Abstract
Purpose of review
The review summarizes our current understanding of the function of the fatty acid translocase, CD36 in lipid metabolism with an emphasis on the influence of CD36 genetic variants and their potential contribution to obesity related complications.
Recent findings
Studies in rodents implicate CD36 in a number of metabolic pathways with relevance to obesity and its associated complications. These include pathways related to fat utilization such as taste perception, intake, intestinal processing and storage in adipose tissue. Dysfunction in these pathways, coupled with the ability of CD36 to transduce intracellular signals that initiate inflammation in response to excess fat supply, promotes metabolic pathology. In the last few years, the relevance of discoveries in rodents to humans has been highlighted by genetic studies, which identified common CD36 variants that influence circulating lipid levels and cardiometabolic phenotypes.
Summary
Recent genetic studies suggest that CD36 plays an important role in lipid metabolism in humans and may be involved in obesity related complications. These findings may accelerate the translation of CD36 metabolic functions determined in rodents to humans. Importantly, these studies highlight the potential utility of assessing CD36 expression and common SNP genotypes.
Keywords: CD36, polymorphisms, triglyceride, cholesterol, obesity
Introduction
Obesity is a major health burden that can contribute to the risk of life threatening metabolic complications. Its pathogenesis involves the dysregulation of energy balance as a result of altered complex interactions between environmental and genetic factors. It is well established that there is a broad range of inter-individual variability in serum lipid profiles and that this variability impacts susceptibility to diseases such as the metabolic syndrome, diabetes or coronary artery disease. Recent research on the scavenger receptor CD36 has highlighted its contribution to individual differences in serum lipids and to some of the metabolic complications of obesity. CD36 functions in the uptake of long chain fatty acids (LCFA), and oxidized low-density lipoproteins (oxLDL). Although early studies suggested that CD36 does not transfer lipid from native lipoproteins, this assumption has been revised based on recent findings implicating CD36 in the metabolism of LDL [1, 2] and HDL [3]. Signal transduction triggered by CD36 ligand binding involves the protein in cellular pathways relevant to some of the metabolic complications of obesity such as insulin resistance, inflammation, atherosclerosis and thrombosis as previously reviewed [4–10].
CD36 is a class B (two transmembrane regions) scavenger receptor originally identified as a platelet glycoprotein in a thrombocytopenic patient with refractoriness to HLA-matched platelet transfusion [11]. The protein is ubiquitously expressed on a variety of cell types, including adipocytes, myocytes, monocytes, macrophages, platelets, hepatocytes, vascular endothelial cells and intestinal enterocytes [9, 10]. Deficiency in CD36 occurs in 3–10% of African and Asian populations and is less common in populations of European descent[12, 13]. CD36 deficiency has been characterized as type I or II on the basis of platelet and/or monocytes expression (Type I, both monocytes and platelets lack CD36, type II, platelet-negative and monocyte-positive for CD36) [14] and little is known about how expression in these cells relates to other tissues. Historically, the relationship between CD36 expression and metabolic health has been unclear [15–19]. CD36 deficient individuals have been reported to exhibit a defect in myocardial uptake of long chain FA, slow clearance of plasma FA after an oral meal, and abnormalities of chylomicron formation. Studies in a small number of deficient subjects reported high LDL cholesterol concentrations [15–19]. While these findings suggest that CD36 deficiency is metabolically detrimental, a number of studies propose an opposing view by showing that a reduction of CD36 level is beneficial and may be atheroprotective [20]. Consistently, increased concentrations of a circulating soluble form of CD36 or high CD36 levels on monocyte have been associated with insulin resistance and with elevated serum levels of inflammation markers [21–23]. Some clarification of these apparently contradictory findings has been provided by genetic studies as highlighted in following sections.
