Abstract
The title compound {systematic name: (6aR,9S)-N-[(2R,5S,10aS,10bS)-5-benzyl-10b-hydroxy-2-methyl-3,6-dioxooctahydro-8H-oxazolo[3,2-a]pyrrolo[2,1-c]pyrazin-2-yl]-7-methyl-4,6,6a,7,8,9-hexahydroindolo[4,3-fg]quinoline-9-carboxamide}, C33H35N5O5, was formed by an epimerization reaction of ergotamine. The non-aromatic ring (ring C of the ergoline skeleton) directly fused to the aromatic rings is nearly planar [maximum deviation = 0.317 (4) Å] and shows an envelope conformation, whereas ring D, involved in an intramolecular N—H⋯N hydrogen bond exhibits a slightly distorted chair conformation. The structure displays chains running approximately parallel to the diagonal of bc plane that are formed through N—H⋯O hydrogen bonds.
Related literature
Ergotaminine is an ergot alkaloid formed by, among others, the fungus Claviceps purpurea on cereal grains and grasses during the growth process; see: Crews et al. (2009 ▶); Müller et al. (2009 ▶). For investigations of the biologically inactive C8-(S)-isomer ergotaminine, see: Pierri et al. (1982 ▶); Komarova & Tolkachev (2001 ▶). For the crystal structure of ergotamine tartrate ethanol solvate, see: Pakhomova et al. (1995 ▶). For the crystal structure of ergometrinine, another C8-(S)-configured ergotalkaloid, see: Merkel et al. (2010 ▶). For the solubility of ergotaminine, see: Stoll (1945 ▶).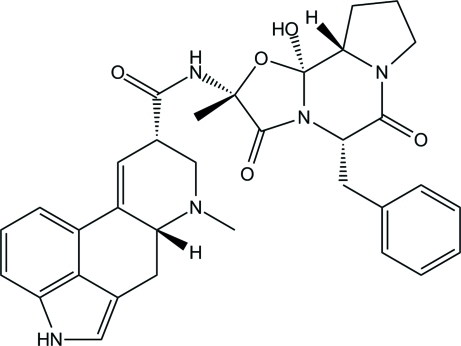
Experimental
Crystal data
C33H35N5O5
M r = 581.66
Monoclinic,

a = 10.974 (3) Å
b = 9.662 (2) Å
c = 14.450 (4) Å
β = 105.059 (15)°
V = 1479.5 (7) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 296 K
0.2 × 0.1 × 0.06 mm
Data collection
Bruker APEX CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2001 ▶) T min = 0.879, T max = 0.986
20196 measured reflections
2781 independent reflections
2240 reflections with I > 2σ(I)
R int = 0.087
Refinement
R[F 2 > 2σ(F 2)] = 0.047
wR(F 2) = 0.093
S = 1.12
2781 reflections
390 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.15 e Å−3
Δρmin = −0.19 e Å−3
Absolute structure: determined from the synthesis
Data collection: SMART (Bruker, 2001 ▶); cell refinement: SAINT (Bruker, 2001 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶) and ORTEPIII (Burnett & Johnson, 1996 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812003674/ds2173sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812003674/ds2173Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812003674/ds2173Isup3.mol
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯N3 | 0.86 | 2.53 | 2.955 (4) | 112 |
| N4—H3⋯O5i | 0.86 | 2.17 | 2.981 (5) | 157 |
Symmetry code: (i)  .
.
supplementary crystallographic information
Comment
The fungus Claviceps purpurea is distributed worldwide through various climatic zones and produces a broad range of ergot alkaloids on grasses and cereal grains during the growth process whereas six epimeric pairs are predominantly formed. One of these main ergot alkaloids is ergotaminine. Contamination of flour and cereal based foods with ergot alkaloids including ergotaminine has previously been reported (Crews et al., 2009; Müller et al., 2009). The biologically inactive C8-(S)-isomer ergotaminine (Pierri et al., 1982) can be converted to the biologically active C8-(R)-isomer ergotamine and vice versa (Komarova & Tolkachev, 2001). The molecule crystallizes in the monoclinic space group P21. The molecular structure of the compound and the atom-labeling scheme are shown in Fig. 1. The absolute configuration could not be defined confidently based on the single-crystal diffraction data. It was however established based on liquid chromatography data that confirmed the epimeric purity of the obtained ergotaminine crystals. Besides the intramolecular hydrogen bonds between N2—H2 and N3 (see Table 1; not shown in Fig. 2), each molecule is connected to two adjacent molecules via intermolecular hydrogen bonds (see Table 1; see dashed green bonds in Fig. 2). As a result adjacent chains run along the [011] and [011] direction in an oppositely slanted fashion and with an inlined angle of 69.4°.
Experimental
Ergotamine tartrate was obtained from Sigma–Aldrich (Taufkirchen, Germany). The stereoselective conversion of ergotamine to ergotaminine was carried out as follows: 12.4 mg ergotamine tartrate were dissolved in a solution of 5 ml methanol and 0.5 ml water. For epimerization reaction the resulting mixture was stored in a sealed vial in darkness at ambient temperature for two weeks. As a result of the slow crystallization colorless crystals of the title compound were formed, because of a substantial solubility difference between ergotamine and ergotaminine (as reported by Stoll (1945)). The isomeric purity (98%) of ergotaminine was proved by HPLC-FLD.
Refinement
In the absence of significant anomalous dispersion effects, Friedel pairs were merged.
The N—H and O—H H atoms were located in difference maps and fixed in their found positions (AFIX 3) with Uiso(H) = 1.2 of the parent atom Ueq or 1.5 Ueq(Cmethyl, O).
Figures
Fig. 1.
