Abstract
Seed dormancy prevents seeds from germinating under environmental conditions unfavourable for plant growth and development and constitutes an evolutionary advantage. Dry storage, also known as after-ripening, gradually decreases seed dormancy by mechanisms not well understood. An Arabidopsis thaliana DOF transcription factor gene (DOF6) affecting seed germination has been characterized. The transcript levels of this gene accumulate in dry seeds and decay gradually during after-ripening and also upon seed imbibition. While constitutive over-expression of DOF6 produced aberrant growth and sterility in the plant, its over-expression induced upon seed imbibition triggered delayed germination, abscisic acid (ABA)-hypersensitive phenotypes and increased expression of the ABA biosynthetic gene ABA1 and ABA-related stress genes. Wild-type germination and gene expression were gradually restored during seed after-ripening, despite of DOF6-induced over-expression. DOF6 was found to interact in a yeast two-hybrid system and in planta with TCP14, a previously described positive regulator of seed germination. The expression of ABA1 and ABA-related stress genes was also enhanced in tcp14 knock-out mutants. Taken together, these results indicate that DOF6 negatively affects seed germination and opposes TCP14 function in the regulation of a specific set of ABA-related genes.
Keywords: ABA1, abscisic acid, after-ripening, DOF6/DOF3.2, germination, heat shock proteins, seeds, TCP14, transcription factors
Introduction
The seed is an important organ for plant survival and species dispersion. At the end of development, mature seeds may enter a dormant state that prevents germination even under favourable conditions (Bewley, 1997), an important adaptation and a commercial trait (Gubler et al., 2005). In Arabidopsis thaliana, as in many other species, both dormancy and germination potential are determined by the interaction between genetic and environmental factors encountered during seed development, storage, and imbibition (Finkelstein et al., 2002; Donohue et al., 2005; Liu et al., 2005; Penfield et al., 2005; Finch-Savage and Leubner-Metzger, 2006; Holdsworth et al., 2008; Matakiadis et al., 2009; Piskurewicz et al., 2009; Rodriguez-Gacio et al., 2009; Yano et al., 2009; Barrero et al., 2010; Josse et al., 2011). These processes are mediated mainly by the ratio of two antagonistic hormones: abscisic acid (ABA) and gibberellins (GAs). Dry storage gradually reduces dormancy, a process called after-ripening. Upon imbibition of after-ripened (AR) seeds, a dramatic quick decay of ABA levels is concomitant with a gradual increase in GAs, which allows germination to occur. In contrast, dormant non-germinating seeds have higher levels of ABA upon imbibition when compared to AR seeds (Ogawa et al., 2003; Kushiro et al., 2004; Millar et al., 2006). Accordingly, genetic alterations of genes involved in ABA biosynthesis, catabolism, or signalling have a profound effect on the germination potential of seeds (Debeaujon and Koornneef, 2000; Lefebvre et al., 2006; Penfield and King, 2009; Lee et al., 2010).
The mechanism by which dormancy is regulated by after-ripening and other environmental signals is not well understood. Dry seeds contain mRNAs from the embryogenesis and maturation phases of seed development, some of them required during the early stages of germination (Holdsworth et al., 2008). During after-ripening, changes in the abundance of specific mRNAs and proteins take place as a result of active transcription and translation (Bove et al., 2005; Leubner-Metzger, 2005; Chibani et al., 2006; Iglesias-Fernández and Matilla, 2009), although major changes in gene expression occur upon seed imbibition (Ogawa et al., 2003; Yamauchi et al., 2004; Nakabayashi et al., 2005; Cadman et al., 2006; Carrera et al., 2008; Kimura and Nambara, 2010). Whereas AR imbibed seeds show a large increase in RNAs encoding proteins associated with protein translation and degradation, reserve mobilization, and cell-wall modification, imbibed dormant seeds show an up-regulation of stress-related genes, many of which are expressed during seed maturation under the control of ABA signalling (Cadman et al., 2006).
Transcriptomic analyses of dry and germinating seeds have shown the overlapping nature of late seed development and germination programmes. Members of several transcription factor (TF) families accumulate their mRNAs in dry seeds and play a pivotal role in triggering and maintaining gene expression during early stages of imbibition (Kimura and Nambara, 2010), or are differentially expressed between dormant and AR imbibed seeds (Yano et al., 2009; Barrero et al., 2010). In barley, TFs belonging to different families have been shown to regulate gene expression both during seed maturation and at early post-germinative phases and, in some cases, as occurs with the DOF TFs BPBF and HvDOF19, play opposite roles (Mena et al., 1998, 2002; Moreno-Risueño et al., 2007a). In the A. thaliana genome, the DOF family is composed of 36 different members, mainly associated with plant-specific phenomena (Yanagisawa, 2002; Lijavetzky et al., 2003; Moreno-Risueño et al., 2007b). Two of them, DAG1 and DAG2 were shown to influence, with opposite effects, the germination of Arabidopsis seeds (Papi et al., 2000; Gualberti et al., 2002; Gabriele et al., 2010).
An A. thaliana DOF TF (At3g45610; DOF3.2/DOF6; Yanagisawa, 2002; Moreno-Risueño et al., 2007b), hereafter DOF6, has been characterized as a negative regulator of seed germination. DOF6 transcripts accumulate in dry seeds and decay gradually upon imbibition and during after-ripening. While constitutive over-expression of DOF6 produced aberrant growth and sterility in the plant, inducible over-expression (IOEX) of DOF6 upon seed imbibition triggered delayed germination, ABA-hypersensitive phenotypes, and increased expression of genes involved in ABA biosynthesis and stress. These altered responses were under the control of the AR status of the seed. A search for DOF6 protein partners identified TCP14, a TF previously identified as a positive regulator of germination potential (Tatematsu et al., 2008). The same ABA-related genes were up-regulated in ioexDOF6 and tcp14 knock-out (KO) mutants. These results indicate that DOF6 negatively affects seed germination and that DOF6 and TCP14 play opposite roles on this process.
Materials and methods
Plant material and growth conditions
A. thaliana Columbia accession (Col-0) was used as the wild type (WT) in this study. A dof6-1 single mutant (SALK_010732) was identified in the Salk T-DNA insertion database (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al., 2003). tcp14-1 and tcp14-3 mutants were previously described in Tatematsu et al. (2008). dof6-1 and tcp14-3 mutant seeds were obtained from NASC (University of Nottingam, UK) and tcp14-1 was provided by Prof. Lucia Colombo (Università di Milano, Italy). Plants were grown either on Petri dishes containing half-strength Murashige and Skoog (MS) medium buffered with 2 mM MES (2-N-morpholino ethanesulfonic acid), pH 5.7, and 0.7% (w/v) agar, or in soil, and grown to maturity at 16 h light at 22 °C /8 h dark at 20 °C and 60% relative humidity. Seeds were harvested when plants had ceased flowering and siliques were starting to dehisce and stored in the dark at 22 °C and 30% relative humidity.
