Abstract
Purpose
Chromosomal rearrangements involving the ROS1 receptor tyrosine kinase gene have recently been described in a subset of non–small-cell lung cancers (NSCLCs). Because little is known about these tumors, we examined the clinical characteristics and treatment outcomes of patients with NSCLC with ROS1 rearrangement.
Patients and Methods
Using a ROS1 fluorescent in situ hybridization (FISH) assay, we screened 1,073 patients with NSCLC and correlated ROS1 rearrangement status with clinical characteristics, overall survival, and when available, ALK rearrangement status. In vitro studies assessed the responsiveness of cells with ROS1 rearrangement to the tyrosine kinase inhibitor crizotinib. The clinical response of one patient with ROS1-rearranged NSCLC to crizotinib was investigated as part of an expanded phase I cohort.
Results
Of 1,073 tumors screened, 18 (1.7%) were ROS1 rearranged by FISH, and 31 (2.9%) were ALK rearranged. Compared with the ROS1-negative group, patients with ROS1 rearrangements were significantly younger and more likely to be never-smokers (each P < .001). All of the ROS1-positive tumors were adenocarcinomas, with a tendency toward higher grade. ROS1-positive and -negative groups showed no difference in overall survival. The HCC78 ROS1-rearranged NSCLC cell line and 293 cells transfected with CD74-ROS1 showed evidence of sensitivity to crizotinib. The patient treated with crizotinib showed tumor shrinkage, with a near complete response.
Conclusion
ROS1 rearrangement defines a molecular subset of NSCLC with distinct clinical characteristics that are similar to those observed in patients with ALK-rearranged NSCLC. Crizotinib shows in vitro activity and early evidence of clinical activity in ROS1-rearranged NSCLC.
INTRODUCTION
Recent advances with targeted therapies have led to a major paradigm shift in oncology. The success of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), such as erlotinib and, more recently, the ALK/MET TKI crizotinib, highlights the need to match targeted therapies to the appropriate genetically defined patient population.1–4 Thus, rapid and efficient identification of key driver genes in non–small-cell lung cancer (NSCLC) is becoming increasingly important.5 Clinical screening efforts have revealed that the most common mutations in lung cancer specimens involve KRAS and EGFR, along with 10 other genes that show a prevalence of mutation in 5% or less of tumors. The ALK gene is rearranged in 3% of patients with NSCLC and has been the focus of intense basic and clinical research over the last 3 years.6–8 Even with large-scale genotyping efforts, 40% of NSCLCs do not have an identifiable driver mutation.
Our interest in exploring chromosomal rearrangements other than ALK as potential drivers in lung cancer has led us to study ROS1, a gene recently shown to be involved in chromosomal translocations in lung cancer.9 ROS1 is a receptor tyrosine kinase of the insulin receptor family. Chromosomal rearrangements involving the ROS1 gene were originally described in glioblastomas, where ROS1 (chromosome 6q22) is fused to the FIG gene (chromosome 6q22 immediately adjacent to ROS1),10–12 and have been shown to be transforming in transgenic mice.13 More recently, ROS1 fusions were identified as potential driver mutations in an NSCLC cell line (HCC78; SLC34A2-ROS1) and an NSCLC patient sample (CD74-ROS1).9 These fusions lead to constitutive kinase activity and are associated with sensitivity in vitro to TKIs. Our prior studies of the activity of the ALK inhibitor TAE684 in a large panel of cancer cell lines showed that the HCC78 cell line is among the 10 most sensitive cell lines.14 Because the other nine lines have ALK abnormalities (translocations or amplifications), the data suggest that ROS1 is inhibited as an off-target effect of TAE684. Currently there are no ROS1-specific agents in clinical trial. The clinical characteristics of patients with ROS1-rearranged NSCLC have not yet been described.
Here, we determine the prevalence of ROS1 rearrangements in NSCLC and define the clinicopathologic characteristics of ROS1-positive tumors, with the identification of 18 patients with ROS1-rearranged NSCLC (approximately 2% of screened patients). Our data indicate that ROS1-positive NSCLCs arise in young never-smokers with adenocarcinoma, which interestingly is a profile similar to patients with ALK-rearranged NSCLCs. In vitro studies suggest that the ALK/MET inhibitor crizotinib may effectively inhibit the growth of ROS1-positive tumors.
PATIENTS AND METHODS
Study Population
The study included an institutional review board–approved retrospective analysis of a series of 1,073 patients with NSCLC seen at Massachusetts General Hospital (MGH) Cancer Center (n = 574), Vanderbilt University Medical Center (n = 443), University of California Irvine Medical Center (n = 52), and Fudan University Shanghai Cancer Center (n = 4). For all patients, medical records were reviewed to extract data on clinicopathologic characteristics, including age, sex, stage, histology, overall survival (OS), and smoking history. OS was measured from the date of diagnosis until the date of death; patients lost to follow-up, deaths unrelated to NSCLC, or patients alive and well were censored. For the patients in the crizotinib trial, responses were classified by using standard Response Evaluation Criteria in Solid Tumors (RECIST), version 1.0.