Genetic studies have provided valuable insight into the potential physiological role of CD36 in humans. Earlier studies showed that mutations in the gene (located on chromosome 7q) associated with insulin resistance, low adiponectin level and type 2 diabetes [24]. Using quantitative trait loci mapping, the CD36 gene region (~300-kb) was linked to circulating lipid concentrations [25–27]. Among the early single nucleotide polymorphism (SNP) association studies, Ma X et al., was the most extensive analysis of CD36 variants in any population [28] and provided a reference for subsequent work. Interest in the genetics of CD36 has recently increased and this review will summarize the knowledge gained regarding the metabolic influence of CD36 genetic variants and their potential contribution to obesity related complications.
CD36 role in dietary lipid perception, absorption and storage
The followings section will first present a brief overview of what is known about CD36’s role in lipid metabolism emphasizing aspects that have been consistent with the genetic discoveries.
Processing of dietary fat
A fascinating research area of late relates to the function of CD36 in fat taste perception. CD36 is localized on lingual taste bud cells in the circumvallate papillae where it was shown to mediate FA perception and the initiation of the cephalic phase of digestion [29–31]. Zhang et al., reported a significant reduction of CD36 expression in circumvallate taste buds in high-fat diet induced obese rats when compared to controls [32**]. They suggested that decreased expression of CD36 mRNA and protein in this cell type diminishes sensitivity to fat taste, in turn increasing the intake of fatty foods as a compensatory response. In contrast to reduced CD36 levels, complete deficiency appears to diminish fat intake possibly as a result of postingestive effects of delayed absorption [31]. In humans, greater preference for fatty foods and their increased consumption have been documented in obese subjects [33]. It has been suggested that the increased fat intake may reflect an attenuation of oral and gastrointestinal FA sensitivity in obesity [34]. Little is known about whether CD36 functions in fat taste perception in humans. Recently, the protein was identified by immunohistochemistry in human taste buds of the circumvallate papillae [35]. Work currently underway suggests that common CD36 SNPs that were shown to influence CD36 expression may influence fat taste perception (Pepino MY, personal communication).
Great progress has been generated related to the proteins that function in intestinal processing of dietary lipids, but the mechanisms underlying dietary lipid absorption remain incompletely understood as recently reviewed elsewhere [36, 37]. As dietary fats are ingested, chylomicrons (~82% triglyceride, 9% cholesterol, 7% phospholipid, 2% protein) are synthesized in absorptive enterocytes to transport fat, cholesterol and fat-soluble vitamins from the villi of the small intestine into the bloodstream [38]. In the proximal small intestine, CD36 is abundantly expressed on villus enterocytes of rodents where it facilitates FA and cholesterol uptake but its contribution to net absorption is small and predominantly manifested in a shift of fat absorption to more distal parts of the intestine [39].
CD36 may however be required in coordinating the incorporation of FA and cholesterol into triglycerides for chylomicron production via its ability for signal transduction [39] and/or by facilitating assembly and ER budding of the prechylomicron transport vesicles [40]. There is evidence for its role in chylomicron synthesis from both CD36 deficient rodents [41] and humans [42] where the production of smaller chylomicrons and prolonged postprandial hypertriglyceridemia were documented. In line with this, two common single nucleotide polymorphisms (SNPs) were identified that associated with reduced plasma concentrations of vitamin E, which is secreted by enterocytes into chylomicrons (SUpplementation en VItamines et Minéraux AntioXydants, n = 621, and the adolescent cross-sectional HELENA; Healthy Lifestyle in Europe by Nutrition in Adolescence, n = 993) (Table 1). CD36 was also shown in rodents to mediate the effect of glucagon-like peptide-2 (GLP-2) on chylomicron formation [60] but whether the same applies to humans remains to be determined. In addition to its role in chylomicron synthesis, CD36 in the intestine is required for transport of the oleic acid needed for generation of oleoylethanolamide (OEA), a lipid messenger that reduces food intake [61]. OEA administration to mice reduces fat deposition and the main effect appears related to a reduction of lipid transport possibly via CD36 [62].
Table 1. Reported trait-associated CD36 SNPs.
