellular proteins were separated by SDS–PAGE and subjected to immunoblot analysis using antisera in 1:200 to 1:1000 dilutions in TTBS buffer [100 mM TRIS-HCl, pH 7.5, 150 mM NaCl; 0.2% Tween-20; 0.1% bovine serum albumin (BSA)]. Non-specific signals were blocked by 1–5% skim milk powder and 0.03–0.12% BSA. Secondary anti-rabbit IgG alkaline phosphatase and anti-rabbit IgG horseradish peroxidase antibodies (Sigma-Aldrich) were diluted 1:30 000 and 1:8000, respectively. Blots were either stained for alkaline phosphatase reaction with 0.3 mg ml−1 nitroblue tetrazolium (NBT) and 0.16 mg ml−1 BCIP in 100 mM TRIS pH 9.5, 100 mM NaCl, 5 mM MgCl2 or for enhanced chemoluminescence (ECL; with horseraddish peroxidase) for 1 min in the dark with 0.5% (w/v) luminol, 0.22% (w/v) coumaric acid, 0.009% (v/v) H2O2 in 100 mM TRIS-HCl pH 8.5. Luminescence was monitored by X-ray films (Kodak Biomax MR; Perkin Elmer).
Promoter–GUS staining
For promoter–GUS analysis, 1501 bp and 729 bp regions upstream of the designated start codons of At-OEP16.1 and At-OEP16.2, respectively, were PCR amplified on genomic DNA by gene-specific primers (Supplementary Table S1 at JXB online). The resulting products were subcloned into the binary pKGWFS7 vector (Karimi et al., 2005). Generation of stable transformants in Arbidopsis Col-0 and histochemical GUS analysis were performed as described in Goetze et al. (2006).
Metabolite analysis
Carbohydrates and free amino acids of ∼50 mg of seed and seedling tissue were extracted and analysed as described previously (Kogel et al., 2010). Amino acid analysis on total seed proteins was performed by Genaxxon Bioscience (Ulm, Germany).
Results
Structure and topology of OEP16 in the outer envelope membrane of plastids
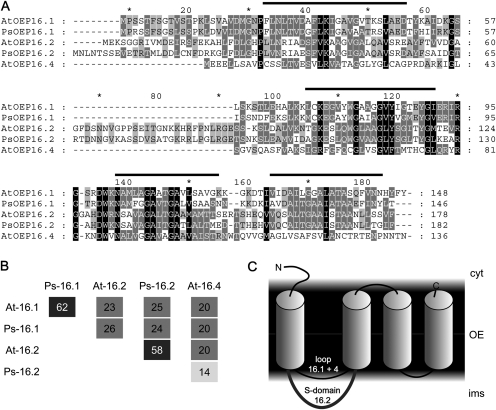
For isolation of Ps-OEP16.2, degenerated oligonucleotide primers were used to PCR-amplify a fragment on pea leaf cDNA. Subsequently, a cDNA, which contained an 546 bp long open reading frame as well as a 5′ untranslated region (UTR; 121 bp) and a 3′UTR (150 bp), was identified by RACE (rapid amplification of cDNA ends)-PCR (GenBank accession no. JN859826). The corresponding Ps-OEP16.2 protein is 183 amino acids long (Fig. 1A) and has a predicted molecular mass of 19.2 kDa. When compared with the Arabidopsis orthologue At-OEP16.2 (179 amino acids, 18.8 kDa), Ps-OEP16.2 shows 58% amino acid identity (Fig. 1B), while the closest relative can be found in M. truncatula (GenBank accession no. BI312211, 86% identity with Ps-OEP16.2). As defined for Ps-OEP16.1 (Linke et al., 2004) and described for all members of the PRAT superfamily (Murcha et al., 2007; Pudelski et al., 2010), four membrane-spanning α-helical domains are likely for Ps-OEP16.2 (Fig. 1A, C). Further, Ps-OEP16.2 contains an extended interhelical loop region between the first and second membrane helix, which is believed to be specifically conserved and inherited throughout evolution of angiosperm seed plants and therefore designated as the S-domain (Drea et al., 2006).
Fig. 1.
Structure and membrane topology of OEP16 proteins. (A) Amino acid sequence alignment of OEP16 isoforms from Arabidopsis and pea. Bars indicate the positions of the four membrane-spanning α-helices as defined for Ps-OEP16.1 (Linke et al., 2004). Identical and similar amino acids shared by all (100%), three or more (≥60%), and two (≥40%) of the sequences are shaded in black, dark grey, and light grey, respectively. (B) Percentage of identical amino acids between the proteins aligned in A. Identities among distinct OEP16.1 and OEP16.2 isoforms of different species are >58% (dark grey squares). (C) Topology model of OEP16 proteins in the outer envelope membrane (OE) of plastids. α-Helical membrane domains (cylinders) are depicted as defined for Ps-OEP16.1 (Linke et al., 2004). Please note that only the N-terminus and the extended interhelical loop region between the first and second helix (compare with alignment in A), which defines the S-domain of OEP16.2 (Drea et al., 2006), most probably are not embedded in the lipid bilayer environment. Cyt, cytosol; ims, intermembrane space.
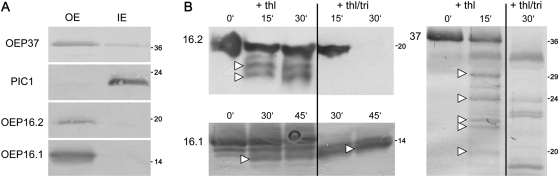
Generation of a specific antibody against Ps-OEP16.2 allowed localization of the 19 kDa protein in outer envelope membranes of very young chloroplasts from pea seedlings (Fig. 2A). By protease treatment of these envelope vesicles and subsequent immunoblot analysis (Fig. 2B), it was possible to determine the orientation of OEP16.1 and OEP16.2 proteins within the membrane (see Fig. 1C). In general, it has been shown that purified outer envelope membranes from isolated pea chloroplasts form vesicles with right-side-out orientation (Keegstra and Youssif 1986; Waegemann et al., 1992). Thus, the protease thermolysin is able to proteolyse only those peptide stretches which are exposed to the cytosol (i.e. at the outside of the envelope vesicles). The proteolytic pattern of the control protein OEP37 (Schleiff et al., 2003) confirmed vesicle orientation and protease function (Fig. 2B). Thermolysin treatment of Ps-OEP16.2 produced two distinct protein fragments that were ∼2.5 kDa and 3 kDa smaller than the signal of untreated samples (19 kDa). Since Ps-OEP16.1 showed a similar proteolytic pattern, it can be concluded that the N- and C-terminus of OEP16.1 and OEP16.2 point to the outer face of the vesicles, which means in vivo to the cytosol. Because the C-terminus of both proteins directly ends with the fourth membrane helix, thermolysin can only cut at the N-terminal end, which is 3.1 kDa for Ps-OEP16.2 and 2.4 kDa for Ps-OEP16.1 (compare Fig. 1A and C). The interhelical loop region between the second and third helix is very short (10 amino acids) and thus most probably was not be accessible to the protease. Experiments on isolated intact Arabidopsis chloroplasts (Li and Philippar, unpublished results) displayed the same pattern for At-OEP16.1 and confirmed results for pea envelopes. In general, both α-helical OEP16 proteins are tightly folded, which is indicated by their resistance to proteolysis even when membranes were solubilized by Triton (Fig. 2B). In particular, the thermolysin fragment of Ps-OEP16.1 that lacks the N-terminus was extraordinarily stable. Ps-OEP16.2 was somewhat less protease resistant than Ps-OEP16.1, probably due to the extended, interhelical S-domain between the first and second helix (compare Fig. 1). However, a β-barrel protein like OEP37 with several larger soluble loop regions (compare Schleiff et al., 2003) was proteolysed into many more fragments, which were further degraded when membranes were solubilized by detergent (Fig. 2B).
Fig. 2.
Localization of OEP16.1 and OEP16.2 in the plastid outer envelope membrane. For immunodetection of OEP16.2, envelope membranes were prepared from isolated chloroplasts of 7-day-old pea seedlings (all others: 10-day-old plants). Numbers indicate the molecular mass of proteins in kDa. (A) Immunoblot analysis of Ps-OEP16.1 and Ps-OEP16.2 in purified outer (OE) and inner (IE) envelope membranes from pea chloroplasts. Antisera against the proteins Ps-OEP37 (OE) and PIC1 (IE) were used as controls. Equal amounts of proteins were loaded per lane: for detection of OEP37, 5 μg; for PIC1, 20 μg; for OEP16.2, 30 μg; and for OEP16.1, 1 μg. (B) Outer envelope membrane vesicles from pea chloroplasts were protease digested with thermolysin (thl) and subsequently subjected to immunblot analysis for Ps-OEP16.2, Ps-OEP16.1, and Ps-OEP37. Thermolysin was incubated for 0–45 min in a 2:1 ratio (w/w) per μg of envelope proteins. To solubilize membranes, 1% Triton (+thl/tri) was added in control assays. For detection of OEP16.2 (upper panel) 90 μg, and for OEP16.1 (lower panel) and OEP37 (right panel) 2.5 μg of protein were loaded equally in each lane. Arrowheads indicate major digestion fragments. For the control OEP37, fragment sizes correspond to those detected by Schleiff et al. (2003).