Molecular Pathology and Fluorescent In Situ Hybridization
Hematoxylin and eosin staining was performed on 5-μm sections from formalin-fixed paraffin-embedded (FFPE) tumor tissue. All tumors were evaluated by at least two pathologists, classified using WHO criteria, and staged according to updated American Joint Committee on Cancer TNM criteria. We used a break-apart fluorescent in situ hybridization (FISH) approach using BAC clones corresponding to the 5′ (RP11-835I21) and 3′ (RP11-1036C2) sequences flanking the ROS1 gene labeled by nick translation in green and red. FFPE slides were deparaffinized, treated with protease, codenatured with FISH probes using a Hybrite slide processor (Abbott Molecular, Chicago, IL), washed, counterstained, cover-slipped, and analyzed using an Olympus BX61 fluorescence microscope (Olympus, Tokyo, Japan) equipped with red, green, and 4′,6-diamidino-2-phenylindole filters. Images were captured and analyzed using Cytovision software (Genetix, San Jose, CA). Positive cases were defined as tumors harboring more than 15% of cells with split signals.
Reverse Transcriptase Polymerase Chain Reaction
ROS1 breakpoints were mapped using reverse transcriptase polymerase chain reaction (RT-PCR) combined with Sanger sequencing of PCR products. Only two ROS1 rearrangements have been mapped in NSCLC—the SLC34A2-ROS1 fusion in the HCC78 cell line (fusing exon 4 of SLC34A2 to exon 32 of ROS1) and the CD74-ROS1 fusion in an NSCLC patient sample (fusing exon 6 of CD74 to exon 34 of ROS1). Thus, we performed analysis of RNA from 14 ROS1-positive samples with sufficient tissue with a panel of PCR primers including SLC34A2 exon 4 and CD74 exon 6 forward primers paired each with a ROS1 exon 32 and exon 34 reverse primer. RNA was extracted from FFPE tissue using the Agencourt Formapure method (Agencourt Biosciences, Beverly, MA) and reverse transcribed using the Superscript III cDNA synthesis kit (Invitrogen, Carlsbad, CA). The following primers were used to determine SLC34A2-ROS1 or CD74-ROS1 breakpoints: SLC34A2 exon 4 forward, TCGGATTTCTCTACTTTTTCGTG; CD74 exon 6 forward, CTCCTGTTTGAAATGAGCAGG; ROS1 exon 32 reverse, GGAATGCCTGGTTTATTTGG; and ROS1 exon 34 reverse, TGAAACTTGTTTCTGGTATCCAA. PCR amplifications were performed in an Eppendorf Mastercycler Gradient (Eppendorf, Hamburg, Germany), using Platinum Taq polymerase (Invitrogen) under standard conditions with 40 ng of cDNA. cDNA was sequenced using the BigDye 3.0 kit (Life Technologies, Carlsbad, CA).
Cell Line Crizotinib Sensitivity Testing
Human NSCLC lines PC9, HCC827, MGH006, NCI-H3122, and HCC-78 cells (obtained from DSMZ, Braunschweig, Germany) were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (RPMI 1640 growth medium). Crizotinib purchased from ChemieTek (Indianapolis, IN), was dissolved in dimethyl sulfoxide for cell culture experiments. For 72-hour drug treatments, 3,000 cells were plated in replicates of six into 96-well plates. After drug treatments, cells were incubated with CellTiter-Glo assay reagent (Promega, Fitchburg, WI) for 10 minutes, and luminescence was measured using a Centro LB 960 microplate luminometer (Berthold Technologies, Oak Ridge, TN). For transfection experiments, the HEK 293 cell line (ATCC, Manassas, VA) was cultivated in DMEM (Mediatech, Manassas, VA) supplemented with 10% FBS. Two hundred ninety-three cells were transfected with CD74-ROS1 or EML4-ALK E13;A20 cDNA constructs using Superfect reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Western Blots
Cells were resuspended in lysis buffer (20 mmol/L Tris, 150 mmol/L NaCl, 1% Nonidet P-40, 10% glycerol, 1 mmol/L EDTA, 1 mmol/L ethyleneglycoltetracetic acid, and protease and phosphatase inhibitors), incubated on ice for 10 minutes, and centrifuged for 5 minutes (15,000 rpm). Protein concentration determination and immunoblotting were performed as previously described.15 The phospho-ROS1, ROS1, phospho-ALK (Y1604), and ALK antibodies were obtained from Cell Signaling Technology (Danvers, MA). The actin antibody was purchased from Sigma (St Louis, MO).
Expression Constructs
The 3FLAG-EML4-ALK E13;A20 (variant 1) construct has been described previously.16 cDNA for CD74-ROS1 was synthesized by Geneart (Regensburg, Germany). The cDNAs were subcloned into pcDNA3.1+ (Invitrogen).