SNPs are listed based on chromosomal position. Average minor allele frequencies (MAF), Average r2 and captured SNPs were generated in TagSNP. “Tag Associated phenotypes” refers to parameters reported to associate with the particular SNP used as a Tag (listed in the Tag SNP column). TagSNP associated trait refers to phenotypes reported to associate with the SNPs in the “Captured SNPs” column. For example, LDL-associated TagSNP rs2151916 is in linkage disequilibrium with the “captured SNP” rs3211816, which was reported to associate with BMI, WC and BMI-dependent insulin sensitivity [49]. Abbreviations: MetS-metabolic syndrome; TAG-triacylglycerol; DHA-dehydroascorbc acid; WC-waist circumference, MI-myocardial infarction. HapMap populations (ASW- African ancestry in Southwest USA; CEU- Utah residents with Northern and Western European ancestry from the CEPH collection; JPT-Japanese in Tokyo, Japan).
| Tag SNP | Avg MAF | Avg r2 | Captured SNP list by population | tag SNP associated trait | Captured SNP associated trait | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| ASW | CEU | JPT | ||||||
| rs1761663 | 0.41 | 0.93 | rs1761663 | rs1761663 rs2151916 rs3211816 |
rs1761663 rs3211816 |
Left Ventricular Mass, BMI | [43] | |
| rs1984112 | 0.23 | 1.00 | TAG, FFA, TAG response to DHA | [28, 44] | ||||
| rs1761667 | 0.42 | 1.00 | FFA, HDL, monocyte CD36 expression, TAG response to DHA, platelet CD36 expression, plasma a-tocopherol, Macular pigment optical density | [20, 28, 44–47] | ||||
| rs2151916 | 0.33 | 0.95 | rs1761663 rs2151916 rs3211816a |
rs2151916 | LDL | BMIa, WCa, BMI dependent insulin sensitivitya | [48, 49] | |
| rs9784998 | 0.15 | 0.94 | rs3173798b rs9784998 |
rs3173798 rs3211867c rs3211883 rs9784998 |
rs3211867 rs9784998 |
HDL, BMI, WC, BMI dependent insulin sensitivity, platelet CD36 expression, | MetSb, Macular Degenerationb, obesity, platelet CD36 expressionc | [45, 49–52] |
| rs1334513 | 0.45 | 1.00 | rs1334513 | rs3211909 rs3211913 |
MetS | [52] | ||
| rs1527479 | 0.38 | 0.94 | rs1527479 | rs1049654 rs10499859d rs1054516e rs1358337 rs1527479 rs3211842 rs3211849f rs3211870 |
rs1049654 rs10499859 rs13438282 rs1527479 rs3211849 |
Type 2 diabetes, insulin resistance, reduced plasma a-tocopherol | Left Ventricular Massd, HDLd,e,f platelet CD36 expressiond,f | [45, 46, 52–54] |
| rs1049654 | 0.39 | 0.94 | rs1049654 | rs1049654 rs10499859 rs1054516 rs1358337 rs1527479 rs3211842 rs3211849 rs3211870 |
rs1049654 rs10499859 rs1527479 rs3211849 |
HDL, MetS, platelet CD36 expression | [45, 52] | |
| rs3211842 | 0.46 | 0.93 | rs1049654 rs10499859 rs1054516 rs1358337 rs1527479 rs3211842 rs3211849 rs3211870 |
rs1054516 rs3211842 |
HDL, platelet CD36 expression | [45, 52] | ||
| rs3211870 | 0.44 | 0.93 | rs3211870 | rs1049654 rs10499859 rs1054516 rs13438282 rs1358337 rs1527479 rs3211842 rs3211849 rs3211870 |
rs3211870 | HDL, platelet CD36 expression | [45, 52] | |
| rs1358337 | 0.43 | 0.92 | rs1358337 | rs1049654 rs10499859 rs1054516 rs13438282 rs1358337 rs1527479 rs3211842 rs3211849 rs3211870 |
rs1358337 | Alzheimer’s disease, HDL, monocyte CD36, platelet CD36 | [20, 45, 52, 55] | |