In summary, the data demonstrate that the N- and C-termini of the membrane-integral proteins OEP16.1 and OEP16.2 in the plastid outer envelope are localized in the cytosol. The OEP16.2-specific interhelical S-domain points to the intermembrane space (see Fig. 1C).
Specific expression of OEP16 isoforms during seed development and germination
Public microarray data sets of Arabidopsis development (Honys and Twell, 2003; Schmid et al., 2005) confirmed that OEP16.1 is the major leaf and stem isoform, while OEP16.2 is expressed strongly and exclusively in embryos, seeds, and pollen tissue (Supplementary Fig. S2 at JXB online). OEP16.4 transcripts can be found ubiquitously, but at a low level—usually about >4 times less than OEP16.1 and/or OEP16.2. In roots, OEP16.1 and OEP16.4 transcripts are present in equal but low amounts (Supplementary Fig. S2A). Of particular interest, however, was the strict regulation of OEP16.2 expression. After seed germination is completed, OEP16.2 is absent in leaves, roots, and stems of all developmental stages. In mature plants, transcripts first appear in flowers, where expression is restricted to stamen and pollen grains, and peaks in early pollen development (Supplementary Fig. S2B, S2C). In addition, elevated expression of OEP16.2 in pollen was confirmed by promoter–GUS studies (not shown, and Drea et al., 2006) and identification of the protein in Arabidopsis pollen proteome mapping (Holmes-Davis et al., 2005).
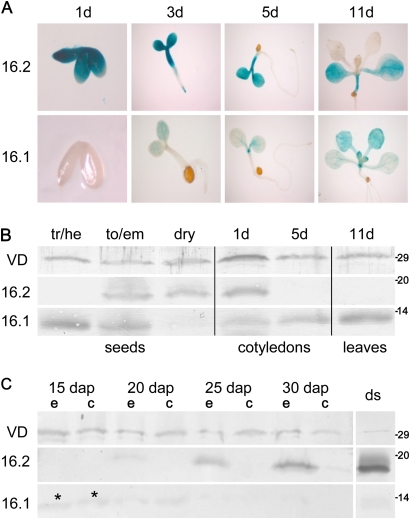
To follow the OEP16 expression pattern and protein levels during seed development and germination, At-OEP16.1 and At-OEP16.2 promoter–GUS signals were studied and immunoblot analysis was performed on Arabidopsis tissues (Fig. 3A, B). Interestingly, OEP16.1 and OEP16.2 show alternating and complementing expression patterns. Whereas OEP16.1 is present until torpedo stage embryos, resumes in cotyledons, and is highest in the first true leaves of the seedling, OEP16.2 fills the gap in late embryonic stages, and peaks in embryos of dry and germinating seeds. Further, OEP16.2 promoter activity was detectable in early cotyledon/hypocotyl tissue and the germinating root tip. Thus, a previous analysis on alternating protein levels of OEP16 isoforms in seedlings (OEP16.1 in first true leaves, OEP16.2 in cotyledons; compare Philippar et al., 2007) could be confirmed and this principle could be expanded for seed development and germination. In addition, the same complementary OEP16.1/OEP16.2 pattern was detected in dissected developing pea seeds, where weak signals of Ps-OEP16.1 proteins are found in early embryo and seed coats, but Ps-OEP16.2 was restricted to late embryos and was highest in dry seeds (Fig. 3C).
Fig. 3.
OEP16.1 and OEP16.2 expression during seed development and germination. (A) Histochemical detection of GUS activity directed by the At-OEP16.1 and At-OEP16.2 promoter in transgenic Arabidopsis plants. GUS expression was monitored after germination for 1, 3, 5, and 11 d. For the 1-day-old tissue (1d) the seed coat was removed manually. Pictures show representative results for at least three different transgenic lines. (B) Immunoblot analysis of At-OEP16.1 and At-OEP16.2 in protein extracts from developing seeds (tr/he, transition/heart stage; to/em, torpedo/embryo stage), dry seeds, cotyledons (1–5 d old), and leaves (11 d old) of seedlings. For seeds 10 μg, and for seedlings 20 μg of protein were loaded in each lane. (C) Immunoblot of Ps-OEP16.2 and Ps-OEP16.1 in protein extracts (4 μg each) from developing and dry (ds) pea seeds. Seed stages are given in days after pollination (dap). During development seeds were separated into embryo (e) and seed coat (c) tissue, the latter containing endosperm layers. Asterisks indicate weak OEP16.1 signals in early embryo and seed coat (15 dap). (B and C) Antiserum against the marker protein VDAC (outer membrane of mitochondria) was used as loading control. Numbers indicate the molecular mass of proteins in kDa.
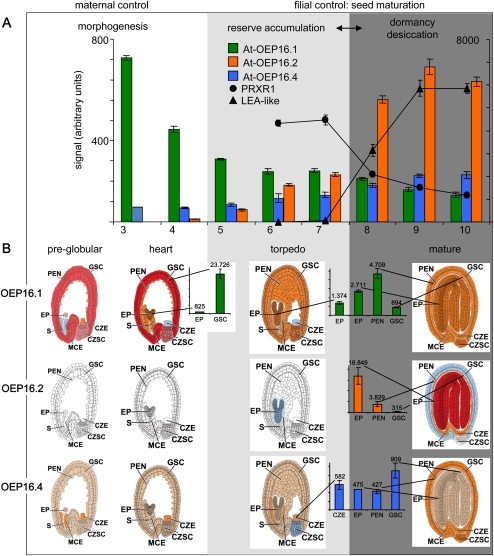
Publicly available Arabidopsis transcript data sets of seed development (Schmid et al., 2005) and laser capture microdissected seed organs (Harada-Goldberg Arabidopsis microarrays: ‘Gene Networks in Seed Development’) allowed further insight into the seed-specific expression pattern of OEP16 isoforms (Fig. 4). Here OEP16.2 expression constantly increases during seed maturation and peaks in late stages when desiccation tolerance and dormancy are acquired (Fig. 4A). OEP16.2 transcripts are exceptionally high during the late desiccation phase, confirming the high protein levels found in dry seeds (compare Fig. 3B and C). Further dissected seed and embryo organs revealed that OEP16.2 transcripts exclusively locate to the embryo and endosperm of mature seeds (Fig. 4B; Supplementary Fig. S3 at JXB online). Because the transition from the phase of reserve accumulation to seed desiccation is accompanied by a distinct switch in gene expression and accumulation of specific free metabolites (Angelovici et al., 2010), two genes were selected from the microarray study by Angelovici et al. (2009) as transcriptional markers for this maturation–desiccation switch: At4g21960, encoding a peroxidase (PRXR1) and up-regulated in late maturation stages but down-regulated upon desiccation, and At2g41280, similar to late embryogenesis abundant (LEA) proteins and expressed only during the seed desiccation phase (see Fig. 4A). In comparison with these two marker genes, OEP16.2 followed the expression pattern of At2g4128, and thus groups to the LEA genes, which are up-regulated during desiccation only. In contrast, OEP16.1 mRNA is increased in early stages of embryo morphogenesis and drops upon seed maturation (Fig. 4A). Interestingly, strong OEP16.1 expression in early morphogenesis is associated with the maternally derived seed coat, but in mature seeds OEP16.1 is present in all seed compartments at a lower level (Fig. 4B; Supplementary Fig. S3). OEP16.4 transcripts, however, are ∼30–40 times less than those for OEP16.1 in seed coats of heart stage embryos or for OEP16.2 in embryos of mature seeds. In the latter, OEP16.4 is restricted to endosperm and seed coat (Fig. 4B; Supplementary Fig. S3).
Fig. 4.
