Abstract
Purpose
Mutations of the PIK3CA gene may predict response to phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) inhibitors. Concomitant mutations in the mitogen-activated protein kinase (MAPK) pathway may mediate resistance.
Patients and Methods
Tumors from patients with breast, cervical, endometrial, and ovarian cancer referred to the Clinical Center for Targeted Therapy (Phase I Program) were analyzed for PIK3CA, KRAS, NRAS, and BRAF mutations. Patients with PIK3CA mutations were treated, whenever feasible, with agents targeting the PI3K/AKT/mTOR pathway.
Results
Of 140 patients analyzed, 25 (18%) had PIK3CA mutations, including five of 14 patients with squamous cell cervical, seven of 29 patients with endometrial, six of 29 patients with breast, and seven of 60 patients with ovarian cancers. Of the 25 patients with PIK3CA mutations, 23 (median of two prior therapies) were treated on a protocol that included a PI3K/AKT/mTOR pathway inhibitor. Two (9%) of 23 patients had stable disease for more than 6 months, and seven patients (30%) had a partial response. In comparison, only seven (10%) of 70 patients with the same disease types but with wild-type PIK3CA treated on the same protocols responded (P = .04). Seven patients (30%) with PIK3CA mutations had coexisting MAPK pathway (KRAS, NRAS, BRAF) mutations (ovarian cancer, n = 5; endometrial cancer, n = 2), and two of these patients (ovarian cancer) achieved a response.
Conclusion
PIK3CA mutations were detected in 18% of tested patients. Patients with PIK3CA mutations treated with PI3K/AKT/mTOR inhibitors demonstrated a higher response rate than patients without mutations. A subset of patients with ovarian cancer with simultaneous PIK3CA and MAPK mutations responded to PI3K/AKT/mTOR inhibitors, suggesting that not all patients demonstrate resistance when the MAPK pathway is concomitantly activated.
INTRODUCTION
Activating oncogenic mutations are attractive drug targets in many malignancies.1–5 Mutations in the p110α subunit of PI3K, called PIK3CA, are often responsible for activation of the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway and have been reported in various human cancers.6 PIK3CA mutations can cause neoplastic transformation and promote cancer progression.7,8 The PI3K/AKT/mTOR pathway is often dysregulated in gynecologic and breast cancers, and PIK3CA mutations have been reported in approximately 18% of breast,9 17% to 33% of cervical,10,11 39% of endometrial,12 and 12% of ovarian cancers.9 Preclinical studies suggested that PIK3CA mutations could predict response to PI3K and mTOR inhibitors, although mutations in the mitogen-activated protein kinase (MAPK) pathway (KRAS, NRAS, BRAF) might mediate resistance.13–15
We investigated PIK3CA mutation status, and when enough tissue permitted, we also assessed the MAPK pathway (KRAS, NRAS, BRAF) mutation status of patients with advanced breast, cervical, endometrial, and ovarian cancers referred to the Phase I Clinical Trials Program clinic (known as the Clinical Center for Targeted Therapy). When feasible, the results of molecular matching were used for treatment selection, and in those cases, patients with PIK3CA mutations were offered treatment targeting the PI3K/AKT/mTOR pathway.
PATIENTS AND METHODS
Patients
Patients with advanced breast, cervical, endometrial, and ovarian cancers who experienced treatment failure with standard therapy and who had tissue available for mutation analysis were eligible. The study was carried out in the Department of Investigational Cancer Therapeutics (Phase I Clinical Trials Program) at The University of Texas MD Anderson Cancer Center (MD Anderson). The registration of patients in the database, pathology assessment, and mutation analysis were performed at MD Anderson. Eligible patients were those referred for phase I clinical trials for targeted therapeutic agents. The study and all treatments were conducted in accordance with the guidelines of the MD Anderson Institutional Review Board.
