Abstract
7365AB, a recessive genetic male sterility system, is controlled by BnMs3 in Brassica napus, which encodes a Tic40 protein required for tapetum development. However, the role of BnMs3 in rapeseed anther development is still largely unclear. In this research, cytological analysis revealed that anther development of a Bnms3 mutant has defects in the transition of the tapetum to the secretory type, callose degradation, and pollen-wall formation. A total of 76 down-regulated unigenes in the Bnms3 mutant, several of which are associated with tapetum development, callose degeneration, and pollen development, were isolated by suppression subtractive hybridization combined with a macroarray analysis. Reverse genetics was applied by means of Arabidopsis insertional mutant lines to characterize the function of these unigenes and revealed that MSR02 is only required for transport of sporopollenin precursors through the plasma membrane of the tapetum. The real-time PCR data have further verified that BnMs3 plays a primary role in tapetal differentiation by affecting the expression of a few key transcription factors, participates in tapetal degradation by modulating the expression of cysteine protease genes, and influences microspore separation by manipulating the expression of BnA6 and BnMSR66 related to callose degradation and of BnQRT1 and BnQRT3 required for the primary cell-wall degradation of the pollen mother cell. Moreover, BnMs3 takes part in pollen-wall formation by affecting the expression of a series of genes involved in biosynthesis and transport of sporopollenin precursors. All of the above results suggest that BnMs3 participates in tapetum development, microspore release, and pollen-wall formation in B. napus.
Keywords: BnMs3, Brassica napus, exine development, gene expression, microspore release, recessive genetic male sterility, suppression subtractive hybridization, tapetum
Introduction
The recessive genetic male sterility (RGMS) system plays an important role in hybrid seed production due to its amazing stability in maintaining sterility (complete male sterility) with no negative cytoplasmic effects. 7365AB, a RGMS system controlled by the BnMs3 gene in Brassica napus, was identified by Huang et al. (2007). Presently, it has been widely used for heterosis in China because of its advantages in obtaining a 100% sterile population using a temporary maintainer line, in which there is no need to remove 50% of the fertile plants during commercial hybrid seed production of the F1 generation (Xiao et al., 2008). The cause of RGMS is attributed to the abnormal development of anthers. In Arabidopsis, anther development is a complicated process, which consists of two phases divided into 14 stages (Sanders et al., 1999; Ma, 2005). During phase I, from stages 1 to 8, cells divide and differentiate to form anther tetra lobes. Each lobe contains five cellular layers from the exterior to the interior: the epidermis, endothecium, middle layer, tapetum, and microspore mother cell (Sanders et al., 1999; Ma, 2005). Tapetal differentiation and microspore separation involved in this phase are essential for anther development. During phase II, after the microspores undergo mitosis to develop into pollen grains, mature pollen grains are released to the surface of anther by its dehiscence. Tapetal PCD (programmed cell death) and pollen-wall formation play important roles in phase II.
B. napus and Arabidopsis are both members of the Brassicaceae and they share a high level of similarity in exon sequences (85.64%) and genomic synteny regions (Cavell et al., 1998). It has been reported that Arabidopsis genes and their orthologues in Brassica have approximately equivalent functions (Auger et al., 2009; Zhang et al., 2009). Therefore, studying gene functions during anther development of B. napus by using the public information resources from Arabidopsis is a valid strategy.
Several genes that are crucial for early anther development have been elucidated in Arabidopsis. AGAMOUS acts in the specification of stamen and carpels (Yanofsky et al., 1990; Ito et al., 2004). SPOROCYTELESS (SPL)/NOZZLE and ROXY control the archesporial cell differentiation (Schiefthaler et al., 1999; Yang et al., 1999; Xing and Zachgo, 2008). Tapetal/microsporocyte determination is regulated by the interaction of TPD1 and EMS1 (Canales et al., 2002; Zhao et al., 2002; Jia et al., 2008). DYSFUNCTIONAL TAPETUM 1 (DYT1), ABORTED MICROSPORE (AMS), TAPETAL DEVELOPMENT FUNCTION 1 (TDF1), AtMYB103, and MALE STERILITY 1 (MS1) play vital roles in tapetal development and PCD (Wilson et al., 2001; Sorensen et al., 2003; Zhang et al., 2006, 2007; Zhu et al., 2008). Moreover, according to several global transcriptome data for spl, ems1, and ms1 mutants, models of the gene regulatory network for early anther development have been proposed in Arabidopsis (Wijeratne et al., 2007; Yang et al., 2007).
During anther development, the timely degradation of the tetrad inner wall (callose) and the outer wall (the primary cell wall of pollen mother cell) is critical for microspore release from tetrads (Rhee and Somerville, 1998). Callose, which is mainly composed of β-1,3-glucans, is degraded by callase (β-1,3-glucanase) (Frankel et al., 1969; Stieglitz and Stern, 1973; Stieglitz, 1977). Several tapetum-specific expressed β-1,3-glucanase genes have been identified, such as Tag1 from tobacco and A6 from B. napus and Arabidopsis (Hird et al., 1993; Bucciaglia and Smith, 1994). Premature expression of a β-1,3-glucanase using a tapetum-specific promoter in tobacco resulted in male sterility (Worrall et al., 1992). In addition to callose degradation, that of the primary wall of pollen mother cell (PMC) is also required for microspore separation (Preuss et al., 1994). The three quartet mutants (qrt1, qrt2, and qrt3) in Arabidopsis are defective in the PMC primary wall degradation and generate tetrad pollen (Rhee et al., 2003; Francis et al., 2006; Ogawa et al., 2009).
The biopolymer sporopollenin is the major component of exine (Piffanelli et al., 1998). It has been thought that sporopollenin is a complex polymer primarily made of a mixture of fatty acids and phenolic compounds (Piffanelli et al., 1998). Therefore, biosynthesis of fatty acids and phenols is essential for exine formation. Recent studies have demonstrated that FACELESS POLLEN 1 (FLP1), NEF1, MS2, Acyl-CoA Synthetase 5 (ACOS5), CYP703A2, and CYP704B1 are involved in the biosynthesis of lipid molecules during anther development (Aarts et al., 1997; Ariizumi et al., 2003, 2004; Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009; Shi et al., 2011). LESS ADHESIVE POLLEN 5 and LESS ADHESIVE POLLEN 6, encoding anther-specific chalcone synthase (CHS), play a role in the biosynthesis of both fatty acids and phenols (Dobritsa et al., 2010). Apart from its biosynthesis, the export process of the sporopollenin precursor is also required for exine formation. In rice, it is likely that Osc6 encodes a lipid transfer protein (LTP) to transfer lipid molecules from the tapetal cells to the pollen wall (Zhang et al., 2010).
BnMs3, encoding a tapetally expressed Tic40 protein, has been isolated by a map-based cloning approach (Dun et al., 2011). The Bnms3 mutant shows abnormal tapetal cell enlargement and aborted microspores. The terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay has revealed that the pattern of PCD in the tapetal cells of the Bnms3 mutant is delayed (Dun et al., 2011).
This study reports the functional significance of BnMs3 in anther development of B. napus. Semi-thin sections reveal that the principal cause of male sterility in the Bnms3 mutant is a defect in the transition of the tapetum to the secretory type. The results of cytochemical staining for the callose wall suggest that callose cannot be degraded in time. Subsequently, suppression subtractive hybridization (SSH) and macroarray analysis are used to identify differentially expressed genes between the Bnms3 mutant and the wild-type plant, a few of which are associated with tapetum development, callose degradation, and pollen-wall biosynthesis. Furthermore, the information on Arabidopsis genes involved in the regulatory network of anther development is used to further understand the role of BnMs3 in anther development and it reveals that BnMs3 takes part in tapetal differentiation and PCD, microspore separation, and exine development by affecting the expression of a series of key genes involved in these pathways. The functional importance of the three differentially expressed genes in the Bnms3 mutant is further analysed by reverse genetics using orthologues of B. napus genes in Arabidopsis, which indicates that AtMSR02, encoding an ATP-binding cassette (ABC) protein, is only required for exine development but not for cuticle formation in the anther. The work provides deeper insight into the role of BnMs3 in tapetal function and pollen development in B. napus.
Materials and methods
Plant materials
This work used the RGMS two-type line (7365AB) as materials. The 7365AB line was maintained by full sib-mating (7365A×7365B), so that line 7365A (Bnms3Bnms3), which is male sterile, and line 7365B (BnMs3Bnms3), which is male fertile, are near-isogenic lines (NILs) differing only in the fertility trait. Seeds were sown in the rapeseed research field of Huazhong Agricultural University, Wuhan, China. Bud lengths ranging from <1, 1–2, and 2–3 mm were collected separately for cDNA library construction. Anthers from bud lengths <3 mm were dissected for macroarray analysis. To investigate the validity of the cDNA macroarray analysis, buds <1, 1–2, 2–3, 3–4, 4–5, and >5 mm in size were harvested for Northern blot analysis. All the harvested samples were immediately deep frozen in liquid nitrogen and stored at –80 °C.
Light microscopy of 7365AB
Morphological observations of anther semi-thin sections were performed as described by Chen et al. (2009). Aniline blue staining was conducted as described by Zhang et al. (2007). Micrographs of fluorescence expression of callose were taken using a fluorescence microscope (Nikon Eclipse 80i) with appropriate filter under ultraviolet light.