Generation of transgenic lines
The pDOF6::GUS plasmid was generated using oligonucleotides pDOF6-fw and pDOF6-rv (Supplementary Table S1, available at JXB online) to amplify a 1.2 kb fragment containing the putative promoter region of DOF6 from A. thaliana genomic DNA. The amplified fragment was digested with SalI and BamHI, cloned into the pENTR3C vector (Invitrogen, USA) and transferred by Gateway LR recombination (Invitrogen) into the destination vector pMDC163, containing the β-glucuronidase (GUS) reporter gene (Curtis and Grossniklaus, 2003). The amiRNA DOF6 construct was designed using the resource at www.weigelworld.org and the oligonucleotides used were DOF6-I-miR-fw, DOF6-II-miR-rv, DOF6-III-miR*-fw, and DOF6-IV-miR*-rv (Supplementary Table S1). Cloning procedure was as described in www.weigelworld.org and the amiRNA sequence was digested with XhoI and SpeI and cloned into the pER8 vector (Zuo et al., 2000). A RNAi DOF6 construct (CATMA3a38625) from the Agrikola collection, together with the pSOUP vector, was provided by NASC (Hilson et al., 2004). The p35S::DOF6 and ioexDOF6 constructs were generated using the oligonucleotides DOF6-fw and DOF6-rv (Supplementary Table S1) to amplify the DOF6 open reading frame (ORF) from A. thaliana cDNA. The amplified fragment was digested with SalI and EcoRI, and cloned into the pENTR3C vector (Invitrogen) and introduced by LR Gateway reaction into the destination vectors pEarleyGate201 (Earley et al., 2006) and pMDC7 (Curtis and Grossniklaus, 2003). All constructs were introduced into Agrobacterium tumefaciens strain C58C1 GV3101 by electroporation (in the case of RNAi DOF6, together with pSOUP vector) and transformed into Arabidopsis by the floral dip method (Clough and Bent, 1998).
Seed germination assays
WT and mutant seeds were collected at the same time and obtained from plants grown in the same conditions. Storage conditions were 22 °C and 30% relative humidity in the dark for 1 week for freshly harvested (FH) seeds and for 3 months for AR seeds. For each genotype, approximately 50 seeds were placed onto filter papers (Whatman No. 3, UK) moistened with 3 ml of sterile water in 6 cm diameter Petri dishes. Plates were sealed with Micropore tape (Micropore 3M, USA) and incubated at 22 °C under 16/8 h light/dark conditions. Germination was scored as radicle emergence through the endosperm and testa every 24 h. All germination assays were carried out in triplicate with at least two independent seed batches.
estradiol treatments
To induce gene expression in ioexDOF6 and amiDOF6 lines in germination assays, 17-β-estradiol (Sigma Aldrich, Spain) was diluted directly in the imbibition water to a final concentration of 50 μM. For continuous induction on agar plates, drops of 50 μM estradiol were directly added to each plant every 2 days. For induction on rosette leaves, 50 μM estradiol containing 0.02% (v/v) Tween 20 was sprayed on the leaves and samples were harvested after 16 h.
Gene expression analyses
Total RNA was isolated from seeds and other tissues, as described by Oñate-Sánchez and Vicente-Carbajosa (2008). Seeds (15 mg) from each genotype were germinated on moistened filter paper and samples were taken at different hours after imbibition (hai). The Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics, Mannheim, Germany) was used to synthesize cDNA. For quantitative reverse transcription (RT)-PCR, 8–16 ng cDNA was used as template together with 0.5 μM of forward and reverse specific oligonucleotides (Supplementary Table S1) and Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Cycling conditions (ABI Prism 7300; Applied Biosystems) were as follows: 10 min at 95 °C and 50 cycles of 15 s at 95 °C and 60 s at 60 °C, linked to a default dissociation stage programme to detect non-specific amplification. Three technical and at least two biological replicates were included in every experiment and the ubiquitin gene UBC21 (At5g25760) was used to normalize expression levels. Therefore, gene-specific mRNA levels shown in all figures except Fig. 5 are relative to UBC21.
Fig. 5.
mRNA levels of ABA-related genes in germinating freshly harvested (FH) ioexDOF6 seeds. Expression of genes involved in ABA metabolism and signalling (A) and ABA-inducible genes (B) was compared by quantitative RT-PCR between wild-type (WT) and ioexDOF6 FH seeds 48 hours after imbibition in 50 μM estradiol. Gene-specific mRNA levels are relative to UBC21. All the UBC21 normalized expression values are relative to the values obtained for each gene in the WT sample.
ABA measurements
Seeds were collected 48 hai in 50 μM estradiol and processed as described in Durgbanshi et al. (2005).
In situ hybridization and histochemical GUS analyses
In situ hybridization was performed as described by Iglesias-Fernández et al. (2011). Specific oligonucleotides (DOF6-insitu-fw and DOF6-insitu-rv; Supplementary Table S1) were used to amplify a 200 bp fragment from the DOF6 3′-non-coding region. Sense and antisense digoxigenin (DIG)-labelled RNA probes were synthesized with the DIG RNA labelling mix (Roche Diagnostics). Probes were hybridized at 52 °C overnight.
GUS staining was performed using the protocol described by Jefferson et al. (1987). After 3 h of seed imbibition, testa and endosperm were separated from the embryos and were incubated with the staining solution at 37 °C until blue colour was visible. Analyses were performed using at least eight independent lines.
Yeast two-hybrid assays
A BD-DOF6 construct was obtained by the LR Gateway recombination reaction between the entry vector pENTR3C, harbouring the DOF6-ORF cassette, and the destination vector pDEST32 (Invitrogen) and was transformed into the Saccharomyces cerevisiae pJ694α strain (James et al., 1996). Yeast two-hybrid screening of an arrayed TF yeast library containing c. 1200 A. thaliana TFs was performed as described by Castrillo et al. (2011). The plates used in the screening to select the positive interactions contained 30 mM 3-amino-1,2,4-triazole (3-AT; Sigma Aldrich). Yeast transformation was done by the polyethylene glycol method. Quantification of β-galactosidase (LacZ) activity in liquid culture in the S. cerevisiae SFY526 strain was calculated using Miller’s formula as described by Lara et al. (2003).
Bimolecular fluorescent complementation
TCP14 ORF was amplified from A. thaliana cDNA using the oligonucleotides TCP14-GW-fw and TCP14-GW-rv (Supplementary Table S1) and cloned into the pDONR221 plasmid by the BP Gateway reaction. Both DOF6 and TCP14 were fused in frame with the N- and C-terminal fragments of yellow fluorescent protein (YFP), respectively, by LR Gateway recombination with the destination vectors pE-SPYNE-GW and pE-SPYCE-GW (Weltmeier et al., 2006). Co-bombardment experiments were done in inner epidermal layers of fresh onions (Allium cepa) using a biolistic Helium gun device (DuPont PDS-1000; BioRad Laboratories, Hercules, CA, USA). The fluorescence emission was observed after 24 h of incubation at 22 °C in the dark under a fluorescence Zeiss Axiophot microscope (Carl Zeiss, Germany) with the following filter set: excitation, 450–490 nm; emission, 520 nm. Images were captured with a CCD colour Leica DFC300FX camera and processed with the Leica Application Suite 2.8.1 build 1554 software (Leica, http://www.leica.com). Each bombardment was performed in four independent plates and complementation was confirmed in two independent assays. Transformation efficiencies were estimated by bombardment with a p35S::GFP construct.