Statistical Analysis
Statistical Analysis consisted of the Fisher's exact test (association of genotype with dichotomous factors), χ2 test, or t test (comparison of means). The Kaplan-Meier method was used to estimate OS, and differences between genotypes were compared using the log-rank test. Data analysis was conducted using Prism 5.0b (GraphPad Software, San Diego, CA), and significance was defined as P < .05. All P values were two-tailed.
Crizotinib Trial
Our study also includes preliminary data of clinical response in one patient (MGH, Boston, MA) enrolled onto an open-label, multicenter trial of the ALK/MET TKI crizotinib (Pfizer, La Jolla, CA; ClinicalTrials.gov identifier: NCT00585195). The trial was conducted in accordance with the Declaration of Helsinki and was approved by the ethics committee at each participating institution; patients on the crizotinib trial were required to give written informed consent before enrolling onto that study. The crizotinib phase I study was sponsored by Pfizer.
RESULTS
Development of ROS1 FISH and Patient Screening
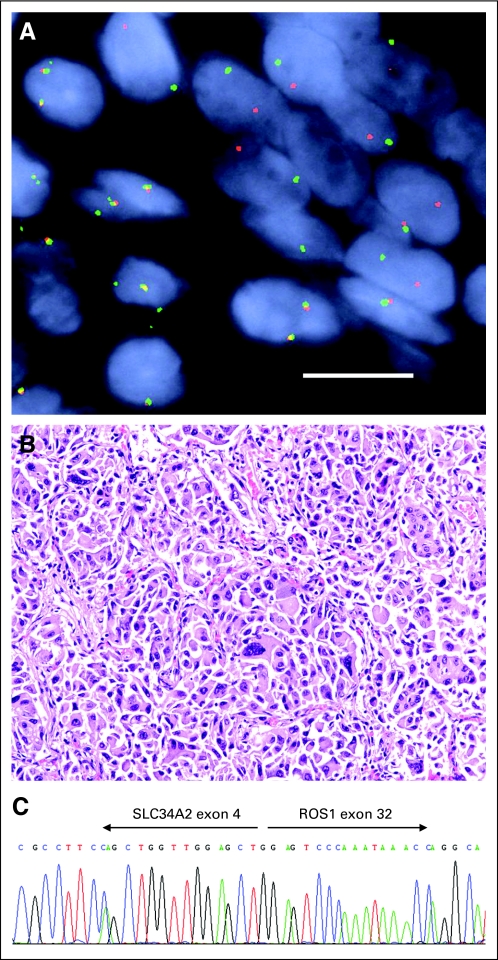
Because only two cases of chromosomal rearrangements involving ROS1 in NSCLC have been published to date (including one patient sample and one cell line, HCC78), little is known about the natural history of these tumors, and their clinical characteristics have not been established. To identify such patients from our clinical archives, we established a split-apart FISH assay to detect ROS1 rearrangements, using two bacterial artificial chromosome clones flanking the ROS1 gene, one labeled red and the other labeled green. Using this assay, we have confirmed the presence of the ROS1 rearrangement in the HCC78 cell line and have performed retrospective analysis of archived lung cancer specimens (Fig 1A). We observed 18 ROS1 FISH-positive NSCLCs in 1,073 tumors analyzed, for a prevalence rate of 1.7%, and these rearrangements are mutually exclusive from ALK rearrangement (ALK rearrangement was observed in 31 other tumors). Although our FISH approach allowed us to determine the presence of a ROS1 translocation within a sample, it provides no information about the translocation partner, which may have important consequences. Therefore, we used RT-PCR and sequencing to confirm the presence of known ROS1 rearrangements and identify the translocation partner in specimens with sufficient tissue to obtain RNA. CD74 was found as the partner in five specimens, SLC34A2 was found as the partner in one specimen, and no partner was identified in eight specimens (Fig 1C). Four specimens were not tested as a result of insufficient tissue.
Fig 1.
(A) A break-apart fluorescent in situ hybridization probe reveals separation of the 5′ ROS1 probe (green) from the 3′ ROS1 probe (red) in a non–small-cell lung cancer formalin-fixed paraffin-embedded specimen. Nuclei are stained with 4′,6-diamidino-2-phenylindole. Size bar = 10 μm. (B) Representative histologic appearance of a ROS1-rearranged tumor, stained with hematoxylin and eosin, showing a solid subtype of adenocarcinoma with highly atypical cytologic features. (C) Sanger sequencing of a reverse transcriptase polymerase chain reaction product from a tumor harboring a SLC34A2-ROS1 rearrangement.
