Abstract
In the title thione, C26H36N6OS, the 1,2,4-triazole ring is planar (r.m.s. deviation = 0.020 Å) and the benzene ring is twisted out of this plane [dihedral angle = 62.35 (12)°]. Supramolecular zigzag chains feature in the crystal packing. These are sustained by O—H⋯N(piperazine) hydrogen bonds, and are connected into the three-dimensional crystal structure by C—H⋯S and C—H⋯O interactions. The crystal studied was a racemic twin.
Related literature
For the biological activity of adamantyl derivatives, see: Vernier et al. (1969 ▶); El-Emam et al. (2004 ▶); Kadi et al. (2007 ▶, 2010 ▶); Al-Omar et al. (2010 ▶). For related adamantane structures, see: Al-Tamimi et al. (2010 ▶); Kadi et al. (2011 ▶).
Experimental
Crystal data
C26H36N6OS
M r = 480.67
Monoclinic,
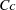
a = 16.1006 (6) Å
b = 14.2182 (5) Å
c = 11.4641 (4) Å
β = 92.440 (4)°
V = 2622.00 (16) Å3
Z = 4
Mo Kα radiation
μ = 0.15 mm−1
T = 100 K
0.25 × 0.20 × 0.15 mm
Data collection
Agilent SuperNova Dual diffractometer with Atlas detector
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011 ▶) T min = 0.963, T max = 0.977
12526 measured reflections
4840 independent reflections
4374 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.039
wR(F 2) = 0.100
S = 1.03
4840 reflections
312 parameters
3 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.25 e Å−3
Δρmin = −0.18 e Å−3
Absolute structure: Flack (1983 ▶), with 1811 Friedel pairs
Flack parameter: 0.06 (7)
Data collection: CrysAlis PRO (Agilent, 2011 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶) and DIAMOND (Brandenburg, 2006 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812005107/hg5175sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812005107/hg5175Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812005107/hg5175Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1O⋯N6i | 0.85 (1) | 1.86 (1) | 2.699 (3) | 170 (5) |
| C13—H13B⋯S1ii | 0.99 | 2.72 | 3.700 (2) | 170 |
| C22—H22⋯O1iii | 0.95 | 2.33 | 3.149 (3) | 145 |
Symmetry codes: (i) 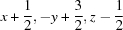 ; (ii)
; (ii) 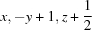 ; (iii)
; (iii) 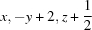 .
.
Acknowledgments
The financial support of the Deanship of Scientific Research and the Research Center of the College of Pharmacy, King Saud University is greatly appreciated. We also thank the Ministry of Higher Education (Malaysia) for funding structural studies through the High-Impact Research Scheme (grant No. UM.C/HIR/MOHE/SC/12).
supplementary crystallographic information
Comment
Derivatives of adamantane have long been known for their diverse biological activities including anti-viral activity against the influenza (Vernier et al., 1969) and HIV viruses (El-Emam et al., 2004). Moreover, adamantane derivative were recently reported to exhibit marked anti-bacterial and anti-inflammatory activities (Kadi et al., 2007, 2010). In continuation of our interest in the chemical and pharmacological properties of adamantane derivatives, and as a part of on-going structural studies of adamantane derivatives (Kadi et al., 2011; Al-Tamimi et al., 2010) we synthesized the title compound (I) as potential chemotherapeutic agent.
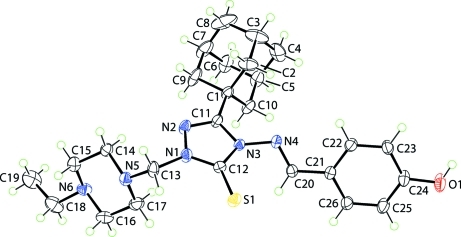
The structure determination of (I), Fig. 1, confirms the presence of the thione in the solid-state. The central 1,2,4-triazole ring is planar [r.m.s. deviation = 0.020 Å] and the benzene ring is twisted out of this plane [dihedral angle = 62.35 (12)°]. Globally, the plane of the five-membered ring bisects the adamantyl group with the benzene and piperazine substituents lying to one side.
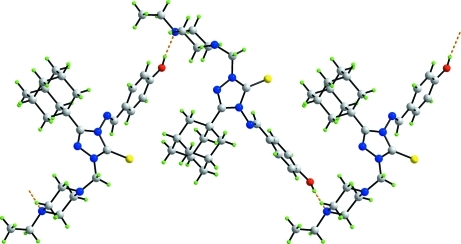
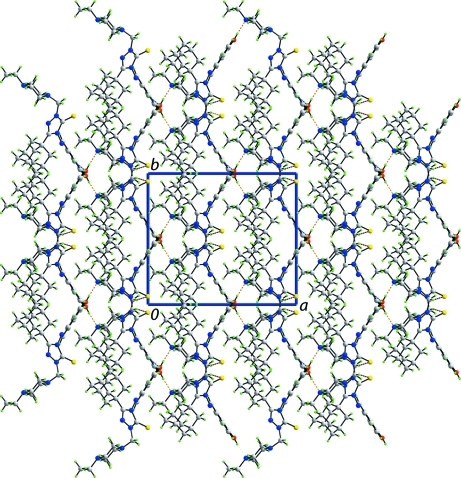
In the crystal packing, supramolecular zigzag chains are formed via O—H···N(piperazine) hydrogen bonds, Fig. 2 and Table 1. These are connected into the three-dimensional crystal structure by C—H···S and C—H···O interactions, Fig. 3 and Table 1.
