Abstract
Purpose
Recent genome-wide screens have identified genetic variations in ARID5B associated with susceptibility to childhood acute lymphoblastic leukemia (ALL). We sought to determine the contribution of ARID5B single nucleotide polymorphisms (SNPs) to racial disparities in ALL susceptibility and treatment outcome.
Patients and Methods
We compared the association between ARID5B SNP genotype and ALL susceptibility in whites (> 95% European genetic ancestry; 978 cases and 1,046 controls) versus in Hispanics (> 10% Native American ancestry; 330 cases and 541 controls). We determined the relationships between ARID5B SNP genotype and ALL relapse risk in 1,605 children treated on the Children's Oncology Group (COG) P9904/9905 clinical trials.
Results
Among 49 ARID5B SNPs interrogated, 10 were significantly associated with ALL susceptibility in both whites and Hispanics (P < .05), with risk alleles consistently more frequent in Hispanics than in whites. rs10821936 exhibited the most significant association in both races (P = 8.4 × 10−20 in whites; P = 1 × 10−6 in Hispanics), and genotype at this SNP was highly correlated with local Native American genetic ancestry (P = 1.8 × 10−8). Multivariate analyses in Hispanics identified an additional SNP associated with ALL susceptibility independent of rs10821936. Eight ARID5B SNPs were associated with both ALL susceptibility and relapse hazard; the alleles related to higher ALL incidence were always linked to poorer treatment outcome and were more frequent in Hispanics.
Conclusion
ARID5B polymorphisms are important determinants of childhood ALL susceptibility and treatment outcome, and they contribute to racial disparities in this disease.
INTRODUCTION
Acute lymphoblastic leukemia (ALL) is the most common type of cancer in children.1 Substantial racial differences exist in both the incidence and treatment outcome of childhood ALL. Among major racial/ethnic groups in the United States, the incidence of ALL during childhood and adolescence is significantly higher in Hispanics than in other groups (incidence rates: blacks < whites and Asians < Hispanics).2–5 Treatment outcome of ALL also varies by race/ethnicity: Hispanic and black children fare worse than white and Asian children with the same disease.6–8 Although there is some evidence that the survival disparity in black children can be mitigated by contemporary risk-adapted ALL therapy,9 the outcome differences between Hispanics and whites have persisted.6,7,10,11 Because populations with different geographic ancestry (ie, racial/ethnic groups) are well distinguished by genetic polymorphisms,12 it is conceivable that these ancestry-related genomic variations may contribute to racial disparities in the incidence and outcome of ALL, in conjunction with environmental/socioeconomic factors.13,14 In fact, the higher risk of ALL relapse in Hispanic children is at least partly attributable to genomic variations characteristic of Native American (NA) genetic ancestry.10
Until recently, the role of inherited genetic variation in ALL predisposition remained controversial because prior genetic studies focusing on candidate genes (eg, those involved in the folate pathway and xenobiotic metabolism/transport) have not demonstrated consistent evidence of association with these genetic polymorphisms.15–17 In contrast, in genome-wide association studies, we and others have independently identified single nucleotide polymorphisms (SNPs) in the ARID5B gene that are strongly associated with ALL susceptibility.18,19 This finding has been repeatedly confirmed in subsequent studies.20–24 The question then arises: What is the contribution of ARID5B genetic polymorphisms to racial differences in ALL incidence? Interestingly, ARID5B genetic variation has also been linked to interpatient variability in antileukemic drug (methotrexate) metabolism,18 arguing for its possible effects on ALL treatment outcome (and racial differences) as well.
In this study, we evaluated 49 common germline SNPs in the ARID5B gene and compared their association with childhood ALL susceptibility and treatment outcome in white versus Hispanic children.
PATIENTS AND METHODS
ALL Cases and Genotyping
The ALL cases investigated comprised 1,605 children with newly diagnosed B-precursor ALL who were treated on the Children's Oncology Group (COG) P9904/P9905 clinical trials25 whose germline DNA was successfully genotyped by using the Affymetrix SNP Array 6 (Affymetrix, Santa Clara, CA).10 Genotype calls (coded as 0, 1, and 2 for AA, AB, and BB genotypes) were determined by the Birdseed algorithm.26 SNPs with a minor allele frequency less than 1% and/or a call rate less than 95% were excluded.10 In subsequent genotype-phenotype association studies, we included 49 SNPs within 10 Kb upstream or downstream of the ARID5B gene. This study was approved by the institutional review boards, and informed consent was obtained from parents, guardians, or patients, as appropriate. Patients included in the genetic association analyses represented 85.3% (1,605 of 1,882) of total enrolled participants on the COG P9904/9905 treatment protocols (Data Supplement).