Results
DOF6 is expressed in A. thaliana seeds
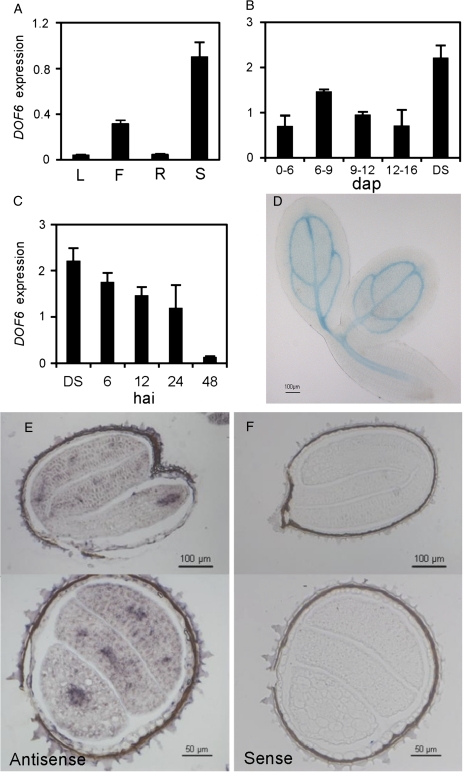
Since several DOF proteins have been strongly linked to the regulation of gene expression during seed development, it was decided to explore whether other phylogenetically related DOFs could have a regulatory role in this organ. DOF6 was selected for further studies since quantitative RT-PCR analyses revealed that its mRNA was preferentially expressed in siliques, although it was also found in leaves, roots, and flowers (Fig. 1A). During silique development, DOF6 was expressed throughout the maturation phase, reaching its maximum expression levels in dry seeds (Fig. 1B). Upon imbibition of FH seeds, DOF6 mRNA levels gradually decreased, being 20 times less abundant at 48 hai than in dry seeds (Fig. 1C). In order to localize the expression of DOF6 in seeds, A. thaliana Col-0 plants were transformed with the GUS reporter gene under the control of the DOF6 promoter. GUS activity was detected in the vascular tissues of the embryo at 3 hai, an expression pattern also observed at 24 hai (Fig. 1D). To confirm this pattern, mRNA in situ hybridization experiments were performed in seeds imbibed for 24 h. As shown in Fig. 1E, DOF6 transcripts were observed in the embryo, mainly in the vascular tissues, and could not be detected in the samples hybridized with a control sense probe (Fig. 1F).
Fig. 1.
DOF6 expression patterns. DOF6 mRNA levels relative to UBC21 in: (A) wild-type plants in different plant organs (L, adult rosette leaves; F, flowers; R, roots; S, pool of developing siliques from 4 to 15 days after pollination (dap); DS, dry seeds); (B) siliques at different dap and (C) seeds at different hours after imbibition (hai). Means and standard errors for three replicates are shown. (D) Expression of the GUS reporter gene driven by the DOF6 promoter in seeds imbibed for 3 h. Seed covers were removed before incubating with the staining solution. (E, F) In situ hybridization of DOF6 transcripts in Col-0 seed sections. Seeds were imbibed for 24 h and hybridized using antisense (E) and sense (F) DOF6 probes. Gene-specific mRNA levels are relative to UBC21. Bars, 100 μm (D), 100 and 50 μm (E and F).
DOF6 over-expression causes severe growth defects in Arabidopsis
To analyse DOF6 function in the plant, T-DNA insertion mutants were searched in public A. thaliana KO collections. Only one line was found to contain an insertion in the DOF6 genomic region (SALK_010732). Homozygous plants for this insertion line did not show alterations in DOF6 expression levels, probably due to the position of the T-DNA located at 900 bp upstream of the initiation codon (Supplementary Fig. S1). Therefore, to reduce DOF6 mRNA levels, transgenic plants carrying constructs for constitutive or inducible expression of DOF6-specific ihpRNAs or amiRNAs, respectively, were generated. DOF6 mRNA levels were not reduced in any of the 15 ihpRNA or eight amiRNA transgenic lines analysed (Supplementary Fig. S1).
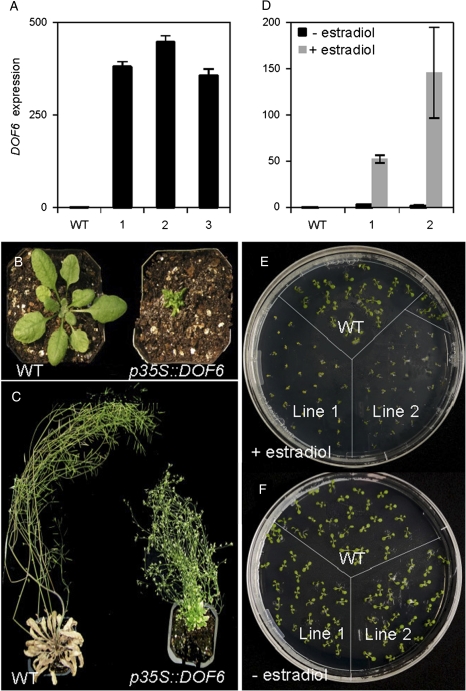
To study the effect of DOF6 over-expression, Arabidopsis plants were transformed with the DOF6 ORF under the control of the CaMV 35S promoter (p35S::DOF6). Eight independent transgenic lines were selected with higher DOF6 mRNA levels than the WT. The expression of DOF6 in three of these lines is shown in Fig. 2A. All these lines showed a stunted phenotype and severe growth defects (Fig. 2B). Only one line was able to develop flowers, although producing very few non-viable seeds (Fig. 2C; Supplementary Fig. S2).
Fig. 2.
DOF6 over-expression produces growth and developmental defects on the plant. (A) DOF6 mRNA levels in rosette leaves of wild-type (WT) and p35S::DOF6 transgenic plants. Representative independent transgenic lines are shown. (B and C) Three- and eight-week-old plants over-expressing DOF6 (p35S::DOF6), respectively and show a dwarf phenotype compared to the WT and are unable to produce seeds. (D) DOF6 mRNA levels in WT and two representative ioexDOF6 lines, before and 16 h after spraying with 50 μM estradiol. (E and F) WT and two representative ioexDOF6 lines germinated on half-strength Murashige and Skoog agar with 50 μM estradiol added every 2 days (E) and without estradiol (F); pictures were taken 3 weeks after sowing. Gene-specific mRNA levels are relative to UBC21.
Since constitutive over-expression of DOF6 mRNA levels had deleterious effects to the plant, an IOEX strategy was adopted. Arabidopsis plants were transformed with the DOF6 ORF under the control of an estradiol-inducible promoter (Curtis and Grossniklaus, 2003). The transgenic lines obtained, hereafter ioexDOF6, increased DOF6 mRNA expression after 16 h of being sprayed with 50 μM estradiol, while no significant differences with WT levels were observed when sprayed with a mock solution (Fig. 2D). ioexDOF6 plants developed normally when grown in the absence of estradiol but, when the induction was maintained by adding fresh estradiol to the growing media every 2 days, seedling growth was delayed, producing a stunted phenotype similar to that observed for the constitutive over-expression lines (Fig. 2E and 2F).