Experimental
A mixture of 709 mg (2 mmol) of 3-(1-adamantyl)-4-(4-hydroxybenzylideneamino)-4H-1,2,4-triazole-5-thiol (Al-Omar et al., 2010), 1-ethylpiperazine (228 mg, 2 mmol) and 37% formaldehyde solution (1 ml), in ethanol (8 ml) was heated under reflux for 15 min when a clear solution was obtained. Stirring was continued for 12 h at room temperature and the mixture was allowed to stand overnight. Cold water (5 ml) was added and the mixture was stirred for 20 min. The precipitated crude product were filtered, washed with water, dried, and crystallized from ethanol to yield 425 mg (44%) of the title compound (I) as colourless crystals. m.p. 436–438 K. 1H NMR (CDCl3): δ 1.18 (t, 3H, CH3, J = 6.5 Hz), 1.69–1.76 (m, 6H, adamantane-H), 1.95 (s, 6H, adamantane-H), 2.03 (s, 3H, adamantane-H), 2.56 (q, 2H, CH2CH3, J = 6.5 Hz), 2.85–3.90 (m, 8H, piperazine-H), 5.18 (s, 2H, CH2), 6.75 (d, 2H, Ar—H, J = 8.0 Hz), 7.67 (d, 2H, Ar—H, J = 8.0 Hz), 9.21 (s, 1H, CH═N). 13C NMR: δ 11.11 (CH3), 27.84, 35.09, 36.48, 38.39 (adamantane-C), 49.50 (CH2CH3), 52.29, 58.43 (piperazine-C), 68.56 (CH2), 116.53, 123.49, 130.87, 161.53 (Ar—C), 154.94 (triazole C-3), 163.34 (CH═N), 164.65 (C═ S).
Refinement
Carbon-bound H atoms were placed in calculated positions [C—H = 0.95 to 1.00 Å, Uiso(H) = 1.2 to 1.5 Ueq(C)] and were included in the refinement in the riding model approximation. The hydroxy H atom was located in a difference Fourier map, and was refined with a distance restraint of O—H = 0.84±0.01 Å, and Uiso was refined.
Owing to poor agreement a number of reflections, i.e. (11 1 4), (4 0 6), (8 4 2), (15 1 2), (10 2 1), (9 5 1), (13 1 1), (14 4 2), (13 3 2) and (14 2 1), were omitted from the final refinement.
The crystal is a racemic twin; the Flack parameter was explicitly refined.
There is a significant Hirshfeld rigid-bond alert for the C7—C8 bond. Their displacement factors are somewhat large, indicating some disorder. However, no model for the disorder was resolved.
Figures
Fig. 1.
The molecular structure of (I) showing the atom-labelling scheme and displacement ellipsoids at the 50% probability level.
Fig. 2.
A view of the zigzag supramolecular chain along [100] in (I). The O—H···N hydrogen bonds are shown as orange dashed lines.
Fig. 3.
A view in projection down the c axis of the unit-cell contents for (I). The O—H···N, C—H···S and C—H···O interactions are shown as orange, brown and blue dashed lines, respectively.