Clinical Characteristics of Patients WithROS1-Rearraranged Tumors
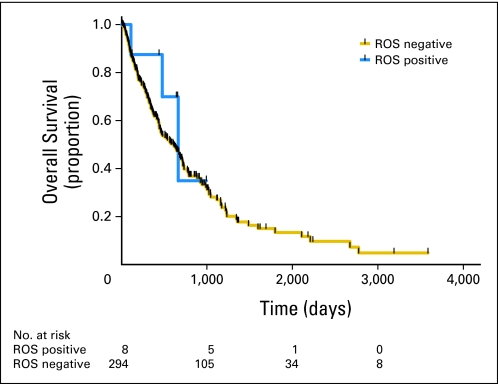
Analysis of the clinicopathologic characteristics of this cohort has revealed that ROS1-positive patients define a new and important genetic subtype of NSCLC (Table 1; the clinicopathologic details of each of the 18 ROS1-positive patients are listed in Table 2). There is significant overlap with ALK-positive NSCLC because ROS1-positive patients tend to be younger (median age, 49.8 years) never-smokers with a histologic diagnosis of adenocarcinoma.7 There is also an overrepresentation of Asians (P < .001) and patients presenting with stage IV disease, with the realization that those conclusions are based on small numbers. Detailed histologic analysis of ROS1-positive NSCLCs did not reveal a clear correlation with a subtype of adenocarcinoma. The presence of signet ring cells, a common feature of ALK-rearranged NSCLCs,17 was not common in ROS1-rearranged tumors. In fact, there was a broad distribution of tumor grade; however, eight of the tumors were poorly differentiated with highly atypical infiltrating tumor cells (Fig 1B and Appendix Fig A1, online only). Kaplan-Meier survival analysis of patients with metastatic NSCLC treated at one of our institutions (MGH) reveals similar OS in patients with and without ROS1 rearrangements (median OS, 663 days in ROS1-positive patients and 607 days ROS1-negative patients; P = .42; Appendix Fig A2, online only). The long survival in the MGH ROS1-negative cohort likely reflects a bias toward patients with NSCLC subtypes with better prognosis (eg, EGFR-mutant tumors) in our practice. This is supported by the observation that the median OS of the ROS1-negative, ALK-negative, EGFR mutation–negative population is 472 days (data not shown).
Table 1.
Demographics and Clinical Characteristics of Patients With ROS1-Positive NSCLC
| Demographic or Clinical Characteristic | All Patients (n = 1,073) |
ROS1 Positive (n = 18) |
ALK Positive (n = 31) |
ROS1 Negative (n = 1,055) |
P (ROS1 positive v ROS1 negative) | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | ||
| Age, years | |||||||||
| Median | 62.0 | 49.8 | 51.6 | 62.3 | < .001 | ||||
| Range | 32-87 | 32-79 | 29-73 | 32-87 | |||||
| Sex | |||||||||
| Male | 523 | 49 | 7 | 39 | 17 | 55 | 516 | 49 | .480 |
| Female | 550 | 51 | 11 | 61 | 14 | 45 | 539 | 51 | |
| Smoking history | |||||||||
| Never-smoker | 239 | 22 | 14 | 78 | 13 | 42 | 225 | 21 | < .001 |
| Light smoker | 62 | 6 | 1 | 6 | 1 | 3 | 61 | 6 | |
| Smoker | 695 | 65 | 2 | 11 | 3 | 10 | 693 | 66 | |
| NA | 77 | 7 | 1 | 6 | 14 | 45 | 76 | 7 | |
| Ethnicity | |||||||||
| Asian | 45 | 4 | 5 | 28 | 2 | 6 | 40 | 4 | < .001 |
| Non-Asian | 942 | 88 | 13 | 72 | 18 | 58 | 929 | 88 | |
| NA | 86 | 8 | 0 | 0 | 11 | 35 | 86 | 8 | |
| Pathology | |||||||||
| Adenocarcinoma | 694 | 65 | 18 | 100 | 16 | 52 | 676 | 64 | .019 |
| Squamous | 200 | 19 | 0 | 0 | 1 | 3 | 200 | 19 | |
| NSCLC, NOS | 59 | 5 | 0 | 0 | 0 | 0 | 59 | 6 | |
| Adenosquamous | 10 | 1 | 0 | 0 | 0 | 0 | 10 | 1 | |
| Other | 38 | 4 | 0 | 0 | 0 | 0 | 38 | 4 | |
| NA | 72 | 7 | 0 | 0 | 14 | 45 | 72 | 7 | |
| Stage | |||||||||
| IA | 218 | 20 | 1 | 6 | 1 | 3 | 217 | 21 | NS |
| IB | 140 | 13 | 1 | 6 | 1 | 3 | 139 | 13 | NS |
| IIA | 44 | 4 | 1 | 6 | 2 | 6 | 43 | 4 | NS |
| IIB | 87 | 8 | 0 | 0 | 1 | 3 | 87 | 8 | NS |
| IIIA | 139 | 13 | 2 | 11 | 5 | 16 | 137 | 13 | NS |
| IIIB | 73 | 7 | 2 | 11 | 2 | 6 | 71 | 7 | NS |
| IV | 327 | 30 | 11 | 61 | 12 | 39 | 316 | 30 | .010 |
| NA | 45 | 4 | 0 | 0 | 7 | 23 | 45 | 4 | |
Abbreviations: NA, not available; NOS, not otherwise specified; NS, not significant; NSCLC, non–small-cell lung cancer.