SSH library construction and differential screening
The library was constructed using a SMART PCR cDNA Synthesis kit (Clontech, USA) combined with the PCR-Select cDNA Subtraction kit (Clontech), according to the manufacturer’s recommendations. The cDNA inserts were amplified using the nested PCR primers 1 and 2R provided in the PCR-select cDNA Subtraction kit and 2 μl bacterial overnight culture as the template. PCR products of the expected lengths (8 μl) were evaluated on 1.2% agarose gel, purified by ethanol precipitation, dissolved in 20 μl distilled deionized water, transferred into 384-well microplates and dotted onto Hybond-N+ nylon membranes (Amersham, UK) using a Biomek 2000 Laboratory Automation Workstation (Beckman Coulter, USA). The membranes (8×12cm) were arrayed into grids of 384 dots that contained triplicate dots. The membranes were denatured in 0.2 M NaOH for 15 min, neutralized in 0.5 M TRIS-HCl (pH 7.5) for 5 min, and rinsed in distilled water twice for 2 min. The cDNAs were permanently fixed onto the membranes by baking at 80 °C for 2 h and the filters were stored at –20 °C. The anther cDNAs from line 7365A or line 7365B were used as probes for membrane screening to isolate the differentially expressed genes. Probes were added to the hybridization solution, which contained 0.5 M NaH2PO4 buffer, 1 mM EDTA (pH8.0), 7% sodium dodecyl sulphate (SDS), and 0.6% bovine serum albumin. Hybridization was carried out overnight at 68 °C. The membranes were washed twice with 2×standard sodium citrate (SSC) and 0.1% SDS for 15 min at room temperature and then washed with 0.5×SSC and 0.1% SDS for 5 min at 68 °C.
The hybridized membranes were exposed using the Phosphor Screen system (FUJIFILM, Japan) for 5 h. The signals from the Phosphor plate were obtained using a FLA-5000 Plate/Fluorescent Image Analyser (FUJIFILM) and analysed with the software Multi Gauge version 3.0. On the basis of the results of the dot-blot hybridization, differentially expressed clones were selected for sequencing.
Sequencing and bioinformatic analysis
Differentially expressed colonies were picked from the subtractive cDNA library for sequence analysis using a ABI 3730 Genetic Analyzer (AuGCT Biotechnology, Beijing, China). Unigenes were obtained using SeqMan software from DNASTAR (Madison, WI). Similarity searches were performed using BLASTN in The Arabidopsis Information Resource (TAIR). Unigenes with BLASTN expectation values (E-values) >10−5 were designated as having insignificant similarity and were analysed using the GenBank database with BLASTN at the NCBI dbEST network service (http://www.ncbi.nlm.nih.gov/BLAST/). Functional categories of unigenes were assigned according to the Munich Information Center for Protein Sequences (MIPS) catalogue of the Arabidopsis thaliana genome (Mewes et al., 2008). If an individual protein was allotted to multiple functional categories, the most probable category (lowest P-value) was chosen (Table 1 and Supplementary Table S1, available at JXB online).
Table 1.
Co-down-regulated expressed genes clustered according to different assembly in the Bnms3 mutant in B. napus and the Atms1, Atspl, and/or Atems1 mutants in A. thaliana
| Cluster | Unigene no. | Locus | Description | Fold change in ms1 | Fold change in ems1/spl |
| Cluster A | BnMSR72 | AT1G67990 | Caffeoyl-CoA 3-O-methyltransferase | −8.2 | −9.4 |
| BnMSR73 | AT3G52160 | Beta-ketoacyl-CoA synthase family protein | −13.9 | −15.3 | |
| BnMSR20 | AT1G71160 | Fatty acid elongase 3-ketoacyl-CoA synthase 7 | −4.4 | −39.0 | |
| BnMSR45 | AT1G75940 | Beta-glucosidase protein | −21.3 | −5.9 | |
| BnMSR42 | AT5G65205 | Short-chain dehydrogenase/reductase (SDR) family protein | −8.2 | −2.0 | |
| BnMSR69 | AT1G76470 | Cinnamoyl-CoA reductase family | −6.6 | −18.7 | |
| BnMSR39 | AT2G03740 | Late embryogenesis abundant domain-containing protein | −7.3 | −5.4 | |
| BnMSR12 | AT3G10410 | Serine carboxypeptidase-like 49 precursor | −2.5 | −2.5 | |
| BnMSR23 | AT3G26125 | Cytochrome p450 monooxygenase 86c2 | −7.8 | −2.1 | |
| BnMSR04 | AT3G51590 | Lipid transfer protein 12 | −51.9 | −11.4 | |
| BnMSR09 | AT1G06260 | Cysteine proteinase | −33.2 | −2.5 | |
| BnMSR38 | AT4G28395 | Lipid transfer protein | −10.8 | −8.3 | |
| BnMSR58 | AT1G75910 | Family II extracellular lipase 4 | −43.1 | −2.3 | |
| BnMSR48 | AT4G37900 | Glycine-rich protein | −22.3 | −2.4 | |
| BnMSR53 | AT5G62320 | MYB99 | −2.9 | −2.5 | |
| BnMSR13 | AT1G61110 | NAC domain containing protein 25 | −11.5 | −2.8 | |
| BnMSR37 | AT5G13380 | Auxin-responsive GH3 family protein | −8.2 | −3.7 | |
| BnMSR63 | AT1G68875 | Expressed protein | −109.9 | −15.4 | |
| BnMSR65 | AT5G48210 | Expressed protein | −16.6 | −5.2 | |
| Cluster B | BnMSR59 | AT1G54540 | Expressed protein | – | −2.7 |
| BnMSR54 | AT1G33430 | Galactosyltransferase family protein | – | −47.0 | |
| BnMSR50 | AT5G15960 | Cold and ABA inducible protein kin1 | – | −2.4 | |
| BnMSR07 | AT5G07230 | Lipid transfer protein family protein | – | −50.6 | |
| BnMSR52 | AT4G31500 | Cytochrome p450 monooxygenase 83b1 | – | −2.5 | |
| BnMSR11 | AT4G20420 | Tapetum-specific protein-related protein | – | −33.9 | |
| BnMSR66 | AT3G23770 | Glycosyl hydrolase family 17 protein | – | −28.5 | |
| BnMSR29 | AT3G15400 | ATA20 | – | −21.6 | |
| BnMSR02 | AT3G13220 | ABC transporter family protein | – | −52.2 | |
| Cluster C | BnMSR57 | AT2G23800 | Geranylgeranyl pyrophosphate synthase 2 | −8.8 | – |
| BnMSR76 | AT2G19070 | Spermidine hydroxycinnamoyl transferase | −14.7 | – | |
| BnMSR75 | AT1G30020 | Expressed protein | −22.1 | – |
Data for ms1 were taken from Yang et al. (2007); data for ems1/spl were taken from Wijeratne et al. (2007). –, No data.
Northern blot analysis
Total RNA (20 μg) were separated by electrophoresis on a 1.2% formaldehyde agarose gel in 1×MOPS buffer, subsequently transferred onto Hybond-N+ nylon membranes, finally fixed by baking at 80 °C for 2 h, and hybridized (Sambrook and Russell, 2001). The selected cDNA inserts were amplified using nested PCR primers 1 and 2R. About 25 ng of each purified PCR product was labelled using the Prime-a-Gene Labeling system (Promega, USA). The results were obtained using the Phosphor Screen system.
Real-time PCR analysis
Total RNA isolated from four bud stages (<0.5, 0.5–1, 1–2, 2–3 mm) for the lines 7365A and 7365B were used for real-time PCR analysis. First-strand cDNA was generated from 2 μg total RNA from each sample using MMLV reverse transcriptase (K1631, Fermentas) according to the manufacturer’s instructions and the products were diluted 100-fold with sterilized ddH2O and amplified by PCR using the SYBR Green Real-time PCR Master Mix (QPK-201, Toyobo) and in triplicate with the CFX96 Real-time system (Bio-Rad). The results were analysed using BnACTIN7 (EV220887.1) as a control to normalize the expression data and with CFX Manager software according to the 2−ΔΔCt method (Livak and Schmittgen, 2001). The primer sequences for real-time PCR assay are shown in Table 2 and Supplementary Table S2.
Table 2.
Genes and corresponding primers used for real-time PCR analysis in B. napus based on the information for genes involved in A. thaliana tapetal development and function (Zhu et al., 2008)
| Gene name | AGI accession | Length of in silico gene (bp) | Identity | E-value | Forward sequence (5′–3′) | Reverse sequence (5′–3′) | Length of products (bp) |
| BnSPL | AT4G27330 | 942 | 86% | 5e−48 | ACTTCAACGAGGCGACAAATCTTAC | CGGGAAAAATTCGTACTCCTTCA | 101 |
| BnROXY2 | AT5G14070 | 413 | 90% | e−122 | GAGCTCGACCTCCACCCTCAT | GAAGTGCCCCCGGAGAAGT | 100 |
| BnEMS1 | AT5G07280 | 3702 | 82% | e−138 | CTTGGTTGGTTGGGTTGTTCAGA | ACGTAGCATGGCCTGTTTGAAA | 100 |
| BnTPD1 | AT4G24972 | 546 | 86% | 2e−74 | AGGCAGCGACCGAACCTATG | TGACGTGGATCCTCGAGATGTT | 99 |
| BnDYT1 | AT4G21330 | 651 | 86% | e−148 | TGTGCCTGTTGGGATTTGAGA | AGTCACCATCACATAGTCCCTGAAGT | 108 |
| BnTDF1 | AT3G28470 | 972 | 92% | e−166 | GTGAAGAACCACTGGAACACGAA | TGAATTCCGTAAGGACCTGAGAAAC | 100 |
| BnAMS | AT2G16910 | 511 | 88% | e−135 | CGAGATGGTGCCAGCTGAAC | GGATTGAACCAGTTGCCATCTG | 101 |
| BnMYB103 | AT5G56110 | 963 | 91% | 0.0 | CCACTACACTCAATCCTCCTCAAGTC | TCAACACGTTTCTTGGTGAGCAA | 100 |
| BnMS1 | AT5G22260 | 1489 | 89% | 0.0 | ATGCCTCCACAAGAATGC | TCCCATCTAACACCAATCC | 190 |
| BnACTIN7 | AT5G09810 | 734 | 90% | 0.0 | CGCGCCTAGCAGCATGAA | GTTGGAAAGTGCTGAGAGATGCA | 101 |
BnACTIN7 was used as a control.