Crystal data
| C26H36N6OS | F(000) = 1032 |
| Mr = 480.67 | Dx = 1.218 Mg m−3 |
| Monoclinic, Cc | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: C -2yc | Cell parameters from 4991 reflections |
| a = 16.1006 (6) Å | θ = 2.5–27.5° |
| b = 14.2182 (5) Å | µ = 0.15 mm−1 |
| c = 11.4641 (4) Å | T = 100 K |
| β = 92.440 (4)° | Prism, colourless |
| V = 2622.00 (16) Å3 | 0.25 × 0.20 × 0.15 mm |
| Z = 4 |
Data collection
| Agilent SuperNova Dual diffractometer with Atlas detector | 4840 independent reflections |
| Radiation source: SuperNova (Mo) X-ray Source | 4374 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.036 |
| Detector resolution: 10.4041 pixels mm-1 | θmax = 27.6°, θmin = 2.5° |
| ω scans | h = −20→16 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2011) | k = −18→18 |
| Tmin = 0.963, Tmax = 0.977 | l = −14→14 |
| 12526 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.039 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.100 | w = 1/[σ2(Fo2) + (0.0544P)2 + 0.5109P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 4840 reflections | Δρmax = 0.25 e Å−3 |
| 312 parameters | Δρmin = −0.18 e Å−3 |
| 3 restraints | Absolute structure: Flack (1983), with 1811 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.06 (7) |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.49872 (4) | 0.56121 (4) | 0.50028 (4) | 0.02603 (13) | |
| O1 | 0.57958 (13) | 1.01838 (13) | 0.12630 (14) | 0.0397 (5) | |
| H1O | 0.602 (3) | 1.0657 (19) | 0.160 (4) | 0.103 (16)* | |
| N1 | 0.38211 (12) | 0.55968 (12) | 0.66504 (16) | 0.0249 (4) | |
| N2 | 0.32689 (12) | 0.61698 (13) | 0.71956 (16) | 0.0268 (4) | |
| N3 | 0.38731 (11) | 0.69230 (12) | 0.57892 (15) | 0.0201 (4) | |
| N4 | 0.41828 (11) | 0.77076 (12) | 0.51938 (14) | 0.0210 (4) | |
| N5 | 0.32453 (12) | 0.40404 (13) | 0.71060 (16) | 0.0257 (4) | |
| N6 | 0.16089 (12) | 0.32789 (13) | 0.70910 (17) | 0.0292 (4) | |
| C1 | 0.28270 (14) | 0.78354 (15) | 0.69923 (18) | 0.0244 (5) | |
| C2 | 0.34125 (17) | 0.85912 (18) | 0.7529 (2) | 0.0387 (6) | |
| H2A | 0.3724 | 0.8331 | 0.8219 | 0.046* | |
| H2B | 0.3818 | 0.8788 | 0.6952 | 0.046* | |
| C3 | 0.2896 (2) | 0.9442 (2) | 0.7892 (3) | 0.0545 (9) | |
| H3 | 0.3275 | 0.9931 | 0.8245 | 0.065* | |
| C4 | 0.2445 (2) | 0.98457 (18) | 0.6805 (3) | 0.0486 (8) | |
| H4A | 0.2134 | 1.0418 | 0.7014 | 0.058* | |
| H4B | 0.2853 | 1.0021 | 0.6221 | 0.058* | |
| C5 | 0.18436 (17) | 0.91073 (17) | 0.6291 (2) | 0.0350 (6) | |
| H5 | 0.1535 | 0.9376 | 0.5593 | 0.042* | |
| C6 | 0.12260 (17) | 0.88242 (19) | 0.7202 (2) | 0.0409 (6) | |
| H6A | 0.0906 | 0.9382 | 0.7439 | 0.049* | |
| H6B | 0.0829 | 0.8356 | 0.6864 | 0.049* | |
| C7 | 0.1689 (2) | 0.8407 (2) | 0.8263 (2) | 0.0474 (8) | |
| H7 | 0.1281 | 0.8216 | 0.8853 | 0.057* | |
| C8 | 0.2276 (2) | 0.9136 (3) | 0.8790 (3) | 0.0678 (12) | |
| H8A | 0.2579 | 0.8870 | 0.9483 | 0.081* | |
| H8B | 0.1956 | 0.9688 | 0.9045 | 0.081* | |