Table 2.
Clinical Details of 18 Patients With ROS1-Positive NSCLC
| Patient No. | Age (years) | Sex | Ethnicity | Smoking (No. of pack-years) | RT-PCR | Stage | Histology | Subtype |
|---|---|---|---|---|---|---|---|---|
| 1 | 42.0 | Male | Asian | 0 | Negative | 1A | AdCA | Acinar (50%), solid (40%); high grade |
| 2 | 37.0 | Female | Asian | 0 | Negative | 1B | AdCA | BAC, mucinous (90%), acinar (10%) |
| 3 | 53.0 | Male | White | 0 | Positive, CD74 exon 6, ROS exon 34 | IIA | AdCA | Acinar (60%), papillary (30%), BAC nonmucinous (10%); high grade |
| 4 | 39.0 | Female | White | 0 | Negative | IIIA | AdCA | Papillary (60%), acinar (40%); high grade |
| 5 | 32.0 | Female | White | 0 | Negative | IIIA | AdCA | Acinar (100%); high grade |
| 6 | 39.0 | Female | White | 0 | ND | IIIB | AdCA | Acinar (100%) |
| 7 | 51.0 | Female | Asian | 0 | Positive, CD74 exon 6, ROS exon 34 | IIIB | AdCA | Papillary (60%), acinar (40%) |
| 8 | 71.0 | Female | White | 0 | Positive, SLC34A2 exon 4., ROS exon 32 | IV | AdCA | Solid (100%); high grade |
| 9 | 43.0 | Male | Asian | 0 | Negative | IV | AdCA | Papillary (60%), acinar (40%) |
| 10 | 79.0 | Male | White | 75 | Negative | IV | AdCA | Solid (100%) |
| 11 | 68.0 | Male | White | 0 | Negative | IV | AdCA | Solid (100%) |
| 12 | 55.0 | Female | White | 23 | Positive, CD74 exon 6, ROS exon 34 | IV | AdCA | Papillary (60%), acinar (40%) |
| 13 | 65.0 | Female | White | 10 | ND | IV | AdCA | Acinar (90%), papillary (10%); high grade |
| 14 | 47.0 | Male | White | 0 | Negative | IV | AdCA | Acinar (100%) |
| 15 | 39.0 | Male | Asian | 0 | ND | IV | AdCA | Papillary (100%); high grade |
| 16 | 44.0 | Female | White | Positive, CD74 exon 6, ROS exon 34 | IV | AdCA | Solid (100%) | |
| 17 | 35.0 | Female | White | 0 | ND | IV | AdCA | Solid (100%); high grade |
| 18 | 57.0 | Female | White | 0 | Positive, CD74 exon 6, ROS exon 34 | IV | AdCA | Acinar (60%), papillary (30%), BAC nonmucinous (10%) |
Abbreviations: AdCA, adenocarcinoma; BAC, bronchioloalveolar carcinoma; ND, not determined; NSCLC, non–small-cell lung cancer; RT-PCR, reverse transcriptase polymerase chain reaction.
Crizotinib Inhibits ROS1 Activity and Cell Growth In Vitro
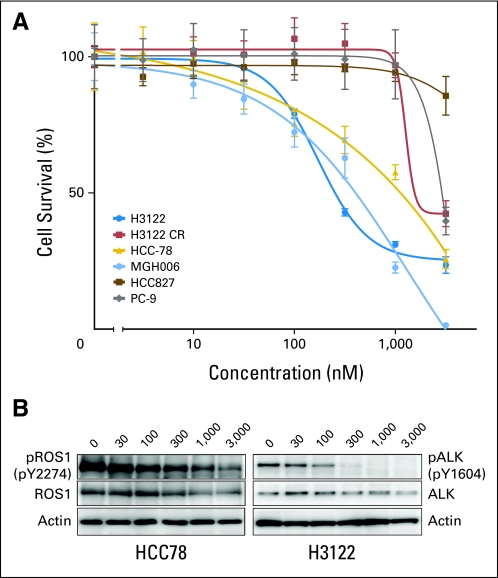
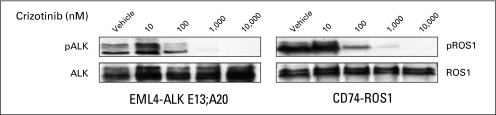
Because we previously found that an experimental compound TAE684 (primarily an ALK kinase inhibitor) effectively inhibited the growth of the ROS1-rearranged NSCLC line HCC78, we analyzed whether crizotinib (primarily an ALK and MET inhibitor) could also inhibit HCC78 growth. Our data indicate that crizotinib is moderately effective at inhibiting HCC78 cells (Fig 2A), with a growth inhibition curve between that of the ALK-rearranged H3122 and MGH006 lines and the ALK- and ROS1-negative lines PC-9 (EGFR mutant) and HCC827 (EGFR mutant) and a crizotinib-resistant H3122 line. Inhibition of ROS1 phosphorylation by crizotinib in the HCC78 cell line was moderate, supporting evidence that ROS1 is likely the target of crizotinib in this cell line (Fig 2B). To confirm this activity, we showed that crizotinib also inhibits ROS1 phosphorylation in HEK 293 cells transfected with a CD74-ROS1 fusion gene expression construct (Fig 3). This inhibition was at concentrations similar to that at which crizotinib inhibited ALK phosphorylation in HEK 293 cells transfected with an EML4-ALK fusion gene.