Tissue Samples and Mutation Analyses
PIK3CA, KRAS, NRAS, and BRAF mutations were investigated in archival formalin-fixed, paraffin-embedded tissue blocks or material from fine-needle aspiration biopsy obtained from diagnostic and/or therapeutic procedures. All histologies were centrally reviewed at MD Anderson. PIK3CA, KRAS, NRAS, and BRAF mutation testing was performed in the Clinical Laboratory Improvement Amendment–certified Molecular Diagnostic Laboratory within the Division of Pathology and Laboratory Medicine at MD Anderson. DNA was extracted from micro-dissected, paraffin-embedded tumor sections and analyzed using a polymerase chain reaction–based DNA sequencing method for PIK3CA mutations in codons [c]532 to [c]554 of exon 9 (helical domain) and c1011 to c1062 of exon 20 (kinase domain), which included the mutation hotspot region of the PIK3CA proto-oncogene by Sanger sequencing after amplification of 276– and 198–base pair amplicons, respectively, using primers designed by the MD Anderson Molecular Diagnostic Laboratory. Whenever possible, in addition to PIK3CA, mutation analysis was done for KRAS and NRAS c12, c13, and c61 mutations of exons 1 and 2 and BRAF c595 to c600 mutations of exon 15 using pyrosequencing as previously described.16
Treatment and Evaluation
Starting in October 2008, consecutive patients (N = 140) with advanced breast, cervical, endometrial, and ovarian cancers were studied. Patients with PIK3CA mutations were enrolled, whenever possible, onto clinical trials containing inhibitors of the PI3K/AKT/mTOR pathway. These clinical trials included temsirolimus, bevacizumab, and liposomal doxorubicin17 (ClinicalTrials.gov identifier: NCT00761644); single-agent temsirolimus (ClinicalTrials.gov identifier: NCT00877773); temsirolimus and bevacizumab (ClinicalTrials.gov identifier: NCT00610493); sirolimus and docetaxel (ClinicalTrials.gov identifier: NCT01054313); and PX86618 (ClinicalTrials.gov identifier: NCT00726583). Treatment continued until disease progression or unacceptable toxicity occurred. Treatment was carried out according to the specific requisites in the treatment protocols selected.
Assessments, including history, physical examination, and laboratory evaluations, were performed as specified in each protocol, typically before the initiation of therapy, weekly during the first cycle, and then, at a minimum, at the beginning of each new treatment cycle. Efficacy was assessed using computed tomography scans and/or magnetic resonance imaging at baseline before treatment initiation and then every two cycles (6 to 8 weeks). All radiographs were read in the Department of Radiology at MD Anderson and reviewed in the Department of Investigational Cancer Therapeutics tumor measurement clinic. Responses were categorized per Response Evaluation Criteria in Solid Tumors (RECIST) version 1.0 and were reported as best response.19 In brief, complete response (CR) was defined as the disappearance of all measurable and nonmeasurable disease. Partial response (PR) was defined as at least a 30% decrease in the sum of the longest diameter of measurable target lesions. Progressive disease was defined as at least a 20% increase in the sum of the longest diameter of measurable target lesions, unequivocal progression of a nontarget lesion, or the appearance of a new lesion. Stable disease (SD) was defined as neither sufficient shrinkage of tumor burden to qualify as a PR nor sufficient increase in tumor volume to qualify as progressive disease. A confirmation of CR/PR required repeat imaging at least 28 days after the initial response assessment.
Statistical Analysis
Statistical analysis was verified by our statistician (J.J.L.). The Fisher's exact test was used to assess the association among categorical variables and PIK3CA mutation status. The Wilcoxon rank sum test assessed the association between age and PIK3CA mutation status. Time to progression (TTP) was defined as the time interval from the start of therapy to the first observation of disease progression or death, whichever occurred first. All tests were two-sided, and P < .05 was considered statistically significant. All statistical analyses were carried out using SPSS 17 software (SPSS, Chicago, IL).
RESULTS
Patients
A total of 140 patients with advanced breast, cervical, endometrial, and ovarian cancers were analyzed for the presence of PIK3CA mutations. Their median age was 56.5 years (range, 25 to 91 years), and 113 patients (81%) were white, 11 (8%) were African American, seven (5%) were Hispanic, and nine (6%) were Asian. Sixty patients (43%) had ovarian cancer, 29 (21%) had endometrial cancer, 29 (21%) had breast cancer, 14 (10%) had squamous cell cervical cancer, and eight (5%) had cervical adenocarcinoma. Detailed patient characteristics are listed in Table 1.