Non-ALL Controls
We obtained genome-wide SNP genotypes from four non-ALL cohorts: the Genetic Association Information Network (GAIN) schizophrenia cohort (database of Genotypes and Phenotypes [dbGAP] phs000017.v3),27,28 unrelated Mexican samples in HapMap III (MEX), the Human Variation Panel (HD100MEX), and Mexican participants in the Genetics of Asthma in Latino Americans (GALA) study.29 All samples were genotyped by using the Affymetrix SNP Array 6. Because of the extremely low incidence of survivors of childhood ALL in the general population (< 1:10,000), these groups were used as non-ALL controls.18,19
Genetic Ancestry-Based Race Classification
Genetic ancestry was determined by using STRUCTURE (version 2.2.3),10,30 on the basis of genotypes at 30,000 SNPs randomly selected from the Affymetrix array. HapMap samples from descendants of Northern Europeans (CEU, n = 90), West Africans (YRI, n = 90), East Asians (CHB/JPT, n = 90), and NA references (n = 105)31 were used to represent European, African, Asian, and NA ancestries, respectively. We assumed that these four ancestries sum to 100% in each patient. For all analyses, whites were defined as having more than 95% European genetic ancestry and Hispanics were defined as having more than 10% NA genetic ancestry, respectively.
Association Between ARID5B SNP Genotype and ALL Susceptibility
Case-control association analyses were performed for two racial/ethnic groups as defined by genetic ancestry: whites (978 children with ALL and 1,046 non-ALL controls [GAIN]) and Hispanics (330 Hispanic children with ALL and 541 Hispanic controls [HapMap MEX, HD100MEX, and GALA]). For each of the 49 ARID5B SNPs, association between SNP genotype and ALL status was determined by comparing genotype frequencies between ALL cases and controls in each race group by logistic regression, with genetic ancestry as a covariate to avoid population stratification.10 To determine whether the SNP genotypes were independently associated with ALL susceptibility, SNPs related to ALL in univariate analyses (P < .1) were subjected to forward selection–based multivariate analyses32 in each population: the SNP with the strongest association would enter the model first, followed by additional SNPs that remained associated with ALL in the presence of polymorphisms already in the model. For association analyses combining rs10821936 and rs7915732, we first grouped Hispanic individuals on the basis of the number of risk alleles at these two SNPs: those carrying more than two copies of risk alleles (homozygous for the risk allele for at least one SNP or heterozygous at both SNPs), those carrying one copy of the risk allele (heterozygous at either one of the two SNPs but not both), and those carrying zero copies of risk alleles (homozygous for the nonrisk allele at both SNPs, as reference). The odds ratio (OR) for developing ALL was estimated by comparing each of the two risk allele-carrying groups with the reference group.
ALL genetic subtype was defined on the basis of chromosomal abnormalities in leukemic blasts: ETV6–RUNX1 fusion, TCF3–PBX1 fusion, and hyperdiploidy (DNA index ≥ 1.16). No patient in the COG P9904/9905 studies had the BCR-ABL fusion or MLL rearrangements,25 and patients without any known chromosomal abnormalities were grouped as “B other.” Associations between germline SNP genotype and ALL subtype, sex, and age at diagnosis (≥ 10 years v < 10 years) were determined by the χ2 test for all 49 ARID5B SNPs.
Relation of ARI5B SNP Genotype to NA Ancestry
We examined the relationship between ARID5B SNP genotypes and NA ancestry in two analyses. First, we estimated local ancestry (European, African, or NA) at each ARID5B SNP in 330 Hispanic patients by using Local Ancestry in Admixed Populations (LAMP),33 with HapMap CEU, YRI, and NA references as reference ancestral populations. The association between genotype and the inferred percentage of NA ancestry at each SNP was assessed by Spearman rank correlation. Second, to confirm the population differences in genotype frequency at rs10821936, we also genotyped this SNP in five indigenous NA populations from Guatemala (Kaqchikel and Kiche) and Mexico (Zapotec, Mixtec, and Mixe) by TaqMan assay (Applied Biosystems, Foster City, CA).
Association Between ARID5B SNP Genotype and Treatment Outcome in the COG P9904/9905 Studies
Relapse was defined as bone marrow and/or extramedullary relapse. Lineage switch, second malignancy, and death during remission were considered as competing events. Association of SNP genotype with relapse risk was determined by using the Fine and Gray model,34 stratifying by treatment arms A, B, C, and D in the COG P9904/9905 regimens. An additive genetic model was used (ie, relapse risk is proportionally associated with the number of copies of the risk allele at an SNP),10,35 and genetic ancestry was included as a covariate to control for population stratification.10 In multivariate analyses, the association of SNP genotype with relapse was evaluated after adjusting for minimal residual disease (MRD) status (at least 0.01% v less than 0.01%) at the end of remission induction therapy (day 29).25 Associations between SNP genotype and overall survival were determined by the Cox regression test.
RESULTS
Association of ARID5B SNP Genotype With ALL Susceptibility in Whites and Hispanics
To identify genetic variations associated with ALL in each population, we first compared genotype frequencies at 49 ARID5B SNPs between ancestry-matched ALL cases and controls: between 978 white children with ALL and 1,046 white controls (> 95% European genetic ancestry); between 330 Hispanic children with ALL and 541 Hispanic controls (> 10% NA genetic ancestry). After adjusting genetic ancestry to control for population stratification, we observed that 10 SNPs were significantly associated with ALL in both populations, and five and three SNPs were significant only in whites and Hispanics, respectively (Table 1 and Data Supplement). In both race groups, rs10821936 was most significantly associated with ALL (P = 8.4 × 10−20 in whites and P = 1 × 10−6 in Hispanics). Interestingly, the frequency of the ALL risk allele (allele C) at rs10821936 was higher in Hispanics (47%) than in whites (33%), consistent with the higher incidence of ALL in Hispanics.3 In fact, for all 10 SNPs significant in both populations, the risk alleles were consistently more common in Hispanics than in whites (Table 1). After correcting for multiple tests (12 independent SNP clusters with pair-wise r2 < 0.1; Data Supplement), five SNPs remained significantly associated with ALL susceptibility in both races, and three were specific to whites (nominal P < .004).