| C9 | 0.21912 (18) | 0.75416 (19) | 0.7898 (2) | 0.0408 (7) | |
| H9A | 0.1810 | 0.7059 | 0.7556 | 0.049* | |
| H9B | 0.2486 | 0.7266 | 0.8592 | 0.049* | |
| C10 | 0.23419 (15) | 0.82507 (16) | 0.5923 (2) | 0.0281 (5) | |
| H10A | 0.2735 | 0.8437 | 0.5323 | 0.034* | |
| H10B | 0.1961 | 0.7770 | 0.5578 | 0.034* | |
| C11 | 0.33174 (14) | 0.69771 (15) | 0.66707 (18) | 0.0231 (4) | |
| C12 | 0.42254 (13) | 0.60297 (14) | 0.58011 (18) | 0.0205 (4) | |
| C13 | 0.39656 (14) | 0.46264 (15) | 0.7065 (2) | 0.0273 (5) | |
| H13A | 0.4365 | 0.4323 | 0.6551 | 0.033* | |
| H13B | 0.4232 | 0.4655 | 0.7859 | 0.033* | |
| C14 | 0.27018 (14) | 0.42163 (15) | 0.80734 (18) | 0.0240 (5) | |
| H14A | 0.2378 | 0.4799 | 0.7926 | 0.029* | |
| H14B | 0.3039 | 0.4297 | 0.8809 | 0.029* | |
| C15 | 0.21187 (14) | 0.33867 (16) | 0.8179 (2) | 0.0278 (5) | |
| H15A | 0.2445 | 0.2806 | 0.8333 | 0.033* | |
| H15B | 0.1754 | 0.3489 | 0.8841 | 0.033* | |
| C16 | 0.21480 (17) | 0.31573 (17) | 0.6094 (2) | 0.0353 (6) | |
| H16A | 0.1800 | 0.3122 | 0.5363 | 0.042* | |
| H16B | 0.2457 | 0.2558 | 0.6184 | 0.042* | |
| C17 | 0.27601 (17) | 0.39628 (17) | 0.6011 (2) | 0.0323 (5) | |
| H17A | 0.3134 | 0.3847 | 0.5362 | 0.039* | |
| H17B | 0.2456 | 0.4557 | 0.5850 | 0.039* | |
| C18 | 0.10554 (18) | 0.24398 (19) | 0.7131 (3) | 0.0436 (7) | |
| H18A | 0.1404 | 0.1868 | 0.7211 | 0.052* | |
| H18B | 0.0729 | 0.2393 | 0.6381 | 0.052* | |
| C19 | 0.04676 (18) | 0.2464 (2) | 0.8111 (3) | 0.0496 (7) | |
| H19A | 0.0136 | 0.1886 | 0.8101 | 0.074* | |
| H19B | 0.0098 | 0.3009 | 0.8014 | 0.074* | |
| H19C | 0.0785 | 0.2514 | 0.8857 | 0.074* | |
| C20 | 0.42424 (13) | 0.75687 (15) | 0.40960 (18) | 0.0211 (4) | |
| H20 | 0.4039 | 0.7001 | 0.3753 | 0.025* | |
| C21 | 0.46203 (13) | 0.82785 (14) | 0.33739 (18) | 0.0213 (4) | |
| C22 | 0.50449 (14) | 0.90456 (15) | 0.38680 (19) | 0.0250 (5) | |
| H22 | 0.5069 | 0.9122 | 0.4692 | 0.030* | |
| C23 | 0.54296 (15) | 0.96938 (15) | 0.31823 (19) | 0.0267 (5) | |
| H23 | 0.5711 | 1.0216 | 0.3534 | 0.032* | |
| C24 | 0.54077 (15) | 0.95855 (17) | 0.19726 (19) | 0.0284 (5) | |
| C25 | 0.49716 (16) | 0.88341 (18) | 0.1466 (2) | 0.0325 (6) | |
| H25 | 0.4938 | 0.8768 | 0.0640 | 0.039* | |
| C26 | 0.45868 (15) | 0.81838 (16) | 0.21584 (18) | 0.0275 (5) | |
| H26 | 0.4297 | 0.7668 | 0.1806 | 0.033* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0246 (3) | 0.0275 (2) | 0.0267 (3) | 0.0066 (2) | 0.0095 (2) | 0.0022 (2) |
| O1 | 0.0465 (12) | 0.0445 (10) | 0.0276 (9) | −0.0220 (9) | −0.0034 (8) | 0.0134 (8) |
| N1 | 0.0260 (10) | 0.0223 (9) | 0.0272 (9) | 0.0052 (8) | 0.0092 (8) | 0.0052 (7) |
| N2 | 0.0282 (10) | 0.0265 (9) | 0.0266 (9) | 0.0083 (8) | 0.0129 (8) | 0.0063 (8) |
| N3 | 0.0206 (9) | 0.0206 (8) | 0.0196 (8) | 0.0011 (7) | 0.0060 (7) | 0.0006 (7) |
| N4 | 0.0205 (9) | 0.0217 (8) | 0.0212 (9) | −0.0005 (7) | 0.0042 (7) | 0.0046 (7) |
| N5 | 0.0257 (10) | 0.0230 (9) | 0.0288 (9) | 0.0039 (8) | 0.0056 (8) | 0.0029 (8) |