Table 1.
Demographics and Clinical Characteristics of All Enrolled Patients With Breast, Cervical, Endometrial, and Ovarian Cancers (N = 140)
| Demographic or Clinical Characteristic | Wild-Type PIK3CA (n = 115)* |
Mutant PIK3CA (n = 25)* |
||
|---|---|---|---|---|
| No. of Patients | % | No. of Patients | % | |
| Age, years | ||||
| Median | 58 | 52 | ||
| Range | 25-91 | 35-72 | ||
| < 50 | 36 | 31.3 | 11 | 44.0 |
| 50-70 | 67 | 58.3 | 13 | 52.0 |
| > 70 | 12 | 10.4 | 1 | 4.0 |
| Ethnicity | ||||
| White | 95 | 82.6 | 18 | 72.0 |
| African American | 7 | 6.1 | 4 | 16.0 |
| Hispanic | 6 | 5.2 | 1 | 4.0 |
| Asian | 7 | 6.1 | 2 | 8.0 |
| Tumor type, histology | ||||
| Breast | 23 | 20.0 | 6 | 24.0 |
| Cervical | 17† | 14.8 | 5‡ | 20.0 |
| Endometrial | 22 | 19.1 | 7 | 28.0 |
| Ovarian | 53 | 46.1 | 7 | 28.0 |
| Site of mutation analysis | ||||
| Primary tumor | 54 | 47.0 | 15 | 60.0 |
| Metastatic tumor | 61 | 53.0 | 10 | 40.0 |
| No. of prior therapies | ||||
| Median | 4 | 3 | ||
| Range | 1-14 | 1-12 | ||
| ≤ 2 | 31 | 27.0 | 11 | 44.0 |
| > 2 | 84 | 73.0 | 14 | 56.0 |
There was no significant difference in any of the listed characteristics between patients with wild-type and mutant PIK3CA.
Adenocarcinoma, n = 8; squamous cell carcinoma, n = 9.
All squamous cell carcinomas.
PIK3CA Mutations
PIK3CA proto-oncogene mutations were detected in 25 (18%) of the 140 study patients (Table 2). The most frequent mutation was H1047R (a mutation in c1047 of PIK3CA that changes the encoded amino acid from histidine to arginine) detected in 11 patients (Table 3). PIK3CA mutations were detected in five (36%) of 14 patients with squamous cell cervical cancer, seven (24%) of 29 patients with endometrial cancer, six (21%) of 29 patients with breast cancer, and seven (12%) of 60 patients with ovarian cancer. PIK3CA mutation status was not significantly associated with age, disease type, or ethnicity.
Table 2.
Distribution of PIK3CA, KRAS, NRAS, and BRAF Mutations
| Oncogene | Mutated |
Total Tested (No.) | P | |
|---|---|---|---|---|
| No. | % | |||
| PIK3CA | 25 | 18 | 140 | NA |
| KRAS | 10 | 10 | 98 | NA |
| NRAS | 2 | 4 | 53 | NA |
| BRAF | 2 | 2 | 84 | NA |
| KRAS/NRAS or BRAF | 14 | 17 | 81 | NA |
| KRAS in mutant PIK3CA | 5 | 23 | 22 | .04 |
| KRAS in wild-type PIK3CA | 5 | 7 | 76 | |
| KRAS/NRAS or BRAF in mutant PIK3CA | 7* | 35 | 20 | .04 |
| KRAS/NRAS or BRAF in wild-type PIK3CA | 7 | 11 | 61 | |
Abbreviation: NA, not applicable.
Simultaneous mutations in PIK3CA and KRAS (n = 5), PIK3CA and BRAF (n = 1), or PIK3CA and NRAS (n = 1).
Table 3.