DOF6 over-expression delays seed germination in freshly harvested but not in after-ripened seeds
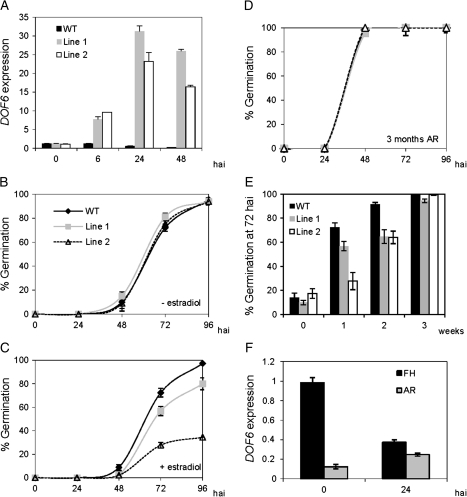
To study the effect of DOF6 over-expression on germination, FH ioexDOF6 seeds were imbibed in the presence or absence of 50 μM estradiol. As shown in Fig. 3A, DOF6 mRNA levels were similar in ioexDOF6 and in WT dry seeds, indicating the absence of significant leaky expression from the inducible promoter. Upon imbibition with estradiol, DOF6 mRNA levels increased up to 30 times in ioexDOF6 seeds. The germination ability of WT seeds was similar in the absence or presence of estradiol and undistinguishable from that observed for the ioexDOF6 lines in the absence of the inducer (Fig. 3B and 3C). However, in the presence of estradiol, FH ioexDOF6 seeds showed delayed germination when compared to WT seeds (Fig. 3C). Although the induction of DOF6 mRNA levels varied between three selected ioexDOF6 lines, and even between descendants of the same plant, the delayed germination phenotype was consistently observed in the three independent transgenic lines, and two showing lower germination percentages were chosen for further studies (Supplementary Fig. S3).
Fig. 3.
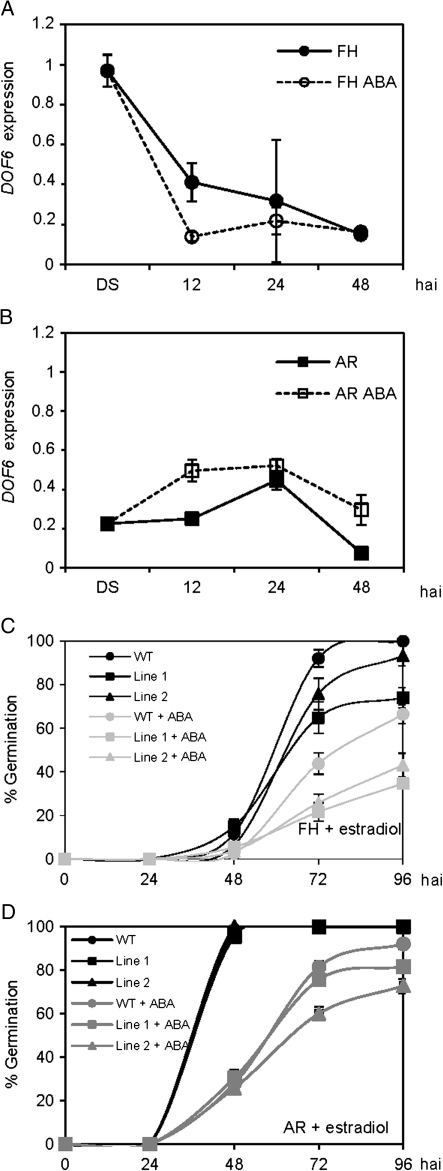
Germination of ioexDOF6 seeds and DOF6 expression in freshly harvested (FH) and after-ripened (AR) seeds. (A) DOF6 expression levels in FH wild type (WT) and two representative ioexDOF6 lines upon imbibition in 50 μM estradiol. (B and C) Germination of FH WT and ioexDOF6 seeds imbibed in 50 μM estradiol (C) and controls without estradiol (B). (D) Germination of AR WT and ioexDOF6 seeds in 50 μM estradiol. (E) Germination percentage of WT and ioexDOF6 seeds at 72 hours after imbibition (hai) in 50 μM estradiol after different weeks of dry storage. (F) DOF6 mRNA levels in FH and in AR WT dry seeds and 24 hai. Gene-specific mRNA levels are relative to UBC21.
To assess if this delayed germination phenotype is influenced by the after-ripening status of the seed, a similar germination experiment was done with AR seeds (3 months of storage at 22 °C; Fig. 3D). In this case, the germination kinetics were identical in WT and in ioexDOF6 seeds in the presence of estradiol. When the experiment was repeated with seeds that had been after-ripened for different periods, the delayed germination of FH ioexDOF6 seeds disappeared gradually with the dry storage time (Fig. 3E). Suppression of the delayed phenotype was also observed in FH ioexDOF6 seeds when stratified (4 °C, 48 h) or imbibed at 17°C (Supplementary Fig. S4). To investigate whether DOF6 endogenous transcripts were being affected by after-ripening, DOF6 mRNA levels were quantified in dry and in imbibed FH and AR seeds. While DOF6 mRNA levels were similar at 24 hai in FH and AR seeds, FH dry seeds contained nearly ten times more transcripts than AR dry seeds (Fig. 3F). These results indicate that the ioexDOF6-delayed germination phenotype is influenced by the after-ripening and dormancy status of the seed and that DOF6 mRNA levels decay during after-ripening and upon imbibition of FH seeds.
DOF6 over-expression increases ABA1 mRNA in freshly harvested seeds
Since DOF6 mRNA levels in seeds decreased during after-ripening and upon seed imbibition and increased during seed maturation, a pattern that resembled ABA dynamics in seeds (Kushiro et al., 2004; Vicente-Carbajosa and Carbonero, 2005; Millar et al., 2006), it seemed appropriate to quantify DOF6 mRNA levels in seeds in response to ABA. FH and AR WT seeds were imbibed in the presence or absence of ABA, and DOF6 transcripts were quantified during seed imbibition. As shown in Fig. 4A and B, DOF6 had different expression patterns in FH and AR imbibed seeds, both with or without ABA during the first hours of imbibition, but the transcript levels were not significantly altered by the presence of ABA after 24 h.
Fig. 4.
ABA effect on DOF6 expression and ioexDOF6 seed germination. (A and B) DOF6 expression by quantitative RT-PCR in freshly harvested (FH; A) and after-ripened (AR; B) Col-0 seeds imbibed in water with 2 and 5 μM ABA, respectively. (C and D) Germination of FH (C) and AR (D) wild-type (WT) and ioexDOF6 seeds in water supplemented with 50 μM estradiol and 0.5 and 1 μM ABA, respectively. Germinating percentages are represented as the mean±standard error from three replicates. Germination patterns were confirmed in two different seed batches. Gene-specific mRNA levels are relative to UBC21.