Fig 2.
(A) Dose-response cell survival curves of ROS1-rearranged cell line (HCC78) and ROS1-negative cell lines in response to crizotinib (nM); ALK-positive lines H3122 and MGH006 are positive controls for crizotinib response, and the PC-9, HCC827, and crizotinib-resistant H3122 CR lines are negative controls. (B) Western blot reveals a three-fold reduction of phospho-ROS1 in HCC78 cells at the same concentration (300 nmol/L) of crizotinib that results in near-complete reduction of phospho-ALK in H3122 cells. Total ROS1 and ALK, as well as actin, are shown as controls. The concentration of crizotinib is indicated above each lane (in nM).
Fig 3.
Two hundred ninety-three cells were transfected with cDNAs encoding EML4-ALK E13;A20 or CD74-ROS1. At approximately 45 hours after transfection, cells were treated with increasing amounts of crizotinib for 2 hours. Lysates were subjected to immunoblotting with antibodies specific for the indicated proteins.
Crizotinib Demonstrates Marked Antitumor Activity in a Patient With Advanced ROS1-Positive NSCLC
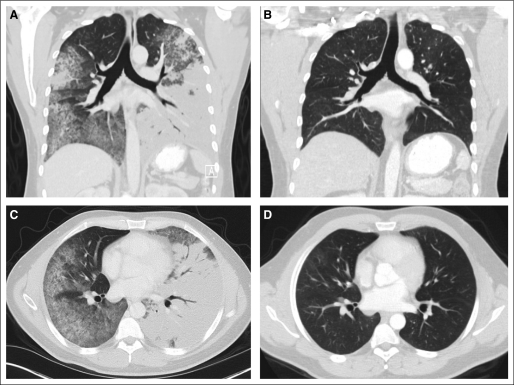
To determine whether ROS1 rearrangement confers sensitivity to ROS1 inhibition in patients, we enrolled a ROS1-positive patient with advanced NSCLC into an expansion cohort of the early phase study of crizotinib.1 Details of this trial, including design and eligibility requirements, have been reported previously.1 The patient is a 31-year-old male never-smoker diagnosed with multifocal bronchioloalveolar carcinoma in August 2010. Genetic testing of his tumor demonstrated no EGFR mutation or ALK rearrangement. He was treated at an outside institution with first-line erlotinib with no response. As a result of progressively worsening symptoms and hypoxia, he was referred to MGH for additional genetic testing and was found to be ROS1 positive. On April 20, 2011, the patient was started on crizotinib at the standard dose of 250 mg twice daily. In less than 1 week, he noted a significant improvement in symptoms, and by 2 weeks, his hypoxia had resolved. Restaging scans at 8 weeks demonstrated near complete resolution of his multifocal lung tumor, which was subsequently confirmed at 12 weeks (Fig 4). At the time of this report (6 months), the patient continues on crizotinib with no evidence of recurrence. This case suggests that patients with ROS1-positive NSCLC may be exquisitely sensitive to therapeutic ROS1 inhibition.
Fig 4.
Response of an ROS1-positive patient with advanced non–small-cell lung cancer to crizotinib. Computed tomography scans of the chest were obtained (A and C) at baseline and (B and D) after 12 weeks of crizotinib. Shown are (A and B) coronal reconstructions and (C and D) axial slices.
DISCUSSION
We have screened our NSCLC tissue archives and have found that approximately 2% of NSCLCs harbor ROS1 rearrangements. With an estimated 200,000 new cases of lung cancer per year in the United States, we extrapolate that there are 4,000 new ROS1-rearranged tumors per year, approximately half as common as ALK rearrangements in NSCLC.1,6–8,18 Chromosomal rearrangements involving the ROS1 receptor tyrosine kinase gene were originally described in glioblastoma, before their identification in NSCLC and more recently in cholangiocarcinoma, suggesting that ROS1 may have roles in other tumor types as well.10–12,19 Because our preclinical work has indicated that ROS1-rearranged tumors are sensitive to a subset of kinase inhibitors, we expect that the identification of ROS1-rearranged tumors will build on the ALK model for rapid validation of an emerging biomarker that will have a long-term impact on diagnosis and treatment of lung cancer.