PIK3CA, KRAS, NRAS, and BRAF Mutations
| Patient No. | Histology | PIK3CA Mutation | KRAS Mutation | NRAS Mutation | BRAF Mutation |
|---|---|---|---|---|---|
| 1 | Breast: lobular ER positive/PR positive/HER2 negative | H1047R | None | None | None |
| 2 | Breast: metaplastic, triple negative | H1047R | None | Not done | None |
| 3 | Breast: ductal, ER positive/PR positive/HER2 negative | E542K | None | None | None |
| 4 | Breast: ductal, ER negative/PR negative/HER2 positive | H1047R | None | Not done | Not done |
| 5 | Breast: ductal, ER positive/PR positive/HER2 positive | M1043I | Not done | Not done | Not done |
| 6 | Breast: ductal, ER positive/PR positive/HER2 negative | H1047R | None | None | Not done |
| 7 | Breast: ductal, ER positive/PR positive/HER2 negative | None | Q61L | None | None |
| 8 | Cervix: squamous | E545K | None | None | None |
| 9 | Cervix: squamous | E545K/D549H | None | None | None |
| 10 | Cervix: squamous | E545K | None | None | None |
| 11 | Cervix: squamous | E542K | None | Not done | None |
| 12 | Cervix: squamous | E545K | None | Not done | None |
| 13 | Cervix: squamous | None | G12D | None | None |
| 14 | Endometrial: clear cell | H1047R | Not done | Not done | None |
| 15 | Endometrial: endometrioid | H1047R | None | None | None |
| 16 | Endometrial: endometrioid | G1049R | Not done | Not done | Not done |
| 17 | Endometrial: papillary | H1047R | None | G13D | None |
| 18 | Endometrial: endometrioid | E545G | G12A | None | None |
| 19 | Endometrial: endometrioid | H1047L | None | Not done | None |
| 20 | Endometrial: endometrioid | H1047R | None | None | None |
| 21 | Endometrial: endometrioid | None | G13D | Not done | None |
| 22 | Endometrial: clear cell | None | Q61L | Not done | Not done |
| 23 | Endometrial: endometrioid | None | None | Q61L | Not done |
| 24 | Ovarian: endometrioid | Q546K | Q61H | None | None |
| 25 | Ovarian: high-grade serous | E542K | None | None | None |
| 26 | Ovarian: clear cell | G1049R | None | None | None |
| 27 | Ovarian: clear cell | H1047R | None | None | V600E |
| 28 | Ovarian: endometrioid | H1047R | G12D | None | None |
| 29 | Ovarian: high-grade serous | H1047R | G13D | None | None |
| 30 | Ovarian: endometrioid | M1043V | G12V | None | None |
| 31 | Ovarian: clear cell | None | Q61H | Not done | Not done |
| 32 | Ovarian: low-grade serous | None | None | None | V600E |
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Simultaneous RAS and PIK3CA Mutations
KRAS mutations in exons 1 or 2 were assessed in 98 patients who had enough tissue available for mutation analysis and were identified in 10 patients (10%; Table 2). The mutations were most frequent in codon 12 (G12D [changes the encoded amino acid from glycine to aspartic acid], n = 2; G12A [changes the encoded amino acid from glycine to alanine], n = 1; G12V [changes the encoded amino acid from glycine to valine], n = 1; Table 3). The presence of KRAS mutations was significantly associated with PIK3CA mutations. Indeed, 23% of patients (five of 22 patients) with a PIK3CA mutation (who had enough tissue for KRAS mutation analysis) also had a KRAS mutation, whereas only 7% of patients (five of 76 patients) without a PIK3CA mutation (who were also tested for KRAS) harbored a KRAS mutation (P = .04; Table 2). Of the 10 patients with KRAS mutations, five (50%) had simultaneous PIK3CA mutations. In contrast, of the 88 patients without a KRAS mutation, only 17 (19%) had a PIK3CA mutation (P = .04). Of the five patients with simultaneous PIK3CA and KRAS mutations, four had ovarian cancer, and one had endometrial cancer (Table 3).
NRAS mutations (G13D [changes the encoded amino acid from glycine to aspartic acid] in codon 13 and Q61L [changes the encoded amino acid from glutamine to leucine] in codon 61) were detected in two patients (4%) of 53 tested who had enough tissue available for NRAS mutation analysis (Table 2), and one of those two patients had a simultaneous PIK3CA mutation (endometrial cancer; Table 3).