| rs3211883 | 0.16 | 0.85 | rs3173798 rs3211883 rs9784998 |
rs3211883 | BMI, WC, %BF, platelet CD36 expression | [45, 49] | ||
| rs3211909 | 0.22 | 1.00 | rs3211913 rs1334513 |
HDL, monocyte and platelet CD36 expression | [20, 52] | |||
| rs3211913 | 0.35 | 1.00 | rs3211909 | HDL, monocyte and platelet CD36 expression | [20, 52] | |||
| rs3211928 | 0.39 | 0.97 | rs3211928 rs3211931 |
rs13230419 rs13246513 rs3173804 rs3211928 rs3211931 rs3211960 rs7755 |
rs3211928 rs3211931 rs7755 |
Total Stroke | [56] | |
| rs3211938 | 0.09 | 1.00 | rs3211938 | -- | -- | Monocyte and Platelet CD36 expression, MetS, HDL | [52] | |
| rs3211956 | 0.16 | 0.98 | rs1527483 rs3211956 |
rs1527483 rs3211956 |
BMI, WC, Acute MI | [49, 57] | ||
| rs7755 | 0.39 | 0.96 | rs7755 | rs13230419 rs13246513 rs3173804 rs3211928 rs3211931 rs3211960 rs7755 |
rs3211928 rs3211931 rs7755 |
Total cholesterol, Stroke, MetS | [52, 56, 58] | |
| rs1049673 | 0.36 | 1.00 | rs1049673 | TAG response to DHA, FFA in men, MetS | [28, 44, 59] | |||
| rs13246513 | 0.39 | 0.96 | rs13230419g rs13246513 rs3173804h rs3211928 rs3211931 rs3211960 rs7755 |
rs13230419 rs13246513 rs3173804 rs3211960 |
HDLh, total strokeg,h, MetS, plasma luten | [52, 56] | ||
There is recent evidence in humans to support a role of CD36 in storage of FFAs in adipose tissue. FA storage is an advantageous mechanism that prevents excessive circulating FFA levels, which can have adverse physiological effects [63, 64] and lead to pro-inflammatory processes that impair insulin receptor-mediated signaling in adipose, liver and skeletal muscle [65, 66]. In rodents, significant contributions of CD36 to FA storage in adipose tissue has been shown [67] but no data were available in humans, until recently. A new study using labeled palmitate tracer, examined FFA storage rates in the postabsorptive state in 25 men and 49 premenopausal women [68]. When adjusted by adipocyte number, abdominal CD36 protein levels (determined by ELISA) positively correlated with upper body subcutaneous fat storage rates in women (r=0.66, p=0.0001) along with two other lipogenic factors that were tested (acyl-CoA synthetase, and diacylglycerol acyltransferase). However, only abdominal CD36 independently predicted upper body subcutaneous FFA storage similar to plasma palmitate concentrations. These data suggested a role of CD36 in abdominal FFA storage in the postprandial state and a potential impact of CD36 expression on adiposity in that region [68**].
CD36 expression associates with obesity related complications
Obesity is an independent risk factor for atherosclerosis and cardiovascular disease [69]. A key early event in the development of atherosclerosis is thought to be altered vascular endothelial cell function. The link between obesity and atherosclerosis involves the secretory function of adipose tissue to produce adipocytokines, which affect the integrity of vascular endothelial cells and mediate adipose tissue inflammatory macrophage infiltration [70]. Lipid accumulation in macrophages contributes to foam cell formation and to proinflammatory cytokine secretion, events critical to atherosclerotic plaque formation.