OEP16 expression profiles during Arabidopsis seed development. Data used to create the digital northern blots were obtained from the respective experiments deposited at the NASCArrays website (http://affy.arabidopsis.info/narrays/experimentbrowse.pl) and from the Harada-Goldberg Arabidopsis miccoarray data set of laser capture microdissected seeds (‘Gene Networks in Seed Development’ at http://estdb.biology.ucla.edu/seed/). Mean signal intensities (arbitrary units ±SD) were averaged from 2–3 replicates. Morphogenesis, reserve accumulation, and desiccation/dormancy phases are indicated by white, light grey, and dark grey background, respectively. (A) Embryo and seed development (AtGenExpress siliques and seeds, NASCARRAYS-154). The seed stages 3–10 are defined according to embryo development as follows: 3, mid-globular to early heart; 4, early heart to late heart; 5, late heart to mid-torpedo; 6, mid-torpedo to late torpedo; 7, late torpedo to early walking stick; 8, walking stick to early curled cotyledons; 9, curled cotyledons to early green cotyledons; 10, green cotyledons. Note that the seed stages 3–5 include silique tissue. Signals for At-OEP16.1 (green) and for At-OEP16.4 (blue) correspond to the right y-axis, while the left y-axis was scaled for At-OEP16.2 (orange) and the controls PRXR1 (dots) and LEA-like (triangles). Differential expression of the latter two genes defines the transcriptional switch from reserve accumulation to the acquirement of desiccation tolerance and dormancy. (B) Tissue-specific expression pattern of OEP16 isoforms in pre-globular, heart, torpedo, and mature seed stages (Harada-Goldberg Arabidopsis LCM Gene-Chip Data Set). Different tissues are defined as follows: CZE, chalazal endosperm; CZSC, chalazal seed coat; EP, embryo proper; GSC, general seed coat; MCE, micropylar endosperm; PEN, peripheral endosperm; S, suspensor. Signal values (arbitrary units) for At-OEP16.1 (green), At-OEP16.2 (orange), and At-OEP16.4 (blue) are presented in tissues with the highest expression (for complete data sets see Supplementary Fig. S3 and Supplementary Table S2 at JXB online). Seed tissues are coloured according to transcript density for signals that are absent (white), insufficient (blue), <500 (beige), 500–5000 (orange), 5000–10 000 (purple), and >10 000 (dark red).
Because in the analysis of pea seeds (Fig. 3C) prepared seed coats contained attached endosperm layers, OEP16 expression could not be distinguished between maternal seed coat and filial endosperm. However, expression profiles of OEP16 orthologues in dissected soybean seeds (Harada-Goldberg data set of Glycine max microarrays: ‘Gene Networks in Seed Development’) allowed comparison of the two legumes and Arabidopsis. As found in pea and Arabidopsis, the OEP16.1 isoform in soybean is highly expressed in seed coat, endosperm, and embryo tissues of early (globular) and late maturation stages, whereas OEP16.2 again is restricted to filial compartments (endosperm/cotyledons and embryo) in the mature seed only (Supplementary Fig. S4, at JXB online). OEP16.2 thereby can be unequivocally allocated to those proteins associated with the seed desiccation process, which are specifically expressed during late embryogenesis, in dry seeds, and during early germination. These findings are further substantiated since At-OEP16.2 represents one of the most abundant mRNAs, which are stored in dry seeds for subsequent germination (Nakabayashi et al., 2005).
As a summary of all expression data, OEP16.2 function appears to be closely linked to desiccation-tolerant grain tissues, which are represented by pollen and in particular seeds, where OEP16.2 is restricted to filial embryo and endosperm. Further, OEP16.2 appears to provide a functional link for bridging the dormant and desiccation-tolerant status during seed maturation and the germination potential of mature seeds. In contrast, a corresponding role might be performed by OEP16.1 during maternally controlled early morphogenesis events and by accompanying reserve accumulation in the seed coat, endosperm, and embryo.
Specific control of OEP16.2 expression by the phytohormone ABA in seeds
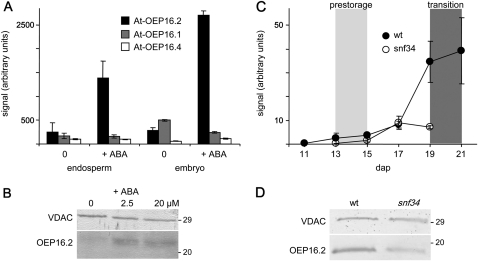
Since OEP16.2 expression specifically correlates with seed desiccation and dormancy processes controlled by the phytohormone ABA, OEP16 transcript and protein content was monitored in germinating Arabidopsis seeds that were treated with ABA. In the analysis by Penfield et al. (2004), At-OEP16.2 transcripts were specifically and strongly increased upon germination for 24 h in the presence of ABA (Fig. 5A). In endosperm tissue, the induction was ∼5-fold, and in embryos even 9-fold. In contrast, At-OEP16.1 and At-OEP16.4 were not regulated in endosperm but responded slightly to ABA in embryos, showing ∼2-fold down-regulation (OEP16.1) and ∼2-fold up-regulation (OEP16.4). When testing for protein levels, it was found that after 48 h of ABA treatment during germination, OEP16.2 protein content also substantially increased (Fig. 5B).
Fig. 5.
Specific ABA induction of OEP16.2 expression in seeds. (A) Digital northern blot of At-OEP16.2 (black), At-OEP16.1 (grey), and At-OEP16.4 (white) expression (arbitrary units) in Arabidopsis endosperm and embryo tissue. Prior to dissection, seeds were germinated for 24 h without (0) and with 20 μM ABA (described in Penfield et al., 2004). Data used to create the expression profile were obtained from the analysis by Penfield et al. (2004) using NASCArrays database (http://affy.arabidopsis.info/narrays/experimentbrowse.pl), experiment NASCARRAYS 386. Signal intensities were averaged from three biological replicates (n=3, ±SD). (B) Western blot analysis of At-OEP16.2 in protein extracts (7.5 μg each) from Arabidopsis seeds, germinated for 48 h on medium containing 0, 2.5, and 20 μM ABA. (C) Transcript content of Ps-OEP16.2 (n=2, ±SD, arbitrary units) in developing pea seeds of the wild type (black) and the Vf-SnRK1-antisense line snf34 (white). The age of seeds is given in days after pollination (dap). According to the definition by Radchuk et al. (2006), delayed down-regulated genes are highly expressed in the pre-storage phase 13–15 dap (light grey area), and delayed up-regulated genes are continuously increased during seed maturation, starting in the transition phase at 19–22 dap (dark grey area). (D) Immunoblot analysis of Ps-OEP16.2 in protein extracts (4 μg each) from wild-type and VfSnRK1-antisense (snf34) pea seeds, isolated 20 dap. (B and D) Antiserum against the marker protein VDAC (outer membrane of mitochondria) was used as a loading control. Numbers indicate the molecular mass of proteins in kDa.
Isolation of the cDNA sequence of OEP16.2 from pea further allowed tracking of Ps-OEP16.2 expression in several studies during pea seed development. By analysis of embryos from antisense lines with decreased expression of sucrose non-fermenting-1-related protein kinase (SnRK1), the expressed sequence tag (EST) for Ps-OEP16.2 (PSC23A24, table II in Radchuk et al., 2006) was found to be grouped to those genes that showed a delayed up-regulation in the transition stage of seed development. In wild-type embryos Ps-OEP16.2 transcripts strongly accumulated during the transition phase 19–22 days after pollination (dap; ∼10-fold when 21 dap are compared with 15 dap; Fig. 5C). As described for Arabidopsis (see above), this transition state during pea seed maturation is associated with a transcriptional and metabolic switch (Radchuk et al., 2006, and references therein). Interestingly, in SnRK1 antisense seeds of line snf34 (corresponding to Vic-34 in Radchuk et al., 2006), Ps-OEP16.2 up-regulation was absent (Fig. 5C), reflecting the fact that SnRK1 antisense seeds contain reduced levels of ABA (Radchuk et al., 2010b). When regulation on the protein level (Fig. 5D) was tested, it was found that compared with the wild type, snf34 embryos at 20 dap contain only ∼70% Ps-OEP16.2 proteins (73.9±12.6%, n=3, ±SD). Further, ABA control of Ps-OEP16.2 expression in pea seeds was confirmed by decreased transcripts in embryos with reduced levels of free ABA, which were generated by immuno modulation (‘anti-ABA’ seeds, see table 1 in Radchuk et al., 2010a). With 69.82±10.05% (n=4, ±SD) of wild-type OEP16.2 proteins, the decrease in anti-ABA embryos at 22 dap was in the same range as for SnRK1 antisense lines.
In conclusion, OEP16.2 expression during late seed development and early germination is specifically controlled and strongly stimulated by the dormancy and desiccation tolerance-inducing hormone ABA.
Loss of OEP16 proteins provokes ABA hypersensitivity during seed germination
To document the function of OEP16 proteins during seed development and germination, all combinations of oep16 triple and double knockout mutants were generated in Arabidopsis. Therefore, the double mutant oep16.1-1/oep16.4-2 was crossed with the single mutant oep16.2-1, both representing knockout lines for At-OEP16.1, At-OEP16.4, and At-OEP16.2, respectively (Philippar et al., 2007). In the F3 generation of the crossed lines, the homozygous oep16 triple mutant (1/2/4), a corresponding triple wild-type line (wt), and the respective double mutants 1/2, 1/4, and 2/4 were segregated (for details, see the Materials and methods). The loss of the respective OEP16 isoforms was documented on the transcript as well as on the protein level (Supplementary Fig. S1 at JXB online). Because OEP16.1 and OEP16.2 function is most probably implicated in seed development and germination, mutants were analysed for visible defects during these processes. What was found was, that in comparison with wild-type plants, none of the oep16 knockout mutants showed a phenotype during germination, growth, and development under standard conditions (compare also Philippar et al., 2007; Pudelski et al., 2009), or any macroscopic defects of embryo and seed maturation (e.g. arrested or distorted embryos, smaller siliques, or differences in seed weight or amount).