Simultaneous BRAF and PIK3CA Mutations
BRAF exon 15 mutations were assessed in 84 patients (Table 2) who had enough tissue available for BRAF mutation analysis. Two patients (2%) had a V600E mutation (a mutation in c600 of BRAF that changes the encoded amino acid from valine to glutamic acid), and one of those two patients (ovarian cancer) had a simultaneous PIK3CA mutation (Table 3).
Simultaneous MAPK Pathway (KRAS, NRAS, BRAF) Mutations and PIK3CA Mutations
We analyzed associations between PIK3CA mutations and all tested MAPK pathway mutations (KRAS, NRAS, BRAF). This analysis included patients tested for RAS (KRAS or NRAS) and BRAF mutations. Not all patients could be tested for all mutations because of the limited amount of available tumor tissue. Mutations in KRAS, NRAS, and BRAF are considered mutually exclusive20; therefore, patients with mutations in RAS (KRAS or NRAS) or BRAF were included, even if they were not tested for both RAS and BRAF. In total, 81 patients were included in this analysis. RAS (KRAS or NRAS) or BRAF mutations were identified in 14 (17%) of these 81 patients. The presence of RAS (KRAS or NRAS) or BRAF mutations was significantly associated with PIK3CA mutations. Thirty-five percent of patients (seven of 20 patients) with a PIK3CA mutation also had a RAS (KRAS or NRAS) or BRAF mutation, whereas only 11% of patients (seven of 61 patients) without a PIK3CA mutation harbored a RAS (KRAS or NRAS) or BRAF mutation (P = .04).
Response in Patients With PIK3CA Mutations Treated With PI3K/AKT/mTOR Inhibitors
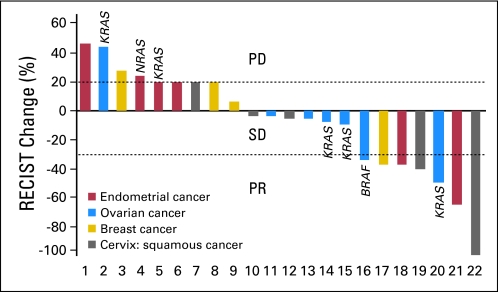
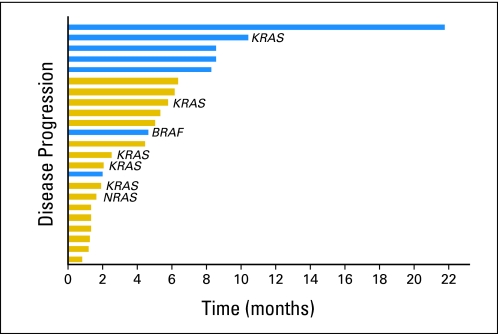
Twenty-three (92%) of 25 patients with an underlying PIK3CA mutation were enrolled onto clinical trials that included a PI3K/AKT/mTOR inhibitor.17,18,21 Two patients were not enrolled because of eligibility or logistical reasons. These 23 patients were refractory to a median of two prior therapies (range, one to 12 prior therapies). Of the 23 patients, seven had ovarian cancer, six had endometrial cancer, five had breast cancer, and five had squamous cell cervical cancer. A response (six confirmed PRs, one unconfirmed PR) was observed in seven patients (30%; 95% CI, 16% to 51%; Figs 1 and 2). Duration of response in the seven responders was 2.0, 4.6, 8.2, 8.4, 8.5, 10.3, and 21.6 months (Fig 3). Two patients (9%; 95% CI, 2% to 27%) who did not have a PR experienced prolonged SD, which lasted for more than 6 months (Fig 3). In total, nine patients (39%) achieved either SD for more than 6 months or a PR. Of the seven patients who responded, six received a combination of a PI3K/AKT/mTOR inhibitor and a cytotoxic drug (liposomal doxorubicin). The responders had a median of two prior therapies (range, one to 12 prior therapies); five of the responders received prior platinum-based, five received prior taxane-based, and three received prior doxorubicin-based therapy and experienced progression. In comparison, only seven (10%; 95% CI, 5% to 19%) of 70 patients with the same disease types with wild-type PIK3CA treated on the same protocols responded (PR or CR; P = .04). Of those 70 patients, 68 received combination therapies, most commonly with cytotoxic drugs such as liposomal doxorubicin or docetaxel. In patients with PIK3CA mutations, there were no associations among response and other patient characteristics, such as age, race, number of prior therapies (> v ≤ two therapies), time from tumor sample collection to mutation analysis, and site of mutation analysis (primary tumor v metastasis). Patients with an H1047R PIK3CA mutation experienced a response rate of 44% (four of nine patients; 95% CI, 19% to 73%) compared with a response rate of 21% (three of 14 patients; 95% CI, 8% to 48%) in patients with other PIK3CA mutations (P = .36). Patients treated with combinations of agents demonstrated a trend to a higher response rate of 44% (seven of 16 patients; 95% CI, 23% to 67%) compared with a rate of 0% (zero of seven patients; 95% CI, 0% to 35%) in patients treated with single-agent therapies (P = .06). Response rates per tumor type were as follows: breast, 20% (one of five patients); squamous cell cervical, 40% (two of five patients); endometrial, 33% (two of six patients); and ovarian, 29% (two of seven patients; Fig 1).