ORTEP representation of the title compound with atomic labeling shown with 30% probability displacement ellipsoids.
Fig. 2.
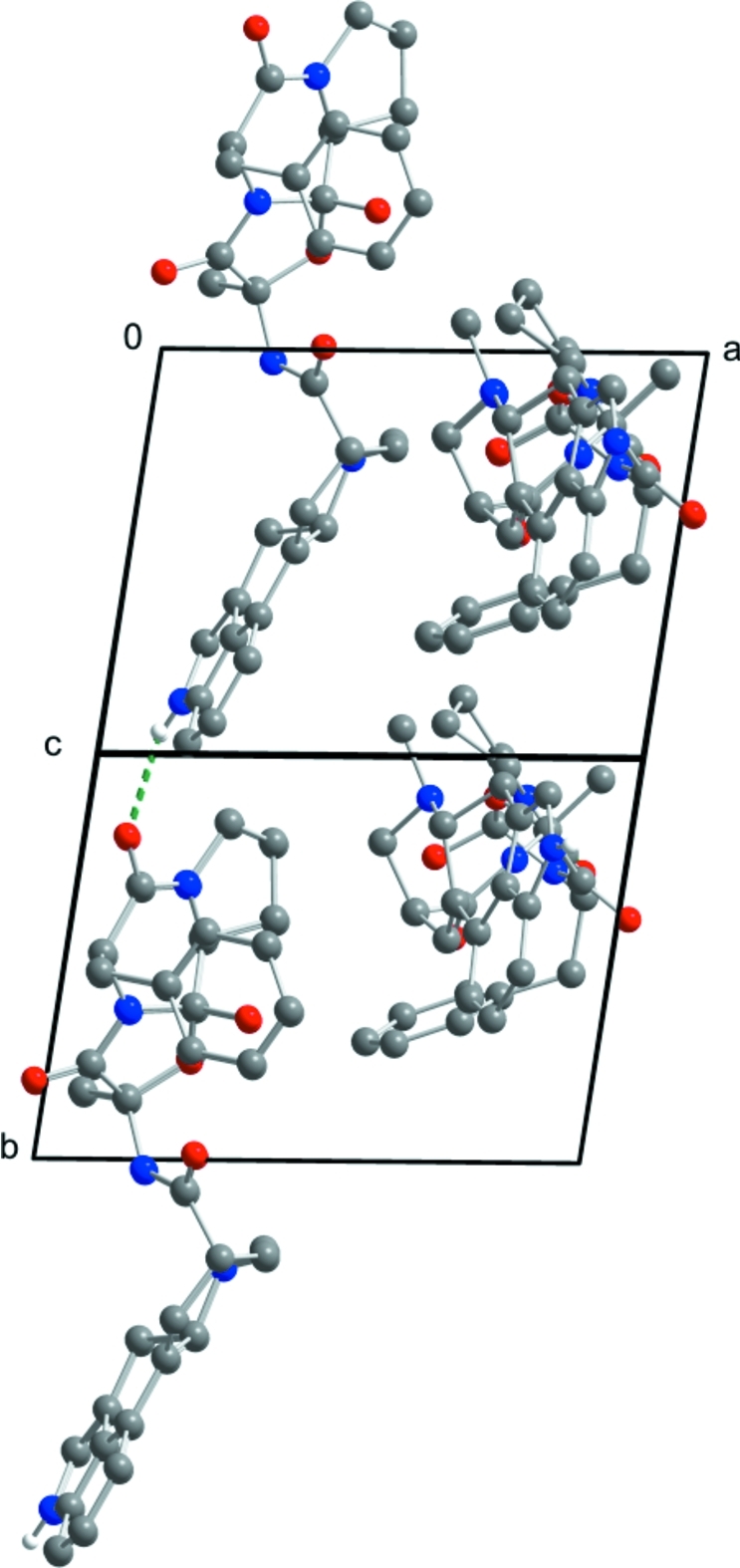
View of the unit cell of the title compound, showing the hydrogen-bonded chains are running approximately parallel to the diagonal of b-c plane. Hydrogen bonds are drawn as dashed green lines. H atoms are omitted for clarity.