Table 1.
Association Between ARID5B SNP Genotype and ALL Susceptibility in Whites and Hispanics
| SNP ID | Location*(chromosome 10) | Risk Allele | Whites† |
Hispanics† |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk Allele Frequency |
P | OR | 95% CI | Risk Allele Frequency |
P | OR | 95% CI | |||||
| Control | Case | Control | Case | |||||||||
| Significant in both whites and Hispanics | ||||||||||||
| rs10821936 | 63723577 | C | 0.33 | 0.48 | 8.38× 10−20 | 1.83 | 1.60 to 2.08 | 0.47 | 0.62 | 1.00 × 10−6 | 1.82 | 1.43 to 2.31 |
| rs7896246 | 63724390 | A | 0.33 | 0.48 | 1.03× 10−19 | 1.83 | 1.60 to 2.08 | 0.47 | 0.61 | 5.45× 10−6 | 1.74 | 1.37 to 2.20 |
| rs10821938 | 63724773 | A | 0.42 | 0.54 | 1.47× 10−14 | 1.64 | 1.45 to 1.87 | 0.54 | 0.68 | 2.96× 10−6 | 1.80 | 1.40 to 2.30 |
| rs7923074 | 63723440 | T | 0.42 | 0.54 | 1.5× 10−13 | 1.60 | 1.41 to 1.82 | 0.53 | 0.68 | 4.69× 10−7 | 1.88 | 1.47 to 2.41 |
| rs10994982 | 63710104 | A | 0.50 | 0.58 | 2.66× 10−7 | 1.39 | 1.23 to 1.58 | 0.57 | 0.70 | 1.35× 10−5 | 1.70 | 1.34 to 2.16 |
| rs2893881 | 63688672 | C | 0.14 | 0.18 | .001 | 1.33 | 1.12 to 1.57 | 0.30 | 0.34 | .0423 | 1.29 | 1.01 to 1.65 |
| rs6479778 | 63689077 | T | 0.14 | 0.18 | .0029 | 1.29 | 1.09 to 1.53 | 0.29 | 0.33 | .0296 | 1.32 | 1.03 to 1.69 |
| rs4948488 | 63685154 | C | 0.16 | 0.20 | .0043 | 1.26 | 1.08 to 1.48 | 0.41 | 0.46 | .0345 | 1.28 | 1.02 to 1.61 |
| rs4948487 | 63669865 | C | 0.51 | 0.55 | .0096 | 1.18 | 1.04 to 1.34 | 0.56 | 0.59 | .0457 | 1.28 | 1.00 to 1.62 |
| rs6479779 | 63695048 | C | 0.38 | 0.41 | .013 | 1.17 | 1.04 to 1.33 | 0.40 | 0.46 | .0364 | 1.27 | 1.02 to 1.59 |
| Significant in Hispanics only | ||||||||||||
| rs2393732 | 63767229 | T | 0.03 | 0.03 | .205 | 1.28 | 0.88 to 1.86 | 0.23 | 0.27 | .0126 | 1.40 | 1.08 to 1.83 |
| rs17215180 | 63688728 | C | 0.53 | 0.55 | .251 | 1.08 | 0.95 to 1.22 | 0.70 | 0.74 | .0181 | 1.37 | 1.05 to 1.77 |
| rs2393782 | 63670859 | G | 0.12 | 0.13 | .088 | 1.17 | 0.98 to 1.41 | 0.27 | 0.30 | .029 | 1.32 | 1.03 to 1.70 |
| Significant in whites only | ||||||||||||
| rs7087125 | 63773039 | A | 0.46 | 0.53 | 9.22× 10−6 | 1.32 | 1.17 to 1.50 | 0.43 | 0.45 | .555 | 1.07 | 0.85 to 1.35 |
| rs9415636 | 63826186 | C | 0.91 | 0.93 | .0015 | 1.45 | 1.15 to 1.82 | 0.95 | 0.96 | .18 | 1.46 | 0.84 to 2.54 |
| rs7922394 | 63666691 | T | 0.53 | 0.58 | .0025 | 1.21 | 1.07 to 1.38 | 0.59 | 0.61 | .19 | 1.17 | 0.92 to 1.49 |
| rs10994983 | 63712827 | C | 0.91 | 0.94 | .0031 | 1.44 | 1.13 to 1.84 | 0.97 | 0.97 | .41 | 1.33 | 0.68 to 2.61 |
| rs6415872 | 63660689 | G | 0.50 | 0.54 | .0049 | 1.20 | 1.06 to 1.36 | 0.55 | 0.57 | .149 | 1.19 | 0.94 to 1.51 |
NOTE. P values < .05 (significant) are shown in bold.
Abbreviations: ALL, acute lymphoblastic leukemia; ID, identification; OR, odds ratio; SNP, single nucleotide polymorphism.
All chromosomal locations are based on hg19.
Whites are defined as > 95% European genetic ancestry and Hispanics are defined as > 10% Native American genetic ancestry.