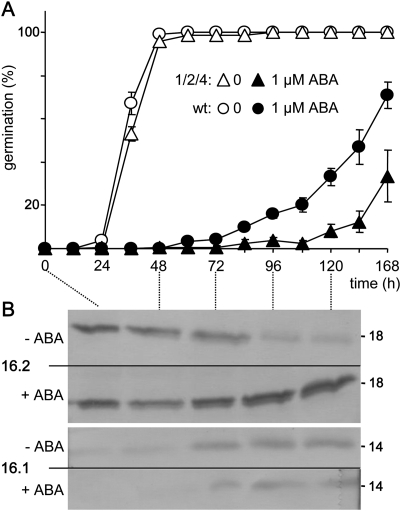
However, when mutant seeds were germinated in the presence of ABA, the oep16 triple mutant displayed ABA hypersensitivity that was characterized by delayed germination when compared with the corresponding wild-type seeds (Fig. 6A; Supplementary Fig. S5 at JXB online). Whereas without ABA, germination rates of all seeds analysed were similar, upon ABA addition it took ∼24 h longer to reach the same number of germinated seeds in the oep16 triple mutant than in the wild type. When seeds were germinated on agar containing 2.5 μM ABA, this delay was maximal after 72 h (Supplementary Fig. S5). Further, germination rates of oep16 double mutants after 72 h incubation with ABA (Supplementary Fig. S5) and germination tests with the single mutant lines (not shown) demonstrated that most probably the loss of OEP16.2 has the strongest effect on ABA-hypersensitive seed germination. However, in all experiments, the ABA-controlled germination delay was strongest in subsequent generations of the oep16 triple mutant. The triple mutant and the corresponding wild type were therefore chosen for all further experiments and analysis of metabolic signatures during ABA-controlled germination. Therefore, seeds were germinated on nylon filters in the presence of 1 μM ABA (Fig. 6A). When compared with experiments on agar, seed germination on nylon filters in general and in the presence of ABA was delayed by ∼12 h and 48 h, respectively (compare Supplementary Fig. S5). However, ABA hypersensitivity of the oep16 triple mutant could be confirmed to appear in a time window between 3 d and 6 d and again revealed an ABA-induced germination delay of ∼24 h. Parallel immunoblot analysis in addition showed that expression of OEP16.2 proteins disappeared 3 d after germination, while upon ABA addition high OEP16.2 levels were maintained throughout the germination process (Fig. 6B). In contrast, expression of OEP16.1, which was initiated ∼3 d after germination, was not responsive to ABA.
Fig. 6.
ABA hypersensitivity during germination of OEP16 triple mutants. (A) Germination rates of oep16 triple mutant (1/2/4, triangles) and corresponding wild-type seeds (circles). Seed germination on nylon filters was followed for 168 h in the absence and presence of 1 μM ABA (open and filled symbols, respectively). Data represent mean values ±SE of n=4 independent experiments. (B) Immunoblot analysis of At-OEP16.2 (upper panel) and At-OEP16.1 (lower panel) in protein extracts (25 μg each) from wild-type Arabidopsis seeds, germinated as described in A. Numbers indicate the molecular mass of proteins in kDa. (–), no ABA; (+), addition of 1 μM ABA.
Thus, ABA hypersensitivity of germinating oep16 mutant seeds is most probably due to the loss of the ABA-induced isoform OEP16.2, which is predominant during seed maturation, in dry seeds, and in early germination. It cannot, however, be excluded that the lack of OEP16.1 and OEP16.4 in the triple mutant might have additional effects on germination as well.
Developing and germinating seeds of the oep16.1/2/4 mutant show changes in metabolic signatures
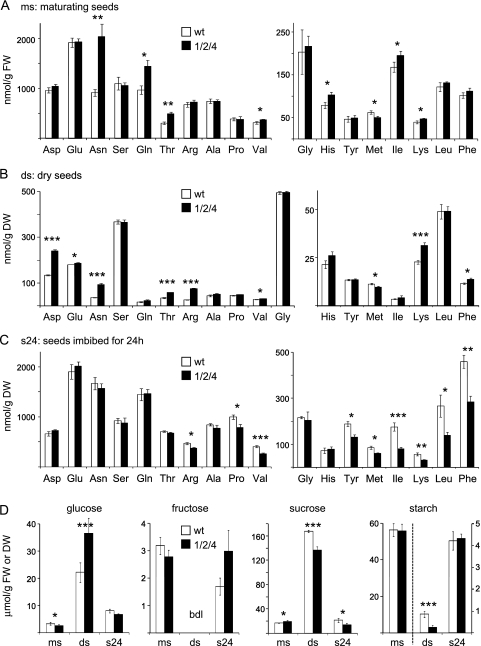
To analyse the impact of OEP16 on the content of seed metabolites, amino acids and sugars in oep16 triple mutants and the corresponding wild type were determined during maturation and germination of seeds (Fig. 7; Supplementary Table S3 at JXB online). In general, free amino acids during late maturation (Fig. 7A) were at a high level, decreased in dry seeds (Fig. 7B), and increased again after initiation of germination by 24 h imbibition (Fig. 7C). Glycine, however, behaved in the opposite manner and was highest in dry seeds. During seed development, the oep16 triple mutant was characterized by a significant increase in amino acids when compared with the wild type: asparagine (Asn), glutamine (Gln), threonine (Thr), valine (Val), histidine (His), isoleucine (Ile), and lysine (Lys) in maturing seeds; aspartate (Asp), glutamate (Glu), Asn, Thr, arginine (Arg), Val, Lys, and phenylalanine (Phe) in dry seeds (Fig. 7A, B). Interestingly, this switched to a decrease in oep16.1/2/4 when seeds were germinated [Arg, proline (Pro), Val, tyrosine (Tyr), methionine (Met), Ile, Lys, leucine (Leu), and Phe; Fig. 7C]. The only exception was Met, which was lower in the oep16 triple mutant in all three stages analysed. However, although statistically significant, the latter reduction was only marginal (∼1.2- to 1.4-fold). Major changes in amino acids of the oep16 triple mutant were found for Asn, Gln, and Thr in maturing seeds as well as for Asp, Asn, Thr, and Arg in dry seeds (2.2-, 1.5-, 1.6-, and 1.8-, 2.6-, 1.7-, and 2.8-fold increase, respectively; see Supplementary Table S3). Furthermore, in imbibed seeds, the decrease of Val (1.5), Ile (2.2), Lys (1.8), Leu (1.9), and Phen (1.6) was >1.5-fold.
Fig. 7.
Amino acid and sugar content during seed maturation of the oep16 triple mutant and wild type. Metabolites were determined in maturing seeds (ms), dry seeds (ds), and seeds imbibed for 24 h (s24) of the oep16.1/2/4 triple mutant and corresponding wild type (black and white bars, respectively). Maturing seeds correspond to stages 7–10 as depicted in Fig. 4A. Data represent mean values of independent biological replicates (n=3–4, ±SD). Please note that while concentrations for maturing seeds are given per gram of tissue fresh weight (FW), they are related to dry weight (DW) for dry and imbibed seeds. Significance analysis was performed by a Student’s t-test with P-values <0.05 (*); <0.01 (**); and <0.005 (***), respectively. For complete data sets see Supplementary Table S3 at JXB online. (A–C) Amino acid concentration (nmol/g FW or DW). Tryptophan and cysteine were not determined. (D) Glucose, fructose, sucrose, and starch (μmol/g FW or DW). Fructose values in dry seeds were below the detection limit (bdl) of the analysis. For starch, the right y-axis is scaled for concentrations in dry and imbibed seeds.
As expected, glucose and sucrose, which have to be mobilized for subsequent germination, accumulated in dry seeds of the mutant and wild type. In contrast, starch breakdown occurred during seed desiccation and thus starch content was highest in late seed maturation (Fig. 7D). Differences in sugar content between the oep16.1/2/4 mutant and the wild type were observed in dry seeds, whereas in the oep16 triple mutant glucose levels were increased 1.65-fold and sucrose and starch decreased by 1.2- and 3.2-fold, respectively. Further, in imbibed mutant seeds, sucrose content was still 1.5-fold less than in the wild type (Fig. 7D; Supplementary Table S3 at JXB online).