Fig 1.
Waterfall plot of patients with PIK3CA mutations treated with phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin inhibitors. The overall response rate was 30%. Five patients with ovarian cancer and two patients with endometrial cancer had simultaneous PIK3CA and MAPK pathway (KRAS, NRAS, or BRAF) mutations. One patient with breast cancer who was never evaluated for response is not depicted. PD, progressive disease; PR, partial response; RECIST, Response Evaluation Criteria in Solid Tumors; SD, stable disease.
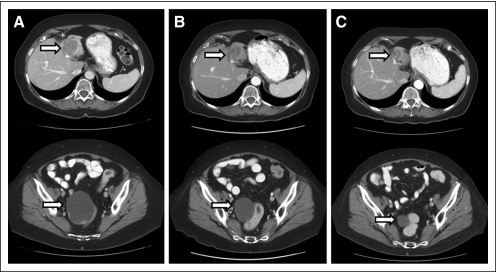
Fig 2.
Computed tomography scans of a responding patient (previously treated with cisplatin/gemcitabine, paclitaxel, carboplatin, liposomal doxorubicin, and cetuximab) with endometrial cancer demonstrating response on therapy with temsirolimus, bevacizumab, and liposomal doxorubicin. (A) Pretreatment scans of liver metastasis and pelvic mass. (B) Restaging scans after two cycles (6 weeks) demonstrating response in liver and pelvic mass. (C) Restaging scans after six cycles (18 weeks) demonstrating continuing response in liver and pelvic mass.
Fig 3.
Time to progression (TTP). Seven patients (30%), of whom five had a partial response (blue bars), experienced TTP longer than 6 months. All patients were off therapy at the time of analysis (19 experienced progression, and four withdrew consent).
DISCUSSION
We detected helical and kinase domain PIK3CA mutations in 18% of 140 patients with advanced breast, cervical, endometrial, and ovarian cancers. The prevalence was highest in squamous cell cervical cancer (36%), followed by endometrial cancer (24%), breast cancer (21%), and ovarian cancer (12%). Although the small number of patients precludes definitive conclusions regarding absolute difference in mutation rates, our results are consistent with those in the literature.9,11,12 Eligible patients received treatments, if feasible, containing a PI3K/AKT/mTOR inhibitor. In 23 PIK3CA-mutant patients with breast and gynecologic cancers who experienced treatment failure with standard therapies, we observed a response rate of 30%. This response rate is favorable compared with a 10% response rate in patients with advanced breast, cervical, endometrial, and ovarian cancers and wild-type PIK3CA treated on the same protocols (P = .04). The latter is comparable to the response rate of 4% to 11% reported by our group and others when patients were treated on phase I trials without molecular selection.22–25 It is also conceivable that the response rate of 10% in unselected patients was on the high end compared with response rates reported previously from other phase I clinics,22,23 because even when mutation status was negative, physicians tended to select patients for PI3K/AKT/mTOR inhibitor studies based on the known frequency of other mutations in the PI3K/AKT/mTOR pathway not detected by our assay (eg, PIK3R1, PTEN, AKT).26 Previously published oncogene-driven clinical trials with a BRAF inhibitor in BRAF-mutant melanoma, epidermal growth factor receptor (EGFR) inhibitors in EGFR-mutant non–small-cell lung cancer, and an ALK inhibitor in non–small-cell lung cancer with an underlying EML4-ALK fusion also show a relatively high response rate, even in the phase I setting.4,5,27 In our study, most observed responses were durable, with five (70%) of seven responses lasting longer than 8 months.