To test the ABA effects on the ioexDOF6 germination phenotype, FH and AR ioexDOF6 and WT seeds were imbibed with estradiol in the absence or presence of ABA and their germination scored. When FH seeds were used, the germination delay of ioexDOF6 seeds compared to WT was enhanced by the presence of ABA and an overall reduction in germination efficiency was detected for all the genotypes tested (Fig. 4C). The AR ioexDOF6 seeds, which had the same germination kinetics as the WT in the absence of ABA, showed a delayed germination in its presence (Fig. 4D). These data indicate that the ioexDOF6 lines are hypersensitive to exogenous ABA both in FH and in AR seeds.
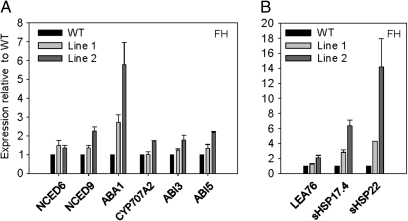
FH ioexDOF6 seeds germinated in the presence of estradiol were used to quantify mRNA levels of key genes involved in ABA metabolism and signalling pathways: ABA1, NCED6, and NCED9, encoding a zeaxanthin epoxidase and two 9-cis-epoxycarotenoid dioxygenases, respectively, involved in ABA biosynthesis (Frey et al., 1999; Audran et al., 2001; Lefebvre et al., 2006); CYP707A2, encoding an ABA 8′-hydroxylase involved in ABA degradation (Okamoto et al., 2006); and ABI3 and ABI5, encoding TFs involved in ABA signalling (Giraudat et al., 1992; Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002). ABA1 was significantly induced in ioexDOF6 lines compared to the WT (more than three times in one line and more than five in the other; Fig. 5A). Such differences were not found in the expression levels of NCED6, NCED9, CYP707A2, ABI3, and ABI5 between the WT and the ioexDOF6 lines, although NCED9 and ABI5 were induced more than two-fold in one of the ioexDOF6 lines (Fig. 5A). Then, the expression of seed ABA-inducible genes was analysed, such as those encoding late embryogenesis abundant (LEA) and small heat shock proteins (sHSPs; Kotak et al., 2007; Holdsworth et al., 2008). It was found that sHSP17.4 and sHSP22 were induced up to ten times in the ioexDOF6 lines compared with the WT (Fig. 5B). None of these genes showed an increased expression when AR seeds were used for the analyses (data not shown).
DOF6 interacts with TCP14, a transcription factor that regulates embryonic growth potential during seed germination.
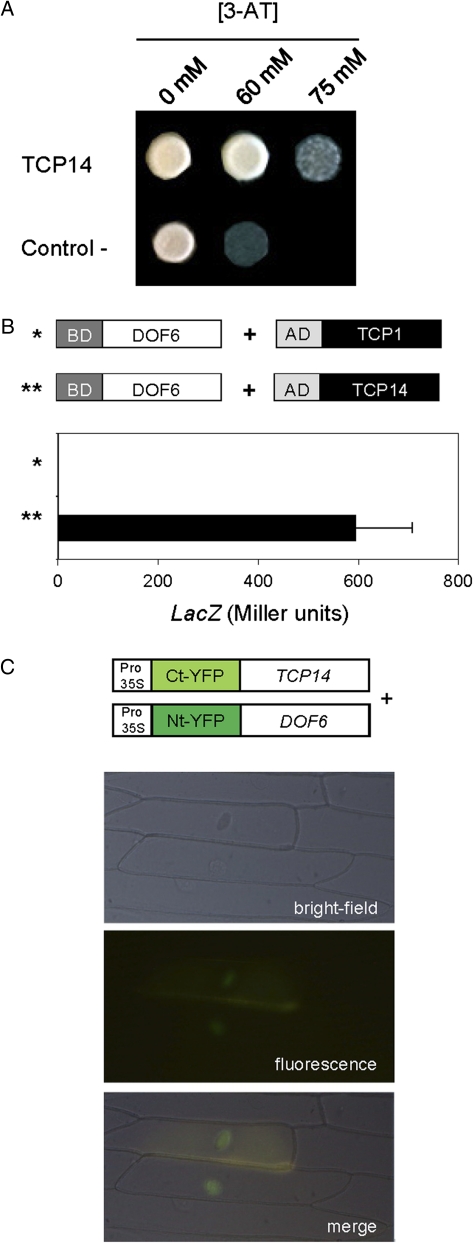
Since transcriptional regulation is a combinatorial process, possible interactors of DOF6 were looked for. To identify such interacting proteins, a yeast two-hybrid screening was performed, using as prey an arrayed library of c. 1200 A. thaliana TFs fused to the GAL4 activation domain (AD-TF). The DOF6 ORF was fused to the GAL4 DNA-binding domain (BD-DOF6) and used as a bait against each of the library TFs by a high-throughput mating assay (Castrillo et al., 2011). A positive interaction with TCP14 was identified and confirmed by growing the mated cells on the appropriate auxotrophic media with increasing concentrations of 3-AT (Fig. 6A). This interaction was quantified using a different yeast strain (S. cerevisiae SFY526) that contains a LacZ reporter gene under the control of GAL4BD binding sites. The SFY526 strain was transformed with BD-DOF6 and AD-TCP14 or with BD-DOF6 and AD-TCP1 constructs. TCP1 was used as a negative control since it is a member of the TCP family that did not interact with DOF6 in the screening. The LacZ activity of the yeast strain carrying the BD-DOF6 and AD-TCP14 constructs was several orders of magnitude above that of the control, indicating that this is a strong interaction (Fig. 6B). Moreover, these results confirm that this interaction is independent of yeast ploidy or genotype.
Fig. 6.
DOF6 interaction with TCP14. (A) Diploid cells carrying BD-DOF6 and AD-TCP14 (TCP14) or BD-DOF6 and AD empty vector (control –) were grown on the appropriate selection media with increasing concentrations of 3-amino-1,2,4-triazole (3-AT). (B) Quantification of the DOF6–TCP14 interaction in yeast using the β-galactosidase reporter gene. An AD-TCP1 construct was used as a negative control. (C) Bimolecular fluorescent complementation assays in onion epidermal cells by particle bombardment. TCP14 and DOF6 were fused to the C- and N-terminal fragments, respectively, of yellow fluorescent protein (YFP) and co-bombarded over epidermal onion cells. Images were obtained with a fluorescence microscope.
To validate the DOF6–TCP14 interaction in planta, bimolecular fluorescent complementation experiments were carried out. DOF6 and TCP14 ORFs were translationally fused to the N- and C-terminal fragments of YFP, respectively, and these constructs were used for transient expression by bombardment of epidermal onion cells. When N-YFP-DOF6 and C-YFP-TCP14 constructs were co-bombarded, YFP fluorescence was observed in the nucleus, indicating that DOF6 and TCP14 proteins interact in planta (Fig. 6C). As expected, no reconstruction of YFP activity was achieved when different combinations of TF and/or empty vectors were co-bombarded (data not shown).
