Abstract
The raffinose family of oligosaccharides (RFOs) serve as transport carbohydrates in the phloem, storage compounds in sink tissues, and putative biological agents to combat both abiotic and biotic stress in several plant species. To investigate further the functional roles of this class of compounds in trees, two cDNAs encoding galactinol synthase (GolS, EC 2.4.1.123), which catalyses the first step in the biosynthesis of RFOs, were identified and cloned from hybrid poplar (Populus alba×grandidentata). Phylogenetic analyses of the Populus GolS isoforms with other known GolS proteins suggested a putative role for these enzymes during biotic or abiotic stress in hybrid poplar. The predicted protein sequences of both isoforms (Pa×gGolSI and Pa×gGolSII) showed characteristics of GolS proteins from other species, including a serine phosphorylation site and the ASAAP pentapeptide hydrophobic domain. Kinetic analyses of recombinant Pa×gGolSI and Pa×gGolSII resulted in Km values for UPD-galactose of 0.80 and 0.65 mM and Vmax values of 657.5 and 1245 nM min−1, respectively. Pa×gGolSI inherently possessed a broader pH and temperature range when compared with Pa×gGolSII. Interestingly, spatial and temporal expression analyses revealed that Pa×gGolSII transcript levels varied seasonally, while Pa×gGolSI did not, implying temperature-regulated transcriptional control of this gene in addition to the observed thermosensitivity of the respective enzyme. This evidence suggested that Pa×gGolSI may be involved in basic metabolic activities such as storage, while Pa×gGolSII is probably involved in seasonal mobilization of carbohydrates.
Keywords: galactinol synthase, GolS, raffinose family of oligosaccharides, hybrid poplar, abiotic stress, biotic stress
Introduction
Plants have the innate ability to store and translocate a portion of the fixed carbon derived from photosynthesis, affording them the plasticity required to grow and survive in a variety of environments and under a myriad of conditions. Typically, they do this by synthesizing starch for storage and translocating sucrose for metabolism and the synthesis of integral structural moieties. However, other sources of translocatable soluble carbohydrate exist in the plant kingdom, with the raffinose family of oligosaccharides (RFOs) being the most prominent (Keller and Pharr, 1996). Chemically, the RFOs are natural extensions of sucrose to which varying numbers of α-galactosyl residues are attached. As non-reducing carbohydrates, they are good storage compounds that can accumulate in large quantities without affecting primary metabolic processes (Peters, 2007). The potential role of RFOs in stress tolerance has been studied intensively in seeds, mainly with respect to desiccation tolerance and longevity in the dehydrated state (Downie et al., 2003). Additionally, RFO accumulation has commonly been associated with abiotic stress conditions such as cold, heat, or drought in several plant species. In Populus tremuloides, endogenous raffinose and stachyose levels have been shown to increase as temperatures drop in early winter and to diminish as temperatures rise in spring. Furthermore, raffinose accumulation has been shown to be strongly dependent on low temperatures during acclimation (Cox and Stushnoff, 2001). In addition, in Populus balsamifera clones, galactinol synthase were among the genes with the most significant differences when trees were subjected to drought treatment (Wilkins et al., 2009)
RFOs also appear to play a prominent role in the response of plants to biotic stresses. For example, a recent report (Philippe et al., 2010) demonstrated the accumulation of galactinol and raffinose in hybrid poplar (Populus trichocarpa×deltoides) trees subjected to simulated feeding by the forest tent caterpillar, Malacosoma disstria. Transcriptome analysis revealed a strong induction of numerous GolS genes. Correspondingly, expression analyses by quantitative PCR in source and sink leaves showed differential transcript accumulation for the various isoforms examined.
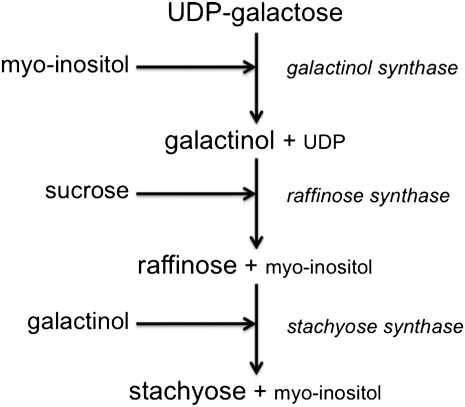
Galactinol synthase (GolS, EC 2.4.1.123) catalyses the first step in the biosynthesis of RFOs (Fig. 1), by reversibly synthesizing galactinol from UDP-D-galactose and myo-inositol (Saravitz et al., 1987; Keller and Pharr, 1996). The only known function for galactinol is as a substrate for the formation of the larger soluble oligosaccharides: raffinose, stachyose, and verbascose (Saravitz et al., 1987). GolS has primarily been characterized biochemically from legume seeds and cucurbit leaves (Keller and Pharr, 1996). However, this enzyme has also been isolated and described in other plants, including common bugle (Ajuga reptans) (Bachmann et al., 1994), zucchini squash (Cucurbita pepo) (Smith et al., 1991), kidney bean (Phaseolus vulgaris) (Liu et al., 1995), soybean (Glycine max) (Ribeiro et al., 2000), and cucumber (Cucumis sativus) (Wakiuchi et al., 2003). Based on these findings, it appears that the enzymes have a pH optimum between 7 and 8, a temperature optimum between 35 and 50 °C, and require manganese ions and dithiothreitol (DTT). In addition to the native expression studies, there have also been studies employing recombinant GolS protein. For example, Taji et al., (2002) expressed three Arabidopsis thaliana GolS genes in Escherichia coli and demonstrated that AtGolS1, -2 and -3 all encoded GolS proteins; however, enzyme kinetics were not discussed. Biologically, it was shown that the transcript abundance of AtGolS1 and -2 were induced by drought and increasing salinity but not by cold. Alternatively, AtGolS3 was induced by cold but not by drought or high salinity. Furthermore, overexpression of AtGolS2 increased the pools of galactinol and raffinose in Arabidopsis and improved the overall drought tolerance of Arabidopsis plants.