| N6 | 0.0253 (11) | 0.0304 (10) | 0.0319 (10) | 0.0012 (8) | 0.0030 (8) | −0.0066 (8) |
| C1 | 0.0272 (12) | 0.0259 (11) | 0.0206 (10) | 0.0077 (9) | 0.0068 (9) | 0.0033 (9) |
| C2 | 0.0395 (15) | 0.0363 (13) | 0.0396 (14) | 0.0138 (12) | −0.0057 (12) | −0.0131 (12) |
| C3 | 0.0535 (19) | 0.0411 (16) | 0.068 (2) | 0.0176 (14) | −0.0071 (17) | −0.0274 (15) |
| C4 | 0.0480 (18) | 0.0238 (12) | 0.075 (2) | 0.0142 (12) | 0.0164 (15) | 0.0012 (13) |
| C5 | 0.0321 (14) | 0.0366 (12) | 0.0369 (14) | 0.0157 (11) | 0.0105 (11) | 0.0127 (11) |
| C6 | 0.0364 (15) | 0.0418 (14) | 0.0459 (15) | 0.0190 (12) | 0.0185 (12) | 0.0127 (12) |
| C7 | 0.0552 (19) | 0.0547 (17) | 0.0344 (14) | 0.0306 (15) | 0.0245 (13) | 0.0088 (13) |
| C8 | 0.083 (3) | 0.082 (2) | 0.0383 (16) | 0.057 (2) | 0.0067 (17) | −0.0153 (16) |
| C9 | 0.0424 (16) | 0.0485 (15) | 0.0334 (13) | 0.0243 (13) | 0.0248 (12) | 0.0166 (12) |
| C10 | 0.0256 (12) | 0.0369 (12) | 0.0223 (10) | 0.0081 (10) | 0.0054 (9) | 0.0047 (10) |
| C11 | 0.0249 (11) | 0.0261 (10) | 0.0186 (10) | 0.0036 (9) | 0.0059 (8) | 0.0043 (9) |
| C12 | 0.0196 (11) | 0.0219 (10) | 0.0200 (10) | 0.0016 (8) | 0.0005 (8) | −0.0004 (9) |
| C13 | 0.0249 (12) | 0.0245 (10) | 0.0332 (12) | 0.0062 (9) | 0.0083 (10) | 0.0106 (10) |
| C14 | 0.0236 (12) | 0.0257 (10) | 0.0226 (10) | 0.0018 (9) | 0.0012 (9) | 0.0021 (9) |
| C15 | 0.0223 (12) | 0.0295 (11) | 0.0318 (12) | 0.0020 (9) | 0.0035 (10) | 0.0037 (10) |
| C16 | 0.0367 (15) | 0.0345 (12) | 0.0348 (13) | 0.0057 (11) | 0.0028 (11) | −0.0121 (11) |
| C17 | 0.0391 (15) | 0.0321 (12) | 0.0261 (11) | 0.0082 (10) | 0.0043 (10) | −0.0020 (10) |
| C18 | 0.0300 (15) | 0.0367 (13) | 0.0640 (18) | −0.0040 (11) | 0.0005 (13) | −0.0110 (13) |
| C19 | 0.0297 (15) | 0.0518 (16) | 0.067 (2) | −0.0095 (13) | 0.0024 (14) | −0.0083 (15) |
| C20 | 0.0181 (11) | 0.0201 (9) | 0.0252 (11) | 0.0008 (8) | 0.0032 (8) | 0.0020 (8) |
| C21 | 0.0189 (11) | 0.0234 (10) | 0.0217 (10) | 0.0012 (8) | 0.0027 (8) | 0.0042 (9) |
| C22 | 0.0260 (12) | 0.0276 (10) | 0.0216 (10) | −0.0014 (9) | 0.0018 (9) | 0.0015 (9) |
| C23 | 0.0282 (12) | 0.0243 (10) | 0.0274 (11) | −0.0080 (9) | 0.0001 (10) | 0.0027 (9) |
| C24 | 0.0262 (12) | 0.0323 (12) | 0.0265 (11) | −0.0065 (9) | −0.0002 (10) | 0.0103 (9) |
| C25 | 0.0362 (15) | 0.0405 (13) | 0.0206 (10) | −0.0119 (11) | −0.0007 (10) | 0.0054 (10) |
| C26 | 0.0286 (13) | 0.0293 (11) | 0.0246 (11) | −0.0078 (10) | 0.0020 (10) | 0.0002 (9) |
Geometric parameters (Å, º)
| S1—C12 | 1.670 (2) | C7—H7 | 1.0000 |
| O1—C24 | 1.349 (3) | C8—H8A | 0.9900 |
| O1—H1O | 0.850 (10) | C8—H8B | 0.9900 |
| N1—C12 | 1.343 (3) | C9—H9A | 0.9900 |
| N1—N2 | 1.376 (3) | C9—H9B | 0.9900 |
| N1—C13 | 1.475 (3) | C10—H10A | 0.9900 |
| N2—C11 | 1.300 (3) | C10—H10B | 0.9900 |
| N3—C11 | 1.380 (3) | C13—H13A | 0.9900 |
| N3—C12 | 1.391 (3) | C13—H13B | 0.9900 |
| N3—N4 | 1.410 (2) | C14—C15 | 1.516 (3) |