Preclinical experiments suggested that PIK3CA mutations render tumors sensitive to PI3K and/or mTOR inhibitors, whereas simultaneous mutations in the MAPK pathway (KRAS, NRAS, BRAF) can mediate resistance to therapy.13–15 We demonstrated that mutations in the MAPK pathway (KRAS, NRAS, BRAF) are more frequent in patients with PIK3CA mutations compared with patients with wild-type PIK3CA (35% v 11%, respectively; P = .04). Of interest, two of our patients with ovarian cancer and coexisting PIK3CA and MAPK pathway mutations responded to PI3K/AKT/mTOR inhibitors.
Conceptually, the use of molecular profiling in early-phase clinical trials has the potential to accelerate the development of new therapies. A decade elapsed before implementation of molecular profiling to identify patients with advanced lung cancer who demonstrated benefit from EGFR tyrosine kinase inhibitors.28 However, development of other targeted therapies, such as ALK inhibitors in lung cancer with an EML4-ALK fusion and BRAF inhibitors in BRAF-mutated melanoma, was more streamlined, with molecular profiling successfully incorporated in the clinical study designs, including phase I trials, which demonstrated striking responses.4,5
Other examples of successful implementation of molecular profiling include imatinib mesylate (a KIT and BCR-ABL kinase inhibitor), which demonstrated response rates greater than 50% in patients with GI stromal tumors (a disorder characterized by KIT kinase mutations) and BCR-ABL–positive chronic myelogenous leukemia.1,29 The response rate in our study is lower than the rates described earlier. Future larger analyses should examine other variables such as coexisting RAS or RAF mutations and/or specific types of PIK3CA mutations to assess factors attenuating responsiveness. In addition, our patients were treated on different dose levels of several early-phase clinical trials. It is plausible that some patients may have received doses or drugs that did not adequately inhibit the pathway.
Finally, all responses in our study were observed with combination, but not single-agent, therapies (44% v 0%, respectively; P = .06). This is in agreement with preclinical data, which suggested that single-agent PI3K/AKT/mTOR pathway inhibition may not always be sufficient to induce a response because PIK3CA mutations often coexist with other concurrent molecular aberrations.13,14 This observation is potentially important for further development of PI3K/AKT/mTOR inhibitors.
In conclusion, we have shown that PIK3CA mutations occur in a significant proportion of patients with advanced breast, cervical, endometrial, and ovarian cancers and that, even in a patient population that has experienced failure with standard therapies, they are associated with response to treatments that include PI3K/AKT/mTOR inhibitors. Such responses have also been anecdotally reported in patients with other PIK3CA-mutant cancers, and we are now analyzing a larger group of such patients.30 Because the number of patients in our series was small and no random assignment occurred, these data must be interpreted cautiously. However, it seems that screening for PIK3CA mutations warrants further investigation in the application of targeted PI3K/AKT/mTOR inhibitors in the clinic, especially in gynecologic and breast cancers, where these mutations are common.
Acknowledgment
We thank Joann Aaron for editorial assistance and Vanda Stepanek for assistance with data collection.
Footnotes
See accompanying editorial on page 765
Supported by Grant No. RR024148 from the National Center for Research Resources, a component of the National Institutes of Health Roadmap for Medical Research.