Crystal data
| C33H35N5O5 | F(000) = 616 |
| Mr = 581.66 | Dx = 1.306 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2yb | Cell parameters from 86 reflections |
| a = 10.974 (3) Å | θ = 4–29° |
| b = 9.662 (2) Å | µ = 0.09 mm−1 |
| c = 14.450 (4) Å | T = 296 K |
| β = 105.059 (15)° | Plate, colourless |
| V = 1479.5 (7) Å3 | 0.2 × 0.1 × 0.06 mm |
| Z = 2 |
Data collection
| Bruker APEX CCD area-detector diffractometer | 2781 independent reflections |
| Radiation source: fine-focus sealed tube | 2240 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.087 |
| ω/2θ scans | θmax = 25.1°, θmin = 1.5° |
| Absorption correction: multi-scan (SADABS; Bruker, 2001) | h = −12→12 |
| Tmin = 0.879, Tmax = 0.986 | k = −11→11 |
| 20196 measured reflections | l = −14→17 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.047 | H-atom parameters constrained |
| wR(F2) = 0.093 | w = 1/[σ2(Fo2) + (0.0345P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.12 | (Δ/σ)max < 0.001 |
| 2781 reflections | Δρmax = 0.15 e Å−3 |
| 390 parameters | Δρmin = −0.19 e Å−3 |
| 1 restraint | Absolute structure: syn |
| Primary atom site location: structure-invariant direct methods |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.2567 (2) | 0.1765 (2) | 0.57682 (16) | 0.0564 (7) | |
| O2 | 0.3529 (2) | 0.2000 (2) | 0.45142 (16) | 0.0526 (6) | |
| H1 | 0.3525 | 0.2846 | 0.4559 | 0.079* | |
| O3 | −0.0202 (3) | 0.3535 (3) | 0.45451 (19) | 0.0753 (8) | |
| O4 | 0.2943 (3) | 0.4688 (3) | 0.52044 (19) | 0.0787 (9) | |
| O5 | 0.0781 (3) | −0.0063 (3) | 0.21039 (18) | 0.0726 (8) | |
| N1 | 0.1306 (2) | 0.2059 (3) | 0.42571 (18) | 0.0458 (7) | |
| N2 | 0.2054 (3) | 0.3898 (3) | 0.6360 (2) | 0.0589 (8) | |
| H2 | 0.1851 | 0.4118 | 0.6877 | 0.071* | |
| N3 | 0.3774 (3) | 0.4339 (3) | 0.8282 (2) | 0.0594 (8) | |
| N4 | 0.1372 (4) | 0.7970 (5) | 1.0691 (3) | 0.0918 (13) | |
| H3 | 0.1068 | 0.8344 | 1.1123 | 0.110* | |
| N5 | 0.2018 (2) | −0.0396 (3) | 0.36023 (19) | 0.0478 (7) | |
| C1 | 0.2503 (3) | 0.1457 (3) | 0.4785 (2) | 0.0438 (8) | |
| C2 | 0.0775 (3) | 0.2859 (4) | 0.4814 (3) | 0.0529 (9) | |
| C3 | 0.1537 (3) | 0.2655 (4) | 0.5853 (2) | 0.0513 (9) | |
| C4 | 0.2866 (4) | 0.4728 (4) | 0.6038 (3) | 0.0606 (10) | |
| C5 | 0.3716 (4) | 0.5658 (4) | 0.6793 (3) | 0.0626 (11) | |
| H4 | 0.4299 | 0.6130 | 0.6487 | 0.075* | |
| C6 | 0.4514 (4) | 0.4756 (4) | 0.7608 (3) | 0.0710 (12) | |
| H5 | 0.4799 | 0.3936 | 0.7338 | 0.085* | |
| H6 | 0.5253 | 0.5270 | 0.7951 | 0.085* | |
| C7 | 0.3456 (3) | 0.5530 (4) | 0.8811 (3) | 0.0563 (10) | |
| H7 | 0.4227 | 0.5813 | 0.9287 | 0.068* | |
| C8 | 0.2471 (4) | 0.5103 (4) | 0.9358 (3) | 0.0649 (11) | |
| H8 | 0.1736 | 0.4707 | 0.8913 | 0.078* | |
| H9 | 0.2830 | 0.4407 | 0.9834 | 0.078* | |
| C9 | 0.2085 (4) | 0.6349 (5) | 0.9840 (3) | 0.0637 (11) | |
| C10 | 0.1623 (4) | 0.6591 (6) | 1.0624 (3) | 0.0858 (14) | |
| H10 | 0.1499 | 0.5914 | 1.1048 | 0.103* | |
| C11 | 0.1678 (4) | 0.8663 (5) | 0.9949 (3) | 0.0732 (12) | |
| C12 | 0.2133 (3) | 0.7655 (4) | 0.9420 (3) | 0.0573 (10) | |
| C13 | 0.2523 (3) | 0.7954 (4) | 0.8593 (3) | 0.0532 (9) | |
| C14 | 0.3004 (3) | 0.6764 (4) | 0.8136 (2) | 0.0487 (9) | |
| C15 | 0.3054 (3) | 0.6754 (4) | 0.7218 (3) | 0.0548 (9) | |
| H11 | 0.2654 | 0.7467 | 0.6822 | 0.066* | |