Animal models have demonstrated increased monocyte derived macrophage recruitment to adipose tissue during obesity [71]. CD36 is proposed to mediate crosstalk between adipocytes and macrophages in obese mice models via its proinflammatory function in response to oxLDL uptake and its role in cytokine secretion by macrophages [72]. Using various in vivo and in vitro strategies, Kennedy et al recently demonstrated that adipose tissue from CD36 knockout mice was more insulin sensitive and had lower levels of inflammatory markers (i.e. IFN-g, MCP-1) as compared to WT mice. Similarly, residential peritoneal macrophages from CD36KO mice had a reduced inflammatory profile (lower IFN-g, MCP-1, MIP1a, and TNF-a) basally and in response to oxLDL. Additionally oxLDL activation of c-Jun N-terminal kinase (JNK), a stress-activated protein kinase related to metabolic deterioration [73], was shown to be CD36 dependent. Finally they showed a significantly diminished interaction between CD36KO derived macrophages and adipocytes in vitro [72*], suggesting that the lack of CD36 expression results in an anti-inflammatory state and is metabolically protective.
CD36 genetic variants in risk of obesity and related complications
Recent genetic association studies have yielded differing results related to the contribution to obesity of common variants in the CD36 gene. Bokor et al. identified four CD36 SNPs (rs3211867, rs3211883, rs3211908, and rs1527483) with association to measures of obesity [50*]. These SNPs could not be confirmed in a large meta-analysis study by Choquet et al 2011 [74**] in 9,973 European subjects. However, as shown in Table 1, other CD36 SNPs associated with waist circumference and BMI in another study by Hemi et al of European subjects (n=1790) at increased risk to type 2 diabetes [49]. Interestingly, in the same study, associations between CD36 SNPs and insulin sensitivity (derived from hyperinsulinemic euglycemic clamp, n=523) were identified and were BMI dependent consistent with a primary effect on BMI [49*].
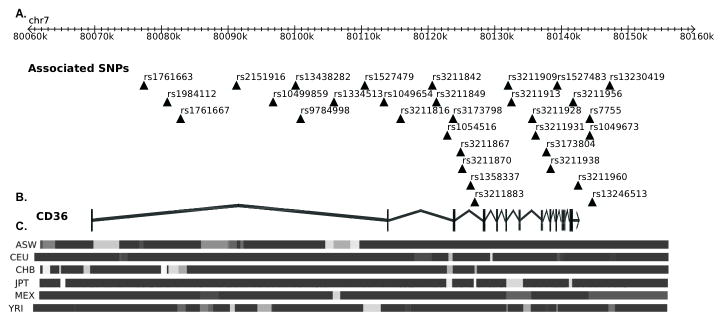
A number of common CD36 SNPs (minor allele frequencies >5%) have been identified to associate with various metabolic traits and obesity related complications. Many of these SNPs are in strong linkage disequilibrium as we show in Figure 1 and Table 1. Using available HapMap genotype data (release 27), we analyzed the trait-associated CD36 SNPs in TagSNP (http://manticore.niehs.nih.gov/) to summarize correlations between variants and associated phenotypes (Table 1).
Figure 1. Chromosome 7q position of CD36 trait-associated SNPs and linkage disequibrium (LD) structure across the gene.
(A) The position of each trait-associated SNP along the gene is indicated by the triangles next to the corresponding rs number. The positions of the SNPs on chromosome 7q are based on HapMap data release 27. (B) Schematic diagram of the CD36 gene (vertical bars representing exons). (C) A graphical representation of the LD block structure of a 100-kb region across 7q21 (80.060– 80.160 Mb) in 6 HapMap populations as indicated on the left (ASW: African ancestry in Southwest USA, CEU: Utah residents with Northern and Western European ancestry from the CEPH collection, CHB: Han Chinese in Beijing, China, JPT: Japanese in Tokyo, Japan, MEX: Mexican ancestry in Los Angeles, California, YRI: Yoruban in Ibadan, Nigeria). Intensity of the block colors is proportional to the strength of the LD (based on pairwise marker to marker r2 values with black =r2 > 0.80 to grey= r2 < 0.80).