Fig. 1.
Schematic representation of the biosynthetic pathway of RFOs.
This study characterized the spatial and temporal transcript expression of three hybrid poplar GolS homologues, both diurnally and annually. Furthermore, the phylogenetic relationships of the Populus genes are described and compared with all known GolS proteins. Two GolS genes were expressed heterologously in Pichia pastoris and their enzyme kinetics were analysed. These results expand our knowledge about the physiological roles of these enzymes in hybrid poplar.
Materials and methods
Phylogenetic analysis
Amino acid sequences obtained from GenBank and Phytozome were used for multiple sequence alignment using the MAFFT program, available at http://www.ebi.ac.uk/. The evolutionary history was inferred using the neighbour-joining method (Saitou and Nei, 1987). The optimal trees with the sum of branch length equal to 0.79719871 and 2.99473095 are shown in Fig. 3A and B, respectively. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic trees. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and bars indicate the number of amino acid substitutions per site. The analysis involved 10 and 35 aa sequences, respectively. All positions containing gaps and missing data were eliminated. There were a total of 323 and 266 positions, respectively, in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). Amino acid percentage similarity was calculated in ClustalW.
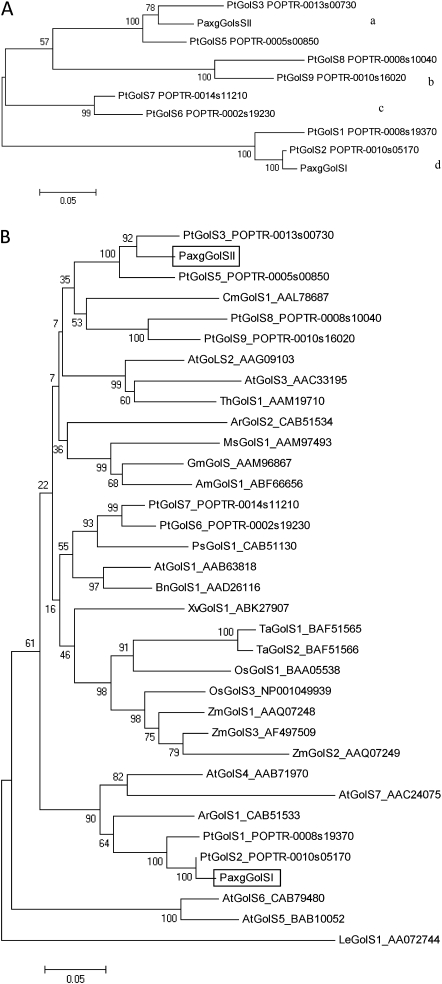
Fig. 3.
Phylogenetic analysis using the neighbour-joining method and based on predicted amino acid sequences of confirmed and putative GolS proteins. (A) GolS proteins from Populus grouped in four clades (a–d). (B) GolS proteins from different plant species. Phytozome accession numbers are provided for the Populus trichocarpa GolS proteins and GenBank accession number are provided for the remaining proteins. The two GolS proteins cloned from hybrid poplar (P. alba×grandidentata) are denoted in rectangles in (B). Bootstrap values are based on 1000 replicates. Bars, 0.05 amino acid substitutions per site.
Cloning GolS from hybrid poplar (Populus alba×grandidentata)
Expressed sequence tags obtained from the Treenomix project of Genome BC (P. trichocarpa, WS02549_C18, WS01812_P18, WS02432_H16, WS01212_F01, WS0141_H15, WS02543_G16, WS0231_K19, WS0189_A11, WS0146_B17, and WS01710_L22) were sequenced and used to design primer sets that amplified the full-length coding region of the GolS genes from hybrid poplar. The primers used to amplify GolSI from leaf cDNA were: GolSLGX.Fw (5′-ATGGCTCCAGG AGTGCCCATGGA-3′) and GolSLGX.Rv (5′-TTAAGCAGCAGATGGCGCAGA-3′), while GolSII was amplified with GoLSLGXIII.Fw (5′-ATGGCTCCTCATATTACAACTGC-3′) and GolSLGXIII.Rv (5′-CTAAGCGGCGGATG GGGCGG-3′). The 1014 bp products amplified by PCR were cloned into a ZERO Blunt vector (Invitrogen Canada Inc., Burlington, ON). Plasmids containing both GolS genes were then used for heterologous expression studies.
Heterologous expression of GolS in Pichia pastoris
The GolSI gene was modified by PCR to include two restriction sites, SnaBI and AvrII, using the primer pair P39GolSI.SnaBI (5′-TCATACGTAATGGCTCCAGGAGTG-3′) and P39GolSI.AvRII (5′-TCCCCTAGGTTAAGCAGCAGATGG-3′) (restriction sites underlined). The 1022 bp product, amplified by PCR, was cloned into pCR-Blunt II-TOPO, (Invitrogen Canada Inc.). The fragment spanning the SnaBI and AvrII sites was then subcloned in frame with the secretion signal (α factor) into the pPIC9K expression vector (Original Pichia Expression Kit; Invitrogen Canada Inc.) and then transformed into E. coli. Colonies were screened using the 5′AOX1 and 3′AOX1 primers (following the manufacturer’s instructions) and sequenced with the 3′AOX primer to confirm that the GolSI sequence was in frame with the α-factor secretion signal, thereby including the Kex2 cleavage site. The pPIC9K-GolSI plasmid was harvested from E. coli, linearized with SalI, and transformed into electrocompetent P. pastoris strain GS115 according to the manufacturer’s specifications with the following modifications: 15 μg linearized DNA was transformed into 80 μl electrocompetent P. pastoris cells using an Electroporator 2510 (Eppendorf, Mississauga, ON) at 1500 V and 5 ms. Transformed colonies were selected on minimal dextrose medium plates and screened for positive GolSI inserts by PCR with total genomic DNA using the 5′AOX1 and 3′AOX1 primers.