| C16 | 0.2457 (4) | 0.9325 (4) | 0.8304 (3) | 0.0661 (11) | |
| H12 | 0.2718 | 0.9580 | 0.7765 | 0.079* | |
| C17 | 0.1988 (4) | 1.0347 (4) | 0.8837 (3) | 0.0831 (14) | |
| H13 | 0.1944 | 1.1262 | 0.8631 | 0.100* | |
| C18 | 0.1594 (4) | 1.0026 (6) | 0.9656 (3) | 0.0841 (15) | |
| H14 | 0.1286 | 1.0705 | 0.9991 | 0.101* | |
| C19 | 0.4491 (5) | 0.3282 (5) | 0.8935 (3) | 0.0904 (14) | |
| H15 | 0.4720 | 0.2545 | 0.8567 | 0.136* | |
| H16 | 0.3980 | 0.2920 | 0.9326 | 0.136* | |
| H17 | 0.5240 | 0.3690 | 0.9336 | 0.136* | |
| C20 | 0.0706 (4) | 0.1934 (5) | 0.6393 (3) | 0.0718 (11) | |
| H18 | 0.0020 | 0.2532 | 0.6426 | 0.108* | |
| H19 | 0.1195 | 0.1718 | 0.7030 | 0.108* | |
| H20 | 0.0378 | 0.1095 | 0.6066 | 0.108* | |
| C21 | 0.0681 (3) | 0.1685 (4) | 0.3258 (2) | 0.0502 (9) | |
| H21 | −0.0208 | 0.1523 | 0.3235 | 0.060* | |
| C22 | 0.1174 (3) | 0.0328 (4) | 0.2951 (3) | 0.0507 (9) | |
| C23 | 0.2471 (3) | −0.0094 (4) | 0.4643 (2) | 0.0470 (8) | |
| H22 | 0.1881 | −0.0499 | 0.4973 | 0.056* | |
| C24 | 0.3714 (4) | −0.0875 (4) | 0.4936 (3) | 0.0651 (11) | |
| H23 | 0.4400 | −0.0325 | 0.4822 | 0.078* | |
| H24 | 0.3914 | −0.1134 | 0.5607 | 0.078* | |
| C25 | 0.3467 (4) | −0.2143 (4) | 0.4293 (3) | 0.0805 (13) | |
| H25 | 0.3059 | −0.2862 | 0.4573 | 0.097* | |
| H26 | 0.4252 | −0.2505 | 0.4202 | 0.097* | |
| C26 | 0.2613 (4) | −0.1660 (4) | 0.3349 (3) | 0.0577 (10) | |
| H27 | 0.3097 | −0.1456 | 0.2891 | 0.069* | |
| H28 | 0.1984 | −0.2355 | 0.3080 | 0.069* | |
| C27 | 0.0694 (4) | 0.2867 (4) | 0.2538 (3) | 0.0658 (11) | |
| H29 | 0.0173 | 0.2584 | 0.1917 | 0.079* | |
| H30 | 0.0296 | 0.3669 | 0.2737 | 0.079* | |
| C28 | 0.1947 (4) | 0.3310 (4) | 0.2411 (2) | 0.0551 (10) | |
| C29 | 0.2512 (5) | 0.2625 (5) | 0.1781 (3) | 0.0706 (12) | |
| H31 | 0.2115 | 0.1859 | 0.1443 | 0.085* | |
| C30 | 0.3658 (5) | 0.3069 (5) | 0.1651 (3) | 0.0863 (15) | |
| H32 | 0.4021 | 0.2597 | 0.1229 | 0.104* | |
| C31 | 0.4258 (5) | 0.4192 (6) | 0.2138 (4) | 0.0921 (15) | |
| H33 | 0.5020 | 0.4494 | 0.2041 | 0.110* | |
| C32 | 0.3729 (5) | 0.4871 (5) | 0.2769 (3) | 0.0867 (13) | |
| H34 | 0.4143 | 0.5624 | 0.3113 | 0.104* | |
| C33 | 0.2586 (4) | 0.4445 (4) | 0.2899 (3) | 0.0667 (11) | |
| H35 | 0.2234 | 0.4927 | 0.3323 | 0.080* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0583 (15) | 0.0628 (17) | 0.0461 (14) | 0.0085 (13) | 0.0098 (12) | −0.0074 (12) |
| O2 | 0.0457 (13) | 0.0436 (13) | 0.0695 (15) | −0.0059 (11) | 0.0167 (12) | 0.0008 (12) |
| O3 | 0.0698 (18) | 0.077 (2) | 0.0795 (18) | 0.0295 (17) | 0.0199 (15) | −0.0057 (16) |
| O4 | 0.127 (3) | 0.0594 (17) | 0.0620 (17) | −0.0228 (17) | 0.0460 (17) | −0.0118 (14) |
| O5 | 0.0844 (19) | 0.0737 (18) | 0.0501 (15) | 0.0090 (16) | 0.0004 (14) | −0.0174 (14) |
| N1 | 0.0429 (15) | 0.0467 (16) | 0.0466 (16) | 0.0038 (14) | 0.0095 (13) | −0.0067 (14) |
| N2 | 0.078 (2) | 0.0510 (18) | 0.0569 (19) | −0.0101 (16) | 0.0347 (17) | −0.0146 (15) |
| N3 | 0.0624 (19) | 0.0521 (19) | 0.0624 (19) | 0.0094 (16) | 0.0140 (16) | −0.0056 (16) |
| N4 | 0.107 (3) | 0.116 (4) | 0.056 (2) | 0.025 (3) | 0.027 (2) | −0.015 (2) |
| N5 | 0.0486 (16) | 0.0409 (16) | 0.0509 (16) | 0.0013 (13) | 0.0077 (13) | −0.0074 (13) |
| C1 | 0.0418 (19) | 0.0470 (19) | 0.0424 (19) | −0.0020 (16) | 0.0107 (16) | −0.0030 (15) |