Microscopy and analysis of Atmsr02
Seeds of the mutants Atmsr02 (BnMSR02 orthologue in Arabidopsis), Atmsr42 (BnMSR42 orthologue in Arabidopsis), and Atmsr53 (BnMSR53 orthologue in Arabidopsis) were taken from the SALK insertional mutant lines SALK_062317, SALK_144641, and SALK_121737, respectively. Seeds were grown in soil at 22 °C under a 16/8 h light/dark regime. PCR genotyping was used to identify homozygous insertion lines. Alexander staining was used to test pollen viability (Alexander, 1969). Transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were performed as described by Yi et al. (2010).
Results
Phenotypic characteristics of the Bnms3 mutant
Semi-thin sections were used for both the Bnms3 mutant and the wild type to observe anther defects in mutant and to identify the relationship between bud lengths and anther developmental stages. As a result, anthers from bud lengths <0.5, 0.5–1, and 1–1.5 mm represented the PMC stage (Fig. 1A,G), the meiosis stage (Fig. 1B,H), and the tetrad stage (Fig. 1C,I), respectively. Buds with lengths 1.5–2.0 mm showed microspore release from the tetrad (Fig. 1D), while anthers with bud lengths 2.0–4.5 mm only exhibited the uninuclear microspore stage (Fig. 1D,E). Bicellular microspores were examined in anthers with bud lengths 4.0–4.5 mm (Fig. 1F).
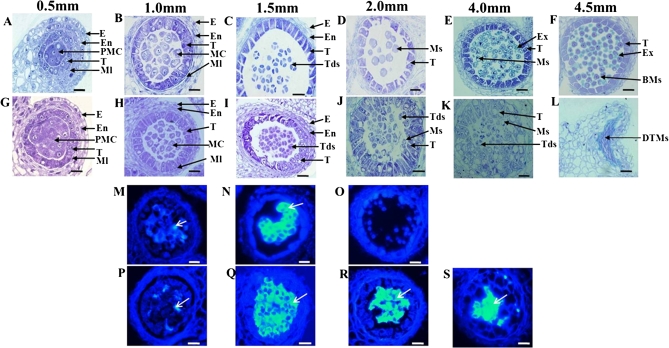
Fig. 1.
Defects in Bnms3 mutant anther development of B. napus in buds of different lengths (0.5, 1.0, 1.5, 2.0, 4.0, and 4.5 mm). (A–L) Comparison of anther development between the wild type (A–F) and the Bnms3 mutant (G–L) by semi-thin sections, revealing that the principal cause of abnormal anther development in the Bnms3 mutant is due to a defect in the transition of the tapetum to the secretory type, which is followed by abnormalities in microspore release and in pollen-wall formation in the few microspores that are released. (M–S) Cytochemical staining for callose wall in the wild type (M–O) and the Bnms3 mutant (P–S) with aniline blue, with the fluorescence expression of callose under ultraviolet light (arrows), showing that callose persists longer in the Bnms3 mutant than in the wild type and leads to the disruption of timely microspore release. BMs, bicellular microspore; DTMs, degenerated tapetum and microspore; E, epidermis; En, endothecium; Ex, exine ; MC, meiotic cell; Ml, middle layer; Ms, microspore; PMC, pollen mother cell; T, tapetum; Tds, tetrads. Bars, 25 μm. (This figure is available in colour at JXB online.)
At the meiosis stage in microspore mother cells, the wild type and the Bnms3 mutant showed no obvious differences in the anthers and the tapetal cells had two nuclei and were vacuolated (Fig. 1B,H). However, some differences in anther development were observed between the mutant and the wild type after meiosis. In the wild type, when the PMCs had finished meiosis and had entered the tetrad stage, the tapetal cell walls were degraded and the tapetal cells shrank and were deeply stained with toluidine blue, which revealed that they had transformed into the secretory type (Fig. 1C). In contrast, the mutant tapetal cell walls remained intact and visible, and the tapetal cells continued to enlarge and vacuolate with reduced staining, meaning that the tapetal cells were unable to transform into the secretory type at any time (Fig. 1I,J,K). During the uninucleate pollen stage, microspores were released from the tetrads in the wild-type anthers (Fig. 1D). Subsequently, microspore exine was formed (Fig. 1E). However, at this stage, the blockage of degradation of the callose surrounding the tetrads resulted in most of the microspores not being released, and the few that were released did not form normal microspore exine (Fig. 1J,K). During the bicellular pollen stage, the tapetal cells and microspores were degenerated instantly in the Bnms3 mutant (Fig. 1L).
In order to further understand the abnormalities of microspore release in the Bnms3 mutant, aniline blue staining was utilized to check the degradation of callose in both the wild-type and the mutant anthers. At the meiosis stage, callose fluorescence appeared in the locules of the wild type and the mutant (Fig. 1M,P) and became the clearest during the tetrad stage (Fig. 1N,Q). After the tetrad stage, callose fluorescence was still observed in the Bnms3 mutant anthers but not detected in the wild type (Fig. 1O,R,S). This result exhibited that callose was not degraded in time in the Bnms3 mutant.
Genes differentially expressed between lines 7365A and 7365B
To identify differentially expressed genes controlled by BnMs3, the subtracted library was constructed using SSH. For the subtracted library, tester cDNA was isolated from the wild type and driver cDNA was from the mutant. Furthermore, 768 clones were randomly chosen from the SSH library for amplification. Macroarray analysis was carried out to screen differentially expressed cDNAs (Supplementary Fig. S1). After scanning, a total of 120 clones showing obviously differential expression were selected for further analysis, 115 clones of which being successfully sequenced, with lengths from 130 bp to 1131 bp. Using SeqMan, 76 unique sequences were obtained from all of the expressed sequence tag (EST) assemblies.
Gene sequences were analysed using BLAST against sequences in TAIR. The 76 unigenes could be classified into two groups. The first group consisted of 68 (89.5%) unigenes with high similarity to genes in Arabidopsis (BLASTN; E-values <10−5 for nucleic acids). The second group included eight unknown unigenes (10.5%) which showed no high similarities with any sequences in the TAIR database. However, by performing BLASTN of the eight sequences in NCBI, similar sequences expressed in anthers or flower buds were identified (Supplementary Table S1). These results reveal that a few novel genes are specifically expressed in the anther of B. napus and are affected by BnMs3.
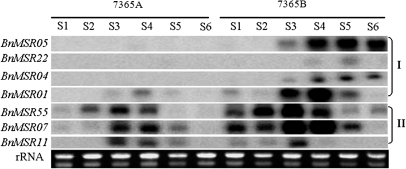
To ascertain the validity of the cDNA macroarray analysis, the expression patterns of seven randomly picked unigenes which were differentially expressed in lines 7365A and 7365B were compared using Northern blot analysis of buds of different lengths. The results were consistent with those from the macroarray analysis and indicated that there were two differential expression types at corresponding stages in the NILs (Fig. 2). Type I included four genes that were expressed only in line 7365B or showed a lower corresponding expression level in the mutant. Type II had three genes expressed in the buds of both NILs, but their expressions were delayed or repressed at the early stage of anther development in line 7365A (Fig. 2).
Fig. 2.
Verification of macroarray results by Northern blot analysis in B. napus. Line 7365A represents the male sterile plant (the Bnms3 mutant) and line 7365B represents the fertile plant (the wild type). S1–S6 represent the bud lengths <1, 1–2, 2–3, 3–4, 4–5, and >5 mm, respectively. I, type I expression type, genes expressed only in line 7365B or at a lower level in the mutant; II, type II expression type, genes expressed in the buds of both lines but expression was delayed or repressed at the early stage of anther development in line 7365A.
Functional categories of differentially expressed genes
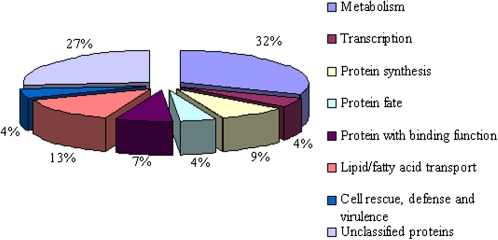
To gain further insights into the functions of the differentially expressed genes, the 68 annotated down-regulated unigenes in the Bnms3 mutant were classified into eight functional categories based on the information from MIPS (Fig. 3; Supplementary Table S1). The major functional groups of down-regulated unigenes were concerned with metabolism, lipid/fatty acid transport, and protein synthesis. The first main group comprised 20 unigenes related to metabolism, encoding enzymes such as cytochrome P450 monooxygenase, glyceraldehyde-3-phosphate dehydrogenase, fatty acid elongase 3-ketoacyl-CoA synthase 7, and caffeoyl-CoA 3-O-methyltransferase. The second main group included nine unigenes annotated as one ABC transport protein and eight LTPs involved in lipid/fatty acid transport. Moreover, six unigenes were associated with protein synthesis, such as translation elongation factor 2-like proteins and ribosomal proteins. The other unigenes were related to transport, protein fate, and stress-responsive proteins.