The positive GS115-pPIC9K-GolSI colony was grown at 28 °C and 250 r.p.m. in minimal dextrose medium overnight, and then for 4 d in minimal methanol induction medium with the addition of 500 μl (0.5%, v/v) of methanol each day. On d 4, the culture was centrifuged and the supernatant containing the secreted proteins was concentrated using an Amicon Ultra-15 filter device (Millipore, Billerica, MA) and partially purified by size-exclusion chromatography using an Econo-Pac 10DG desalting column (Bio-Rad Laboratories Canada Ltd., Mississauga, ON). The desalting buffer contained 50 mM Hepes (pH 7), 10 mM DTT and 0.2% (v/v) BSA. The desalted enzyme solution was used for all enzyme kinetic assays.
The GolSII gene was similarly amplified by PCR to include the SnaBI and AvrII restriction sites into the amplicon, and a 6×His tag at the 3′ end using the primers P39GolSIISnaBI (5′-TTCTACGTAATGGCTCCTCATATTACAACTACCCTTGCTAAC-3′) and P39GolSIIAvRIIhist (5′-TGGCCTAGGCTAATGATGATGATGATGATGAGCGGCGGATGG-3′) (restriction sites and 6×His tag underlined). The PCR product was cloned as for GolSI and ligated in frame with the α-factor of the pPIC9K vector. Pichia pastoris GS115 was transformed with the construct as described previously for GolSI.
Enzyme assays and kinetics
Enzyme assays were based on the method of Ribeiro et al. (2000), with minor modifications: the reaction contained 30 μl GolS desalted enzyme solution, with either 50 mM Hepes/NaCl, Mes, or Tris to achieve different pHs (5.5–7.5), 50 mM myo-inositol, 3 mM DTT, 2 mM MnCl2, and 10 mM UDP-galactose. The reaction was allowed to proceed overnight at various temperatures (30–60 °C) and terminated by placing the tubes in boiling water for 2 min. To each tube, 500 μl water, 10 μl potato apyrase (0.3 U; Sigma, St Louis, MO), 150 μl apyrase reaction mixture [250 mM Tris/HCl (pH 7.5), 25 mM KCl, 7.5 mM CaCl2, 0.5 mM Na2EDTA, and 50 mM glucose] was added and the tube was incubated for 10 min at 37 °C. The apyrase reaction was stopped by the addition of 60 μl 75% (v/v) trichloroacetic acid. The tubes were cooled on ice for 10 min, centrifuged at 3000 g for 10 min and the amount of Pi in the supernatant was determined using a method modified from that of Fiske and Subbarow (1925). Briefly, to each tube was added 100 μl 2.5% (w/v) ammonium molybdate dissolved in 2 M HCl and 100 μl Fiske–Subbarow reducer, and after 2 min (at room temperature) 40 μl 34% (w/v) sodium citrate.2H2O solution was added and the absorbance was measured at 660 nm. The steady-state kinetics (Vmax and Km) were calculated using the Michaelis–Menten non-linear regression model of GraphPad Prism 5.00 program (GraphPad, San Diego, CA). A standard curve was constructed measuring the Pi from KH2PO4.
Expression profile of GolS isoforms
Hybrid poplar trees were planted in the University of British Columbia (Vancouver, BC, Canada) greenhouse in July 2006. After 1 year of growth, they were moved outdoors to experience seasonal conditions, and samples were collected monthly from September 2007 to August 2008. All sampling was done before sunrise between 5:30 and 7:30 a.m. Leaf, phloem (including bark and phloem tissue), lateral apical, and young stem were cut and immediately placed in liquid nitrogen and stored at –80 °C until further use. Total RNA was isolated following the method of Kolosova et al. (2004). A TURBO DNA-free kit (Ambion Inc., Life Technologies Corp., Carlsbad, CA) was used to eliminate contaminating DNA. RNA treated with DNase was quantified and 1 μg was used to generate cDNA using an iScript cDNA Synthesis kit (Bio-Rad). The resulting cDNA was stored at –20 ° C. The same trees were used for diurnal profiling: the samples were taken hourly in July 2009 and cDNA was generated according to the protocol described previously.
Reverse transcriptase PCR
PCR reactions contained 1 μl cDNA, 10 pmol forward and reverse primers, 2mM dNTPs, and 1 μl Taq DNA polymerase with Thermo buffer (New England Biolabs, Ipswich, MA) in a total reaction volume of 20 μl. The primer sequences were as described by Philippe et al. (2010) and corresponded to PtGolS1.2 (same clade as Pa×gGolSI), PtGolS3.1 (homologue of Pa×gGolSII), and PtGolS6.1 (homologue of Pa×gGolSIII). PCR was performed using a PTC200 thermocycler (MJ Research, Waltham, MA), with the following conditions: 94 °C for 4 min, followed by 35 cycles of 94 °C for 30 s, 58, 55 or 60 °C (respectively, for the above transcripts) for 1 min and 72 °C for 1 min. An aliquot of 12 μl of each reaction was run on a 2% (w/v) agarose gel in TAE buffer. Gels were visualized by ethidium bromide staining and captured using an AlphaImager 2200 (Alpha Inotech, San Leandro, CA).
Quantitative real-time PCR
Quantitative real-time PCR reactions consisted of 10 μl SsoFast EvaGreen Supermix (Bio-Rad), 5 pmol forward and reverse primers for PtGolS1.2, PtGolS3.1, PtGolS6.1 (described above), and TIF5A genes, 1 μl cDNA (1:5 dilution), and water to a total volume of 20 μl. RT-PCR was performed using a CFX96 Real-Time PCR Detection System (Bio-Rad). The following thermal cycler conditions were used to amplify the 92, 186, and 110 bp fragments of the PtGolS1.2, PtGolS3.1, PtGolS6.1 transcripts, respectively: one cycle of 30 s at 95 °C, 39 cycles of 95 ° C for 5 s, 58, 55 or 60 °C for 5 s (respectively, for the above transcripts), followed by one cycle of 55 to 95 ° C increase for 5 s to calculate the melting curve. Relative expression was calculated using TIF5A as a reference gene (Coleman et al., 2009).