| C2 | 0.053 (2) | 0.046 (2) | 0.062 (2) | 0.0042 (19) | 0.0190 (19) | −0.0042 (18) |
| C3 | 0.057 (2) | 0.047 (2) | 0.053 (2) | 0.0019 (18) | 0.0218 (18) | −0.0087 (17) |
| C4 | 0.080 (3) | 0.051 (2) | 0.058 (2) | −0.008 (2) | 0.031 (2) | −0.013 (2) |
| C5 | 0.071 (3) | 0.056 (2) | 0.071 (3) | −0.014 (2) | 0.037 (2) | −0.018 (2) |
| C6 | 0.059 (2) | 0.065 (3) | 0.093 (3) | 0.000 (2) | 0.026 (2) | −0.021 (3) |
| C7 | 0.054 (2) | 0.056 (2) | 0.054 (2) | 0.0060 (19) | 0.0054 (18) | −0.0090 (18) |
| C8 | 0.069 (3) | 0.064 (3) | 0.061 (2) | 0.001 (2) | 0.016 (2) | 0.004 (2) |
| C9 | 0.061 (2) | 0.082 (3) | 0.047 (2) | 0.008 (2) | 0.012 (2) | −0.001 (2) |
| C10 | 0.095 (3) | 0.109 (4) | 0.054 (3) | 0.013 (3) | 0.022 (3) | 0.004 (3) |
| C11 | 0.073 (3) | 0.093 (4) | 0.047 (2) | 0.014 (3) | 0.005 (2) | −0.017 (2) |
| C12 | 0.053 (2) | 0.070 (3) | 0.044 (2) | 0.005 (2) | 0.0044 (18) | −0.014 (2) |
| C13 | 0.051 (2) | 0.053 (2) | 0.051 (2) | 0.0027 (18) | 0.0046 (17) | −0.0153 (18) |
| C14 | 0.0437 (19) | 0.052 (2) | 0.049 (2) | −0.0028 (17) | 0.0092 (16) | −0.0082 (17) |
| C15 | 0.060 (2) | 0.048 (2) | 0.058 (2) | −0.0043 (18) | 0.0192 (19) | −0.0085 (18) |
| C16 | 0.071 (3) | 0.064 (3) | 0.060 (2) | 0.005 (2) | 0.013 (2) | −0.012 (2) |
| C17 | 0.100 (3) | 0.059 (3) | 0.081 (3) | 0.017 (2) | 0.007 (3) | −0.021 (2) |
| C18 | 0.094 (3) | 0.094 (4) | 0.056 (3) | 0.025 (3) | 0.005 (2) | −0.033 (3) |
| C19 | 0.091 (3) | 0.070 (3) | 0.100 (3) | 0.023 (3) | 0.006 (3) | 0.006 (3) |
| C20 | 0.083 (3) | 0.067 (3) | 0.074 (3) | −0.011 (2) | 0.036 (2) | −0.003 (2) |
| C21 | 0.0438 (19) | 0.050 (2) | 0.053 (2) | 0.0042 (17) | 0.0063 (16) | −0.0035 (17) |
| C22 | 0.047 (2) | 0.051 (2) | 0.052 (2) | 0.0033 (17) | 0.0077 (17) | −0.0039 (17) |
| C23 | 0.0468 (19) | 0.0442 (18) | 0.050 (2) | 0.0003 (16) | 0.0123 (16) | −0.0009 (16) |
| C24 | 0.068 (3) | 0.049 (2) | 0.070 (2) | 0.009 (2) | 0.003 (2) | 0.001 (2) |
| C25 | 0.085 (3) | 0.056 (2) | 0.089 (3) | 0.018 (2) | 0.003 (2) | −0.012 (2) |
| C26 | 0.059 (2) | 0.047 (2) | 0.070 (2) | 0.0059 (19) | 0.021 (2) | −0.0106 (19) |
| C27 | 0.072 (3) | 0.068 (3) | 0.053 (2) | 0.023 (2) | 0.007 (2) | 0.002 (2) |
| C28 | 0.067 (3) | 0.050 (2) | 0.046 (2) | 0.0169 (19) | 0.0103 (19) | 0.0089 (18) |
| C29 | 0.093 (3) | 0.065 (3) | 0.058 (2) | 0.014 (3) | 0.028 (2) | −0.003 (2) |
| C30 | 0.116 (4) | 0.075 (3) | 0.081 (3) | 0.034 (3) | 0.049 (3) | 0.013 (3) |
| C31 | 0.085 (3) | 0.084 (4) | 0.117 (4) | 0.019 (3) | 0.044 (3) | 0.031 (3) |
| C32 | 0.091 (4) | 0.070 (3) | 0.094 (3) | 0.001 (3) | 0.015 (3) | 0.003 (3) |
| C33 | 0.086 (3) | 0.055 (3) | 0.060 (2) | 0.010 (2) | 0.022 (2) | −0.001 (2) |
Geometric parameters (Å, º)
| O1—C1 | 1.435 (4) | C13—C16 | 1.385 (5) |
| O1—C3 | 1.451 (4) | C13—C14 | 1.490 (5) |
| O2—C1 | 1.388 (4) | C14—C15 | 1.340 (5) |
| O2—H1 | 0.8201 | C15—H11 | 0.9300 |
| O3—C2 | 1.229 (4) | C16—C17 | 1.428 (6) |
| O4—C4 | 1.230 (4) | C16—H12 | 0.9300 |
| O5—C22 | 1.246 (4) | C17—C18 | 1.396 (6) |
| N1—C2 | 1.353 (4) | C17—H13 | 0.9300 |
| N1—C1 | 1.458 (4) | C18—H14 | 0.9300 |
| N1—C21 | 1.474 (4) | C19—H15 | 0.9600 |
| N2—C4 | 1.367 (5) | C19—H16 | 0.9600 |
| N2—C3 | 1.444 (4) | C19—H17 | 0.9600 |
| N2—H2 | 0.8600 | C20—H18 | 0.9600 |
| N3—C19 | 1.472 (5) | C20—H19 | 0.9600 |