Fig. 3.
Functional categorization of down-regulated unigenes contained in the Bnms3 mutant according to the Munich Information Center for Protein Sequences classification. A total of 68 unigenes were grouped into eight functional categories and the percentages of gene transcripts in each group were listed. (This figure is available in colour at JXB online.)
Co-down-regulated genes in the Bnms3 and Arabidopsis mutants
Arabidopsis has been used as a model plant to study the molecular mechanisms in anther development (Ma, 2005; Wilson and Zhang, 2009). A large number of genes relevant to this process have been identified through microarray analysis in Arabidopsis, and there are several available transcription profile data of anther-development mutants, including SPL as a regulator of sporogenesis, EXCESS MICROSPOROCYTES 1 (EMS1) involved in the early tapetal cell initiation, and MALE STERILE 1 (MS1) essential for the late tapetal cell development and function (Alves-Ferreira et al., 2007; Wijeratne et al., 2007; Yang et al., 2007). Since the Bnms3 mutant showed abnormal tapetal cells and pollen exine in anther development, the present study compared the Bnms3 mutant data set with that of spl, ems1, and ms1 mutants in Arabidopsis. Consequently, out of 68 down-regulated unigenes in the Bnms3 mutant, 41.2% (28 genes) were co-down-regulated in spl and/or ems1 anthers, 32.4% (22 genes) were co-down-regulated in ms1 anthers, and 19 (27.9%) genes were co-down-regulated in ms1 as well as in spl and/or ems1 anthers (Table 1, cluster A). It was concluded that several pathways common to all of the mutants were affected, such as lipid biosynthesis (fatty acid elongase 3-ketoacyl-CoA synthase 7), lipid transport (lipid transfer protein 12), hormone-responsive proteins (auxin-responsive GH3 family protein), and pollen coat proteins (family II extracellular lipase 4). Furthermore, it also indicated that nine out of 28 unigenes and three out of 22 unigenes, in Atms1 (Table 1, cluster B) and Atspl and/or Atems1 anthers (Table 1, cluster C), respectively, were not obviously down-regulated.
Expression changes of genes related to tapetal development
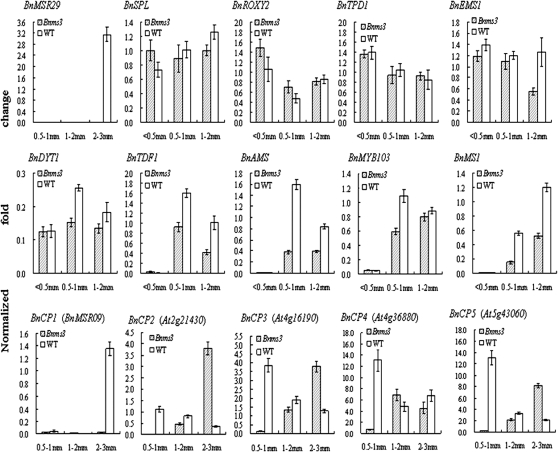
Semi-thin sections revealed that the Bnms3 mutant tapetal cells were unable to transform into the secretory type. Among those down-regulated unigenes, BnMSR29 (ATA20) was tapetum specific (Rubinelli et al., 1998) but was only detected in the buds of the wild type as compared with the Bnms3 mutant (Fig. 4). To date, several putative models for the transcriptional regulation of tapetal development and function have been proposed in Arabidopsis. Therefore, further clarification of the role of BnMs3 in tapetal development was sought using real-time PCR analysis based on the gene information from Arabidopsis. Comparative genomics was used to obtain the homologous Brassica ESTs and genome survey sequences using the full coding sequences of the corresponding key tapetal genes from Arabidopsis by BLASTN in NCBI. Subsequently, according to the conserved sequences of gene-contig assembly, gene-specific primers for real-time PCR were designed to match sequences close to their 3′ untranslated regions (Table 2). The data indicated that the expression levels of DYSFUNCTIONAL TAPETUM 1 and its upstream genes, SPL, ROXY2, TPD1, and EMS1, were unchanged at the PMC stage in the Bnms3 mutant, while those of TDF1, AMS, MYB103, and MS1 were delayed in the Bnms3 mutant buds at the post-meiotic stage of anther development (Fig. 4). These results suggested that the abortion of the Bnms3 mutant emerged at the post-meiotic stage of anther development and BnMs3 might affect tapetal development by affecting the expression of BnTDF1, BnAMS, BnMYB103, and BnMS1, which are associated with tapetal differentiation and/or PCD.
Fig. 4.
Real-time PCR analysis of genes invovled in tapetal development and degradation between the Bnms3 mutant and the wild type, using primers designed for BnSPL, BnROXY2, BnTPD1, BnEMS1, BnDYT1, BnTDF1, BnAMS, BnMYB103, and BnMS1 according to the gene information of regulatory network of Arabidopsis tapetal development and function combined with the expressed sequence tag database in NCBI. BnMSR29 and BnCP1 were isolated from the suppression subtractive hybridization library. Sequences for BnCP2, BnCP3, BnCP4, and BnCP5 (belonging to the cysteine protease family and related to tapetal programmed cell death were obtained using comparative genomics and the Arabidopsis electronic Fluorescent Pictograph browser. The expression levels of BnDYT1 and its upstream genes BnSPL, BnROXY2, BnTPD1, and BnEMS1 were unchanged at the pollen mother cell stage in the Bnms3 mutant, whereas the activities of BnTDF1, BnAMS, BnMYB103, and BnMS1 in the Bnms3 mutant were significantly reduced at the post-meiotic stage of anther development. The expression of five cysteine protease genes was affected in the Bnms3 mutant. Values are means±SD.
Recently, it has been shown that tapetum degeneration in Bnms3 mutants is retarded (Dun et al., 2011). It has been suggested that cysteine proteases may be acting as an effector of PCD in the wild-type tapetum. In the present research, the expression of a unigene encoding cysteine protease was down-regulated in the Bnms3 mutant. In Arabidopsis, there are 30 genes encoding cysteine proteases (Beers et al., 2004), which were analysed using comparative genomics and the Arabidopsis electronic Fluorescent Pictograph browser (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). Four other cysteine protease genes were chosen for real-time PCR analysis, for its orthologues ESTs revealed the presence of corresponding transcripts in anther, flower, and bud cDNA libraries in Brassica. The real-time PCR data showed that the expression patterns of these genes were divided into two clusters. Cluster A involved BnCP2 (At2g21430), BnCP3 (At4g16190), BnCP4 (At4g36880), and BnCP5 (At5g43060), whose expression levels were repressed at the post-meiotic stage when compared with wild type (Fig. 4). However, the expression of BnCP1 (BnMSR09), the one and only member of cluster B, was merely detected at the uninucleate stage of wild-type buds (Fig. 4).
Changes in expression of genes associated with microspore separation
It has been shown that callose cannot be degraded in time in the Bnms3 mutant. In Arabidopsis, A6 (belonging to β-1,3-glucanase family) is likely to be involved in callose dissolution (Zhang et al., 2007; Zhu et al., 2008). Among the down-regulated unigenes, BnMSR66 encodes a β-1,3-glucanase and shares 85% nucleotide identity with At3G23770. Sequences of these genes were aligned using ClustalX version 1.8.3. Bucciaglia first isolated the A6 gene of B. napus (Bucciaglia and Smith, 1994); however, on the basis of the sequences of AT4G14080 and in EST database in NCBI, it lacked five amino acids at the N-terminal. A full-length coding sequence of BnMSR66 was obtained using the analysis of B. napus EST fragments deposited in NCBI and the open reading frame of AT3G23770 in combination with sequencing. BnA6 cDNA contained a 1440-bp open reading frame encoding a polypeptide of 479 amino acids, while that of BnMSR66 was 1431 bp encoding a polypeptide of 476 amino acids. On the basis of the alignment, all sequences contained a classic glycosyl hydrolase family 17 domain (belonging to the β-1,3-glucanase family motif) and an X8 domain, which is well defined as a class of carbohydrate-binding modules responsible for binding β-1,3-glucan (Fig. 5).
Fig. 5.
Sequence analysis of BnA6 and BnMSR66, showing that BnMSR66 and BnA6 are both members of β-1,3-glucanase family and share high sequence identity. The amino acid sequences of BnMSR66 and BnA6 in B. napus and AT3G23770 and AtA6 in Arabidopsis were aligned using ClustalX version 1.8.3 (Jeanmougin et al., 1998). Numbers show the positions of amino acid residues. Black shading indicates conserved residues and grey shading indicates sequence similarity. The putative glycosyl hydrolase family 17 motif sequences are marked by a straight lines. The X8 domain is boxed based on the result of Pfam analysis (http://www.sanger.ac.uk/Software/Pfam/; Finn et al., 2010).
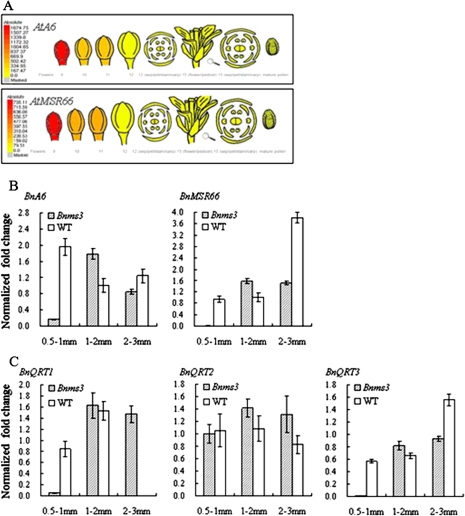
In the publicly available Arabidopsis electronic Fluorescent Pictograph browser, expression patterns of both AtA6 and AtMSR66 (At3G23770) showed high consistency from stages 9 to 11 in the anther, predominantly at stage 9 (Fig. 6A). To further confirm the function of BnMs3 in callose dissolution, both BnA6 and BnMSR66 were selected for real-time PCR analysis. As a result, the expression level of BnA6 was delayed while that of BnMSR66 could not be detected in the Bnms3 mutant at the post-meiotic stage as compared with the wild type (Fig. 6B). Hence, it was demonstrated that this study had found a novel β-1,3-glucanase gene (BnMSR66) which might be responsible for callose degeneration in anther development.