GenBank accession numbers
The sequences of Pa×gGolSI and Pa×gGolSII determined in this study have been deposited in GenBank under accession numbers JF499886 and JF499887, respectively.
Results
Characterization and phylogeny of the GolS proteins
The full-length sequences of two GolS homologues from hybrid poplar (Populus alba×grandidentata) were isolated, cloned, and subsequently named Pa×gGolSI and Pa×gGolSII. The complete coding region of Pa×gGolSI comprised 1014 nt and included an open reading frame encoding a polypeptide of 337 aa. The second gene, Pa×gGolSII, encoded a polypeptide of the same length. The deduced amino acid sequences were aligned to compare consensus regions with other known GolS proteins (Arabidopsis thaliana GolS1 and -5, Oryza sativa GolS1, Cucumis sativus GolS1 and Ajuga reptans GolS1 and -2), which display characteristics common to known GolS proteins (Fig. 2). The MAFFT program was used to align the sequences and the neighbour-joining method was used for phylogenetic analysis. A bootstrap value of 70 was used to define clades. The nine poplar GolS isoforms were positioned in four groups (Fig. 3A). The first group contained Populus trichocarpa GolS3 (PtGolS3; homologue of Pa×gGolSII), which had 89% similarity with PtGolS5. The second group was formed by PtGolS8 and PtGolS9, which shared 88% similarity. The third group was formed by PtGolS7 and PtGolS6, which shared 93% similarity. Finally, PtGolS2 (homologue of Pa×gGolSI) had 92% similarity to PtGolS1 and was found in the fourth group together with PtGolS1. The PtGolS4 gene was not included in the analysis as it is a tandem repeat of PtGolS3 (Philippe et al., 2010).
Fig. 2.
Deduced amino acid sequence alignment of known GolS proteins from Arabidopsis thaliana (AtGolS1 and -5), Ajuga reptans (ArGolS1 and -2), Oryza sativa (OsGolS1), Cucumis melo (CmGolS1) and the two GolS proteins isolated from the P. alba×grandidentata hybrid poplar (Pa×gGolSI and Pa×gGolSII). There is a conserved putative serine phosphorylation site (S) at position 274, and the characteristic hydrophobic pentapeptide (APSAA) is located at the end of the sequence. Identical amino acids are shaded in black and similar residues are shaded in grey.
A second tree grouping all known and putative GolS proteins (Fig. 3B) clustered the proteins into eight main clades. The first clade grouped PtGolS3 and -5 with the hybrid poplar Pa×gGolSII. The second clade included PtGolS8 and -9. The GolS of Cucumis melo (CmGolS1) had a low bootstrap value and could not be considered part of this clade. The third group was formed by ThGolS1 (Thellungiella halophile) and AtGolS2 and -3 (Arabidopsis thaliana). The fourth clade was formed by GmGolS (G. max), AmGolS1 (Ammopiptanthus mongolicus), and MsGolS1 (Medicago sativa), and this group shared 81–85% similarity. ArGolS2 (Ajuga reptans) demonstrated a rather low bootstrap value and was not considered part of this clade. The fifth clade was formed by PtGolS6 and -7, PsGolS1 (Pisum sativum), AtGolS1, and BnGolS1 (Brassica napus). The highest similarity within this group was 93% (the two Populus proteins). The sixth clade was formed primarily by three monocot species, Zea mays (ZmGolS1, -2, and -3 with similarities of 74–90%), Triticum aestivum (TaGolS1 and -2 with 97% similarity) and Oryza sativa (OsGolS1 and OSGolS3). XvGolS1 (Xerophyta viscosa) had a low bootstrap value and was not consider part of this clade. The seventh clade grouped PtGolS1 and -2, Pa×gGolSI, ArGolSI, and AtGolS4 and -7. Pa×gGolSI and ArGolSI shared 80% similarity, while the Arabidopsis proteins shared 72% similarity. Finally, AtGolS5 and -6 clustered in the last clade with 90% similarity. LeGolS1 (Lycopersicon esculentum) stood alone and was used as the root of the tree.
Heterologous expression in Pichia pastoris
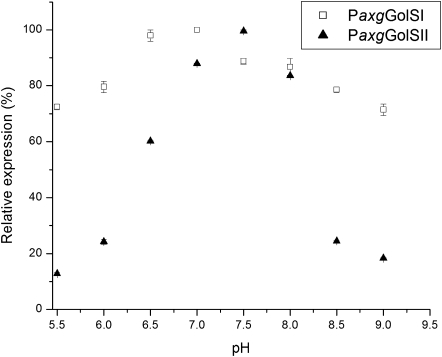
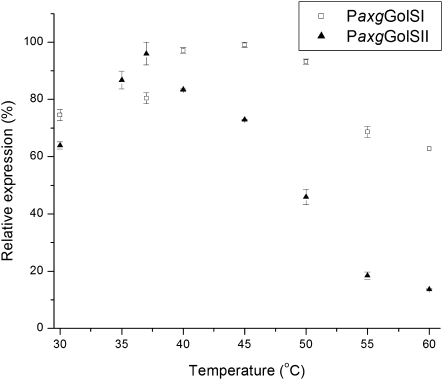
GolSI and GolSII from hybrid poplar were cloned and expressed in the methylotrophic yeast Pichia pastoris GS115. Both isoforms were able to catalyse the formation of galactinol from UDP-galactose and myo-inositol. GolSI displayed a greater pH stability, ranging from 5.5 to 9.0 with an optimum of 7, and losing only a small percentage of its original activity at the extreme pH (Fig. 4). In contrast, GolSII displayed a very narrow pH tolerance, with activity decreasing considerably when the pH deviated from the 7.5 optimum (Fig. 4). A similar pattern was apparent when the enzyme activity was measured at different temperatures: GolSI was more stable at different temperatures than GolSII, showing maximal activity at 45 and 37 °C, respectively (Fig. 5).
Fig. 4.