| N3—C7 | 1.472 (4) | C20—H20 | 0.9600 |
| N3—C6 | 1.477 (5) | C21—C22 | 1.527 (5) |
| N4—C10 | 1.369 (6) | C21—C27 | 1.547 (5) |
| N4—C11 | 1.378 (6) | C21—H21 | 0.9800 |
| N4—H3 | 0.8600 | C23—C24 | 1.519 (5) |
| N5—C22 | 1.334 (4) | C23—H22 | 0.9800 |
| N5—C26 | 1.475 (4) | C24—C25 | 1.519 (6) |
| N5—C23 | 1.485 (4) | C24—H23 | 0.9700 |
| C1—C23 | 1.512 (5) | C24—H24 | 0.9700 |
| C2—C3 | 1.529 (5) | C25—C26 | 1.514 (5) |
| C3—C20 | 1.515 (5) | C25—H25 | 0.9700 |
| C4—C5 | 1.530 (5) | C25—H26 | 0.9700 |
| C5—C15 | 1.503 (5) | C26—H27 | 0.9700 |
| C5—C6 | 1.542 (6) | C26—H28 | 0.9700 |
| C5—H4 | 0.9800 | C27—C28 | 1.495 (5) |
| C6—H5 | 0.9700 | C27—H29 | 0.9700 |
| C6—H6 | 0.9700 | C27—H30 | 0.9700 |
| C7—C14 | 1.539 (5) | C28—C33 | 1.391 (5) |
| C7—C8 | 1.552 (5) | C28—C29 | 1.395 (5) |
| C7—H7 | 0.9800 | C29—C30 | 1.387 (6) |
| C8—C9 | 1.506 (5) | C29—H31 | 0.9300 |
| C8—H8 | 0.9700 | C30—C31 | 1.366 (7) |
| C8—H9 | 0.9700 | C30—H32 | 0.9300 |
| C9—C10 | 1.376 (5) | C31—C32 | 1.370 (6) |
| C9—C12 | 1.407 (6) | C31—H33 | 0.9300 |
| C10—H10 | 0.9300 | C32—C33 | 1.378 (6) |
| C11—C18 | 1.379 (7) | C32—H34 | 0.9300 |
| C11—C12 | 1.406 (5) | C33—H35 | 0.9300 |
| C12—C13 | 1.400 (5) | ||
| C1—O1—C3 | 111.6 (2) | C5—C15—H11 | 118.1 |
| C1—O2—H1 | 109.5 | C13—C16—C17 | 119.8 (4) |
| C2—N1—C1 | 112.7 (3) | C13—C16—H12 | 120.1 |
| C2—N1—C21 | 124.1 (3) | C17—C16—H12 | 120.1 |
| C1—N1—C21 | 122.8 (3) | C18—C17—C16 | 122.5 (4) |
| C4—N2—C3 | 121.4 (3) | C18—C17—H13 | 118.7 |
| C4—N2—H2 | 119.3 | C16—C17—H13 | 118.7 |
| C3—N2—H2 | 119.4 | C11—C18—C17 | 117.5 (4) |
| C19—N3—C7 | 111.6 (3) | C11—C18—H14 | 121.3 |
| C19—N3—C6 | 108.5 (3) | C17—C18—H14 | 121.3 |
| C7—N3—C6 | 112.0 (3) | N3—C19—H15 | 109.5 |
| C10—N4—C11 | 108.9 (4) | N3—C19—H16 | 109.5 |
| C10—N4—H3 | 125.7 | H15—C19—H16 | 109.5 |
| C11—N4—H3 | 125.5 | N3—C19—H17 | 109.5 |
| C22—N5—C26 | 122.0 (3) | H15—C19—H17 | 109.5 |
| C22—N5—C23 | 126.8 (3) | H16—C19—H17 | 109.5 |
| C26—N5—C23 | 111.1 (3) | C3—C20—H18 | 109.5 |
| O2—C1—O1 | 111.5 (3) | C3—C20—H19 | 109.5 |
| O2—C1—N1 | 112.8 (3) | H18—C20—H19 | 109.5 |
| O1—C1—N1 | 104.0 (2) | C3—C20—H20 | 109.5 |
| O2—C1—C23 | 109.2 (3) | H18—C20—H20 | 109.5 |
| O1—C1—C23 | 109.5 (3) | H19—C20—H20 | 109.5 |
| N1—C1—C23 | 109.8 (3) | N1—C21—C22 | 112.7 (3) |
| O3—C2—N1 | 126.2 (3) | N1—C21—C27 | 113.2 (3) |
| O3—C2—C3 | 126.1 (3) | C22—C21—C27 | 111.9 (3) |
| N1—C2—C3 | 107.5 (3) | N1—C21—H21 | 106.1 |
| N2—C3—O1 | 108.9 (3) | C22—C21—H21 | 106.1 |
| N2—C3—C20 | 109.3 (3) | C27—C21—H21 | 106.1 |
| O1—C3—C20 | 110.9 (3) | O5—C22—N5 | 122.3 (3) |
| N2—C3—C2 | 115.7 (3) | O5—C22—C21 | 119.1 (3) |
| O1—C3—C2 | 103.4 (3) | N5—C22—C21 | 118.6 (3) |
| C20—C3—C2 | 108.5 (3) | N5—C23—C1 | 108.8 (3) |
| O4—C4—N2 | 122.1 (3) | N5—C23—C24 | 103.0 (3) |
| O4—C4—C5 | 122.2 (4) | C1—C23—C24 | 117.8 (3) |
| N2—C4—C5 | 115.7 (3) | N5—C23—H22 | 108.9 |
| C15—C5—C4 | 115.8 (3) | C1—C23—H22 | 108.9 |
| C15—C5—C6 | 109.0 (3) | C24—C23—H22 | 108.9 |
| C4—C5—C6 | 109.4 (3) | C25—C24—C23 | 103.0 (3) |
| C15—C5—H4 | 107.4 | C25—C24—H23 | 111.2 |
| C4—C5—H4 | 107.4 | C23—C24—H23 | 111.2 |
| C6—C5—H4 | 107.4 | C25—C24—H24 | 111.2 |
| N3—C6—C5 | 110.9 (3) | C23—C24—H24 | 111.2 |
| N3—C6—H5 | 109.5 | H23—C24—H24 | 109.1 |
| C5—C6—H5 | 109.5 | C24—C25—C26 | 105.8 (3) |