DOF6 and TCP14 participate in the regulation of ABA1 and sHSP genes in imbibed seeds
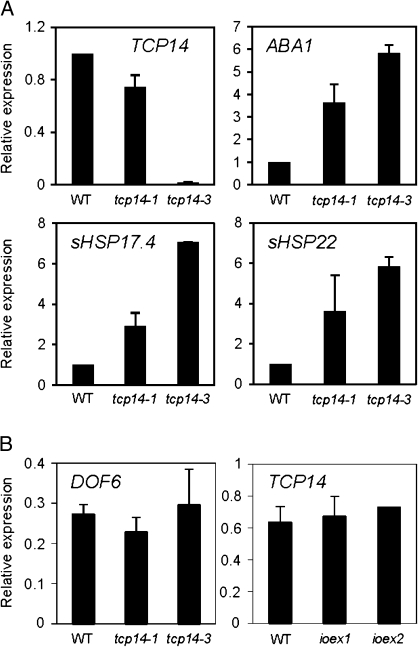
TCP14 had been previously described as a positive regulator of embryonic growth potential during seed germination (Tatematsu et al., 2008) and the phenotypes observed for tcp14 KO seeds resembled those of ioexDOF6 seeds. To test if DOF6 and TCP14 could be involved in the regulation of a common set of genes, the expression of those genes that were found to be induced more than three times during FH seed imbibition in the ioexDOF6 lines were quantified in FH seeds from two previously described tcp14 KO lines (tcp14-1 and tcp14-3; Tatematsu et al. 2008). While the mRNA levels of TCP14 were reduced in both KO lines when compared to the WT, as expected, the expression of ABA1, sHSP17.4, and sHSP22 was enhanced (Fig. 7A), as occurs in the ioexDOF6 lines. These results indicate that DOF6 and TCP14 have opposite effects (activator and repressor, respectively) on the expression of ABA1, sHSP17.4, and sHSP22.
Fig. 7.
mRNA levels of DOF6 and ABA-related genes in germinating tcp14 mutant seeds. (A) Expression of TCP14, ABA1, sHSP17.4, and sHSP22 was compared by quantitative RT-PCR between freshly harvested wild-type (WT) and tcp14 imbibed seeds. WT expression levels are 1 in all cases. (B) mRNA levels of DOF6 and TCP14 were quantified in tcp14 and ioexDOF6 FH seeds, respectively. WT and ioexDOF6 seeds used in the left panel were imbibed for 24 h in 50 μM estradiol. Gene-specific mRNA levels are relative to UBC21.
To analyse a possible cross-regulation between DOF6 and TCP14, their mRNA levels in tcp14 KO and ioexDOF6 germinating seeds, respectively, were quantified. As shown in Fig. 7B, no differences in mRNA levels were detected between the WT and mutant plants, indicating that DOF6 or TCP14 are not regulating each other’s expression.
Discussion
DOF6 is regulated by after-ripening and enhances ABA1 expression
In this study, DOF6 has been demonstrated to negatively affect seed germination, most probably by increasing the expression of ABA-related genes and seed ABA content (Supplementary Fig. S5). This regulation is transient and conditioned by the AR status of the seed. TCP14 has been identified as an interactor of DOF6, and tcp14 seeds displayed phenotypes similar to those observed for ioexDOF6. These similarities extend to the expression of ABA-related genes and could explain a possible regulatory mechanism for their AR-dependent phenotypes and opposing roles.
DOF6 transcripts were abundant in FH dry seeds and decreased upon imbibition and during seed after-ripening. In a systematic study of Arabidopsis TFs, Barrero et al. (2010) identified 39 TF genes that were differentially expressed between FH and AR seeds after short imbibition periods, and suggested that they had different roles in seed dormancy control: genes up-regulated in AR seeds (AR set) would be related to germination and dormancy release, whereas genes up-regulated in FH seeds (FH set) would be involved in dormancy maintenance. Although other DOF genes were highly represented in the AR set, DAG1 among them, DOF6 was not included in any of the sets. However, our data showed that DOF6 was consistently down-regulated by after-ripening and should be, consequently, included in the FH set.
Some DOF TFs have been shown to play dual and/or opposing roles during seed maturation and upon germination in barley: BPBF and HvDOF19 are negative regulators of hydrolase genes expressed in the aleurone of germinating seeds, but are transcriptional activators of protein storage genes during seed maturation (Mena et al., 1998, 2002; Moreno-Risueño et al., 2007a) and SAD and HvDOF17 are an activator and a repressor, respectively, in both phases of seed development (Isabel-LaMoneda et al., 2003; Diaz et al., 2005; Moreno-Risueño et al., 2007a). In Arabidopsis, DAG1 is a DOF TF negatively regulating seed germination (Papi et al., 2000; Gualberti et al., 2002; Gabriele et al., 2010). Despite the phylogenetic closeness between DAG1 and DOF6 and their similar effects on germination, they must fulfil different roles, since DAG1 was up-regulated in AR compared to FH imbibed seeds (Barrero et al., 2010). In addition, dag1 KO seeds did not show altered sensitivity to ABA (Gualberti et al., 2002).
Barrero et al. (2010) analysed loss-of-function mutants for 22 genes belonging to the AR and FH sets. Only two of these mutants, belonging to the AR set, displayed altered dormancy, indicating a high degree of gene redundancy and a high robustness of the dormancy programme. This suggests that gain-of-function approaches, such as the one used in this study, may be more informative when analysing contributions of TFs whose loss-of-function do not show altered phenotypes, or cause lethality. The lack of insertion mutants in public databases together with the impossibility to silence DOF6 by amiRNAs and ihpRNAs, suggested that the reduction in DOF6 mRNA levels may be incompatible with plant viability.
The germination delay of FH ioexDOF6 seeds was accompanied by the upregulation of the ABA biosynthetic gene ABA1. ABA1 encodes a zeaxanthin epoxidase, the first enzyme in ABA biosynthesis, which has also been described to regulate ABA accumulation during seed development (Audran et al., 2001). Mutants deficient on this gene have been reported to show a non-dormant phenotype (Koornneef et al., 1982), whereas ABA1 over-expression affects ABA levels and delays seed germination (Frey et al., 1999; Park et al., 2008). In addition, ABA1 is down-regulated by after-ripening and it has been suggested to play a key role in dormancy maintenance in non-AR seeds (Carrera et al., 2008), whereas mutants deficient in ABA biosynthesis and ABA-signalling pathways show altered dormancy and germination phenotypes (Finkelstein et al., 2002; Rodriguez-Gacio et al., 2009). DOF6 must also play a role in dormancy control since WT germination was restored in FH ioexDOF6 seeds when dormancy-breaking treatments were applied upon imbibition (Supplementary Fig. S4). Other genes up-regulated in ioexDOF6 FH imbibed seeds were sHSPs. These genes are up-regulated by ABA and are normally expressed during seed maturation and have increased expression levels in imbibed dormant seeds (Wehmeyer et al., 1996; Nakabayashi et al., 2005; Cadman et al., 2006; Kotak et al., 2007; Carrera et al., 2008).