Enzymatic activity of the recombinant Pa×gGolSI (homologue of PtGolS2) and Pa×gGolSII (homologue of PtGolS3) in response to pH at 37.5 °C. Relative expression (%) was determined colorimetrically (Ribeiro et al., 2000). Error bars correspond to the standard error of the mean.
Fig. 5.
Enzymatic activity of the recombinant Pa×gGolSI (homologue of PtGolS2) and Pa×gGolSII (homologue of PtGolS3) at different temperatures at optimal pH. Relative expression (%) was determined colorimetrically (Ribeiro et al., 2000). Error bars correspond to the standard error of the mean.
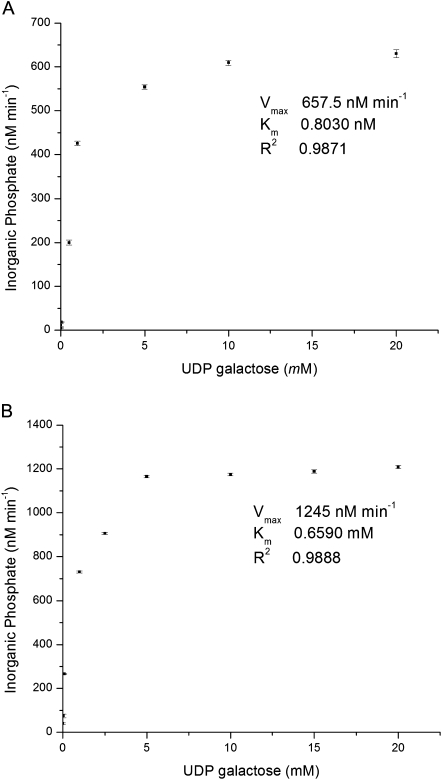
The Vmax and Km values of GolSI for UDP-galactose were 657.5 nM min−1 and 0.80 mM, while GolSII had Vmax and Km values of 1245 nM min−1 and 0.65 mM for the same substrate under their optimum enzyme conditions (Fig. 6). The Vmax, Km, optimal pH, and temperature profiles determined in this study were similar to those reported previously from other plants (Bachmann et al., 1994; Liu et al., 1995; Ribeiro et al., 2000; Wakiuchi et al., 2003).
Fig. 6.
Vmax and Km values for the recombinant GolS enzymes Pa×gGolSI (A) and Pa×gGolSII (B). Error bars represent the standard error of the mean (n=6 and n=4, respectively).
Spatial and temporal expression profile of GolS isoforms
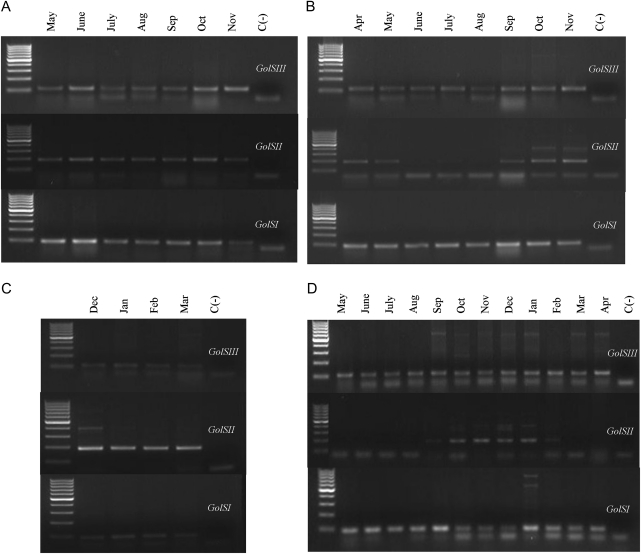
Semi-quantitative and quantitative real-time PCR was used to investigate the expression of three hybrid poplar GolS isoforms, representing three out of four clades containing poplar GolS proteins. Pa×gGolSIII, which is homologous to PtGolS6 (LGII: 15155825–15157384; Philippe et al., 2010) was expressed in source leaves sampled from May to November, showing a slightly higher level of expression during the autumn months. Pa×gGolSII, which is homologous to PtGolS3 (LGXIII: 362675–364038; Philippe et al., 2010), and Pa×gGolSI, which was in the same clade as PtGolS1 (LGVIII: 13321952–13323592; Philippe et al., 2010), were also expressed during these same months. However, the latter showed higher overall expression levels (Fig. 7A). In contrast, in the lateral apical tissue, which includes the apical meristem, small sink leaves and the three first internodes of the stem, differences in expression of the second homologue (Pa×gGolSII) were detected. Specifically, this homologue was expressed during both spring and autumn but was not detectable during the summer. Additionally, of the three genes, Pa×gGolSI had the highest expression level during this same time period (Fig. 7B).
Fig. 7.
Annual expression profiles of hybrid poplar GolS isoforms in different tissues over the course of 1 year of growth. (A) Source leaf, (B) lateral apical stem (first three nodes), (C) branch tip (without buds), (D) phloem. Primers used in the real-time PCRs amplified one member of the following three clades (based on Fig. 3A): clade a Pa×gGolSII, clade c Pa×gGolSIII and clade d Pa×gGolSI. Water was used as the negative control [C(-)].
The profile in the stem (apex without buds) during the winter period showed the highest expression of Pa×gGolSII compared with the other GolS genes surveyed (Fig. 7C). Finally, in the phloem, the expression of Pa×gGolSIII and Pa×gGolSI was similar and consistent for the entire annual cycle, while the expression of Pa×gGolSII was limited to the winter months (Fig. 7D).