To determine which ARID5B SNPs independently contribute to ALL susceptibility, we performed forward selection–based multivariate analyses for SNPs with a trend toward association in univariate analyses (P < .1; Data Supplement). Thus, SNPs with the strongest association signal enter the model first, after which the remaining SNPs compete against each other on the basis of their association significance after adjusting for polymorphisms already in the model (Table 2). In whites, rs10821936 was the first SNP that entered the multivariate model, and none of the remaining SNPs was significant in the presence of rs10821936. In contrast, in Hispanics, one additional ARID5B SNP (rs7915732) was independently associated with ALL susceptibility, even after adjusting for genotype at rs10821936. In addition, Hispanic individuals carrying more than two copies of risk alleles at these two SNPs (homozygous for the risk allele for at least one SNP or heterozygous at both SNPs) were at much higher risk of developing ALL (OR, 5.83; 95% CI, 2.12 to 16.0) than those carrying one copy of the risk allele (heterozygous at one SNP; OR, 2.54; 95% CI, 1.61 to 4.02) or those carrying zero copies of risk alleles (homozygous for nonrisk allele at both SNPs, the reference group), suggesting cumulative effects of these two SNPs on ALL susceptibility. Moreover, these two SNPs were not in linkage disequilibrium with each other (r2 < 0.01), suggesting the possibility of multiple causal variants in the ARID5B gene in Hispanics.
Table 2.
Multivariate Analysis of Association of ARID5B SNPs With ALL Susceptibility
| Whites |
Hispanics |
||||||
|---|---|---|---|---|---|---|---|
| SNP ID | OR | 95% CI | P* | SNP ID | OR | 95% CI | P* |
| rs10821936 | 2.13 | 1.77 to 2.58 | 2.19 × 10−15 | rs10821936 | 1.92 | 1.50 to 2.45 | 2.14 × 10−7 |
| rs7915732† | 2.58 | 1.27 to 3.52 | .0091 | ||||
Abbreviations: ALL, acute lymphoblastic leukemia; ID, identification; OR, odds ratio; SNP, single nucleotide polymorphism.
P values indicate association after adjusting for other SNPs in a forward selection–based multivariate model (see Patients and Methods).
In univariate analyses, rs7915732 was related to ALL susceptibility in Hispanics (P = .059) but not in whites (P = .293).
Association Between ARID5B SNP Genotype and ALL Susceptibility According to ALL Genetic Subtype, Sex, and Age at Diagnosis
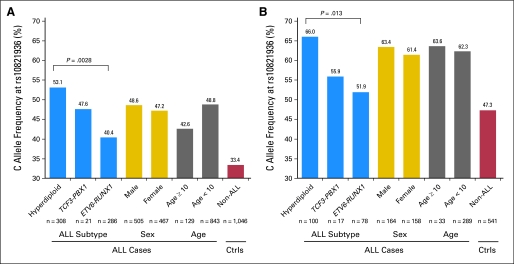
Because ALL consists of multiple subtypes with distinct genetic and prognostic characteristics, we next examined whether ARID5B SNP genotype preferentially predisposes to any specific type of ALL. Patients enrolled on the COG P9904/9905 protocols were divided into four groups on the basis of chromosomal abnormalities: ETV6–RUNX1, TCF3–PBX1, hyperdiploidy (DNA index ≥ 1.16), and B other. Of 18 ALL susceptibility-associated ARID5B SNPs, five were also associated with ALL subtype in both whites and Hispanics, but none was related to age at diagnosis or sex (Data Supplement). For example, in both race groups (Fig 1), the rs10821936 C allele was significantly more common in hyperdiploid ALL than in ALL with the TCF3–PBX1 or ETV6–RUNX1 fusion genes (P = .0028 in whites and P = .013 in Hispanics), similar to previous observations.18,19 Allele frequency did not differ between boys and girls with ALL, arguing against any sex-specific effects of this SNP on ALL susceptibility. Similarly, rs10821936 genotype was not associated with disease onset (age at diagnosis ≥ 10 years or < 10 years).
Fig 1.
rs10821936 genotype frequency by acute lymphoblastic leukemia (ALL) subtype, sex, and age at diagnosis. The risk allele (allele C) at rs10821936 was significantly over-represented in the hyperdiploid subtype of ALL in both whites (A) and Hispanics (B), although no difference was observed by sex or age groups. P values were determined by χ2 tests. Ctrls, controls.
Relationship Between ARID5B SNP Genotype and NA Ancestry
Given the increased allele frequency of ARID5B SNPs in Hispanics (who have a substantial admixture of NA ancestry), we next examined the relationship between ARID5B SNP genotype and NA genetic ancestry. In Hispanics, the number of C alleles (risk allele for ALL) at rs10821936 was positively correlated with the percentage of NA ancestry at this locus: C allele frequency was 51.3%, 63.3%, and 78.9% in patients with 0%, 50%, and 100% NA ancestry at rs10821936, respectively (Data Supplement; n = 322; P = 1.8 × 10−8). We also genotyped rs10821936 in five different indigenous NA populations in Guatemala and Mexico, where the proportion of NA ancestry is high, and we observed substantially higher prevalence of the C allele (52% to 83%) than in whites and Hispanics (Data Supplement). Together, these results indicate that the higher frequency of the risk allele at ARID5B SNPs (eg, rs10821936) in Hispanics is likely to result from the admixture of NA ancestry.