Presented in part at the 13th Biennial Meeting of the International Gynecologic Cancer Society, October 23-26, 2010, Prague, Czech Republic.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00761644.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Apostolia M. Tsimberidou, Baxter (C), Cephalon (C), Caris (C) Stock Ownership: None Honoraria: None Research Funding: Apostolia M. Tsimberidou, National Comprehensive Cancer Network, sanofi-aventis, Celgene Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Filip Janku, Jennifer J. Wheler, Shannon N. Westin, Stacy L. Moulder, Aung Naing, Apostolia M. Tsimberidou, Siqing Fu, Gerald S. Falchook, David S. Hong, Ignacio Garrido-Laguna, Karen H. Lu, Razelle Kurzrock
Administrative support: Filip Janku, Razelle Kurzrock
Provision of study materials or patients: Filip Janku, Jennifer J. Wheler, Shannon N. Westin, Stacy L. Moulder, Aung Naing, Apostolia M. Tsimberidou, Siqing Fu, Gerald S. Falchook, David S. Hong, Ignacio Garrido-Laguna, Rajyalakshmi Luthra, Karen H. Lu, Razelle Kurzrock
Collection and assembly of data: Filip Janku, Ignacio Garrido-Laguna
Data analysis and interpretation: Filip Janku, Ignacio Garrido-Laguna, Rajyalakshmi Luthra, J. Jack Lee, Razelle Kurzrock
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 3.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 4.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 7.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 9.Levine DA, Bogomolniy F, Yee CJ, et al. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–2878. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 10.Janku F, Tsimberidou A, Garrido-Laguna I, et al. PIK3CA, KRAS, and BRAF mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. J Clin Oncol. 2010;28(suppl; abstr 2583):224s. [Google Scholar]
- 11.Miyake T, Yoshino K, Enomoto T, et al. PIK3CA gene mutations and amplifications in uterine cancers, identified by methods that avoid confounding by PIK3CA pseudogene sequences. Cancer Lett. 2008;261:120–126. doi: 10.1016/j.canlet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Hayes MP, Wang H, Espinal-Witter R, et al. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res. 2006;12:5932–5935. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- 13.Ihle NT, Lemos R, Jr, Wipf P, et al. Mutations in the phosphatidylinositol-3-kinase pathway predict for antitumor activity of the inhibitor PX-866 whereas oncogenic Ras is a dominant predictor for resistance. Cancer Res. 2009;69:143–150. doi: 10.1158/0008-5472.CAN-07-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Nicolantonio F, Arena S, Tabernero J, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–2866. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuo Z, Chen SS, Chandra PK, et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22:1023–1031. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 17.Morony JW, Schlumbrecht MP, Helgason T, et al. A phase I trial of liposomal doxorubicin, bevacizumab, and temsirolimus in patients with advanced gynecologic and breast malignancies. Clin Cancer Res. 2011;17:6840–6846. doi: 10.1158/1078-0432.CCR-11-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jimeno A, Herbst RS, Falchook GS, et al. Final results from a phase I, dose-escalation study of PX-866, an irreversible, pan-isoform inhibitor of PI3 kinase. J Clin Oncol. 2010;28(suppl; abstr 3089):255s. [Google Scholar]
- 19.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 20.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 21.Janku F, Tsimberidou AM, Garrido-Laguna I, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitor. Mol Cancer Ther. 2011;10:558–565. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horstmann E, McCabe MS, Grochow L, et al. Risks and benefits of phase 1 oncology trials, 1991 through 2002. N Engl J Med. 2005;352:895–904. doi: 10.1056/NEJMsa042220. [DOI] [PubMed] [Google Scholar]
- 23.Jain RK, Lee JJ, Hong D, et al. Phase I oncology studies: Evidence that in the era of targeted therapies patients on lower doses do not fare worse. Clin Cancer Res. 2010;16:1289–1297. doi: 10.1158/1078-0432.CCR-09-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurzrock R, Benjamin RS. Risks and benefits of phase 1 oncology trials, revisited. N Engl J Med. 2005;352:930–932. doi: 10.1056/NEJMe058007. [DOI] [PubMed] [Google Scholar]
- 25.Moroney J, Wheler J, Hong D, et al. Phase I clinical trials in 85 patients with gynecologic cancer: The M. D. Anderson Cancer Center experience. Gynecol Oncol. 2010;117:467–472. doi: 10.1016/j.ygyno.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): A resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 28.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 29.van Oosterom AT, Judson I, Verweij J, et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: A phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- 30.Janku F, Garrido-Laguna I, Wheler JJ, et al. Screening for PIK3CA mutations, PTEN loss, and RAS/RAF mutations in early-phase protocols with PI3K/mTOR pathway inhibitors. J Clin Oncol. 2011;29(suppl; abstr 10507):631s. [Google Scholar]