DOF6 mRNA levels were down-regulated by after-ripening and this might occur through silencing with an AR-induced miRNA. Reyes and Chua (2007) found that the mRNAs encoding two positive regulators of ABA responses (MYB TFs) were cleaved by the miRNA159 during germination in response to ABA. Regarding DOF6, no miRNA predicted to regulate DOF6 mRNA levels was found by in silico analysis (http://asrp.cgrb.oregonstate.edu/). Increased levels of DOF6 mRNAs have been demonstrated to be sufficient to produce delayed germination and ABA-related gene expression in FH but not in AR seeds.
Biological relevance of the DOF6–TCP14 interaction
A new interaction between TF members of the DOF and TCP families has been uncovered. TCPs are plant-specific TFs that have been associated with promotion of cell growth and proliferation, hormone biosynthesis, and circadian rhythms and they have been shown to interact with armadillo BTB, NAC and TOC1 proteins (Martin-Trillo and Cubas, 2010). The biological significance of the DOF6–TCP14 and its role in after-ripening and ABA biosynthesis pathways is supported by several lines of evidence: (i) interaction of DOF6 and TCP14 was observed in the nuclei of plant cells where TFs usually exert their functions; (ii) both TFs were expressed in dry seeds and in the vascular tissues of the embryo during seed imbibition (this work; Tatematsu et al., 2008); (iii) ioexDOF6 and tcp14 KO mutants had increased expression of specific genes involved in ABA biosynthesis and ABA-mediated stress responses; (iv) ioexDOF6 and tcp14 mutant FH seeds had a delayed germination phenotype which is abolished by after-ripening (this work; Tatematsu et al., 2008); v) ioexDOF6 and tcp14 AR seeds had ABA-hypersensitive germination (this work; Tatematsu et al., 2008); and (vi) p35S:DOF6 plants showed growth retardation and a reduced plant stature as observed in the tcp14 tcp15 double mutant (Kieffer et al., 2011). These results suggest that DOF6 and TCP14 have opposite functions. Since they do not regulate their transcript levels reciprocally (Fig. 7B), and TCP14 mRNA levels do not change during after-ripening (Supplementary Fig. S6), it could be possible that, in FH seeds, DOF6 abundance would exceed that of TCP14 (Supplementary Fig. S7) and DOF6 proteins not ‘sequestered’ in the interaction with TCP14 would be able to promote the expression of ABA-related genes. The balance of this TF abundance would be shifted at the end of after-ripening and would allow TCP14 to promote cell proliferation and germination. Another possibility could be the existence of other factors that may favour or oppose DOF6 function. Good candidates would be those regulated by the AR status of the seed. Out of the 39 TFs identified by Barrero et al. (2010), 29 are present in this study’s TF library (Castrillo et al., 2011). However, no interaction between DOF6 and any of these TFs was observed.
Besides its putative role in seeds, it is possible that DOF6 participates in other developmental and physiological processes, since its mRNA is present in flowers, leaves, and roots (Fig. 1), specifically in the vascular system (Supplementary Fig. S8). This is in agreement with the severe growth defects produced when DOF6 is constitutively over-expressed (Fig. 2). It was also observed that the lack of progeny in p35S::DOF6 plants was due to reduced pollen viability (Supplementary Fig. S2), although this could be an indirect effect. Other TFs involved in seed dormancy and germination are also involved in the regulation of development outside the seed. For instance, SPT is involved in carpel and fruit development (Groszmann et al., 2008), ABI5 is involved in growth-insensitivity to ABA (Lopez-Molina and Chua, 2000), and TCP14 promotes cell division in young internodes (Kieffer et al., 2011). In this report, the inducible approach has allowed the study of DOF6 function in the context of dormancy and germination and isolated from other possible roles in the plant.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Table S1. Oligonucleotide sequences and gene loci.
Supplementary Fig. S1. Strategies used to obtain DOF6 loss-of-function lines.
Supplementary Fig. S2. Constitutive over-expression of DOF6 produces plant sterility.
Supplementary Fig. S3. DOF6 expression levels and germination kinetics in three independent ioexDOF6 lines.
Supplementary Fig. S4. Dormancy-breaking treatments eliminate FH ioexDOF6-delayed germination.
Supplementary Fig. S5. Effect of DOF6 over-expression on ABA levels in imbibed seeds.
Supplementary Fig. S6. TCP14 mRNA levels in FH and AR seeds.
Supplementary Fig. S7. Proposed model of the physiological consequences triggered by the interaction between DOF6 and TCP14 during Arabidopsis seed germination.
Supplementary Fig. S8. Expression of the GUS reporter gene driven by the DOF6 promoter in different plant organs.
Acknowledgments
The authors thank Inmaculada Gude for excellent technical assistance, Dr Wolfgang Dröge-Lasser (Universität Würzburg, Germany) for providing the SPYNE and SPYCE plasmids and Prof Lucia Colombo (Università di Milano, Italy) for providing the tcp14-1 homozygous seeds. Financial support from the Ministerio de Ciencia e Innovación, Spain (project nos. CSD 2007-00057, EUI 2008-03716, and BIO2010-17334) is acknowledged. PR-R is the recipient of a predoctoral fellowship (Formación Personal Investigador, Training Research Personnel) from the Ministerio de Ciencia e Innovación, Spain.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Audran C, Liotenberg S, Gonneau M, North H, Frey A, Tap-Waksman K, Vartanian N, Marion-Poll A. Localisation and expression of zeaxanthin epoxidase mRNA in Arabidopsis in response to drough stress and during seed development. Australian Journal of Plant Physiology. 2001;28:1161–1173. [Google Scholar]
- Barrero JM, Millar AA, Griffiths J, Czechowski T, Scheible WR, Udvardi M, Reid JB, Ross JJ, Jacobsen JV, Gubler F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. The Plant Journal. 2010;61:611–622. doi: 10.1111/j.1365-313X.2009.04088.x. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. The Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove J, Lucas P, Godin B, Oge L, Jullien M, Grappin P. Gene expression analysis by cDNA-AFLP highlights a set of new signaling networks and translational control during seed dormancy breaking in Nicotiana plumbaginifolia. Plant Molecular Biology. 2005;57:593–612. doi: 10.1007/s11103-005-0953-8. [DOI] [PubMed] [Google Scholar]
- Cadman CS, Toorop PE, Hilhorst HW, Finch-Savage WE. Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. The Plant Journal. 2006;46:805–822. doi: 10.1111/j.1365-313X.2006.02738.x. [DOI] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in. Arabidopsis. The Plant Journal. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, Paz-Ares J, Oñate-Sanchez L. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS One. 2011 doi: 10.1371/journal.pone.0021524. 6, e21524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani K, Ali-Rachedi S, Job C, Job D, Jullien M, Grappin P. Proteomic analysis of seed dormancy in Arabidopsis. Plant Physiology. 2006;142:1493–1510. doi: 10.1104/pp.106.087452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiology. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. The Plant Journal. 2005;42:652–662. doi: 10.1111/j.1365-313X.2005.02402.x. [DOI] [PubMed] [Google Scholar]
- Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, Schmitt J. Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution. 2005;59:740–757. [PubMed] [Google Scholar]
- Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho JV, Gomez-Cadenas A. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Journal of Agricultural and Food Chemistry. 2005;53:8437–8442. doi: 10.1021/jf050884b. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. The New Phytologist. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;(14 Suppl):S15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A, Audran C, Marin E, Sotta B, Marion-Poll A. Engineering seed dormancy by the modification of zeaxanthin epoxidase gene expression. Plant Molecular Biology. 1999;39:1267–1274. doi: 10.1023/a:1006145025631. [DOI] [PubMed] [Google Scholar]
- Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. The Plant Journal. 2010;61:312–323. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Paicu T, Smyth DR. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. The Plant Journal. 2008;55:40–52. doi: 10.1111/j.1365-313X.2008.03469.x. [DOI] [PubMed] [Google Scholar]
- Gualberti G, Papi M, Bellucci L, Ricci I, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. The Plant Cell. 2002;14:1253–1263. doi: 10.1105/tpc.010491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV. Dormancy release, ABA and pre-harvest sprouting. Current Opinion in Plant Biology. 2005;8:183–187. doi: 10.1016/j.pbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, et al. Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Research. 2004;14:2176–2189. doi: 10.1101/gr.2544504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. Post-genomics dissection of seed dormancy and germination. Trends in Plant Science. 2008;13:7–13. doi: 10.1016/j.tplants.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Matilla A. After-ripening alters the gene expression pattern of oxidases involved in the ethylene and gibberellin pathways during early imbibition of Sisymbrium officinale L. seeds. Journal of Experimental Botany. 2009;60:1645–1661. doi: 10.1093/jxb/erp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Fernández R, Rodriguez-Gacio MC, Barrero-Sicilia C, Carbonero P, Matilla A. Three endo-beta-mannanase genes expressed in the micropylar endosperm and in the radicle influence germination of Arabidopsis thaliana seeds. Planta. 2011;233:25–36. doi: 10.1007/s00425-010-1257-z. [DOI] [PubMed] [Google Scholar]
- Isabel-LaMoneda I, Diaz I, Martinez M, Mena M, Carbonero P. SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. The Plant Journal. 2003;33:329–340. doi: 10.1046/j.1365-313x.2003.01628.x. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse EM, Gan Y, Bou-Torrent J, et al. A DELLA in disguise: SPATULA restrains the growth of the developing Arabidopsis seedling. The Plant Cell. 2011;23:1337–1351. doi: 10.1105/tpc.110.082594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. The Plant Journal. 2011;68:147–158. doi: 10.1111/j.1365-313X.2011.04674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Nambara E. Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Molecular Biology. 2010;73:119–129. doi: 10.1007/s11103-010-9603-x. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Dellaert LW, van der Veen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutattion Research. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- Kotak S, Vierling E, Baumlein H, von Koskull-Doring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of. Arabidopsis. The Plant Cell. 2007;19:182–195. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. The EMBO Journal. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P, Oñate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicente-Carbajosa J. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. Journal of Biological Chemistry. 2003;278:21003–21011. doi: 10.1074/jbc.M210538200. [DOI] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Tureckova V, Strnad M, Lopez-Molina L. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19108–19113. doi: 10.1073/pnas.1012896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. The Plant Journal. 2006;45:309–319. doi: 10.1111/j.1365-313X.2005.02622.x. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G. Beta-1,3-Glucanase gene expression in low-hydrated seeds as a mechanism for dormancy release during tobacco after-ripening. The Plant Journal. 2005;41:133–145. doi: 10.1111/j.1365-313X.2004.02284.x. [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionary Biology. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Koizuka N, Martin RC, Nonogaki H. The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. The Plant Journal. 2005;44:960–971. doi: 10.1111/j.1365-313X.2005.02588.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. A null mutation in a bZIP factor confers ABA-insensitivity in. Arabidopsis thaliana. Plant and Cell Physiology. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends in Plant Science. 2010;15:31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou JP, Kamiya Y, Nambara E, Truong HN. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiology. 2009;149:949–960. doi: 10.1104/pp.108.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P. A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiology. 2002;130:111–119. doi: 10.1104/pp.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. The Plant Journal. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. The Plant Journal. 2006;45:942–954. doi: 10.1111/j.1365-313X.2006.02659.x. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueño MA, Diaz I, Carrillo L, Fuentes R, Carbonero P. The HvDOF19 transcription factor mediates the abscisic acid-dependent repression of hydrolase genes in germinating barley aleurone. The Plant Journal. 2007a;51:352–365. doi: 10.1111/j.1365-313X.2007.03146.x. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueño MA, Martinez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Molecular Genetics and Genomics. 2007b;277:379–390. doi: 10.1007/s00438-006-0186-9. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. The Plant Journal. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. Gibberellin biosynthesis and response during Arabidopsis seed germination. The Plant Cell. 2003;15:1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiology. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Research Notes. 2008;1:93. doi: 10.1186/1756-0500-1-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M, Sabatini S, Bouchez D, Camilleri C, Costantino P, Vittorioso P. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes and Development. 2000;14:28–33. [PMC free article] [PubMed] [Google Scholar]
- Park HY, Seok HY, Park BK, Kim SH, Goh CH, Lee BH, Lee CH, Moon YH. Overexpression of Arabidopsis ZEP enhances tolerance to osmotic stress. Biochemical and Biophysical Research Communications. 2008;375:80–85. doi: 10.1016/j.bbrc.2008.07.128. [DOI] [PubMed] [Google Scholar]
- Penfield S, Graham S, Graham IA. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochemical Society Transactions. 2005;33:380–383. doi: 10.1042/BST0330380. [DOI] [PubMed] [Google Scholar]
- Penfield S, King J. Towards a systems biology approach to understanding seed dormancy and germination. Proceedings. Biological Sciences. 2009;276:3561–3569. doi: 10.1098/rspb.2009.0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Tureckova V, Lacombe E, Lopez-Molina L. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. The EMBO Journal. 2009;28:2259–2271. doi: 10.1038/emboj.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gacio MC, Matilla-Vazquez MA, Matilla AJ. Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signaling and Behavior. 2009;4:1035–1049. doi: 10.4161/psb.4.11.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. The Plant Journal. 2008;53:42–52. doi: 10.1111/j.1365-313X.2007.03308.x. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. International Journal of Developmental Biology. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- Wehmeyer N, Hernandez LD, Finkelstein RR, Vierling E. Synthesis of small heat-shock proteins is part of the developmental program of late seed maturation. Plant Physiology. 1996;112:747–757. doi: 10.1104/pp.112.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schutze K, Alonso R, Harter K, Vicente-Carbajosa J, Droge-Laser W. Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. The EMBO Journal. 2006;25:3133–3143. doi: 10.1038/sj.emboj.7601206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. The Plant Cell. 2004;16:367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends in Plant Science. 2002;7:555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- Yano R, Kanno Y, Jikumaru Y, Nakabayashi K, Kamiya Y, Nambara E. CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiology. 2009;151:641–654. doi: 10.1104/pp.109.142018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. Technical advance: an estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. The Plant Journal. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.