Fig. 6.
Expression patterns of genes involved in microspore separation in B. napus. (A) Relative expression levels and patterns of AtA6 and AtMSR66 in flowers based on microarray data displayed in the Arabidopsis electronic Fluorescent Pictograph browser, showing that they have similar expression patterns; colour scale shows microarray signal level. (B) Real-time PCR analysis of callase-related genes BnA6 and BnMSR66 in the buds of the Bnms3 mutant and the wild type, showing that expression of both is suppressed in the Bnms3 mutant at the post-meiotic stage. (C) Real-time PCR analysis of BnQRT1, BnQRT2, and BnQRT3 in the buds of the Bnms3 mutant and the wild type, showing that expression of BnQRT1 and BnQRT3 were affected in the Bnms3 mutant at the post-meiotic stage, while that of BnQRT2 was unchanged at the three stages in both the mutant and the wild type. (This figure is available in colour at JXB online.)
Apart from callose dissolution, that of the PMC pectic wall is also important for the microspore separation. There are three genes (AtQRT1, AtQRT2, and AtQRT3) whose mutants fail to undergo microspore separation with the release of viable pollen tetrads (Preuss et al., 1994). To investigate the function of BnMs3 in degrading the PMC wall, BnQRT1, BnQRT2, and BnQRT3 were isolated by means of comparative genomics and chosen for real-time PCR. The down-regulated expression of both BnQRT1 and BnQRT3 was initiated at the post-meiotic stage in the Bnms3 mutant (Fig. 6C) while expression level of BnQRT2 was unchanged at the three stages in both the mutant and the wild type.
Expression changes of genes involved in lipid metabolism
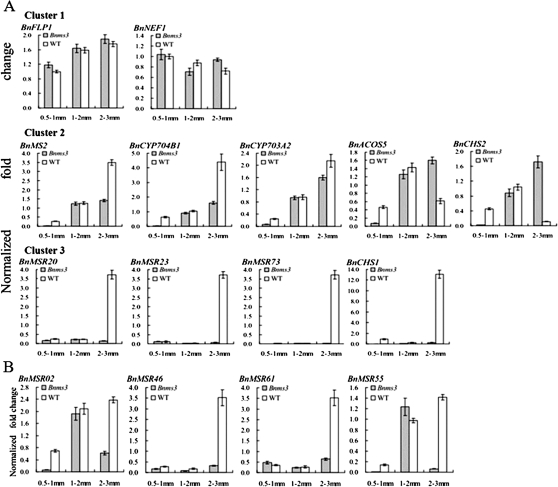
In the Bnms3 mutant, callose dissolution was blocked, but there were still seldom microspores without pollen exine released from the tetrad according to our research. Lipid molecules are a main component of pollen wall. To explore the role of BnMs3 in biosynthesis of lipid molecules, three unigenes obtained from the SSH library and eight ESTs acquired based on the corresponding information from Arabidopsis which participated in this process were analysed by using real-time PCR. According to the different stages at which their differential gene expression initiated, the expression patterns of them could be divided into three clusters. Cluster 1 included BnNEF1 and BnFLP1 which didn’t show any expression changes at the three detected stages between the two materials (Fig. 7A, cluster 1). Cluster 2 was consisted of BnCYP703A2, BnACOS5, BnCYP704B1, BnCHS2 (At4g34850), and BnMS2, showing the initially down-regulated expression in the Bnms3 mutant at the post-meiotic stage (Fig. 7A, cluster 2). The three unigenes (BnMSR20, BnMSR23, and BnMSR73) and BnCHS1 (At4g00040) were attached to cluster 3, which exhibited the down-regulated expression in the Bnms3 mutant initiated at the uninuclear microspore stage (Fig. 7A, cluster 3). Among these differentially expressed genes, BnCYP703A2, BnACOS5, BnCYP704B1, BnMS2, and the three unigenes were relevant to biosynthesis of fatty acids, while BnCHS1 and BnCHS2, which encode chalcone synthase, were involved in biosynthesis of both fatty acids and phenols in Arabidopsis (Dobritsa et al., 2010). These results disclosed that BnMs3 was associated with the biosynthetic pathways of fatty acids and phenolic compounds, which were required for exine development.
Fig. 7.
Real-time PCR analysis of genes related to anther lipid metabolism in the wild type and the Bnms3 mutant in B. napus. (A) According to the different stages at which differential gene expression initiated, the expression patterns of 11 genes involved in biosynthesis of lipid molecules could be divided into three clusters: cluster 1 did not show any gene expression changes at the three detected stages in the mutant and wild type; cluster 2 showed the initially down-regulated gene expression in the Bnms3 mutant at the meiosis stage; cluster 3 exhibited initially down-regulated gene expression in the Bnms3 mutant at the uninuclear microspore stage. (B) Real-time PCR analysis of four genes associated with transport of sporopollenin precursors in both the wild type and the Bnms3 mutant, showing that the expression levels were all affected in the Bnms3 mutant at their initial expression stage.
According to research on wax export from an epidermal cell to the cuticle, wax export consists of two steps of lipid export during pollen-wall formation (Samuels et al., 2008). The first step is the transport of lipid molecules through the plasma membrane (PM) to the extracellular environment, and the second step is transport from the extracellular environment to the surface of microspores, forming pollen exine. BnMSR02, 648 bp in length, belongs to ABC transporter family, which is associated with transmembrane transport of lipid molecules and shares high gene sequence similarity (95.2%) with AtABCG26 (AT3G13220). Real-time PCR analysis showed that the expression of BnMSR02 was much lower in the Bnms3 mutant when compared with that in the wild type during the post-meiotic period (Fig. 7B). It is likely that the anther-expressed LTPs may participate in the transport of sporopollenin precursors (lipid molecules) from the extracellular environment to the microspore during exine deposition (Zhang et al., 2010). In the Bnms3 mutant, the expression levels of eight putative LTPs were found to be altered, namely BnMSR55, BnMSR46, BnMSR24, BnMSR04, BnMSR07, BnMSR10, BnMSR61, and BnMSR38 (Supplementary Table S1). BnMSR46, BnMSR55, and BnMSR61 were analysed by real-time PCR and their corresponding expression levels were reduced in the Bnms3 mutant as compared with the wild-type plant (Fig. 7B). The expression patterns of several LTPs were different, based on the Northern blot and real-time PCR analysis combined with the microarray data from Arabidopsis. BnMSR04 was only expressed in wild-type buds and its expression was initiated at the uninucleate stage (Fig. 2). Expression of BnMSR55 and BnMSR07 was delayed in the Bnms3 mutant at the post-meiotic stage of anther development (Fig. 2 and Fig. 7B). This suggested that lipids synthesized by the tapetal cells might be affected by BnMs3 function and exported in different batches through different LTPs.
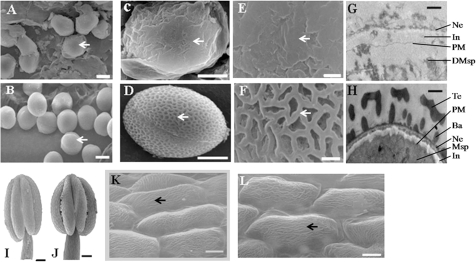
Three genes were selected for further analysis based on gene expression changes in the Bnms3 mutant using transferred-DNA insertion mutants of Arabidopsis to understand their functions in anther development (Supplementary Table S2). As a result, only the Atmsr02 mutant (BnMSR02 orthologue in Arabidopsis) exhibited defects in pollen development (Fig. 8). In this mutant, the insertion occurred at 231 bp upstream of the predicted fifth exon of AtABCG26 (Fig. 8I). The mutant phenotype was indistinguishable from that of the wild type in the heterozygous lines, but homozygous plants of the insertional mutant showed male sterility; hence, the mutation was recessive. The sterile mutant line displayed normal vegetative and floral development but small siliques and no seed yield (Fig. 8E). Pollen grains were absent on the surface of anthers or stigma in the mutant flowers as compared with the wild-type plant (Fig. 8F,G). Moreover, Alexander’s stain showed that mature anthers of Atmsr02 homozygous plants were empty (Fig. 8H). In Arabidopsis, ABCG subfamily transporters involved in the excretion of lipids through the PM have been previously reported (Bird, 2008), and lipids synthesized in the tapetal cells have two destinies, namely biosynthesis of sporopollenin and anther epidermis cutin (Li et al., 2010). To deeply understand BnMSR02 function, the present study used transmission and scanning electron microscopy to observe pollen exine and anther epidermis in the Atmsr02 mutant (Fig. 9). The results indicated that free microspores were visible in the immature anthers of the Atmsr02 mutant (Fig. 9A), but they had an abnormally smooth surface (Fig. 9C,E) and abnormal pollen exine (Fig. 9G). However, it was impossible to differentiate the cytological shape of anther epidermal cutin in the mutant from that of the wild type because of their cuticular ridges in common (Fig. 9K,L). These data suggested that there are two different pathways across the PM of tapetal cells for lipid transport for cuticles and pollen exine materials. MSR02 is required for the transport of lipids related to sporopollenin precursors, forming pollen exine through PM of the tapetal cells, but not for that of cuticular lipid formation.