| N3—C6—H6 | 109.5 | C24—C25—H25 | 110.6 |
| C5—C6—H6 | 109.5 | C26—C25—H25 | 110.6 |
| H5—C6—H6 | 108.0 | C24—C25—H26 | 110.6 |
| N3—C7—C14 | 110.9 (3) | C26—C25—H26 | 110.6 |
| N3—C7—C8 | 110.3 (3) | H25—C25—H26 | 108.7 |
| C14—C7—C8 | 112.1 (3) | N5—C26—C25 | 104.0 (3) |
| N3—C7—H7 | 107.8 | N5—C26—H27 | 111.0 |
| C14—C7—H7 | 107.8 | C25—C26—H27 | 111.0 |
| C8—C7—H7 | 107.8 | N5—C26—H28 | 111.0 |
| C9—C8—C7 | 109.9 (3) | C25—C26—H28 | 111.0 |
| C9—C8—H8 | 109.7 | H27—C26—H28 | 109.0 |
| C7—C8—H8 | 109.7 | C28—C27—C21 | 117.6 (3) |
| C9—C8—H9 | 109.7 | C28—C27—H29 | 107.9 |
| C7—C8—H9 | 109.7 | C21—C27—H29 | 107.9 |
| H8—C8—H9 | 108.2 | C28—C27—H30 | 107.9 |
| C10—C9—C12 | 105.4 (4) | C21—C27—H30 | 107.9 |
| C10—C9—C8 | 136.4 (4) | H29—C27—H30 | 107.2 |
| C12—C9—C8 | 118.1 (3) | C33—C28—C29 | 117.0 (4) |
| N4—C10—C9 | 110.3 (4) | C33—C28—C27 | 121.5 (4) |
| N4—C10—H10 | 124.8 | C29—C28—C27 | 121.5 (4) |
| C9—C10—H10 | 124.8 | C30—C29—C28 | 121.0 (4) |
| N4—C11—C18 | 133.8 (4) | C30—C29—H31 | 119.5 |
| N4—C11—C12 | 106.2 (4) | C28—C29—H31 | 119.5 |
| C18—C11—C12 | 120.0 (4) | C31—C30—C29 | 120.6 (5) |
| C13—C12—C11 | 123.4 (4) | C31—C30—H32 | 119.7 |
| C13—C12—C9 | 127.4 (3) | C29—C30—H32 | 119.7 |
| C11—C12—C9 | 109.2 (4) | C30—C31—C32 | 119.5 (5) |
| C16—C13—C12 | 116.8 (3) | C30—C31—H33 | 120.3 |
| C16—C13—C14 | 127.0 (3) | C32—C31—H33 | 120.3 |
| C12—C13—C14 | 116.2 (3) | C31—C32—C33 | 120.4 (5) |
| C15—C14—C13 | 123.6 (3) | C31—C32—H34 | 119.8 |
| C15—C14—C7 | 122.2 (3) | C33—C32—H34 | 119.8 |
| C13—C14—C7 | 114.2 (3) | C32—C33—C28 | 121.6 (4) |
| C14—C15—C5 | 123.9 (4) | C32—C33—H35 | 119.2 |
| C14—C15—H11 | 118.1 | C28—C33—H35 | 119.2 |
| C3—O1—C1—O2 | 115.5 (3) | C9—C12—C13—C14 | −3.5 (5) |
| C3—O1—C1—N1 | −6.4 (3) | C16—C13—C14—C15 | −23.1 (6) |
| C3—O1—C1—C23 | −123.6 (3) | C12—C13—C14—C15 | 159.1 (3) |
| C2—N1—C1—O2 | −111.3 (3) | C16—C13—C14—C7 | 156.5 (4) |
| C21—N1—C1—O2 | 75.9 (4) | C12—C13—C14—C7 | −21.3 (4) |
| C2—N1—C1—O1 | 9.6 (4) | N3—C7—C14—C15 | −7.1 (5) |
| C21—N1—C1—O1 | −163.2 (3) | C8—C7—C14—C15 | −130.9 (4) |
| C2—N1—C1—C23 | 126.7 (3) | N3—C7—C14—C13 | 173.2 (3) |
| C21—N1—C1—C23 | −46.1 (4) | C8—C7—C14—C13 | 49.5 (4) |
| C1—N1—C2—O3 | 176.7 (4) | C13—C14—C15—C5 | 170.2 (3) |
| C21—N1—C2—O3 | −10.6 (6) | C7—C14—C15—C5 | −9.4 (5) |
| C1—N1—C2—C3 | −8.9 (4) | C4—C5—C15—C14 | 113.1 (4) |
| C21—N1—C2—C3 | 163.8 (3) | C6—C5—C15—C14 | −10.7 (5) |
| C4—N2—C3—O1 | 56.9 (4) | C12—C13—C16—C17 | −1.0 (5) |
| C4—N2—C3—C20 | 178.3 (3) | C14—C13—C16—C17 | −178.8 (4) |
| C4—N2—C3—C2 | −59.0 (5) | C13—C16—C17—C18 | 0.5 (6) |
| C1—O1—C3—N2 | −122.1 (3) | N4—C11—C18—C17 | −178.9 (4) |
| C1—O1—C3—C20 | 117.6 (3) | C12—C11—C18—C17 | −0.8 (7) |
| C1—O1—C3—C2 | 1.5 (4) | C16—C17—C18—C11 | 0.5 (7) |
| O3—C2—C3—N2 | −62.1 (5) | C2—N1—C21—C22 | −154.1 (3) |
| N1—C2—C3—N2 | 123.5 (3) | C1—N1—C21—C22 | 17.9 (4) |
| O3—C2—C3—O1 | 178.9 (4) | C2—N1—C21—C27 | 77.7 (4) |
| N1—C2—C3—O1 | 4.5 (4) | C1—N1—C21—C27 | −110.3 (3) |
| O3—C2—C3—C20 | 61.1 (5) | C26—N5—C22—O5 | 4.5 (5) |
| N1—C2—C3—C20 | −113.3 (3) | C23—N5—C22—O5 | −174.5 (3) |
| C3—N2—C4—O4 | 19.6 (6) | C26—N5—C22—C21 | −175.2 (3) |
| C3—N2—C4—C5 | −157.6 (3) | C23—N5—C22—C21 | 5.7 (5) |
| O4—C4—C5—C15 | 117.7 (4) | N1—C21—C22—O5 | −175.5 (3) |