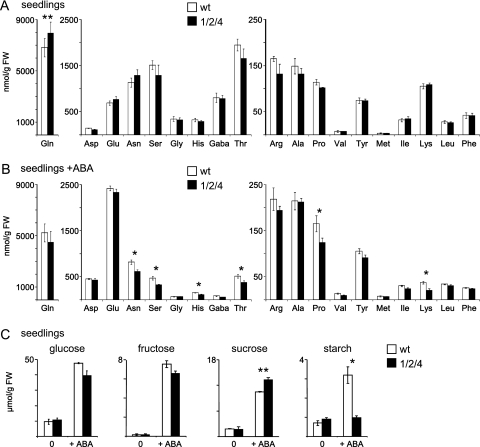
To follow metabolic changes introduced by the observed ABA hypersensitivity of the oep16 triple mutant, amino acids and sugars were analysed in 6-day-old seedlings grown in the absence and presence of 1 μM ABA (Fig. 8; Supplementary Table S3 at JXB online). When oep16 triple mutant seeds were germinated for 6 d, amino acid differences from the wild type that were detected in dry seeds and early germination were almost compensated, except for a 1.2-fold increase in Gln (Fig. 8A). In contrast, in the presence of ABA, down-regulation of amino acids was observed in oep16 triple mutant seedlings (Fig. 8B). Here Asp, serine (Ser), His, Thr, and Pro were slightly decreased (fold change < 1.4) and Lys was down-regulated more pronouncedly (1.8-fold). Thus, with respect to down-regulation of amino acids in the OEP16.1/2/4 mutant, ABA addition to seedlings seems to mimic the status of early germinating, imbibed seeds (compare Fig. 7C, Supplementary Table S3 at JXB online). ABA addition to wild-type and mutant seedlings in general induced an increase in Asp, Glu, Arg, Ala, Pro, Val, and Met, and a decrease in Asn, Ser, Gln, Gly, His, GABA (γ-aminobutyric acid), Thr, Lys, and Phe, while only Ile and Leu remained unchanged (Supplementary Fig. S6 at JXB online). Except for Ile (slight decrease in mutants) this behaviour was the same for wild type and the oep16 triple mutant.
Fig. 8.
Amino acid and sugar content of oep16 triple mutant and wild-type seedlings. (A and B) Amino acid concentrations (nmolg FW, n=3–4 ±SD) in 6-day-old seedlings of the oep16 triple mutant (1/2/4, black bars) and wild type (white bars). Seedlings were grown on nylon filters in the absence (A) and presence of 1 μM ABA (B). Tryptophan and cysteine were not determined. (C) Glucose, fructose, sucrose, and starch concentrations (μmol/g FW, n=3–4, ±SD) in seedling tissue described in A and B. Significance analysis was performed by a Student’s t-test with P-values <0.05 (*); <0.01 (**); and <0.005 (***), respectively. For complete data sets see Supplementary Table S3 at JXB online.
After growth for 6 d in the absence of ABA, significant differences in glucose, fructose, sucrose, and starch were not observed (Fig. 8C). However, as found in dry seeds, in the presence of ABA the starch content in the oep16 triple mutant dropped quite drastically (3.3-fold decrease), but sucrose increased slightly (1.2-fold). In general, dry seeds contained ∼2-fold more glucose and 10-fold more sucrose than 6-day-old seedlings. ABA addition during germination induced an overall increase in sugars (∼5-fold for glucose, sucrose, and starch, and 50-fold for fructose).
In summary, the largest difference for the oep16.1/2/4 mutant compared with the wild type in amino acid and sugar content was found in dry seeds. Whereas from seed maturation to desiccation the mutant is characterized by an increase in amino acids, a switch to decreased amino acids was detected during early germination and a compensation of metabolic differences was revealed after prolonged seedling growth. However, when ABA is present, down-regulation of amino acids and starch is sustained in mutant seedlings, thus mimicking the status of dry seeds.
Discussion
Expression and localization of OEP16 isoforms
Isolation of OEP16.2 cDNA from P. sativum in the current study allowed a comparative description of the membrane topology and tracking of the expressional regulation of the isoforms OEP16.1 and OEP16.2 on the transcript and protein level in the two plant species Arabidopsis and pea. It could be unequivocally demonstrated that OEP16.2 inserts into the outer envelope membrane of plastids and that the N- and C-terminal ends of the OEP16.1 and OEP16.2 proteins are oriented to the cytosol. Previous in vitro studies of protein import into pea chloroplasts, which resulted in a distinct and protease-protected band in the chloroplast fraction for At-OEP16.1 and At-OEP16.2, are in line with the present findings (Murcha et al., 2007).
Whereas previous analyses demonstrated that the isoform OEP16.1 is the major leaf OEP16 (for an overview, see Pudelski et al., 2010), it could be shown here that OEP16.1 plays an additional role in early seed maturation. When seed desiccation and dormancy are induced, OEP16.1 is decreased and thus groups to the so-called ‘maturation class’ of proteins, associated with seed reserve accumulation processes (for overviews, see Holdsworth et al., 2008a; Angelovici et al., 2010). Further, the expression patterns of the OEP16.1 and OEP16.2 isoforms alternate during seed development and germination: when OEP16.1 decreases during mid embryogenesis, OEP16.2 levels rise and are predominant throughout the acquirement of dormancy and desiccation tolerance in dry seeds as well as during early germination. However, although OEP16.2 is still detectable in cotyledons of very young seedlings, the protein is absent in leaves, a tissue where again OEP16.1 expression is strongly induced.
Apart from seeds, OEP16.2 is only present in pollen grains as well as cotyledons and the root tip of germinating seeds. Although OEP16.2 is quite abundant in Arabidopsis pollen plastids, as reflected by its identification in pollen proteome analysis (Holmes-Davis et al., 2005), a phenotype could not be associated with oep16.2 knockout mutants either during in vivo reproduction or by in vitro pollen germination assays (K Philippar, S McCormick, unpublished data). Plastids in pollen and seeds as well as in cotyledons represent metabolic source organelles for germination and growth of pollen tubes, seeds, and seedlings; thus OEP16.2 might function in metabolite export. In root tip plastids, a similar function is likely during initiation of seed germination, in particular when the emerging radicle has to rupture testa and endosperm (see Holdsworth et al., 2008a).
The presence of OEP16.2 in late embryogenesis and early seed germination is exceptionally strong. In dry seeds, OEP16.2 represents one of the most highly abundant mRNAs stored for seed germination (see Nakabayashi et al., 2005) and is closely linked to the seed desiccation process (‘up-regulated only during seed desiccation’, see Angelovici et al., 2010; compare Fig. 4A). Thus, OEP16.2 belongs to the LEA proteins (see Holdsworth et al., 2008a) and is most probably functioning in processes that bridge seed desiccation and germination. In Arabidopsis seeds, OEP16.2 transcripts were found in embryo and endosperm tissue by microarray analysis (Penfield et al., 2004) and in situ hybridization (Drea et al., 2006). In pea and soybean seeds, OEP16.2 protein and transcripts were detected in embryos and cotyledons (Fig. 3C; Supplementary Fig. S4 at JXB online). Thus, unlike OEP16.1, which in addition to the embryo and endosperm can be found in maternally derived seed coats, OEP16.2 is restricted to filial seed tissues.
Localization and orientation of OEP16.1 and OEP16.2 in the outer envelope membrane of plastids could be clearly documented in the current study. For the third isoform OEP16.4, however, the subcellular localization is not completely obvious since in vitro import experiments into chloroplasts and mitochondria as well as immunological studies did not give clear results. However, signals of N-terminal, GFP-tagged At-OEP16.4 in Arabidopsis cell cultures and the expression pattern point to a plastid localization of OEP16.4 (Murcha et al., 2007). Together with its ubiquitous but low expression pattern, it is thus tempting to speculate that OEP16.4 might have a constitutive function in potential oligomeric channels, composed of different OEP16 isoform subunits, for example OEP16.1/OEP16.4 or OEP16.2/OEP16.4 (compare Pudelski et al., 2010).
Regulation of OEP16 expression in seeds: metabolites, ABA, and dormancy
In addition to the data presented, transcriptional regulation of OEP16.1 and OEP16.2 was found in several studies associated with development, metabolite content, and ABA regulation in seeds, summarized in Supplementary Fig. S7 at JXB online and discussed in the following.
Links of OEP16 expression to seed amino acid content
In the so-called Arabidopsis KD mutant, which is characterized by an increased seed Lys content—achieved by seed-specific interruption of Lys biosynthesis feedback inhibition in the background of a Lys catabolism knockout (Angelovici et al., 2009, 2011)—OEP16 expression is de-regulated (Supplementary Fig. S7A at JXB online). First, transcripts of the ‘maturation class’ protein OEP16.1 in KD mutants are low during seed maturation, but adjusted to wild-type levels in dry seeds. Secondly, down-regulation of the LEA protein OEP16.2 is delayed upon germination, linking OEP16.2 with a function in bridging seed desiccation with germination and reflecting the general attenuation of essential germination-associated processes observed in KD mutants (Angelovici et al., 2011). Thus, OEP16 function seems to be linked to the amino acid status of seeds, in particular to that of Lys and other members of the Asp family, which are affected in KD mutants as well (see Angelovici et al., 2011; Galili, 2011). Because in oep16 triple mutant seeds, de-regulated amino acids are mainly Asp derived as well (see below), a function of OEP16 in shuttling these amino acids between the plastid and the cytosol seems possible.