Fig. 8.
Characterization of the Atmsr02 transferred-DNA mutant in A. thaliana. (A) Wild-type plant, with normal silique development (arrows). (E) Atmsr02 transferred-DNA mutant plant, with very small siliques containing no seeds (arrows). (B,C) Wild-type flowers, with many pollen grains displayed on the anther and stigma surfaces (arrows). (D) Wild-type anther by Alexander staining, with viable pollen grains (arrow). (F,G) Atmsr02 transferred-DNA mutant flowers, with no pollen grains present on the surface of anther or stigma (arrows). (H) Atmsr02 transferred-DNA mutant anther by Alexander staining, without viable pollen grains (arrow). (I) Diagrammatic representation of the Atmsr02 transferred-DNA mutant. (This figure is available in colour at JXB online.)
Fig. 9.
Scanning and transmission electron microscopy (SEM, TEM, respectively) of pollen exine and anther epidermis from wild-type and Atmsr02 transferred-DNA mutant A. thaliana. (A–F) SEM micrographs of pollen exine of (A,C,E) Atmsr02 transferred-DNA mutant, showing smooth surfaces of the pollen grains (arrows), and (B,D,F) wild type, displaying a regular reticulate pattern (arrows). (G,H) TEM micrographs of the mature pollen wall of (G) Atmsr02 transferred-DNA mutant, displaying abnormal exine layer on the pollen, and (H) wild type, showing an intact exine structure. (I,J) SEM micrographs of anthers of (I) Atmsr02 transferred-DNA mutant and (J) wild type. (K,L) SEM micrographs of the outmost surface of the anther epidermis in (K) wild type and (L) Atmsr02 transferred-DNA mutant, both exhibiting no apparent differences and having cuticular ridges (arrows). Ba, baculae; DMsp, degenerated microspore; In, intine ; Msp, microspore; Ne, nexine; PM, plasma membrane; Te, tectum. Bars=10 μm (A,B), 2.5 μm (C,D), 1 μm (E,F), 0.5 μm (G,H), 50 μm (I,J), 5 μm (K,L).
Discussion
In this research, cytological defects of the Bnms3 mutant during anther development were investigated and SSH and macroarray approaches were used to identify genes affected by BnMs3 using a NIL (line 7365AB). Results of both the cytological and differential expression analysis exhibited that BnMs3 was associated with tapetal development, callose degradation, and pollen-wall formation. To gain a deeper insight into the function of BnMs3 in these processes, Brassica orthologues of the Arabidopsis genes required for anther development were identified by comparative genomics and analysed by real-time PCR for both the Bnms3 mutant and the wild type.
BnMs3 influences tapetal differentiation and degradation
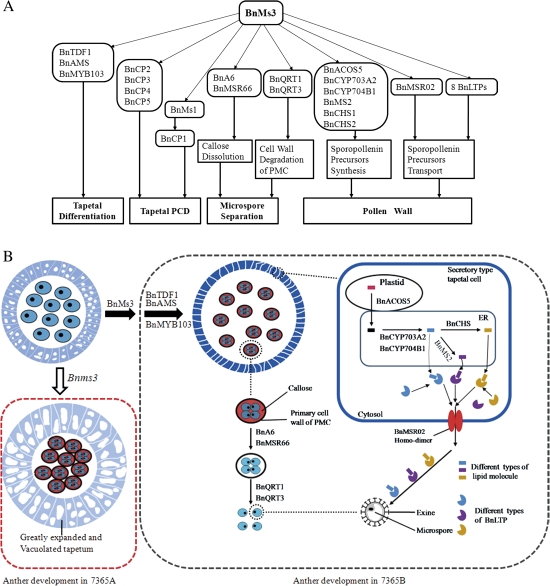
Tapetum is a cell layer adjacent to the anther locule. There are two main tapetal types, designated as the secretory tapetum and the amoeboid tapetum, in the angiosperms (Pacini et al., 1985; Murgia et al., 1991). The secretory tapetum shows cytoplasm and nucleus shrinkage, loss of the cell wall, and reduction in the size of mitochondria (Pacini et al., 1985; Parish and Li, 2010). Tapetal cells transform into the secretory type at the post-meiotic stage to secrete enzymes for the release of microspores from the tetrad and to provide nutrients for pollen development (Pacini et al., 1985; Piffanelli et al., 1998). Once the transition of tapetum is blocked in anther development in Arabidopsis, the tapetal cells become enlarged and abnormally vacuolated, which is followed by aberrant microspore separation and abnormal pollen-wall development (Zhang et al., 2007; Zhu et al., 2008). In Arabidopsis, TDF1, AMS, and MYB103 are involved in this process. AMS encodes a basic helix–loop–helix protein and acts downstream of TDF1 but upstream of MYB103 (Sorensen et al., 2003; Zhang et al., 2007; Zhu et al., 2008). Histological observation revealed that the critical cause of male sterility in the Bnms3 mutant was attributed to a defect in the timely transition of the tapetum to the secretory type, which was similar to that of Attdf1, Atams, and Atmyb103 mutants. Moreover, the anther of B. napus is bigger than that of Arabidopsis, which made it easier to distinguish the secretory type. Real-time PCR analysis demonstrated that the expression levels of BnTDF1, BnAMS, and BnMYB103 were delayed at the post-meiotic stage of anther development in the Bnms3 mutant. These data suggested that BnMs3 shared the same pathway with BnTDF1, BnAMS, and BnMYB103 in tapetal differentiation and might control the transition of the tapetal cells by affecting the expression of BnMYB103 (Fig. 10A,B).
Fig. 10.
Functional model of BnMs3 in anther development of B. napus. (A) The functional network of BnMs3 in tapetum and microspore development in B. napus based on the analysis of suppression subtractive hybridization and gene information from Arabidopsis. (B) Model for the role of BnMs3 in tapetal function and pollen development in B. napus according to cytological observation, suppression subtractive hybridization, and gene information from Arabidopsis. 7365A and 7365B represent the Bnms3 mutant and the wild type, respectively. (This figure is available in colour at JXB online.)
In flowering plants, degeneration of the tapetal cells is required for pollen-wall formation and is considered as the result of PCD (Wu and Cheung, 2000; Vizcay-Barrena and Wilson, 2006). The TUNEL assay has confirmed that tapetal PCD in Atms1, Bncyp704b1, Ostdr (AtAMS orthologue in rice), and Bnms3 mutants was delayed or absent (Li et al., 2006; Vizcay-Barrena and Wilson, 2006; Yi et al., 2010; Dun et al., 2011). Cysteine proteases are induced in PCD (Solomon et al., 1999). OsTDR directly regulates a cysteine protease (OsCP1) associated with PCD (Li et al., 2006). MS1, acting downstream of AMS and CYP704B1, also affects the expression of a cysteine protease gene, At1g06260 (Yang et al., 2007; Chen et al., 2009; Xu et al., 2010). The real-time PCR analysis revealed that the expression levels of BnMS1, BnAMS, and BnCYP704B1 and five cysteine protease genes were affected in the Bnms3 mutant at the corresponding initial expression stages of anther development. These results demonstrated that the Bnms3 mutant exhibited abnormal tapetal degradation due to delayed tapetal PCD, which is affected by BnMs3 by modulating the expression of four cysteine proteases at the tetrad stage and that of BnMS1 to affect the expression of BnCP1 (At1g06260) at the uninucleate stage (Fig. 10A).
BnMs3 is essential for microspore separation
In anther development, the PMC undergoes meiosis to form a tetrad, which is surrounded by the inner wall (the callose secondary cell wall) and the outer wall (the primary wall of PMC) (Rhee and Somerville, 1998). Therefore, degradation of the callose wall and primary wall of the PMC is a critical step during pollen development. The callose wall is made of β-1,3-glucan and is dissolved by callase (β-1,3-glucanase) secreted by the secretory tapetum (Frankel et al., 1969; Stieglitz and Stern, 1973; Stieglitz, 1977). Several tapetum-specific expressed genes encoding β-1,3-glucanase have been cloned, such as Tag1 in tobacco (Bucciaglia and Smith, 1994), Osg1 in rice (Yamaguchi et al., 2002), and A6 from B. napus and Arabidopsis (Hird et al., 1993). Silencing of Osg1 using RNA interference resulted in disruption of callose degradation (Wan et al., 2011). In transgenic tobacco, two pathogenesis-related β-1,3-glucanase genes were fused to the tapetum-specific promoters and the callose wall of the microspores dissolved prematurely, finally leading to male sterility (Worrall et al., 1992; Tsuchiya et al., 1995). These results imply that the timely degradation of the callose wall is crucial for microspore separation and that merely one β-1,3-glucanase is sufficient to form callase for degrading callose. The callose of the Bnms3 mutant in this study was not degraded in time, according to aniline blue staining, which is consistent with results in Attdf1 and Atmyb103 mutants (Zhu et al., 2010). Additionally, reduced expression of A6 in Attdf1 and Atmyb103 mutants may result in abnormal callose degradation (Zhang et al., 2007; Zhu et al., 2008). This study isolated a novel β-1,3-glucanase (BnMSR66) in the Bnms3 mutant, which included a glycosyl hydrolase family 17 motif and an X8 domain and had an expression pattern similar to that of BnA6. Both BnMSR66 and BnA6 showed reduced expression at the post-meiotic stage of anther development in the Bnms3 mutant. Alignment of the sequences exhibited 73% identity at the amino acid level. Therefore, BnMs3 may specifically control several β-1,3-glucanase genes that are likely to be functionally redundant in regulating callase activity in B. napus (Fig. 10A,B). The subsequent degradation of the PMC pectic wall is required for microspore separation (Rhee and Somerville, 1998). In Arabidopsis, QRT1, QRT2, and QRT3 are involved in this process (Preuss et al., 1994). Loss-of-function mutations of any one of the three QRT genes lead to the lack of microspores separation after meiosis and produce mature pollen tetrads (Rhee et al., 2003; Francis et al., 2006; Ogawa et al., 2009). In the Bnms3 mutant, the expression levels of BnQRT1 and BnQRT3 were down-regulated at the post-meiotic stage. These results indicate that BnMs3 is also involved in controlling the degradation of the PMC pectic wall by affecting the expression of BnQRT1 and BnQRT3 (Fig. 10A,B).