| N4—C20 | 1.282 (3) | C14—H14A | 0.9900 |
| N5—C13 | 1.430 (3) | C14—H14B | 0.9900 |
| N5—C17 | 1.455 (3) | C15—H15A | 0.9900 |
| N5—C14 | 1.463 (3) | C15—H15B | 0.9900 |
| N6—C15 | 1.472 (3) | C16—C17 | 1.517 (4) |
| N6—C16 | 1.474 (3) | C16—H16A | 0.9900 |
| N6—C18 | 1.491 (3) | C16—H16B | 0.9900 |
| C1—C11 | 1.508 (3) | C17—H17A | 0.9900 |
| C1—C10 | 1.543 (3) | C17—H17B | 0.9900 |
| C1—C2 | 1.540 (3) | C18—C19 | 1.500 (4) |
| C1—C9 | 1.547 (3) | C18—H18A | 0.9900 |
| C2—C3 | 1.535 (3) | C18—H18B | 0.9900 |
| C2—H2A | 0.9900 | C19—H19A | 0.9800 |
| C2—H2B | 0.9900 | C19—H19B | 0.9800 |
| C3—C8 | 1.527 (6) | C19—H19C | 0.9800 |
| C3—C4 | 1.527 (4) | C20—C21 | 1.455 (3) |
| C3—H3 | 1.0000 | C20—H20 | 0.9500 |
| C4—C5 | 1.529 (4) | C21—C22 | 1.395 (3) |
| C4—H4A | 0.9900 | C21—C26 | 1.399 (3) |
| C4—H4B | 0.9900 | C22—C23 | 1.376 (3) |
| C5—C6 | 1.527 (4) | C22—H22 | 0.9500 |
| C5—C10 | 1.528 (3) | C23—C24 | 1.394 (3) |
| C5—H5 | 1.0000 | C23—H23 | 0.9500 |
| C6—C7 | 1.519 (4) | C24—C25 | 1.392 (3) |
| C6—H6A | 0.9900 | C25—C26 | 1.382 (3) |
| C6—H6B | 0.9900 | C25—H25 | 0.9500 |
| C7—C8 | 1.512 (5) | C26—H26 | 0.9500 |
| C7—C9 | 1.540 (3) | ||
| C24—O1—H1O | 115 (3) | C5—C10—H10B | 109.7 |
| C12—N1—N2 | 113.61 (17) | C1—C10—H10B | 109.7 |
| C12—N1—C13 | 126.00 (18) | H10A—C10—H10B | 108.2 |
| N2—N1—C13 | 120.25 (18) | N2—C11—N3 | 110.08 (19) |
| C11—N2—N1 | 105.06 (18) | N2—C11—C1 | 124.0 (2) |
| C11—N3—C12 | 108.69 (17) | N3—C11—C1 | 125.92 (19) |
| C11—N3—N4 | 124.34 (18) | N1—C12—N3 | 102.45 (18) |
| C12—N3—N4 | 125.08 (18) | N1—C12—S1 | 128.61 (15) |
| C20—N4—N3 | 113.35 (17) | N3—C12—S1 | 128.90 (16) |
| C13—N5—C17 | 114.98 (19) | N5—C13—N1 | 116.01 (18) |
| C13—N5—C14 | 115.93 (18) | N5—C13—H13A | 108.3 |
| C17—N5—C14 | 110.53 (18) | N1—C13—H13A | 108.3 |
| C15—N6—C16 | 110.09 (18) | N5—C13—H13B | 108.3 |
| C15—N6—C18 | 111.71 (19) | N1—C13—H13B | 108.3 |
| C16—N6—C18 | 107.59 (19) | H13A—C13—H13B | 107.4 |
| C11—C1—C10 | 111.59 (18) | N5—C14—C15 | 108.67 (18) |
| C11—C1—C2 | 110.14 (19) | N5—C14—H14A | 110.0 |
| C10—C1—C2 | 109.34 (19) | C15—C14—H14A | 110.0 |
| C11—C1—C9 | 108.30 (18) | N5—C14—H14B | 110.0 |
| C10—C1—C9 | 108.06 (19) | C15—C14—H14B | 110.0 |
| C2—C1—C9 | 109.3 (2) | H14A—C14—H14B | 108.3 |
| C3—C2—C1 | 109.2 (2) | N6—C15—C14 | 109.78 (18) |
| C3—C2—H2A | 109.8 | N6—C15—H15A | 109.7 |
| C1—C2—H2A | 109.8 | C14—C15—H15A | 109.7 |
| C3—C2—H2B | 109.8 | N6—C15—H15B | 109.7 |
| C1—C2—H2B | 109.8 | C14—C15—H15B | 109.7 |
| H2A—C2—H2B | 108.3 | H15A—C15—H15B | 108.2 |
| C8—C3—C4 | 110.8 (3) | N6—C16—C17 | 111.47 (19) |
| C8—C3—C2 | 109.6 (3) | N6—C16—H16A | 109.3 |
| C4—C3—C2 | 108.6 (2) | C17—C16—H16A | 109.3 |
| C8—C3—H3 | 109.3 | N6—C16—H16B | 109.3 |
| C4—C3—H3 | 109.3 | C17—C16—H16B | 109.3 |
| C2—C3—H3 | 109.3 | H16A—C16—H16B | 108.0 |
| C5—C4—C3 | 109.1 (2) | N5—C17—C16 | 109.2 (2) |
| C5—C4—H4A | 109.9 | N5—C17—H17A | 109.8 |
| C3—C4—H4A | 109.9 | C16—C17—H17A | 109.8 |