Studies evaluating CD36 SNPs in association with lipid concentrations and metabolic disease status have been quite consistent in contrast to those related to obesity measures. Previous association studies have related CD36 polymorphisms to abnormal serum FA [28] and low-density lipoproteins (LDL)[48, 58]. CD36 SNPs were correlated with diabetes-associated coronary artery disease (CAD) risk in Caucasians [28] and Korean populations [75]. We identified associations between multiple CD36 SNPs and susceptibility to the metabolic syndrome (MetS) and with serum high-density lipoprotein cholesterol (HDL) in a large African American cohort from the Hypertension Genetic Epidemiology Network (HyperGEN)[52]. The minor allele of nine common CD36 SNPs associated with increased serum HDL and that of seven SNPs associated with reduced HDL. These observations suggested that the role of CD36 in human HDL metabolism might be underappreciated. A role for CD36 in the MetS was also suggested in Puerto Ricans, another population with high prevalence of obesity. The latter study by Noel S et al tested previously published CD36 SNPs for association in a Puerto Rican cohort (n=1178) using multivariate linear regression models. Subjects homozygous for the rs1049673 G-allele (1.89, 95%CI: 1.0–3.5, p=0.04) had an increased risk of MetS and a trend for rs3211931 TT-genotype (1.77, 95% CI: 1.0–3.1, p=0.05) was noted [59*].
Cardiovascular disease and heart function
A potential role for CD36 in cardiovascular disease and in heart function is supported by genetics studies. In a GWA study from the Heart and Aging Research in Genomic Epidemiology consortium, 15 CD36 SNPs tagged by rs3211928 associated with the risk of stroke (p-values of 10−6) [56**]. CD36 SNPs have also been identified to associate with left ventricular mass [53]. The involvement of CD36 in left ventricular function is further supported by a recent association study of 255 hypertensive European families comprised of 1425 individuals [43].
As shown in Table 1, many of the SNPs occur at frequencies >10%, suggesting their potential clinical relevance, unfortunately, few studies have addressed the functional impact of most of these variants. The coding variant rs3211938, which is specific to populations of African ancestry and predicted to result in a truncated CD36 protein, was shown to associate with CD36 deficiency [12, 52]. Two studies to-date have examined the influence of common CD36 SNP on expression of the gene and protein in monocytes and platelets. We determined that the influence of SNPs previously identified to associate with fasting serum HDL-C concentrations in an African American cohort impacted monocyte CD36 protein levels and causality was inferred by mendelian randomization analysis [20**]. We also showed that monocyte CD36 protein level inversely associated with HDL and positively with VLDL. Some of the associations between SNPs and CD36 levels we identified were confirmed in another study, which addressed SNP effects on platelet CD36. This GWA analysis in a Caucasian cohort (n=374) by Ghosh et al., documented that interindividual variability in platelet CD36 surface expression is largely attributed to CD36 variants [45**]. Twenty-three CD36 SNPs met genome wide significance with the strongest association at rs3211870. Many of these SNPs were also significant in previously reported association studies as shown in Table 1. Moreover, Ghosh et al., showed that platelet CD36 expression levels influence the activation response to oxLDL in vitro using platelets derived from healthy donors. Future studies are needed to determine whether genetically determined variability in CD36 expression may influence additional parameters of platelet activation since CD36 was recently shown to be involved in calcium influx leading to release of arachidonic acid and formation of proinflammatory prostaglandins [76].
Conclusion
In summary, although common CD36 genetic variants are not strongly associated with measures of obesity, it is clear that they contribute to individual variability in lipid profiles and to the susceptibility to obesity related complications. Based on their common prevalence CD36 genetic variants may be useful in designing better strategies for prevention and treatment in high-risk individuals. A potentially important conclusion that is suggested by the genetic studies and that should be explored further in future work is that there is a differential effect of partial versus complete CD36 deficiency. Studies with CD36 deficient subjects [15–19] versus those with reduced CD36 levels [20] as a result of common CD36 SNPs indicate that lower CD36 expression is metabolically protective. In contrast complete CD36 deficiency similar to CD36 overexpression is likely to predispose to metabolic complications. This suggests that there may be a “metabolically protective” range or threshold effect of CD36 expression. More studies are needed to determine the contribution of common SNPs to the molecular regulation of CD36 expression and its tissue specific functions. Such information is needed to improve our understanding of how CD36 SNPs may impact specific obesity related phenotypes and complications.