Association of ARID5B SNP Genotype With ALL Treatment Outcome
Finally, we tested whether ARID5B polymorphisms were related to treatment outcome in 1,605 children enrolled on the COG P9904/9905 studies. Of 49 ARID5B SNPs, eight were associated with relapse (P < .05; Table 3 and Data Supplement), all of which were also associated with ALL susceptibility in whites and/or Hispanics (Table 1 and Data Supplement). The alleles related to ALL disease susceptibility were consistently linked to poorer treatment outcome (Table 3). Of eight SNPs associated with ALL relapse, six were associated with MRD status at the end of remission induction therapy, and three remained prognostic after adjusting for MRD (Data Supplement). For example, the T allele at rs6479778 was more frequent in ALL cases than in controls (P = .0029 in whites and P = .0296 in Hispanics), and patients carrying the T allele were also at higher risk of ALL relapse (hazard ratio, 1.48; P = 8.1 × 10−5) and lower overall survival (hazard ratio, 1.73; P = 4.4 × 10−4; Fig 2). Genotype at rs6479778 was also associated with MRD (n = 1,481; P = 1.3 × 10−4; Data Supplement). In a multivariate analysis, rs6479778 exhibited a trend toward association with relapse after adjusting for MRD (P = .05), although this SNP was not prognostic in patients who were negative for MRD. Forward selection–based multivariate analyses indicated that none of the other ARID5B SNPs were prognostic after adjusting for rs6479778. Because the T allele at rs6479778 was more common in Hispanics (29%) than in whites (14%), this polymorphism is likely to contribute to the racial disparity in both the incidence and treatment outcome of ALL.
Table 3.
Association of ARID5B SNP Genotype With Risk of ALL Relapse
| SNP ID | Location*(chromosome 10) | ALL Susceptibility |
ALL Relapse |
Risk Allele Frequency† |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Risk Allele | P (White) | P (Hispanic) | Risk Allele | P | HR | 95% CI | White | Hispanic | ||
| rs6479778 | 63689077 | T | .0029 | .0296 | T | 8.1× 10−5 | 1.48 | 1.22 to 1.80 | 0.14 | 0.29 |
| rs2893881 | 63688672 | C | .001 | .0423 | C | 1.13× 10−4 | 1.45 | 1.20 to 1.75 | 0.14 | 0.30 |
| rs4948488 | 63685154 | C | .0043 | .0345 | C | 2.6× 10−4 | 1.41 | 1.18 to 1.69 | 0.16 | 0.41 |
| rs2393782 | 63670859 | G | .0883 | .029 | G | .0087 | 1.35 | 1.08 to 1.69 | 0.12 | 0.27 |
| rs10821938 | 63724773 | A | 1.47× 10−14 | 2.96× 10−6 | A | .0298 | 1.20 | 1.02 to 1.45 | 0.42 | 0.54 |
| rs7923074 | 63723440 | T | 1.5× 10−13 | 4.69× 10−7 | T | .0391 | 1.20 | 1.01 to 1.43 | 0.42 | 0.53 |
| rs6479779 | 63695048 | C | .0126 | .0364 | C | .0419 | 1.20 | 1.01 to 1.45 | 0.38 | 0.40 |
| rs17215180 | 63688728 | C | .251 | .0181 | C | .0494 | 1.22 | 1.00 to 1.49 | 0.53 | 0.70 |
NOTE. P values < .05 are shown in bold.
Abbreviations: ALL, acute lymphoblastic leukemia; HR, hazard ratio; ID, identification; SNP, single nucleotide polymorphism.
Chromosomal locations are based on hg19.
Allele frequency was determined on the basis of non-ALL controls.
Fig 2.
ARID5B single nucleotide polymorphisms are associated (CC, CT, and TT) with both acute lymphoblastic leukemia (ALL) susceptibility and relapse. (A) Relationship between genotype at ARID5B single nucleotide polymorphism rs6479778 and ALL relapse in the Children's Oncology Group P9904/9905 clinical trials. P value was determined by Fine and Gray's regression model after adjusting for treatment arm and ancestry. (B) Allele T at rs6479778 was more frequent in ALL cases than in controls and more common in Hispanics than in whites. P values were estimated by logistic regression.
DISCUSSION
The underlying causes of racial disparities in childhood ALL are likely to be multifactorial, including both genetic and environmental influences.14 For example, genetic polymorphisms may give rise to racial differences in ALL incidence when the prevalence of a common ALL-predisposing genetic variant differs by race, and/or when genetic variants are associated with ALL in a race-specific manner. Comparison of the association of 49 ARID5B SNPs with ALL in whites versus Hispanics revealed both similarities and differences: the majority of the SNPs most strongly associated with ALL susceptibility (eg, rs10821936) were common to the two races, strongly arguing for shared causal variants at this locus. Interestingly, we previously reported that rs10821936 is also associated with ALL susceptibility in blacks,20 and the risk allele frequency (16%) was significantly lower compared with that in whites (33%) and Hispanics (47%). Taken together, there appears to be a positive correlation between the frequency of the risk allele at this SNP and ALL incidence among race groups (blacks<whites<Hispanics; Fig 3). Several SNPs were associated with ALL in only the white or the Hispanic race (Table 1), suggesting possible population differences in the linkage disequilibrium of these SNPs with the causal variants. Further systematic resequencing of this genomic region in various ancestral groups is warranted to reveal the causative polymorphisms and the exact nature of the racial differences in ALL that are explained by ARID5B. No ARID5B SNPs were significantly associated with genomic loci previously linked to ALL susceptibility (ie, IKZF1; Data Supplement), suggesting that these genes independently contribute to ALL pathobiology.