BnMs3 affects pollen-wall development
The synthesis of pollen exine is a vital step during microspore development (Zinkl et al., 1999). Since polymer sporopollenin largely composed of fatty acids and phenolic compounds are key components of pollen exine (Paxson-Sowders et al., 1997; Piffanelli et al., 1998), lipid metabolism and export are essential for exine formation (Piffanelli et al., 1998; Ahlers et al., 1999). In the Bnms3 mutant, expression levels of five genes encoding putative enzymes involved in lipid metabolism were down-regulated, including two genes (BnMSR20 encoding fatty acid elongase 3-ketoacyl-CoA synthase 7 and BnMSR73 encoding beta-ketoacyl-CoA synthase) concerned with the biosynthesis of long-chain fatty acids, which plays an important role in pollen exine formation (Zhang et al., 2008). Hydroxylation of fatty acids is also required for the exine formation, and several cytochrome P450 enzymes are associated with ω-hydroxylation of fatty acids (Mizutani and Ohta, 2010). It has been shown that CYP703A2 and CYP704B1 catalyse the in-chain hydroxylation of C10 to C14 and C16 to C18 fatty acids during pollen exine formation, respectively (Morant et al., 2007; Dobritsa et al., 2009; Li et al., 2010). In the Bnms3 mutant, the expression levels of both BnCYP703A2 and BnCYP704B1 were decreased. The conversion of fatty acids to fatty alcohols is required for lipid synthesis, which is principally important for pollen exine formation in flowering plants (Aarts et al., 1997). MS2, encoding a fatty acyl carrier protein reductase associated with sporopollenin biosynthesis, is involved in this conversion during pollen development of Arabidopsis and rice (Aarts et al., 1997; Shi et al., 2011). The present study found that the expression of BnMS2 was remarkably decreased at the post-meiotic stage of anther development in the Bnms3 mutant. Moreover, chalcone synthase is able to use medium- to long-chain (C4–C20) fatty acyl-CoA as a substrate for generating phenylpropanoid (an ingredient of sporopollenin precursors), in which At4g00040 (CHS1) and At4g34850 (CHS2) are involved (Dobritsa et al., 2010). The real-time PCR data in this study showed that the expression levels of BnCHS1 and BnCHS2 were affected in the Bnms3 mutant. This suggested that both fatty acid and phenylpropanoid pathways were altered in the Bnms3 mutant.
Apart from playing a role in the formation of pollen-wall materials, BnMs3 also affects the export of lipid molecules from the tapetal cells to the surface of the microspores, which mainly consists of two steps (Samuels et al., 2008). First, the lipid molecules synthesized in the tapetal cells are exported through the PM. ABCG11 and ABCG12 belong to ABC transporter protein family, which uses ATP hydrolysis to transport a variety of substances across biological membranes, and are required for wax transport across the PM in Arabidopsis (Pighin et al., 2004; David et al., 2007; Panikashvili et al., 2007). In the Bnms3 mutant, the expression of BnMSR02 (ABCG26) was changed, and the Atabcg26 mutant (Atmsr02) resulted in a male sterile phenotype due to microspore degradation, which was consistent with the data reported by Quilichini et al. (2010) and Choi et al. (2011). Fatty acids are required for both cuticle and exine formation during anther development; consequently, transmission and scanning electron microscopy was conducted to analyse the cuticle and exine of the Atabcg26 mutant. The cuticle was normal but the pollen exine was abnormal. Therefore it is concluded that ABCG26 was only essential for lipid export of sporopollen precursors through the PM during pollen exine development. The second step of lipid transport is from the extracellular environment to the surface of microspores to form pollen exine (Samuels et al., 2008; Zhang et al., 2010). So far, LTP has been considered as an attractive candidate for this process (DeBono et al., 2009). Expression levels of the eight genes putatively related to LTPs were apparently down-regulated in the Bnms3 mutant. These results suggested that defects in the biosynthesis and transport of lipids, which led to fewer microspores being released from the tetrad and the abnormal pollen exine formation in the Bnms3 mutant, made the pollen abortion more thorough (Fig. 10A,B).
A model for BnMs3 function in anther development
Cytological observation combined with the differential gene expression data from the macroarray has shown that BnMs3 is involved in tapetal function and pollen development. According to putative models for the network of tapetum and pollen development in Arabidopsis and rice (Zhu et al., 2008; Wilson and Zhang, 2009; Xu et al., 2010), a few crucial genes from the models were analysed in this study using real-time PCR, which has provided a deeper insight into the role of BnMs3 during anther development (Fig. 10A,B). All data indicated that BnMs3 plays a specific role in the transition of the tapetal cells to the secretory type. Additionally, it is also required for microspore separation, lipid biosynthesis, export of pollen-wall materials, and pulling the trigger of tapetal PCD by affecting the expression of several key genes in B. napus (Fig. 10A,B).
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Differential screening of the subtracted libraries using macroarray analysis.
Supplementary Table S1. Homology analysis of differentially expressed sequences from the subtracted library.
Supplementary Table S2. Gene-specific primer pairs used in this study.
Acknowledgments
The authors would like to thank the two anonymous reviewers for their helpful comments, and Mayank Gautam for his effort on critical reading and revising the manuscript. This research was supported by funds from the National Natural Science Foundation of China (31130040) and the National ‘863’ High-Tech Project (2006AA10Z146).
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. The Plant Journal. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- Ahlers F, Thom I, Lambert J, Kuckuk R, Rolf W. 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry. 1999;50:1095–1098. [Google Scholar]
- Alexander M. Differential staining of aborted and nonaborted pollen. Biotechnic and Histochemistry. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz EM. Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiology. 2007;145:747–762. doi: 10.1104/pp.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. The Plant Journal. 2004;39:170–181. doi: 10.1111/j.1365-313X.2004.02118.x. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Molecular Biology. 2003;53:107–116. doi: 10.1023/B:PLAN.0000009269.97773.70. [DOI] [PubMed] [Google Scholar]
- Auger B, Baron C, Lucas MO, Vautrin S, Bergès H, Chalhoub B, Fautrel A, Renard M, Nesi N. Brassica orthologs from BANYULS belong to a small multigene family, which is involved in procyanidin accumulation in the seed. Planta. 2009;230:1167–1183. doi: 10.1007/s00425-009-1017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers EP, Jones AM, Dickerman AW. The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry. 2004;65:43–58. doi: 10.1016/j.phytochem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Bird DA. The role of ABC transporters in cuticular lipid secretion. Plant Science. 2008;174:563–569. [Google Scholar]
- Bucciaglia PA, Smith AG. Cloning and characterization of Tag1, a tobacco anther β-1,3-glucanase expressed during tetrad dissolution. Plant Molecular Biology. 1994;24:903–914. doi: 10.1007/BF00014444. [DOI] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Current Biology. 2002;12:1718–1727. doi: 10.1016/s0960-9822(02)01151-x. [DOI] [PubMed] [Google Scholar]
- Cavell AC, Lydiate DJ, Parkin IAP, Dean C, Trick M. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome. 1998;41:62–69. [PubMed] [Google Scholar]
- Chen YN, Lei SL, Zhou ZF, Zeng FQ, Yi B, Wen J, Shen JX, Ma CZ, Tu JX, Fu TD. Analysis of gene expression profile in pollen development of recessive genic male sterile Brassica napus L. line S45A. Plant Cell Reports. 2009;28:1363–1372. doi: 10.1007/s00299-009-0736-9. [DOI] [PubMed] [Google Scholar]
- Choi H, Jin JY, Choi S, Hwang JU, Kim YY, Suh MC, Lee Y. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. The Plant Journal. 2011;65:181–193. doi: 10.1111/j.1365-313X.2010.04412.x. [DOI] [PubMed] [Google Scholar]
- David B, Fred B, Alexandra B, John S, Greer S, Jetter R, Kunst LK, Wu XM, Yephremov A, Samuels L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. The Plant Journal. 2007;52:485–498. doi: 10.1111/j.1365-313X.2007.03252.x. [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ. A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. The Plant Cell. 2009;21:507–525. doi: 10.1105/tpc.108.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuels L. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. The Plant Cell. 2009;21:1230–1238. doi: 10.1105/tpc.108.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Lei ZT, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW. LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiology. 2010;153:937–955. doi: 10.1104/pp.110.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Moller BL, Preuss D. CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiology. 2009;151:574–589. doi: 10.1104/pp.109.144469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dun XL, Zhou ZF, Xia SQ, Wen J, Yi B, Shen JX, Ma CZ, Tu JX, Fu TD. BnaC.Tic40, a plastid inner membrane translocon originating from Brassica oleracea, is essential for tapetal function and microspore development in Brassica napus. The Plant Journal. 2011;68:532–545. doi: 10.1111/j.1365-313X.2011.04708.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Research. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology. 2006;142:1004–1013. doi: 10.1104/pp.106.085274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel R, Izhar S, Nitsan J. Timing of callase activity and cytoplasmic male sterility in Petunia. Biochemical Genetics. 1969;3:451–455. doi: 10.1007/BF00485605. [DOI] [PubMed] [Google Scholar]