Key points.
Recent studies implicate CD36 in dietary fat taste perception, absorption and storage in humans.
Partial versus complete CD36 deficiency may result in differential metabolic effects; protection versus predisposition to disease risk, respectively.
Common CD36 genetic variants contribute to interindividual variability in fasting lipid concentrations and to risk of obesity related complications.
Acknowledgments
Grants from the American Heart Association (L. L-G.) and NIH DK33301 (N.A.A), DK60022 (N.A.A) supported this work. We also thank the Washington University Nutrition and Obesity Research Unit (P30 DK056341) and Diabetes Research Training Centers (P60 DK020579-32).
Footnotes
There are no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Brodeur MR, Brissette L, Falstrault L, et al. Scavenger receptor of class B expressed by osteoblastic cells are implicated in the uptake of cholesteryl ester and estradiol from LDL and HDL3. J Bone Miner Res. 2008;23:326–337. doi: 10.1359/jbmr.071022. [DOI] [PubMed] [Google Scholar]
- 2.Luangrath V, Brodeur MR, Rhainds D, Brissette L. Mouse CD36 has opposite effects on LDL and oxidized LDL metabolism in vivo. Arterioscler Thromb Vasc Biol. 2008;28:1290–1295. doi: 10.1161/ATVBAHA.107.161653. [DOI] [PubMed] [Google Scholar]
- 3.Brundert M, Heeren J, Merkel M, et al. Scavenger receptor CD36 mediates uptake of high density lipoproteins in mice and by cultured cells. J Lipid Res. 2011;52:745–758. doi: 10.1194/jlr.M011981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collot-Teixeira S, Martin J, McDermott-Roe C, et al. CD36 and macrophages in atherosclerosis. Cardiovasc Res. 2007;75:468–477. doi: 10.1016/j.cardiores.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Heilbronn LK, Gregersen S, Shirkhedkar D, et al. Impaired fat oxidation after a single high-fat meal in insulin-sensitive nondiabetic individuals with a family history of type 2 diabetes. Diabetes. 2007;56:2046–2053. doi: 10.2337/db06-1687. [DOI] [PubMed] [Google Scholar]
- 6.Kashyap SR, Ioachimescu AG, Gornik HL, et al. Lipid-induced insulin resistance is associated with increased monocyte expression of scavenger receptor CD36 and internalization of oxidized LDL. Obesity (Silver Spring) 2009;17:2142–2148. doi: 10.1038/oby.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park YM, Febbraio M, Silverstein RL. CD36 modulates migration of mouse and human macrophages in response to oxidized LDL and may contribute to macrophage trapping in the arterial intima. J Clin Invest. 2009;119:136–145. doi: 10.1172/JCI35535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahaman SO, Lennon DJ, Febbraio M, et al. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 2006;4:211–221. doi: 10.1016/j.cmet.2006.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverstein RL, Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal. 2009;2:re3. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su X, Abumrad NA. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol Metab. 2009;20:72–77. doi: 10.1016/j.tem.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda H, Mitani T, Ohnuma M, et al. A new platelet-specific antigen, Naka, involved in the refractoriness of HLA-matched platelet transfusion. Vox Sang. 1989;57:213–217. doi: 10.1111/j.1423-0410.1989.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 12.Aitman TJ, Cooper LD, Norsworthy PJ, et al. Malaria susceptibility and CD36 mutation. Nature. 2000;405:1015–1016. doi: 10.1038/35016636. [DOI] [PubMed] [Google Scholar]
- 13.Yanai H, Chiba H, Morimoto M, et al. Type I CD36 deficiency in humans is not associated with insulin resistance syndrome. Thromb Haemost. 2000;83:786. [PubMed] [Google Scholar]