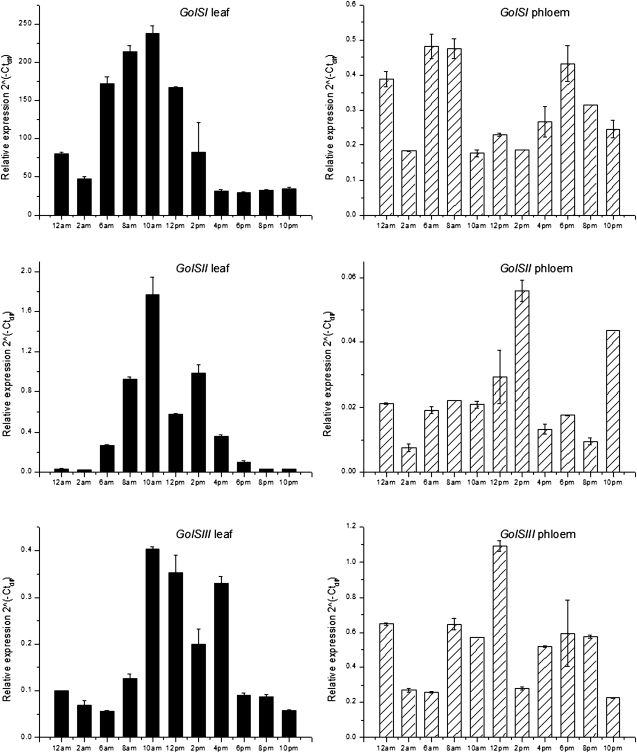
The relative expression levels of the three isoforms were also analysed over the course of a full diurnal cycle to investigate daily changes in expression using quantitative real-time PCR. Two tissues were analysed for this purpose: source leaf and phloem. The levels of the three GolS transcripts in leaves started to increase in the early morning before reaching maximum levels at around 10 a.m. Thereafter, expression decreased in the afternoon to reach a low in the evening (Fig. 8). It was also apparent that expression of the Pa×gGolSI isoform in source leaves was much higher than expression of the other two isoforms (Pa×gGolSII and -III, with differences at the highest relative expression of 139- and 590-fold, respectively). Generally, expression of the transcripts in the phloem was lower than the expression on the leaf for all isoforms during the summer.
Fig. 8.
Diurnal expression profiles of three GolS isoforms in hybrid poplar (P. alba×grandidentata) leaf and phloem tissue. Error bars correspond to standard error of the mean, based on three replicates.
Discussion
GolS proteins from hybrid poplar
The main goal of this study was to identify and characterize the GolS genes from hybrid poplar. Using the Populus trichocarpa genome sequence (Tuskan et al., 2006), nine putative GolS genes were identified. Based on the predicted amino acid sequence, the nine putative GolS genes appear to have evolved from four ancestral genes through genome duplication (Philippe et al., 2010).
Two homologues of the PtGolS genes were cloned from a hybrid poplar (Populus alba×grandidentata) clone. Pa×gGolSI shared 97% similarity at the amino acid level with its homologue PtGolS2, while the second isoform, Pa×gGolSII, had 91.7% similarity to PtGolS3. The predicted protein sequences of both cloned isoforms clearly showed the serine phosphorylation site at position 274 and the ASAAP pentapeptide hydrophobic domain characteristic of all known GolS proteins (Sprenger and Keller, 2000). Furthermore, the predicted isoelectric points for both enzymes were 5.27 and 5.48, respectively, which are similar to that of the GolS purified from zucchini leaves (Smith et al., 1991) and characteristic of other plant GolS proteins.
To confirm the identity of the putative Pa×gGolS isoforms and characterize the enzymatic properties of the proteins, the cloned genes were expressed heterologously in Pichia pastoris. The Pichia pastoris system represents a very useful experimental tool for protein research, as it can produce sufficient amounts of the recombinant protein(s) in soluble form, and offers correct folding and appropriately post-translational modification (Daly and Hearn, 2005). The recombinant GolS Vmax, Km, and optimal pH and temperature profiles were similar to those reported previously from other plants. For example, Smith et al. (1991) determined a pH optimum of 7.5 for purified GolS isolated from mature zucchini (Cucurbita pepo) leaves. In addition, the purified GolS was shown to bind specifically to UDP-galactose (Km= 1.8 mM). Phaseolus vulgaris (kidney beans) GolS has also been purified and characterized (Liu et al., 1995), and was shown to have a pH optimum of 7.0 and a Km of 0.4 mM for UDP-galactose. A crude and partially purified soybean seed GolS activity was shown to exhibit a maximal activity at pH 7.0 and 50 °C. This enzyme showed Km and Vmax values (for UDP-galactose) of 5.2 mM and 195 nmol min−1, respectively (Ribeiro et al., 2000). The Cucumis sativus GolS extracted from leaves has similarly been shown to have an optimal pH of 7–8 and highest activity at 40 °C (Wakiuchi et al., 2003). Based on our results, it was concluded that the two hybrid poplar isoforms encoded true GolS proteins, and functionally displayed similar properties to other known GolS proteins.
Phylogenetic analysis
The predicted amino acid sequences of Pa×gGolS, PtGolS proteins and other confirmed and putative GolS proteins from dicotyledonous and monocotyledonous plant species were used to perform a phylogenetic analysis using the neighbour-joining method. The results showed that the nine GolS proteins from Populus trichocarpa and the seven GolS proteins from Arabidopsis thaliana were distributed among all the clades, demonstrating as much variation within as among species. The Populus trichocarpa GolS proteins were positioned in pairs in four clades, following the pattern of genome duplication of Populus (Tuskan et al., 2006). The two cloned isoforms from hybrid poplar were representative of two different clades. Pa×gGolSI and -II were homologues of PtGolS2 and -3, respectively, with 66% similarity at the amino acid level.
Many GolS genes have been shown to be stress related. For example, the PtGolS genes were recently shown to be highly responsive to biotic stress and showed differential induction with gene-specific patterns in source and sink leaves (Philippe et al., 2010). Similarly, three of the seven Arabidopsis thaliana GolS proteins (GolS1, -2, and -3) were shown previously to be stress-responsive genes (Taji et al., 2002).