- Hird DL, Worrall D, Hodge R, Smartt S, Paul W, Scott R. The anther-specific protein encoded by the Brassica napus and Arabidopsis thaliana A6 gene displays similarity to β-1,3-glucanases. The Plant Journal. 1993;4:1023–1033. doi: 10.1046/j.1365-313x.1993.04061023.x. [DOI] [PubMed] [Google Scholar]
- Huang Z, Chen YF, Yi B, Xiao L, Ma CZ, Tu JX, Fu TD. Fine mapping of the recessive genic male sterility gene (Bnms3) in Brassica napus L. Theoretical and Applied Genetics. 2007;115:113–118. doi: 10.1007/s00122-007-0547-8. [DOI] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature. 2004;430:356–360. doi: 10.1038/nature02733. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends in Biochemical Sciences. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Jia GX, Liu XD, Owen HA, Zhao DZ. Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proceedings of the National Academy of Sciences USA. 2008;105:2220–2225. doi: 10.1073/pnas.0708795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, et al. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. The Plant Cell. 2010;22:173–190. doi: 10.1105/tpc.109.070326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. The Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annual Review of Plant Biology. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- Mewes HW, Dietmann S, Frishman D, et al. MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Research. 2008;36:D196–D201. doi: 10.1093/nar/gkm980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. Diversification of P450 genes during land plant evolution. Annual Review of Plant Biology. 2010;61:291–315. doi: 10.1146/annurev-arplant-042809-112305. [DOI] [PubMed] [Google Scholar]
- Morant M, Jorgensen K, Schaller H, Pinot F, Moller BL, Werck-Reichhart D, Bak S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. The Plant Cell. 2007;19:1473–1487. doi: 10.1105/tpc.106.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M, Charzynska M, Rougier M, Cresti M. Secretory tapetum of Brassica oleracea L.: polarity and ultrastructural features. Sexual Plant Reproduction. 1991;4:28–35. [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. The Plant Cell. 2009;21:216–233. doi: 10.1105/tpc.108.063768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini E, Franchi GG, Hesse M. The tapetum: its form, function, and possible phylogeny in Embryophyta. Plant Systematics and Evolution. 1985;149:155–185. [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Hofer R, Schreiber L, Chory J, Aharoni A. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiology. 2007;145:1345–1360. doi: 10.1104/pp.107.105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish RW, Li SF. Death of a tapetum: a programme of developmental altruism. Plant Science. 2010;178:73–89. [Google Scholar]
- Paxson-Sowders DM, Owen HA, Makaroff CA. A comparative ultrastructural analysis of exine pattern development in wild-type Arabidopsis and a mutant defective in pattern formation. Protoplasma. 1997;198:53–65. [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ. Biogenesis and function of the lipidic structures of pollen grains. Sexual Plant Reproduction. 1998;11:65–80. [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306:702–704. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ. ATP-binding bassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiology. 2010;154:678–690. doi: 10.1104/pp.110.161968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Osborne E, Poindexter PD, Somerville CR. Microspore separation in the quartet 3 mutants of Arabidopsis is Impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiology. 2003;133:1170–1180. doi: 10.1104/pp.103.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. The Plant Journal. 1998;15:79–88. doi: 10.1046/j.1365-313x.1998.00183.x. [DOI] [PubMed] [Google Scholar]
- Rubinelli P, Hu Y, Ma H. Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Molecular Biology. 1998;37:607–619. doi: 10.1023/a:1005964431302. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: a laboratory manual. New York: Cold Spring Harbor Press; 2001. [Google Scholar]
- Samuels L, Kunst L, Jetter R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annual Review of Plant Biology. 2008;59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction. 1999;11:297–322. [Google Scholar]
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA. 1999;96:11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Tan HX, Yu XH, et al. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. The Plant Cell. 2011;23:2225–2246. doi: 10.1105/tpc.111.087528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. The Plant Cell. 1999;11:431–444. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AM, Krober S, Unte US, Huijser P, Dekker K, Saedler H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. The Plant Journal. 2003;33:413–423. doi: 10.1046/j.1365-313x.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- Stieglitz H. Role of β-1,3-glucanase in postmeiotic microspore release. Developmental Biology. 1977;57:87–97. doi: 10.1016/0012-1606(77)90356-6. [DOI] [PubMed] [Google Scholar]
- Stieglitz H, Stern H. Regulation of β-1,3-glucanase activity in developing anthers of Lilium. Developmental Biology. 1973;34:169–173. doi: 10.1016/0012-1606(73)90347-3. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Toriyama K, Yoshikawa M, Ejiri S, Hinata K. Tapetum-specific expression of the gene for an endo-β-1,3-glucanase causes male sterility in transgenic tobacco. Plant and Cell Physiology. 1995;36:487–494. doi: 10.1093/oxfordjournals.pcp.a078784. [DOI] [PubMed] [Google Scholar]
- Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. Journal of Experimental Botany. 2006;57:2709–2717. doi: 10.1093/jxb/erl032. [DOI] [PubMed] [Google Scholar]
- Wan LL, Zha WJ, Cheng XY, et al. A rice β-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta. 2011;233:309–323. doi: 10.1007/s00425-010-1301-z. [DOI] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun YJ, Liu WL, Albert R, Zheng ZQ, Oppenheimer DG, Zhao DZ, Ma H. Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. The Plant Journal. 2007;52:14–29. doi: 10.1111/j.1365-313X.2007.03217.x. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. The Plant Journal. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB. From Arabidopsis to rice: pathways in pollen development. Journal of Experimental Botany. 2009;60:1479–1492. doi: 10.1093/jxb/erp095. [DOI] [PubMed] [Google Scholar]
- Worrall D, Hird DL, Hodge R, Paul W, Draper J, Scott R. Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. The Plant Cell. 1992;4:759–771. doi: 10.1105/tpc.4.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HM, Cheung AY. Programmed cell death in plant reproduction. Plant Molecular Biology. 2000;44:267–281. doi: 10.1023/a:1026536324081. [DOI] [PubMed] [Google Scholar]
- Xiao L, Yi B, Chen YF, Huang Z, Chen W, Ma CZ, Tu JX, Fu TD. Molecular markers linked to Bn;rf: a recessive epistatic inhibitor gene of recessive genic male sterility in Brassica napus L. Euphytica. 2008;164:377–384. [Google Scholar]
- Xing S, Zachgo S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. The Plant Journal. 2008;53:790–801. doi: 10.1111/j.1365-313X.2007.03375.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Yang CY, Yuan Z, Zhang DS, Gondwe MY, Ding ZW, Liang WQ, Zhang DB, Wilson ZA. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. The Plant Cell. 2010;22:91–107. doi: 10.1105/tpc.109.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nakayama K, Hayashi T, Tanaka Y, Koike S. Molecular cloning andcharacterization of a novel β-1,3-glucanase gene from rice. Bioscience, Biotechnology, and Biochemistry. 2002;66:1403–1406. doi: 10.1271/bbb.66.1403. [DOI] [PubMed] [Google Scholar]
- Yang CY, Vizcay-Barrena G, Conner K, Wilson ZA. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. The Plant Cell. 2007;19:3530–3548. doi: 10.1105/tpc.107.054981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes and Development. 1999;13:2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yi B, Zeng FQ, Lei SL, et al. Two duplicate CYP704B1-homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. The Plant Journal. 2010;63:925–938. doi: 10.1111/j.1365-313X.2010.04289.x. [DOI] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yin CS, Zong J, Gu FW, Zhang DB. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiology. 2010;154:149–162. doi: 10.1104/pp.110.158865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Molecular Plant. 2008;1:599–610. doi: 10.1093/mp/ssn028. [DOI] [PubMed] [Google Scholar]
- Zhang JF, Lu Y, Yuan YX, Zhang XW, Geng JF, Chen Y, Cloutier S, McVetty PBE, Li GY. Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Molecular Biology. 2009;69:553–563. doi: 10.1007/s11103-008-9437-y. [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun YJ, Timofejeva L, Chen CB, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by dysfunctional tapetum1 (dyt1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. The Plant Journal. 2007;52:528–538. doi: 10.1111/j.1365-313X.2007.03254.x. [DOI] [PubMed] [Google Scholar]
- Zhao DZ, Wang G, Speal B, Ma H. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes and Development. 2002;16:2021–2031. doi: 10.1101/gad.997902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN. Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. The Plant Journal. 2008;55:266–277. doi: 10.1111/j.1365-313X.2008.03500.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dun XL, Zhou ZF, Xia SQ, Yi B, Wen J, Shen JX, Ma CZ, Tu JX, Fu TD. A separation defect of tapetum cells and microspore mother cells results in male sterility in Brassica napus: the role of abscisic acid in early anther development. Plant Molecular Biology. 2010;72:111–123. doi: 10.1007/s11103-009-9556-0. [DOI] [PubMed] [Google Scholar]
- Zinkl GM, Zwiebel BI, Grier DG, Preuss D. Pollen-stigma adhesion in Arabidopsis: a species-specific interaction mediated by lipophilic molecules in the pollen exine. Development. 1999;126:5431–5440. doi: 10.1242/dev.126.23.5431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.