| N2—C4—C5—C15 | −65.1 (5) | C27—C21—C22—O5 | −46.7 (4) |
| O4—C4—C5—C6 | −118.7 (4) | N1—C21—C22—N5 | 4.2 (4) |
| N2—C4—C5—C6 | 58.5 (4) | C27—C21—C22—N5 | 133.1 (3) |
| C19—N3—C6—C5 | 169.6 (3) | C22—N5—C23—C1 | −33.7 (4) |
| C7—N3—C6—C5 | −66.7 (4) | C26—N5—C23—C1 | 147.2 (3) |
| C15—C5—C6—N3 | 47.1 (4) | C22—N5—C23—C24 | −159.5 (3) |
| C4—C5—C6—N3 | −80.4 (4) | C26—N5—C23—C24 | 21.4 (4) |
| C19—N3—C7—C14 | 166.1 (3) | O2—C1—C23—N5 | −74.8 (3) |
| C6—N3—C7—C14 | 44.2 (4) | O1—C1—C23—N5 | 162.9 (2) |
| C19—N3—C7—C8 | −69.1 (4) | N1—C1—C23—N5 | 49.3 (3) |
| C6—N3—C7—C8 | 169.0 (3) | O2—C1—C23—C24 | 41.9 (4) |
| N3—C7—C8—C9 | −176.1 (3) | O1—C1—C23—C24 | −80.4 (4) |
| C14—C7—C8—C9 | −52.0 (4) | N1—C1—C23—C24 | 166.0 (3) |
| C7—C8—C9—C10 | −154.0 (5) | N5—C23—C24—C25 | −33.9 (4) |
| C7—C8—C9—C12 | 28.8 (5) | C1—C23—C24—C25 | −153.6 (3) |
| C11—N4—C10—C9 | −0.5 (6) | C23—C24—C25—C26 | 35.2 (4) |
| C12—C9—C10—N4 | 0.9 (5) | C22—N5—C26—C25 | −178.9 (3) |
| C8—C9—C10—N4 | −176.5 (4) | C23—N5—C26—C25 | 0.2 (4) |
| C10—N4—C11—C18 | 178.1 (5) | C24—C25—C26—N5 | −22.0 (4) |
| C10—N4—C11—C12 | −0.1 (5) | N1—C21—C27—C28 | 65.2 (4) |
| N4—C11—C12—C13 | 178.8 (3) | C22—C21—C27—C28 | −63.3 (4) |
| C18—C11—C12—C13 | 0.2 (6) | C21—C27—C28—C33 | −96.9 (4) |
| N4—C11—C12—C9 | 0.7 (5) | C21—C27—C28—C29 | 84.6 (4) |
| C18—C11—C12—C9 | −177.8 (4) | C33—C28—C29—C30 | −0.2 (5) |
| C10—C9—C12—C13 | −179.0 (4) | C27—C28—C29—C30 | 178.3 (4) |
| C8—C9—C12—C13 | −1.0 (6) | C28—C29—C30—C31 | −0.1 (6) |
| C10—C9—C12—C11 | −1.0 (4) | C29—C30—C31—C32 | 1.0 (7) |
| C8—C9—C12—C11 | 177.0 (3) | C30—C31—C32—C33 | −1.4 (7) |
| C11—C12—C13—C16 | 0.7 (5) | C31—C32—C33—C28 | 1.1 (6) |
| C9—C12—C13—C16 | 178.4 (4) | C29—C28—C33—C32 | −0.2 (5) |
| C11—C12—C13—C14 | 178.7 (3) | C27—C28—C33—C32 | −178.7 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···N3 | 0.86 | 2.53 | 2.955 (4) | 112 |
| N4—H3···O5i | 0.86 | 2.17 | 2.981 (5) | 157 |
Symmetry code: (i) x, y+1, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: DS2173).
References
- Bruker (2001). SMART, SAINT and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Burnett, M. N. & Johnson, C. K. (1996). ORTEPIII Report ORNL-6895. Oak Ridge National Laboratory, Tennessee, USA.
- Crews, C., Anderson, W. A. C., Rees, G. & Krska, R. (2009). Food Addit. Contam. Part B, 2, 79–85. [DOI] [PubMed]
- Komarova, E. L. & Tolkachev, O. N. (2001). Pharm. Chem. J. 35, 37–45.
- Merkel, S., Köppen, R., Koch, M., Emmerling, F. & Nehls, I. (2010). Acta Cryst. E66, o2275. [DOI] [PMC free article] [PubMed]
- Müller, C., Kemmlein, S., Klaffke, H., Krauthause, W., Preiss-Weigert, A. & Wittkowski, R. (2009). Mol. Nutr. Food Res. 53, 500–507. [DOI] [PubMed]
- Pakhomova, S., Ondráucek, J., Huusák, M., Kratochvíl, B., Jegorov, A. & Stuchlík, J. (1995). Acta Cryst. C51, 308–311.
- Pierri, L., Pitman, I. H., Rae, I. D., Winkler, D. A. & Andrews, P. R. (1982). J. Med. Chem. 25, 937–942. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Stoll, A. (1945). Helv. Chim. Acta, 28, 1283–1308.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812003674/ds2173sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812003674/ds2173Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812003674/ds2173Isup3.mol
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