Another link of OEP16 with the amino acid content in seed tissue can be found in pea mutants where the seed-specific overexpression of a plasma membrane-localized amino acid permease results in a parallel increase of amino acids and Ps-OEP16.1 transcripts in embryos (Weigelt et al., 2008). Further, reduced expression of the oxoglutarate/malate translocator OMT in the inner envelope of pea seed plastids, which affects conversion of carbohydrates from sucrose into amino acids and proteins (Riebeseel et al., 2010), leads to strongly reduced Ps-OEP16.1 and Ps-OEP16.2 transcript content in late pre-storage, transition stage, and early maturation of seeds. Interestingly, the change in amino acid profiles, in particular the increase of free Asp of the OMT antisense pea seeds, resembles that observed in the maturing seeds of the oep16 triple mutant (see below). Thus, expression of OEP16 genes during seed development seems to be closely linked to the metabolic status of the cell, in particular to that of amino acids and sugars.
ABA control of OEP16.2 expression
In the current study it is shown that in seeds OEP16.2 transcript and protein accumulation is under the control of the phytohormone ABA. Previously, Drea et al. (2006) described that the promoter of At-OEP16.2 contains four ABREs, which are induced during acquirement of desiccation tolerance and dormancy of seeds. Further, in seed/silique tissue, OEP16.2 mRNA accumulation is under the control of the transcription factors ABI3 and ABI5 (for ABA INSENSITIVE 3 and 5), which represent central elements of the ABA signal transduction chain and function as key regulators of seed development (Finkelstein et al., 2002). Microarray analysis of seeds from ABA signalling mutants confirms that ABI3 controlled OEP16.2 expression by ∼4.8- (abi3) and 3-fold (fus3) down-regulation when compared with the wild type (Supplementary Fig. S7B at JXB online). In the latter mutant, FUS3 (FUSCA3) represents another central ABA-associated transcription factor in seeds, which is closely linked to ABI3 and controls ABA-dependent seed storage protein expression (Kagaya et al., 2005; Suzuki and McCarthy, 2008). FUS3 control of OEP16.2 is also indirectly confirmed in double mutants of the polycomb group complex proteins CURLY LEAF (CLF) and SWINGER (SWN) (Chanvivattana et al., 2004), where transcript levels of FUS3 are described to be highly up-regulated (Makarevich et al., 2006). Surprisingly OEP16.2 mRNA was found to be drastically induced (∼1700-fold) in clf/swn seedlings as well, while constant expression of OEP16.1 and of the housekeeping protein translocon channel TOC75 indicates a normal number of plastid organelles in these mutants (Supplementary Fig. S7B at JXB online). Thus, via FUS3, the LEA protein OEP16.2 is under the control of polycomb group proteins, which repress flowering and endosperm proliferation.
Expression during dormancy, after-ripening and germination
Through ABA control, OEP16.2 is linked to the acquirement of seed desiccation and further seems to function in bridging dormancy stages with early germination. Therefore, OEP16 transcript content was followed by digital northern analysis of several transcriptome studies associated with seed dormancy, after-ripening, and germination. When transcript content is analysed after seed imbibition in phase two of germination (i.e. while metabolic processes are reinitiated), OEP16.2 is classified as down-regulated by a previous after-ripening phase (Carrera et al., 2007, 2008). Studies on seed dormancy release and cycling in the deep-dormant ecotype Cvi (‘Cape Verde Islands’, Cadman et al., 2006; Finch-Savage et al., 2007), however, clearly show that OEP16.2 down-regulation is induced by imbibition, no matter whether seeds are in the primary dormant stage (i.e. freshly harvested without an after-ripening phase) or non-dormant after a prolonged after-ripening phase [compare OEP16.2 transcript content in PDD and DDL (dry) versus all other seed stages (imbibed) in Supplementary Fig. S7C at JXB online]. As soon as seeds are imbibed, OEP16.2 transcript content drops in non-deep-dormant Col-0 as well as in deep-dormant Cvi (Yamagishi et al., 2009). Sufficient after-ripening (DDL and DL stages in Supplementary Fig. S7C), however, seems to maximize the drop in OEP16.2 expression upon initiation of germination by imbibition [5-fold decrease in DDL to DL, compared with an ∼2fold decrease without sufficient after-ripening (e.g. DDL to PDL)]. For OEP16.1 down-regulation was also found upon imbibition, but in contrast to OEP16.2 in a time-dependent manner (Supplementary Fig. S7C). Interestingly, in seeds which are capable of germinating (non-dormant DL and primary dormant, light and nitrate-treated PDLN, Supplementary Fig. S7C), imbibition-induced OEP16.1 transcript reduction is neutralized. These findings are underlined by a recently established genome-wide network model that captures seed germination (Bassel et al., 2011), where OEP16.1 is categorized as germination up-regulated and the ‘dormancy up’ gene OEP16.2 is grouped only with co-expressed, ABA-induced genes. Thus, in summary, it can be concluded that OEP16.2 expression is clearly linked to the acquirement of dormancy and desiccation tolerance in drying seeds, while after-ripening has only secondary effects. Finally, initiation of germination by imbibition leads to down-regulation of OEP16.2 expression, which is restricted in bridging dormancy with very early germination events. In contrast, OEP16.1 RNA levels are likely to correlate with germination capacity of seeds.
Further, OEP16.1 mRNA is decreased 3-fold in after-ripened and imbibed seed tissue of a knockout mutant of the peroxisomal ATP-binding cassette transporter COMATOSE, which is crucial for reactivation of metabolic processes during transition from dormancy to germination (Carrera et al., 2007). In addition, in imbibed comatose mutant seeds, sucrose mobilization is restricted, as demonstrated by higher sucrose but lower glucose levels than the wild type (Footitt et al., 2002). Thus, since OEP16.1 expression is strongly enhanced (∼75-fold; Gonzali et al., 2006) by the metabolic signalling molecule sucrose, up-regulation in comatose seeds might be mediated by higher sucrose levels. Interestingly, a similar metabolic pattern (i.e. high sucrose but low glucose) is found in maturing seeds of the oep16 triple mutant (compare Fig. 7C and Supplementary Table S3 at JXB online), indicating that the lack of OEP16 during reserve accumulation of seeds affects not only amino acid content (see below) but also carbohydrate metabolism.
ABA hypersensitivity and metabolic imbalance of oep16 triple knockout mutants
Because OEP16.2 expression in seed development is controlled by ABA, germination of OEP16 mutants was followed in the presence and absence of this phytohormone (Fig. 6; Supplementary Fig. S5 at JXB online). Thereby it was possible to detect an ABA hypersensitivity of germination, which is most probably directly linked to the absence of OEP16.2. Whereas without ABA, normally OEP16.2 proteins are absent in growing seedlings, expression and thus function of OEP16.2 is sustained when ABA is present. A similar effect can be found for the metabolic imbalance observed, since oep16.1/2/4 seedlings grown in the presence of ABA show the same changes (i.e. down-regulation of amino acids and starch) as dry mutant seeds, which followed their endogenous dormancy and desiccation programme (without external addition of ABA). Interestingly, in the high Lys KD mutant seeds (Angelovici et al., 2010) an attenuation in the initiation of the germination programme is also correlated with a delayed down-regulation of OEP16.2 expression (see above, Supplementary Fig. S7A at JXB online). Thus, ABA hypersensitivity of germinating oep16 mutant seeds most probably is due to the loss of the ABA-induced isoform OEP16.2, which is predominant during dormancy and desiccation. It cannot, however, be excluded that the lack of OEP16.1 and OEP16.4 in the triple mutant might have additional, mostly ABA-independent effects on germination as well.