The clinical profile of patients with ROS1-rearranged NSCLCs is remarkably similar to that of ALK-rearranged NSCLCs, including young age of onset and nonsmoking history.7 The similar characteristics suggest that the two genetic subtypes may share a common pathogenesis, possibly sharing environmental or genetic risk factors. However, we still have few clues as to the pathogenesis of ALK- or ROS1-positive tumors, as well as what may be predisposing these young nonsmokers to these specific genetic rearrangements. The histologic appearance of ROS1-rearranged tumors is distinct from ALK-positive tumors, with the only recurrent feature being the presence of high-grade highly atypical infiltrating cells in one third of ROS1-positive patients. Although ROS1 rearrangements are not limited to young never-smokers with high-grade histology, knowledge of this clinical profile will help clinicians select patients most likely to harbor this genetic subtype and most likely to benefit from targeted inhibitors such as crizotinib. Of all never-smokers in this study, 6% harbor ROS1 rearrangements. Thus, together with ALK rearrangements and EGFR mutation, ROS1 rearrangements can be added to the growing list of NSCLC genetic subtypes that arise independently from smoking.
We believe that definitive genetic subclassification of NSCLC is increasingly important in patient management.20 FISH is currently the most effective diagnostic technology to detect chromosomal rearrangements in tumor tissue. Other assays include RT-PCR, immunohistochemistry, and next-generation sequencing. However, FISH may not be the optimal biomarker assay because of the cost and need for technical expertise, although RT-PCR failed to detect previously described rearrangements in a substantial number of FISH-positive cases. This suggests that alternative ROS1 partners or other CD74-ROS1 and SLC34A2-ROS1 breakpoints will be uncovered. Our own work has shown that immunohistochemistry is equally sensitive to FISH in the detection of ALK-positive NSCLCs21 but is currently limited by the lack of commercially available ALK antibodies. Currently, there are no effective ROS1 antibodies for the detection of ROS1 rearrangements in paraffin sections.
For the expanded molecularly enriched cohort portion of the phase I trial of crizotinib in advanced-stage NSCLC, greater than 1,500 patients were screened to identify the 82 patients who eventually were enrolled onto the expanded cohort of ALK-positive patients.1 Although that trial is still ongoing, the objective response rate has been consistent over time and is approximately 60%. One of the most exciting lessons we have learned from this trial is that drug development can be accelerated by matching the right gene mutation to the right drug. The discovery of ALK rearrangements in NSCLC was published in late 2007,8 clinical screening for ALK rearrangements began by early 2008, and initial results of the phase I trial were published in 2010. Definitive phase III registration trials of crizotinib in this population have already been initiated, including one with crizotinib as first-line treatment and another trial with crizotinib as second-line treatment, and US Food and Drug Administration approval was received in 2011. Our observation that the ROS1-rearranged cell line HCC78 is at least moderately sensitive to crizotinib and the observation that crizotinib inhibits ROS1 phosphorylation in controlled transfection experiments support ROS1 as a bona fide target of crizotinib. That our ROS1-positive patient has shown a remarkable response to crizotinib therapy indicates that the clinical development of ROS1-specific kinase inhibitors, including crizotinib and others,22 should be accelerated and focused on this subpopulation of NSCLC.
In summary, we have found that approximately 2% of patients with NSCLC harbor ROS1 rearrangements. These patients are typically younger never-smokers who may benefit from crizotinib therapy.
Appendix
Fig A1.
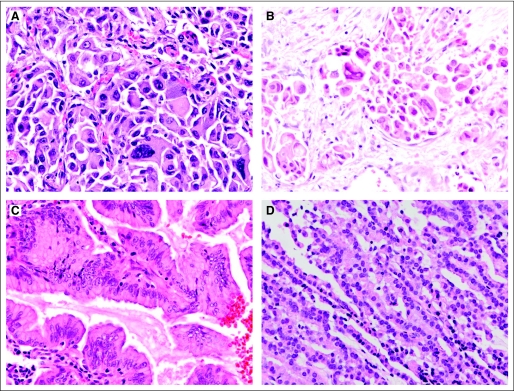
Representative histologic appearances of ROS1-rearranged tumors, stained with hematoxylin and eosin, with (A and B) two tumors harboring adenocarcinoma with highly atypical cytologic features including malignant giant cells (A is a high-magnification image of patient in Fig 1), (C) a mucinous bronchioloalveolar carcinoma, and (D) a low-grade adenocarcinoma forming thin tortuous glandular structures.
Fig A2.
Kaplan-Meier overall survival curves of ROS1-positive and -negative populations.