Fig 3.
Racial differences in the risk allele frequency at rs10821936 and in acute lymphoblastic leukemia (ALL) incidence. (A) Genotype of rs10821936 is associated with ALL susceptibility in all three race groups. (B) Frequency of the risk allele (allele C) increases in order for blacks, whites, and Hispanics, consistent with the racial differences in ALL incidence. *Association of rs10821936 with ALL in blacks is based on a previous report by Yang et al.20 †ALL incidence by race is based on the report by Linabery et al.3
Interestingly, our multivariate analyses identified two ARID5B SNPs that independently contributed to ALL susceptibility in Hispanics, although a single SNP (rs10821936) almost entirely accounted for the association signal at this locus in whites. These observations imply that relationships between genetic variations and ALL susceptibility should be examined in the context of genetic ancestry. The C allele at rs10821936 was significantly over-represented in patients with a high percentage of NA ancestry at this locus and was surprisingly common in indigenous NA populations, raising the question of whether NAs are particularly susceptible to ALL. Although there is a paucity of data specifically assessing the incidence of ALL in NAs, a recent analysis of Surveillance, Epidemiology, and End Results (SEER) data from five states in the United States reported a 1.63-fold greater incidence of ALL in NA children than in whites, although the difference did not reach statistical significance because of small sample size.4 Internationally, one of highest incidence rates of childhood ALL occurs in Costa Rica,36,37 where Mestizos (admixed groups with NA ancestry) constitute a high proportion of the population. These observations are of interest but should be interpreted with caution for several reasons: race/ethnic classification by self-reports, relatively small size of NA samples, and possible incompleteness of the cancer registries (particularly in developing countries). It is intriguing that the population frequency of this allele somewhat parallels the order of human migration out of Africa38: the frequency is lowest in blacks (Africa), intermediate in whites and Asians (Europe and Asia), and highest in Native Americans (the Americas). Whether the ARID5B locus has been subject to selection pressure during human evolution warrants further investigation.39
Finally, one of the most significant findings from this study is that ARID5B germline SNPs related to ALL susceptibility were also associated with ALL outcome. To the best of our knowledge, this is the first report describing the relation between ARID5B and ALL treatment response in the context of a frontline ALL clinical trial. Further examination of ARID5B variation in the context of different ALL treatment regimens is warranted to refine its value as a prognostic marker. Interestingly, we previously observed that ARID5B SNP genotype is associated with leukemic cell accumulation of methotrexate metabolites (ie, polyglutamated methotrexate),18 offering a plausible mechanism by which ARID5B is linked to ALL relapse. Together, these results point to the possibility that leukemogenesis and antileukemic drug response mechanisms may converge on common pathways.
Supplementary Material
Acknowledgment
We thank Jeannette Pullen, MD, for assistance in classification of patients with ALL and the Center for Molecular Medicine for genome-wide genotyping of Children's Oncology Group (COG) P9904/9905 samples. We thank Sharon Naron for manuscript editing, Deepa Bhojwani, MD, for insightful comments, and Raul Ribeiro, MD, and Pedro de Alarcon, MD, for coordinating collaborations with Guatemala.
Appendix
Funding support for the Whole Genome Association Study of Bipolar Disorder was provided by the US National Institute of Mental Health (NIMH), and the genotyping of samples was provided through the Genetic Association Information Network (GAIN). The data sets used for the analyses described in this article were obtained from the database of Genotypes and Phenotypes (dbGaP; accession number phs000017.v3). Samples and associated phenotype data for the Collaborative Genomic Study of Bipolar Disorder were provided by the NIMH Bipolar Disorder Genetics Initiative. Data and biomaterials were collected in four projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1991 to 1998, the principal investigators and co-investigators were Indiana University, Indianapolis, IN, U01 MH46282, J. Nurnberger, M. Miller, and E. Bowman; Washington University, St. Louis, MO, U01 MH46280, T. Reich, A. Goate, and J. Rice; The Johns Hopkins University, Baltimore, MD, U01 MH46274, J.R. DePaulo Jr, S. Simpson, and C. Stine; and the NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, E. Gershon, D. Kazuba, and E. Maxwell. Data and biomaterials were collected as part of 10 projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1999 to 2003, the principal investigators and co-investigators were Indiana University, Indianapolis, IN, R01 MH59545, J. Nurnberger, M.J. Miller, E.S. Bowman, N.L. Rau, P.R. Moe, N. Samavedy, and R. El-Mallakh; University of Louisville, Louisville, KY, H. Manji; Wayne State University, Detroit, MI, D.A. Glitz, E.T. Meyer, C. Smiley, T. Foroud, L. Flury, D.M. Dick, and H. Edenberg; Washington University, St. Louis, MO, R01 MH059534, J. Rice, T. Reich, A. Goate, and L. Bierut; The Johns Hopkins University, Baltimore, MD, R01 MH59533, M. McInnis, J.R. DePaulo Jr, D.F. MacKinnon, F.M. Mondimore, J.B. Potash, P.P. Zandi, D. Avramopoulos, and J. Payne; University of Pennsylvania, Philadelphia, PA, R01 MH59553, W. Berrettini; University of California, Irvine, CA, R01 MH60068, W. Byerley and M. Vawter; University of Iowa, Iowa City, IA, R01 MH059548, W. Coryell and R. Crowe; University of Chicago, Chicago, IL, R01 MH59535, E. Gershon, J. Badner, F. McMahon, C. Liu, A. Sanders, M. Caserta, S. Dinwiddie, T. Nguyen, and D. Harakal; University of California, San Diego, CA, R01 MH59567, J. Kelsoe and R. McKinney; Rush University, Chicago, IL, R01 MH059556, W. Scheftner, H.M. Kravitz, D. Marta, A. Vaughn-Brown, and L. Bederow; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, F.J. McMahon, L. Kassem, S. Detera-Wadleigh, L. Austin, and D.L. Murphy.