| C5—C4—H4B | 109.9 | N5—C17—H17B | 109.8 |
| C3—C4—H4B | 109.9 | C16—C17—H17B | 109.8 |
| H4A—C4—H4B | 108.3 | H17A—C17—H17B | 108.3 |
| C6—C5—C10 | 110.1 (2) | N6—C18—C19 | 113.7 (2) |
| C6—C5—C4 | 109.7 (2) | N6—C18—H18A | 108.8 |
| C10—C5—C4 | 108.8 (2) | C19—C18—H18A | 108.8 |
| C6—C5—H5 | 109.4 | N6—C18—H18B | 108.8 |
| C10—C5—H5 | 109.4 | C19—C18—H18B | 108.8 |
| C4—C5—H5 | 109.4 | H18A—C18—H18B | 107.7 |
| C7—C6—C5 | 109.8 (2) | C18—C19—H19A | 109.5 |
| C7—C6—H6A | 109.7 | C18—C19—H19B | 109.5 |
| C5—C6—H6A | 109.7 | H19A—C19—H19B | 109.5 |
| C7—C6—H6B | 109.7 | C18—C19—H19C | 109.5 |
| C5—C6—H6B | 109.7 | H19A—C19—H19C | 109.5 |
| H6A—C6—H6B | 108.2 | H19B—C19—H19C | 109.5 |
| C8—C7—C6 | 109.2 (3) | N4—C20—C21 | 120.16 (19) |
| C8—C7—C9 | 109.3 (3) | N4—C20—H20 | 119.9 |
| C6—C7—C9 | 109.9 (2) | C21—C20—H20 | 119.9 |
| C8—C7—H7 | 109.5 | C22—C21—C26 | 118.51 (19) |
| C6—C7—H7 | 109.5 | C22—C21—C20 | 121.40 (19) |
| C9—C7—H7 | 109.5 | C26—C21—C20 | 120.05 (19) |
| C7—C8—C3 | 109.9 (2) | C23—C22—C21 | 121.1 (2) |
| C7—C8—H8A | 109.7 | C23—C22—H22 | 119.4 |
| C3—C8—H8A | 109.7 | C21—C22—H22 | 119.4 |
| C7—C8—H8B | 109.7 | C22—C23—C24 | 120.1 (2) |
| C3—C8—H8B | 109.7 | C22—C23—H23 | 120.0 |
| H8A—C8—H8B | 108.2 | C24—C23—H23 | 120.0 |
| C7—C9—C1 | 109.7 (2) | O1—C24—C25 | 118.1 (2) |
| C7—C9—H9A | 109.7 | O1—C24—C23 | 122.5 (2) |
| C1—C9—H9A | 109.7 | C25—C24—C23 | 119.4 (2) |
| C7—C9—H9B | 109.7 | C26—C25—C24 | 120.3 (2) |
| C1—C9—H9B | 109.7 | C26—C25—H25 | 119.9 |
| H9A—C9—H9B | 108.2 | C24—C25—H25 | 119.9 |
| C5—C10—C1 | 109.84 (19) | C25—C26—C21 | 120.6 (2) |
| C5—C10—H10A | 109.7 | C25—C26—H26 | 119.7 |
| C1—C10—H10A | 109.7 | C21—C26—H26 | 119.7 |
| C12—N1—N2—C11 | 0.8 (2) | C10—C1—C11—N3 | 52.9 (3) |
| C13—N1—N2—C11 | 176.73 (19) | C2—C1—C11—N3 | −68.8 (3) |
| C11—N3—N4—C20 | −140.2 (2) | C9—C1—C11—N3 | 171.7 (2) |
| C12—N3—N4—C20 | 57.3 (3) | N2—N1—C12—N3 | −2.5 (2) |
| C11—C1—C2—C3 | −177.9 (2) | C13—N1—C12—N3 | −178.14 (19) |
| C10—C1—C2—C3 | 59.2 (3) | N2—N1—C12—S1 | 175.18 (16) |
| C9—C1—C2—C3 | −59.0 (3) | C13—N1—C12—S1 | −0.5 (3) |
| C1—C2—C3—C8 | 60.0 (3) | C11—N3—C12—N1 | 3.2 (2) |
| C1—C2—C3—C4 | −61.2 (3) | N4—N3—C12—N1 | 168.07 (18) |
| C8—C3—C4—C5 | −57.6 (3) | C11—N3—C12—S1 | −174.48 (16) |
| C2—C3—C4—C5 | 62.8 (3) | N4—N3—C12—S1 | −9.6 (3) |
| C3—C4—C5—C6 | 58.3 (3) | C17—N5—C13—N1 | 55.3 (3) |
| C3—C4—C5—C10 | −62.2 (3) | C14—N5—C13—N1 | −75.8 (2) |
| C10—C5—C6—C7 | 59.2 (3) | C12—N1—C13—N5 | −128.4 (2) |
| C4—C5—C6—C7 | −60.5 (3) | N2—N1—C13—N5 | 56.2 (3) |
| C5—C6—C7—C8 | 60.8 (3) | C13—N5—C14—C15 | −164.70 (17) |
| C5—C6—C7—C9 | −59.0 (3) | C17—N5—C14—C15 | 62.2 (2) |
| C6—C7—C8—C3 | −59.5 (3) | C16—N6—C15—C14 | 57.6 (2) |
| C9—C7—C8—C3 | 60.8 (3) | C18—N6—C15—C14 | 177.0 (2) |
| C4—C3—C8—C7 | 58.6 (3) | N5—C14—C15—N6 | −60.5 (2) |
| C2—C3—C8—C7 | −61.2 (3) | C15—N6—C16—C17 | −55.8 (2) |
| C8—C7—C9—C1 | −59.7 (3) | C18—N6—C16—C17 | −177.7 (2) |