Footnotes
See accompanying article on page 878
Supported by National Institutes of Health Grant No. 1R21CA161590-01 (A.J.I.) and by Pfizer, which supported the crizotinib phase I trial.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Clinical trial information can be found for the following: NCT00585195.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: Keith D. Wilner, Pfizer (C) Consultant or Advisory Role: Alice T. Shaw, Pfizer (C), ARIAD (C); A. John Iafrate, Pfizer (C), Abbott Molecular (C) Stock Ownership: Keith D. Wilner, Pfizer Honoraria: None Research Funding: Alice T. Shaw, AstraZeneca, Novartis; Eunice L. Kwak, Pfizer; Jeffrey W. Clark, Pfizer Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Keith D. Wilner, William Pao, A. John Iafrate
Financial support: Keith D. Wilner, A. John Iafrate
Administrative support: Keith D. Wilner, William Pao, A. John Iafrate
Provision of study materials or patients: Alice T. Shaw, Sai-Hong Ignatius Ou, Pierre P. Massion, Rong Fang, Keith D. Wilner, Jeffrey W. Clark, Hongbin Ji, William Pao, A. John Iafrate
Collection and assembly of data: Kristin Bergethon, Alice T. Shaw, Sai-Hong Ignatius Ou, Ryohei Katayama, Christine M. Lovly, Nerina T. McDonald, Pierre P. Massion, Christina Siwak-Tapp, Adriana Gonzalez, Julie M. Batten, Haiquan Chen, Keith D. Wilner, Eunice L. Kwak, Jeffrey W. Clark, David P. Carbone, Jeffrey A. Engelman, Mari Mino-Kenudson, William Pao, A. John Iafrate
Data analysis and interpretation: Kristin Bergethon, Alice T. Shaw, Sai-Hong Ignatius Ou, Ryohei Katayama, Christine M. Lovly, Nerina T. McDonald, Rong Fang, Eugene J. Mark, Keith D. Wilner, Jeffrey W. Clark, Hongbin Ji, Mari Mino-Kenudson, William Pao, A. John Iafrate
Manuscript writing: All authors
Final approval of manuscript: All authors
Affiliations
Kristin Bergethon, Alice T. Shaw, Ryohei Katayama, Eugene J. Mark, Julie M. Batten, Eunice L. Kwak, Jeffrey W. Clark, Jeffrey A. Engelman, Mari Mino Kenudson, and A. John Iafrate, Massachusetts General Hospital, Boston, MA; Sai-Hong Ignatius Ou and Christina Siwak-Tapp, Chao Family Comprehensive Cancer Center, University of California Irvine Medical Center, Orange; Keith D. Wilner, Pfizer, La Jolla, CA: Christine M. Lovly, Nerina T. McDonald, Pierre P. Massion, Adriana Gonzalez, David P. Carbone, and William Pao, Vanderbilt University Medical Center; Pierre P. Massion, Nashville Veterans Affairs Medical Center, Nashville, TN; Rong Fang and Hongbin Ji, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences; Haiquan Chen, Fudan University Shanghai Cancer Center; and Haiquan Chen, Shanghai Medical College, Fudan University, Shanghai, China.
REFERENCES
- 1.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perner S, Wagner PL, Demichelis F, et al. EML4-ALK fusion lung cancer: A rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 9.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Birchmeier C, O'Neill K, Riggs M, et al. Characterization of ROS1 cDNA from a human glioblastoma cell line. Proc Natl Acad Sci U S A. 1990;87:4799–4803. doi: 10.1073/pnas.87.12.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birchmeier C, Sharma S, Wigler M. Expression and rearrangement of the ROS1 gene in human glioblastoma cells. Proc Natl Acad Sci U S A. 1987;84:9270–9274. doi: 10.1073/pnas.84.24.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charest A, Lane K, McMahon K, et al. Fusion of FIG to the receptor tyrosine kinase ROS in a glioblastoma with an interstitial del(6)(q21q21) Genes Chromosomes Cancer. 2003;37:58–71. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 13.Charest A, Wilker EW, McLaughlin ME, et al. ROS fusion tyrosine kinase activates a SH2 domain-containing phosphatase-2/phosphatidylinositol 3-kinase/ mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2006;66:7473–7481. doi: 10.1158/0008-5472.CAN-06-1193. [DOI] [PubMed] [Google Scholar]
- 14.McDermott U, Iafrate AJ, Gray NS, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68:3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 15.Engelman JA, Janne PA, Mermel C, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovly CM, Heuckmann JM, de Stanchina E, et al. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res. 2011;71:4920–4931. doi: 10.1158/0008-5472.CAN-10-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeuchi K, Choi YL, Soda M, et al. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clin Cancer Res. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- 19.Gu TL, Deng X, Huang F, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One. 6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pao W, Iafrate AJ, Su Z. Genetically informed lung cancer medicine. J Pathol. 2011;223:230–240. doi: 10.1002/path.2788. [DOI] [PubMed] [Google Scholar]
- 21.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Deeb IM, Park BS, Jung SJ, et al. Design, synthesis, screening, and molecular modeling study of a new series of ROS1 receptor tyrosine kinase inhibitors. Bioorg Med Chem Lett. 2009;19:5622–5626. doi: 10.1016/j.bmcl.2009.08.029. [DOI] [PubMed] [Google Scholar]