Morphologically oep16.1/2/4 mutant embryos developed normally and seed weight, total protein as well as N and C content of oep16 triple mutant seeds did not change when compared with the wild type. Further, amino acid composition and content in total seed storage proteins also did not differ from the wild type (Supplementary Table S4 at JXB online). Therefore, the observed variations in seed amino acid concentrations are completely related to free amino acids. The most obvious imbalance observed in oep16.1/2/4 was an increase in amino acids in maturing as well as in dry seeds. Interestingly, this shifted to a decrease when germination was initiated by imbibition and when seedlings were treated with ABA. The only amino acid that was constantly decreased during mutant seed development and imbibition was Met (see Fig. 7A, B, C; Supplementary Table S3). Because Met biosynthesis is localized in plastids only (see Wirtz and Droux, 2005), a Met decrease in oep16.1/2/4 might be due to incomplete Met export via the outer envelope membrane when the OEP16 channel pore is lacking. Further, Met has been shown to be crucial for seed germination and seedling growth (Gallardo et al., 2002) and thus, when reduced, might contribute to the ABA-induced germination delay of the oep16 triple mutant. The most pronounced increase in amino acids in maturing and dry seeds of oep16.1/2/4 was detected for Asn, leading to an increased ratio of Asn to Asp in maturing seeds (Supplementary Table S3). Interestingly, the same can be found for pea seed mutants of the plastidic oxoglutarate/malate translocator OMT (Riebeseel et al., 2010). During seed maturation, Asn (beside Gln) represents an N source, which is loaded via plasma membrane-integral permeases from the maternal tissue to the developing embryo (Tegeder et al., 2000; Miranda et al., 2001). Because released ammonia is primarily fixed via the plastidic glutamine synthetase/glutamate synthase (GS/GOGAT) cycle, it is tempting to speculate that the loss of the amino acid-selective channel OEP16 in the outer envelope membrane of seed plastids results in reduced amino acid transfer to plastids and thus accumulation of Asn and other amino acids in the cytosol of developing seeds. During further germination, seeds completely rely on their endogenous amino acid supply, which now is reduced in the oep16 triple mutant, most probably due to previous incomplete N fixation and amino acid transfer to plastids in seed maturation. This hypothesis is underlined by the finding that amino acids such as Arg, Val, Lys, and Phe, which were up-regulated in dry mutant seeds, are down-regulated after imbibition and initiation of germination. Interestingly, among all amino acids regulated in maturing and dry seeds as well as in ABA-treated seedlings, those that are Asp derived (Asp, Asn, Lys, Met, Ile, and Thr) were over-represented (see Supplementary Table S3), suggesting an OEP16 transport function for the latter.
Whereas sucrose mobilization seems to be restricted in maturing oep16.1/2/4 seeds —as indicated by reduced glucose but elevated sucrose—this pattern is turned around in dry and imbibed mutant seeds where decreased sucrose levels point to a higher demand for carbohydrate catabolism than in the wild type (see Supplementary Table S3 at JXB online). Furthermore, strongly reduced starch levels in dry seeds and ABA-treated seedlings not only demonstrate that ABA addition to oep16 triple mutant seedlings mimics the status of dry seed tissue (see above), but underline increased carbohydrate degradation in oep16.1/2/4 seeds. Interestingly, starch content per seed is also decreased in ABA-deficient pea mutant seeds (Radchuk et al., 2010a), again linking OEP16 function with ABA and sugar signalling as well as with regulation of the metabolic status in seeds.
Conclusion on the function of OEP16
By studying localization and regulation of OEP16 transcripts and proteins as well as ABA hypersensitivity and metabolite distribution of oep16 triple knockout mutants in the current study, a role for the OEP16.1 and OEP16.2 isoforms in seed development and germination can now be assigned. Because in these developmental stages the content of free amino acids is affected by the loss of OEP16 and due to previous in vitro data (Pohlmeyer et al., 1997), it is concluded that OEP16 functions in shuttling amino acids across the outer envelope of plastids. Further, the results on seed development and the phenotype of the oep16 triple mutant clearly contradict a previously proposed role for OEP16 in pre-protein transport of the protochlorophyllide oxidoreductase A (Reinbothe et al., 2004, 2005; Pollmann et al., 2007; Samol et al., 2011a, b; for details and discussion, see Philippar et al., 2007; Pudelski et al., 2009, 2010; Bonneville and Tichtinsky, 2010). Due to the alternating expression pattern in seeds, OEP16.1 is involved in reserve accumulation processes during early seed maturation, but, when a transcriptional and metabolic switch induces the ABA-controlled seed desiccation and dormancy, OEP16.2 comes into play, bridging late seed development with early germination.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Characterization of OEP16 mutant lines.
Figure S2. OEP16 expression profiles during Arabidopsis development.
Figure S3. Tissue-specific expression of OEP16 isoforms in Arabidopsis seed development.
Figure S4. Tissue-specific expression of OEP16 isoforms in soybean seed development.
Fig. S5. Hypersensitivity during germination correlates with the loss of OEP16.2.
Figure S6. ABA-induced amino acid changes in Arabidopsis seedlings.
Figure S7. Regulation of OEP16 expression in a seed high-lysine mutant, within the ABA signalling chain, and during dormancy and germination.
Acknowledgments
We would like to thank Olga Lesina and Ingrid Jeshen (LMU Munich) for initial experiments on ABA immunoblots of OEP16.2, Iryna Ilkavets (LMU Munich) for generation of promoter–GUS lines, as well as Tanja Bender and Alfred Schmiedl (FAU Erlangen) for technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) and CiPSM to JS, as well as within the Bavarian research network FOR PLANTA (projects of US, JS).
Glossary
Abbreviations
- ABA
abscisic acid
- OEP16
outer envelope protein of 16 kDa
References
- Angelovici R, Fait A, Fernie AR, Galili G. A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytologist. 2011;189:148–159. doi: 10.1111/j.1469-8137.2010.03478.x. [DOI] [PubMed] [Google Scholar]
- Angelovici R, Fait A, Zhu X, Szymanski J, Feldmesser E, Fernie AR, Galili G. Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiology. 2009;151:2058–2072. doi: 10.1104/pp.109.145631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Galili G, Fernie AR, Fait A. Seed desiccation: a bridge between maturation and germination. Trends in Plant Science. 2010;15:211–218. doi: 10.1016/j.tplants.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ. Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proceedings of the National Academy of Sciences, USA. 2011;108:9709–9714. doi: 10.1073/pnas.1100958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville JM, Tichtinsky G. Correction for ‘A plant porphyria related to defects in plastid import of protochlorophyllide oxidoreductase’. Proceedings of the National Academy of Sciences, USA. 2010;107:5693. doi: 10.1073/pnas.0610934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Hofmann–Benning S, Weber APM. Comparative proteomics of chloroplast envelopes from C-3 and C-4 plants reveals specific adaptations of the plastid envelope to C-4 photosynthesis and candidate proteins required for maintaining C-4 metabolite fluxes. Plant Physiology. 2008a;148:568–579. doi: 10.1104/pp.108.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Shrestha RP, Whitten D, Wilkerson CG, Carr KM, Froehlich JE, Weber APM. Low-coverage massively parallel pyrosequencing of cDNAs enables proteomics in non-model species: comparison of a species-specific database generated by pyrosequencing with databases from related species for proteome analysis of pea chloroplast envelopes. Journal of Biotechnology. 2008b;136:44–53. doi: 10.1016/j.jbiotec.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Weber APM. Proteomic analysis of the proplastid envelope membrane provides novel insights into small molecule and protein transport across proplastid membranes. Molecular Plant. 2009;6:1247–1261. doi: 10.1093/mp/ssp070. [DOI] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiology. 2011;155:1678–1689. doi: 10.1104/pp.110.168120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. Erratum in: Plant Journal 47, 164. [DOI] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. The Plant Journal. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Peer W, Schmuths H, Footitt S, Theodoulou FL, Holdsworth MJ. Gene expression profiling reveals defined functions of the ATP–binding cassette transporter COMATOSE late in phase II of germination. Plant Physiology. 2007;143:1669–1679. doi: 10.1104/pp.107.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- Clausen C, Ilkavets I, Thomson R, Philippar K, Vojta A, Möhlmann T, Neuhaus E, Fulgosi H, Soll J. Intracellular localization of VDAC proteins in plants. Planta. 2004;220:30–37. doi: 10.1007/s00425-004-1325-3. [DOI] [PubMed] [Google Scholar]
- Drea SC, Lao NT, Wolfe KH, Kavanagh TA. Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. The Plant Journal. 2006;46:723–735. doi: 10.1111/j.1365-313X.2006.02741.x. [DOI] [PubMed] [Google Scholar]
- Drechsel G, Raab S, Hoth S. Arabidopsis zinc-finger protein 2 is a negative regulator of ABA signaling during seed germination. Plant Physiology. 2010;167:1418–1421. doi: 10.1016/j.jplph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Duy D, Soll J, Philippar K. Solute channels of the outer membrane: from bacteria to chloroplasts. Biological Chemistry. 2007;388:879–889. doi: 10.1515/BC.2007.120. [DOI] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugière S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Molecular and Cellular Proteomics. 2003;2:325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. The Plant Journal. 2007;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham I, Baker A, Holdsworth M. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO Journal. 2002;21:2912–2922. doi: 10.1093/emboj/cdf300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14:15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annual Reviews in Plant Biology. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS. Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. Journal of Proteome Research. 2003;2:413–425. doi: 10.1021/pr034025j. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiologia Plantarum. 2002;116:238–247. doi: 10.1034/j.1399-3054.2002.1160214.x. [DOI] [PubMed] [Google Scholar]