The ArGolS1 and -2 genes of Ajuga reptans were also distributed in different clades and have been proposed to possess inherently distinct biological functions (Sprenger and Keller, 2000). ArGolS1 (grouped with Pa×gGolSI) was predicted to be involved in the synthesis of storage RFOs, while ArGolS2 was implicated in the synthesis and transport of RFOs. The GolS from Cucumis melo (with 74% similarity with Pa×gGolSII) has been shown to be present in developing melon seeds during the active formation of raffinose and stachyose (Volk et al., 2003). Additionally, the GolS promoter isolated from Cucumis melo has been shown to drive gene expression in the minor-vein companion cells of both transgenic tobacco (Nicotiana tabacum) and Arabidopsis. Neither of these plants employs galactinol in a phloem-loading process, suggesting that the promoter responds to a minor-vein-specific regulatory cascade that is highly conserved across a broad range of eudicotyledons (Ayre et al., 2003). This is an interesting observation, as this gene was predicted previously to cluster phylogenetically (Zhao et al., 2004) with the Curcubita pepo GolS that is localized in the intermediary cells (Beebe and Turgeon, 1992). Based on phylogenetic clustering and the functional data available, it is indeed possible that the two GolS proteins from hybrid poplar have different biological roles.
The G. max, Ammopiptanthus mongolicus and Medicago sativa GolS proteins clustered together in our analysis. These genes have been shown to be strongly associated with stress tolerance. In Ammopiptanthus mongolicus, a woody species, only one form of the GolS gene (AmGolS) has been found, which was induced significantly by cold, drought, NaCl, and abscisic acid application (Cao et al., 2009). Similarly, the transcript induction of MsGolS of Medicago sativa was higher in winter-hardy alfalfa cultivars (Cunningham et al., 2003).
The GolS from Thellungiella halophile (ThGolS) was found to group together with two genes from Arabidopsis thaliana (ArGolS2 and -3). Similar to the AtGolS genes, which have been shown to be stress related, ThGolS is suggested to be a putative salinity stress-regulated gene (Wang et al., 2004). Among the dicot species, the LeGolS1 (L. esculentum) amino acid sequence was the most divergent, and, interestingly, expression of this transcript was shown to be induced by dehydration in germinating seeds and by dehydration coupled with cold in seedling leaves (Downie et al., 2003). This may suggest a seed-specific role for some GolS isoforms in plant growth and development.
The similarity of the GolS proteins from monocot species was evident from the close association observed in the cluster analysis. In particular, the cluster contained three maize GolS proteins that have been studied during seed development and germination. Two of the three GolS gene family members in maize have also been detected in stressed seeds, while the other GolS was expressed mainly during seed development (Zhao et al., 2004). Another member of this tightly associated clade is the rice GolS, which was found to be expressed under chilling conditions (Takahashi et al., 1994). Additionally, the X. viscosa GolS gene showed an increase in expression in leaf tissue experiencing water deficit (Peters, 2007).
Based on the phylogenetic relationships of the two hybrid poplar GolS isolates and the GolS proteins from other species, we suggest a potential role of these enzymes during biotic or abiotic stresses in hybrid poplar. Furthermore, the distinct grouping of these isoforms implies different roles for these two GolS proteins.
Expression profiles
The raffinose family of oligosaccharides have long been suggested to act as biological agents to combat abiotic (Taji et al., 2002; Cunningham et al., 2003; Panikulangara et al., 2004) and biotic (Philippe et al., 2010) stress. The three different GolS isoforms analysed in this study are homologues of genes that belong to different clades of the Populus trichocarpa GolS proteins. The Pa×gGolSII homologue (PtGolS3) showed seasonal expression and seemed to be temperature regulated based on the abundance of the transcript during the spring and autumn in the shoot tip along with elevated expression during the winter in the phloem. Additionally, expression in branch tips during winter was higher than that of the other genes. The expression differences during day and night in source leaf poplar revealed that the transcript abundance was much higher during the day, reaching a maximum near 10 a.m. and then decreasing in the afternoon, reaching its lowest level in the evening. Similar patterns were also found in Ajuga reptans, where the GolS activity extracted from pre-dawn leaves represented only 30–50% of the GolS activity usually extracted during the day. Bachmann et al. (1994) hypothesized that GolS proteins may obey a light/dark regulation by protein modification similar to sucrose phosphate synthase (Huber, 1983).
Expression of the Pa×gGolSI homologue was constitutively higher in source leaves compared with the other two GolS proteins. Interestingly, the same isoform was present throughout the year in most of the tissues, and it seems that its expression was not altered by cold acclimation. Additionally, when the kinetics of the enzyme was analysed at different pHs and temperatures, the activity was more stable over a wide range of conditions compared with the Pa×gGolSII isoform, which displayed a very narrow temperature tolerance, suggesting indirectly that this enzyme may be highly temperature related. Overall, the variation in seasonal expression of the Pa×gGolS isoforms suggests that each enzyme may play a different physiological role. The persistent expression of the Pa×gGolSI transcript throughout the year could be associated with the synthesis of RFOs for storage, while Pa×gGolSII may be involved with seasonal mobilization of carbohydrates. However, additional studies of the GolS proteins are required to elucidate further the specific roles of this important enzyme group in phloem transport and storage in poplar.
Acknowledgments
The authors acknowledge financial support from the Natural Science and Engineering Research Council of Canada (NSERC) held by SDM.