Footnotes
Supported by Grants No. CA093552, CA78224, CA21765, R21CA158568, CA98543, RC4CA156449, CA114762, CA114766, CA021765-33, U10CA98413, U01GM61393, U01GM92666, and HL088133 from the National Institutes of Health, the National Cancer Institute Intramural Research Program, the American Lebanese Syrian Associated Charities, and CureSearch and by the American Society of Hematology Scholar Award and the Alex Lemonade Stand Foundation for Childhood Cancer Young Investigator Award (J.J.Y.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00005596.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Mary V. Relling, St. Jude Children's Research Hospital (C) Consultant or Advisory Role: None Stock Ownership: None Honoraria: None Research Funding: Mary V. Relling, Sigma Tau Pharmaceuticals Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Cheng Cheng, Naomi J. Winick, Cheryl L. Willman, Gregory H. Reaman, William L. Carroll, Mignon Loh, William E. Evans, Ching-Hon Pui, Stephen P. Hunger, Mary V. Relling, Jun J. Yang
Administrative support: Celeste Eng, Gregory H. Reaman
Provision of study materials or patients: Naomi J. Winick, Paul L. Martin, Cheryl L. Willman, Bruce M. Camitta, Ching-Hon Pui, Stephen P. Hunger
Collection and assembly of data: Meenakshi Devidas, Geoff Neale, Esteban G. Burchard, Dara G. Torgerson, Celeste Eng, Michael Dean, Frederico Antillon, Naomi J. Winick, Paul L. Martin, Cheryl L. Willman, Mignon Loh, Stephen P. Hunger, Jun J. Yang
Data analysis and interpretation: Heng Xu, Cheng Cheng, Deqing Pei, Yiping Fan, Wenjian Yang, Paul Scheet, Michael Dean, Cheryl L. Willman, Bruce M. Camitta, William E. Evans, Jun J. Yang
Manuscript writing: All authors
Final approval of manuscript: All authors
Affiliations
Heng Xu, Cheng Cheng, Deqing Pei, Yiping Fan, Wenjian Yang, Geoff Neale, William E. Evans, Ching-Hon Pui, Mary V. Relling, and Jun J. Yang, St. Jude Children's Research Hospital, Memphis, TN; Meenakshi Devidas, University of Florida, Gainesville, FL; Paul Scheet, The University of Texas MD Anderson Cancer Center, Houston; Naomi J. Winick, University of Texas Southwestern Medical Center, Dallas, TX; Esteban G. Burchard, Dara G. Torgerson, Celeste Eng, and Mignon Loh, University of California at San Francisco, San Francisco, CA; Michael Dean, National Cancer Institute, Frederick, MD; Federico Antillon, Unidad Nacional de Oncologia Pediatrica, Guatemala City, Guatemala; Paul L. Martin, Duke University, Durham, NC; Cheryl L. Willman, University of New Mexico Cancer Center, Albuquerque, NM; Bruce M. Camitta, Medical College of Wisconsin, Milwaukee, WI; Gregory H. Reaman, George Washington University and Children's National Medical Center, Washington, DC; William L. Carroll, New York University Cancer Institute, New York, NY; and Stephen P. Hunger, University of Colorado Denver School of Medicine and The Children's Hospital, Aurora, CO.