- Galili G. The aspartate-family pathway of plants: linking production of essential amino acids with energy and stress regulation. Plant Signaling Behaviour. 2011;6:192–195. doi: 10.4161/psb.6.2.14425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze TA, Philippar K, Ilkavets I, Soll J, Wagner R. OEP37 is a new member of the chloroplast outer membrane ion channels. Journal of Biological Chemistry. 2006;281:17989–17998. doi: 10.1074/jbc.M600700200. [DOI] [PubMed] [Google Scholar]
- Gonzali S, Loreti E, Solfanelli C, Novi G, Alpi A, Perata P. Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. Journal of Plant Research. 2006;119:115–123. doi: 10.1007/s10265-005-0251-1. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annual Review of Plant Biology. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Holdsworth M, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after ripening, dormancy and germination. New Phytologist. 2008a;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends in Plant Science. 2008b;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Holmes-Davis R, Tanaka CK, Vensel WH, Hurkman WJ, McCormick S. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics. 2005;5:4864–1884. doi: 10.1002/pmic.200402011. [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiology. 2003;132:640–652. doi: 10.1104/pp.103.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Okuda R, Ban A, Toyoshima R, Tsutsumida K, Usui H, Yamamoto A, Hattori T. Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant and Cell Physiology. 2005;46:300–311. doi: 10.1093/pcp/pci031. [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends in Plant Science. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Keegstra K, Youssif AE. Isolation and characterization of chloroplast envelope membranes. Methods in Enzymology. 1986;118:316–325. [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Current Biology. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- Kogel KH, Voll LM, Schäfer P, et al. Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific differences. Proceedings of the National Academy of Sciences, USA. 2010;107:6198–6203. doi: 10.1073/pnas.1001945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linka N, Weber APM. Intracellular metabolite transporters in plants. Molecular Plant. 2010;3:21–53. doi: 10.1093/mp/ssp108. [DOI] [PubMed] [Google Scholar]
- Linke D, Frank J, Pope MS, Soll J, Ilkavets I, Fromme P, Burstein EA, Reshetnyak YK, Emelyanenko VI. Folding kinetics and structure of OEP16. Biophysical Journal. 2004;86:1479–1487. doi: 10.1016/S0006-3495(04)74216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA. Plastids unleashed: their development and their integration in plant development. International Journal of Developmental Biology. 2005;49:557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C. Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Reports. 2006;7:947–952. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Heim U, Sauer N, Wobus U, Weber H. Amino acid permeases in developing seeds of Vicia faba L: expression precedes storage protein synthesis and is regulated by amino acid supply. The Plant Journal. 2001;28:61–71. doi: 10.1046/j.1365-313x.2001.01129.x. [DOI] [PubMed] [Google Scholar]
- Murcha MW, Elhafez D, Lister R, Tonti-Filippini J, Baumgartner M, Philippar K, Carrie C, Mokranjac D, Soll J, Whelan J. Characterisation of the preprotein and amino acids transporter gene family in Arabidopsis. Plant Physiology. 2007;134:199–212. doi: 10.1104/pp.106.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. The Plant Journal. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. The Plant Cell. 2004;16:2705–2718. doi: 10.1105/tpc.104.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K, Geis T, Ilkavets I, Oster U, Schwenkert S, Meurer J, Soll J. Chloroplast biogenesis: the use of mutants to study the etioplast–chloroplast transition. Proceedings of the National Academy of Sciences, USA. 2007;104:678–683. doi: 10.1073/pnas.0610062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar K, Soll J. Intracellular transport: solute transport in chloroplasts, mitochondria, peroxisomes and vacuoles, and between organelles. In: Yeo AR, Flowers TJ, editors. Plant solute transport. Oxford: Blackwell Publishing Ltd; 2007. pp. 133–192. [Google Scholar]
- Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R. Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proceedings of the National Academy of Sciences, USA. 1997;94:9504–9509. doi: 10.1073/pnas.94.17.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Springer A, Buhr F, Lahroussi A, Samol I, Bonneville JM, Tichtinsky G, von Wettstein D, Reinbothe C, Reinbothe S. A plant porphyria related to defects in plastid import of protochlorophyllide oxidoreductase A. Proceedings of the National Academy of Sciences, USA. 2007;104:2019–2023. doi: 10.1073/pnas.0610934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pudelski B, Kraus S, Soll J, Philippar K. The plant PRAT proteins—preprotein and amino acid transport in mitochandria and chloroplasts. Plant Biology (Stuttgart) 2010;12(Suppl 1):42–55. doi: 10.1111/j.1438-8677.2010.00357.x. [DOI] [PubMed] [Google Scholar]
- Pudelski B, Soll J, Philippar K. A search for factors influencing etioplast–chloroplast transition. Proceedings of the National Academy of Sciences, USA. 2009;106:12201–12206. doi: 10.1073/pnas.0902145106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R, Conrad U, Saalbach I, Giersberg M, Emery RJ, Küster H, Nunes-Nesi A, Fernie AR, Weschke W, Weber H. Abscisic acid deficiency of developing pea embryos achieved by immunomodulation attenuates developmental phase transition and storage metabolism. The Plant Journal. 2010a;64:715–730. doi: 10.1111/j.1365-313X.2010.04376.x. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschke W, Weber H. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. The Plant Journal. 2010b;61:324–338. doi: 10.1111/j.1365-313X.2009.04057.x. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiology. 2006;140:263–278. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Pollmann S, Springer A, James RJ, Tichtinsky G, Reinbothe C. A role of Toc33 in the protochlorophyllide-dependent plastid import pathway of NADPH:protochlorophyllide oxidoreductase (POR) A. The Plant Journal. 2005;42:1–12. doi: 10.1111/j.1365-313X.2005.02353.x. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Quigley F, Springer A, Schemenewitz A, Reinbothe C. The outer plastid envelope protein Oep16: role as precursor translocase in import of protochlorophyllide oxidoreductase A. Proceedings of the National Academy of Sciences, USA. 2004;101:2203–2208. doi: 10.1073/pnas.0301962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebeseel E, Häusler RE, Radchuk R, et al. The 2-oxoglutarate/malate translocator mediates amino acid and storage protein biosynthesis in pea embryos. The Plant Journal. 2010;61:350–363. doi: 10.1111/j.1365-313X.2009.04058.x. [DOI] [PubMed] [Google Scholar]
- Samol I, Buhr F, Springer A, Pollmann S, Lahroussi A, Rossig C, von Wettstein D, Reinbothe C, Reinbothe S. Implication of the oep16-1 mutation in a flu-independent, singlet oxygen–regulated cell death pathway in Arabidopsis thaliana. Plant and Cell Physiology. 2011a;52:84–95. doi: 10.1093/pcp/pcq176. [DOI] [PubMed] [Google Scholar]
- Samol I, Rossig C, Buhr F, Springer A, Pollmann S, Lahroussi A, von Wettstein D, Reinbothe C, Reinbothe S. The outer chloroplast envelope protein OEP16-1 for plastid import of NADPH:protochlorophyllide oxidoreductase A in Arabidopsis thaliana. Plant and Cell Physiology. 2011b;52:96–111. doi: 10.1093/pcp/pcq177. [DOI] [PubMed] [Google Scholar]
- Schleiff E, Eichacker LA, Eckart K, Becker T, Mirus O, Stahl T, Soll J. Prediction of the plant beta-barrel proteome: a case study of the chloroplast outer envelope. Protein Science. 2003;12:748–759. doi: 10.1110/ps.0237503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Suzuki M, McCarty DR. Functional symmetry of the B3 network controlling seed development. Current Opinion in Plant Biology. 2008;11:548–553. doi: 10.1016/j.pbi.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Steinkamp T, Hill K, Hinnah SC, Wagner R, Rohl T, Pohlmeyer K, Soll J. Identification of the pore-forming region of the outer chloroplast envelope protein OEP16. Journal of Biological Chemistry. 2000;275:11758–11764. doi: 10.1074/jbc.275.16.11758. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW. Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiology. 2000;122:319–326. doi: 10.1104/pp.122.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow IJ, Rawsthorne S, Raines C, Emes MJ. Plastid metabolic pathways. In: Möller SG, editor. Plastids. Oxford: Blackwell Publishing Ltd; 2005. pp. 60–125. [Google Scholar]
- Waegemann K, Eichacker S, Soll J. Outer envelope membranes from chloroplasts are isolated as right-side-out vesicles. Planta. 1992;187:89–94. doi: 10.1007/BF00201628. [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. Molecular physiology of legume seed development. Annual Review of Plant Biology. 2005;56:253–279. doi: 10.1146/annurev.arplant.56.032604.144201. [DOI] [PubMed] [Google Scholar]
- Weigelt K, Küster H, Radchuk R, Müller M, Weichert H, Fait A, Fernie AR, Saalbach I, Weber H. Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. The Plant Journal. 2008;55:909–926. doi: 10.1111/j.1365-313X.2008.03560.x. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Droux M. Synthesis of the sulfur amino acids: cysteine and methionine. Photosynthesis Research. 2005;86:345–362. doi: 10.1007/s11120-005-8810-9. [DOI] [PubMed] [Google Scholar]
- Yamagishi K, Tatematsu K, Yano R, Preston J, Kitamura S, Takahashi H, McCourt P, Kamiya Y, Nambara E. CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant and Cell Physiology. 2009;50:330–340. doi: 10.1093/pcp/pcn201. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008 doi: 10.1371/journal.pone.0001994. 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.