| C6—C7—C9—C1 | 60.1 (3) | C13—N5—C17—C16 | 166.65 (19) |
| C11—C1—C9—C7 | 179.0 (2) | C14—N5—C17—C16 | −59.7 (2) |
| C10—C1—C9—C7 | −59.9 (3) | N6—C16—C17—N5 | 56.4 (2) |
| C2—C1—C9—C7 | 59.0 (3) | C15—N6—C18—C19 | 59.0 (3) |
| C6—C5—C10—C1 | −60.2 (3) | C16—N6—C18—C19 | 179.9 (2) |
| C4—C5—C10—C1 | 60.1 (3) | N3—N4—C20—C21 | −174.54 (17) |
| C11—C1—C10—C5 | 179.1 (2) | N4—C20—C21—C22 | 10.6 (3) |
| C2—C1—C10—C5 | −58.8 (3) | N4—C20—C21—C26 | −171.5 (2) |
| C9—C1—C10—C5 | 60.1 (3) | C26—C21—C22—C23 | −0.5 (3) |
| N1—N2—C11—N3 | 1.3 (2) | C20—C21—C22—C23 | 177.3 (2) |
| N1—N2—C11—C1 | −179.0 (2) | C21—C22—C23—C24 | −0.7 (4) |
| C12—N3—C11—N2 | −3.0 (2) | C22—C23—C24—O1 | −177.9 (2) |
| N4—N3—C11—N2 | −167.98 (19) | C22—C23—C24—C25 | 2.0 (4) |
| C12—N3—C11—C1 | 177.3 (2) | O1—C24—C25—C26 | 177.8 (2) |
| N4—N3—C11—C1 | 12.3 (3) | C23—C24—C25—C26 | −2.1 (4) |
| C10—C1—C11—N2 | −126.8 (2) | C24—C25—C26—C21 | 0.9 (4) |
| C2—C1—C11—N2 | 111.6 (3) | C22—C21—C26—C25 | 0.4 (3) |
| C9—C1—C11—N2 | −8.0 (3) | C20—C21—C26—C25 | −177.5 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O···N6i | 0.85 (1) | 1.86 (1) | 2.699 (3) | 170 (5) |
| C13—H13B···S1ii | 0.99 | 2.72 | 3.700 (2) | 170 |
| C22—H22···O1iii | 0.95 | 2.33 | 3.149 (3) | 145 |
Symmetry codes: (i) x+1/2, −y+3/2, z−1/2; (ii) x, −y+1, z+1/2; (iii) x, −y+2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5175).
References
- Agilent (2011). CrysAlis PRO Agilent Technologies, Yarnton, Oxfordshire, England.
- Al-Omar, M. A., Al-Abdullah, E. S., Shehata, I. A., Habib, E. E., Ibrahim, T. M. & El-Emam, A. A. (2010). Molecules, 15, 2526–2550. [DOI] [PMC free article] [PubMed]
- Al-Tamimi, A.-M. S., Bari, A., Al-Omar, M. A., Alrashood, K. A. & El-Emam, A. A. (2010). Acta Cryst. E66, o1756. [DOI] [PMC free article] [PubMed]
- Brandenburg, K. (2006). DIAMOND Crystal Impact GbR, Bonn, Germany.
- El-Emam, A. A., Al-Deeb, O. A., Al-Omar, M. A. & Lehmann, J. (2004). Bioorg. Med. Chem. 12, 5107–5113. [DOI] [PubMed]
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Kadi, A. A., Al-Abdullah, E. S., Shehata, I. A., Habib, E. E., Ibrahim, T. M. & El-Emam, A. A. (2010). Eur. J. Med. Chem. 45, 5006–5011. [DOI] [PubMed]
- Kadi, A. A., Alanzi, A. M., El-Emam, A. A., Ng, S. W. & Tiekink, E. R. T. (2011). Acta Cryst. E67, o3127. [DOI] [PMC free article] [PubMed]
- Kadi, A. A., El-Brollosy, N. R., Al-Deeb, O. A., Habib, E. E., Ibrahim, T. M. & El-Emam, A. A. (2007). Eur. J. Med. Chem. 42, 235–242. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Vernier, V. G., Harmon, J. B., Stump, J. M., Lynes, T. L., Marvel, M. P. & Smith, D. H. (1969). Toxicol. Appl. Pharmacol. 15, 642–665. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812005107/hg5175sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812005107/hg5175Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812005107/hg5175Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report