REFERENCES
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 2.McNeil DE, Coté TR, Clegg L, et al. SEER update of incidence and trends in pediatric malignancies: Acute lymphoblastic leukemia. Med Pediatr Oncol. 2002;39:554–557. doi: 10.1002/mpo.10161. discussion 552-553. [DOI] [PubMed] [Google Scholar]
- 3.Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992-2004) Cancer. 2008;112:416–432. doi: 10.1002/cncr.23169. [DOI] [PubMed] [Google Scholar]
- 4.Chow EJ, Puumala SE, Mueller BA, et al. Childhood cancer in relation to parental race and ethnicity: A 5-state pooled analysis. Cancer. 2010;116:3045–3053. doi: 10.1002/cncr.25099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horner MJ, Ries LAG, Krapcho M, et al. Bethesda, MD: National Cancer Institute; 2009. SEER Cancer Statistics Review, 1975-2006. [Google Scholar]
- 6.Bhatia S, Sather HN, Heerema NA, et al. Racial and ethnic differences in survival of children with acute lymphoblastic leukemia. Blood. 2002;100:1957–1964. doi: 10.1182/blood-2002-02-0395. [DOI] [PubMed] [Google Scholar]
- 7.Kadan-Lottick NS, Ness KK, Bhatia S, et al. Survival variability by race and ethnicity in childhood acute lymphoblastic leukemia. JAMA. 2003;290:2008–2014. doi: 10.1001/jama.290.15.2008. [DOI] [PubMed] [Google Scholar]
- 8.Pollock BH, DeBaun MR, Camitta BM, et al. Racial differences in the survival of childhood B-precursor acute lymphoblastic leukemia: A Pediatric Oncology Group Study. J Clin Oncol. 2000;18:813–823. doi: 10.1200/JCO.2000.18.4.813. [DOI] [PubMed] [Google Scholar]
- 9.Pui CH, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA. 2003;290:2001–2007. doi: 10.1001/jama.290.15.2001. [DOI] [PubMed] [Google Scholar]
- 10.Yang JJ, Cheng C, Devidas M, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43:237–241. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhatia S. Disparities in cancer outcomes: Lessons learned from children with cancer. Pediatr Blood Cancer. 2011;56:994–1002. doi: 10.1002/pbc.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 13.Bhatia S. Influence of race and socioeconomic status on outcome of children treated for childhood acute lymphoblastic leukemia. Curr Opin Pediatr. 2004;16:9–14. doi: 10.1097/00008480-200402000-00004. [DOI] [PubMed] [Google Scholar]
- 14.Buffler PA, Kwan ML, Reynolds P, et al. Environmental and genetic risk factors for childhood leukemia: Appraising the evidence. Cancer Invest. 2005;23:60–75. [PubMed] [Google Scholar]
- 15.Chokkalingam AP, Buffler PA. Genetic susceptibility to childhood leukaemia. Radiat Prot Dosimetry. 2008;132:119–129. doi: 10.1093/rpd/ncn255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koppen IJ, Hermans FJ, Kaspers GJ. Folate related gene polymorphisms and susceptibility to develop childhood acute lymphoblastic leukaemia. Br J Haematol. 2010;148:3–14. doi: 10.1111/j.1365-2141.2009.07898.x. [DOI] [PubMed] [Google Scholar]
- 17.Vijayakrishnan J, Houlston RS. Candidate gene association studies and risk of childhood acute lymphoblastic leukemia: A systematic review and meta-analysis. Haematologica. 2010;95:1405–1414. doi: 10.3324/haematol.2010.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Treviño LR, Yang W, French D, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W, Treviño LR, Yang JJ, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia. 2010;24:894–896. doi: 10.1038/leu.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han S, Lee KM, Park SK, et al. Genome-wide association study of childhood acute lymphoblastic leukemia in Korea. Leuk Res. 2010;34:1271–1274. doi: 10.1016/j.leukres.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Healy J, Richer C, Bourgey M, et al. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95:1608–1611. doi: 10.3324/haematol.2010.022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad RB, Hosking FJ, Vijayakrishnan J, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115:1765–1767. doi: 10.1182/blood-2009-09-241513. [DOI] [PubMed] [Google Scholar]
- 24.Vijayakrishnan J, Sherborne AL, Sawangpanich R, et al. Variation at 7p12.2 and 10q21.2 influences childhood acute lymphoblastic leukemia risk in the Thai population and may contribute to racial differences in leukemia incidence. Leuk Lymphoma. 2010;51:1870–1874. doi: 10.3109/10428194.2010.511356. [DOI] [PubMed] [Google Scholar]
- 25.Borowitz MJ, Devidas M, Hunger SP, et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia and its relationship to other prognostic factors: A Children's Oncology Group study. Blood. 2008;111:5477–5485. doi: 10.1182/blood-2008-01-132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.International Schizophrenia Consortium. Purcell SM, Wray NR, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burchard EG, Avila PC, Nazario S, et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169:386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 30.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao X, Bigham AW, Mei R, et al. A genomewide admixture mapping panel for Hispanic/Latino populations. Am J Hum Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allison PD. Cary, NC: SAS Institute; 1999. Logistic Regression Using the SAS System: Theory and Application. [Google Scholar]
- 33.Sankararaman S, Sridhar S, Kimmel G, et al. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 35.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santamaría-Quesada C, Vargas M, Venegas P, et al. Molecular and epidemiologic findings of childhood acute leukemia in Costa Rica. J Pediatr Hematol Oncol. 2009;31:131–135. doi: 10.1097/MPH.0b013e31818c919e. [DOI] [PubMed] [Google Scholar]
- 37.Redaelli A, Laskin BL, Stephens JM, et al. A systematic literature review of the clinical and epidemiological burden of acute lymphoblastic leukaemia (ALL) Eur J Cancer Care (Engl) 2005;14:53–62. doi: 10.1111/j.1365-2354.2005.00513.x. [DOI] [PubMed] [Google Scholar]
- 38.Ramachandran S, Deshpande O, Roseman CC, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci U S A. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casto AM, Feldman MW. Genome-wide association study SNPs in the human genome diversity project populations: Does selection affect unlinked SNPs with shared trait associations? PLoS Genet. 2011;7:e1001266. doi: 10.1371/journal.pgen.1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.