Glossary
Abbreviations
- DTT
dithiothreitol
- GolS
galactinol synthase
- RFOs
raffinose family of oligosaccharides
References
- Ayre BG, Blair JE, Turgeon R. Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiology. 2003;133:1229–1239. doi: 10.1104/pp.103.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann M, Matile P, Keller F. Metabolism of the raffinose family oligosaccharides in leaves of Ajuga reptans L. (cold acclimation, translocation, and sink to source transition: discovery of chain elongation enzyme) Plant Physiology. 1994;105:1335–1345. doi: 10.1104/pp.105.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe DU, Turgeon R. Localization of galactinol, raffinose, and stachyose synthesis in Cucurbita pepo leaves. Planta. 1992;188:354–361. doi: 10.1007/BF00192802. [DOI] [PubMed] [Google Scholar]
- Cao P, Song J, Zhou C, Weng M, Liu J, Wang F, Zhao F, Feng D, Wang B. Characterization of multiple cold induced genes from Ammopiptanthus mongolicus and functional analyses of gene. AmEBP1. Plant Molecular Biology. 2009;69:529–539. doi: 10.1007/s11103-008-9434-1. [DOI] [PubMed] [Google Scholar]
- Coleman HD, Yan J, Mansfield SD. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proceedings of the National Academy of Sciences, USA. 2009;106:13118–13123. doi: 10.1073/pnas.0900188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox SE, Stushnoff C. Temperature-related shifts in soluble carbohydrate content during dormancy and cold acclimation in Populus tremuloides. Canadian Journal of Forest Research. 2001;31:730–737. [Google Scholar]
- Cunningham SM, Nadeau P, Castonguay Y, Laberge S, Volenec JJ. Raffinose and stachyose accumulation, galactinol synthase expression, and winter injury of contrasting alfalfa germplasms. Crop Science. 2003;43:562–570. [Google Scholar]
- Daly R, Hearn MTW. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. Journal of Molecular Recognition. 2005;18:119–138. doi: 10.1002/jmr.687. [DOI] [PubMed] [Google Scholar]
- Downie B, Gurusinghe S, Dahal P, Thacker RR, Snyder JC, Nonogaki H, Yim K, Fukanaga K, Alvarado V, Bradford KJ. Expression of a GALACTINOL SYNTHASE gene in tomato seeds is up-regulated before maturation desiccation and again after imbibition whenever radicle protrusion is prevented. Plant Physiology. 2003;131:1347–1359. doi: 10.1104/pp.016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fiske CH, Subbarow Y. The colorimetric determination of phosphorus. J Biol Chem. 1925;66:375–400. [Google Scholar]
- Huber SC. Role of sucrose-phosphate synthase in partitioning of carbon in leaves. Plant Physiology. 1983;71:818–821. doi: 10.1104/pp.71.4.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Miller B, Ralph S, Ellis BE, Douglas C, Ritland K, Bohlmann J. Isolation of high-quality RNA from gymnosperm and angiosperm trees. BioTechniques. 2004;36:821–824. doi: 10.2144/04365ST06. [DOI] [PubMed] [Google Scholar]
- Keller F, Pharr DM. Metabolism of carbohydrates in sinks and sources: galactosyl-sucrose oligosaccharides. In: Zamski E, Schaffer AA, editors. Photoassimilate Distribution in Plants and Crops. Marcel Dekker New York: 1996. pp. 115–184. [Google Scholar]
- Liu JJ, Odegard W, Delumen BO. Galactinol synthase from kidney bean cotyledon and zucchini leaf – purification and N-terminal sequences. Plant Physiology. 1995;109:505–511. doi: 10.1104/pp.109.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikulangara TJ, Eggers-Schumacher G, Wunderlich M, Stransky H, Schoffl F. Galactinol synthase1. A novel heat shock factor target gene responsible for heat-induced synthesis of raffinose family oligosaccharides in Arabidopsis. Plant Physiology. 2004;136:3148–3158. doi: 10.1104/pp.104.042606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S. Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. Journal of Experimental Botany. 2007;58:1947. doi: 10.1093/jxb/erm056. [DOI] [PubMed] [Google Scholar]
- Philippe RN, Ralph SG, Mansfield SD, Bohlmann J. Transcriptome profiles of hybrid poplar (Populus trichocarpa × deltoides) reveal rapid changes in undamaged, systemic sink leaves after simulated feeding by forest tent caterpillar (Malacosoma disstria) New Phytol. 2010;188:787–802. doi: 10.1111/j.1469-8137.2010.03392.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro M, Felix CR, De Paulino Lozzi S. Soybean seed galactinol synthase activity as determined by a novel colorimetric assay. Revista Brasileira de Fisiologia Vegetal. 2000;12:203–212. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saravitz DM, Pharr DM, Carter TE. Galactinol synthase activity and soluble sugars in developing seeds of four soybean genotypes. Plant Physiology. 1987;83:185–189. doi: 10.1104/pp.83.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PT, Kuo TM, Crawford CG. Purification and characterization of galactinol synthase from mature zucchini squash leaves. Plant Physiology. 1991;96:693–698. doi: 10.1104/pp.96.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N, Keller F. Allocation of raffinose family oligosaccharides to transport and storage pools in Ajuga reptans: the roles of two distinct galactinol synthases. The Plant Journal. 2000;21:249–258. doi: 10.1046/j.1365-313x.2000.00671.x. [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. The Plant Journal. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Joshee N, Kitagawa Y. Induction of chilling resistance by water stress, and cDNA sequence-analysis and expression of water stress-regulated genes in rice. Plant Molecular Biology. 1994;26:339–352. doi: 10.1007/BF00039544. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011 doi: 10.1093/molbev/msr121. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Volk GM, Haritatos EE, Turgeon R. Galactinol synthase gene expression in melon. Journal of the American Society for Horticultural Science. 2003;128:8–15. [Google Scholar]
- Wakiuchi N, Shiomi R, Tamaki H. Production of galactinol from sucrose by plant enzymes. Bioscience Biotechnology and Biochemistry. 2003;67:1465–1471. doi: 10.1271/bbb.67.1465. [DOI] [PubMed] [Google Scholar]
- Wang ZI, Li PH, Fredricksen M, Gong ZH, et al. Expressed sequence tags from Thellungiella halophila, a new model to study plant salt-tolerance. Plant Science. 2004;166:609–616. [Google Scholar]
- Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM. Genotype and time of day shape the Populus drought response. The Plant Journal. 2009;60:703–715. doi: 10.1111/j.1365-313X.2009.03993.x. [DOI] [PubMed] [Google Scholar]
- Zhao TY, Thacker R, Corum JW, Snyder JC, Meeley RB, Obendorf RL, Downie B. Expression of the maize GALACTINOL SYNTHASE gene family: (I) Expression of two different genes during seed development and germination. Physiologia Plantarum. 2004;121:634–646. [Google Scholar]
- Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. Academic Press New York; 1965. pp. 97–166. [Google Scholar]