- 14.Yamamoto N, Akamatsu N, Sakuraba H, et al. Platelet glycoprotein IV (CD36) deficiency is associated with the absence (type I) or the presence (type II) of glycoprotein IV on monocytes. Blood. 1994;83:392–397. [PubMed] [Google Scholar]
- 15.Furuhashi M, Ura N, Nakata T, et al. Genotype in human CD36 deficiency and diabetes mellitus. Diabet Med. 2004;21:952–953. doi: 10.1111/j.1464-5491.2004.01248.x. [DOI] [PubMed] [Google Scholar]
- 16.Kamiya M, Nakagomi A, Tokita Y, et al. Type I CD36 deficiency associated with metabolic syndrome and vasospastic angina: a case report. J Cardiol. 2006;48:41–44. [PubMed] [Google Scholar]
- 17.Nagasaka H, Yorifuji T, Takatani T, et al. CD36 deficiency predisposing young children to fasting hypoglycemia. Metabolism. 2011;60:881–887. doi: 10.1016/j.metabol.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Yasunaga T, Koga S, Ikeda S, et al. Cluster differentiation-36 deficiency type 1 and acute coronary syndrome without major cardiovascular risk factors: case report. Circ J. 2007;71:166–169. doi: 10.1253/circj.71.166. [DOI] [PubMed] [Google Scholar]
- 19.Yoshizumi T, Nozaki S, Fukuchi K, et al. Pharmacokinetics and metabolism of 123I-BMIPP fatty acid analog in healthy and CD36-deficient subjects. J Nucl Med. 2000;41:1134–1138. [PubMed] [Google Scholar]
- 20**.Love-Gregory L, Sherva R, Schappe T, et al. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum Mol Genet. 2011;20:193–201. doi: 10.1093/hmg/ddq449. This study demonstrates that common CD36 variants that influence circulating lipid levels associate associate with changes in CD36 expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handberg A, Levin K, Hojlund K, Beck-Nielsen H. Identification of the oxidized low-density lipoprotein scavenger receptor CD36 in plasma: a novel marker of insulin resistance. Circulation. 2006;114:1169–1176. doi: 10.1161/CIRCULATIONAHA.106.626135. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Scavini M, Orlando RA, et al. Increased CD36 Expression Signals Monocyte Activation Among Patients with Type 2 Diabetes Mellitus. Diabetes Care. 2010;33:2065–2067. doi: 10.2337/dc10-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silverstein RL. Inflammation, atherosclerosis, and arterial thrombosis: role of the scavenger receptor CD36. Cleve Clin J Med. 2009;76 (Suppl 2):S27–30. doi: 10.3949/ccjm.76.s2.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepretre F, Linton KJ, Lacquemant C, et al. Genetic study of the CD36 gene in a French diabetic population. Diabetes Metab. 2004;30:459–463. doi: 10.1016/s1262-3636(07)70143-x. [DOI] [PubMed] [Google Scholar]
- 25.Arya R, Blangero J, Williams K, et al. Factors of insulin resistance syndrome--related phenotypes are linked to genetic locations on chromosomes 6 and 7 in nondiabetic mexican-americans. Diabetes. 2002;51:841–847. doi: 10.2337/diabetes.51.3.841. [DOI] [PubMed] [Google Scholar]
- 26.Lehman DM, Arya R, Blangero J, et al. Bivariate linkage analysis of the insulin resistance syndrome phenotypes on chromosome 7q. Hum Biol. 2005;77:231–246. doi: 10.1353/hub.2005.0040. [DOI] [PubMed] [Google Scholar]
- 27.Malhotra A, Elbein SC, Ng MC, et al. Meta-analysis of genome-wide linkage studies of quantitative lipid traits in families ascertained for type 2 diabetes. Diabetes. 2007;56:890–896. doi: 10.2337/db06-1057. [DOI] [PubMed] [Google Scholar]
- 28.Ma X, Bacci S, Mlynarski W, et al. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum Mol Genet. 2004;13:2197–2205. doi: 10.1093/hmg/ddh233. [DOI] [PubMed] [Google Scholar]
- 29.Laugerette F, Passilly-Degrace P, Patris B, et al. CD36 involvement in orosensory detection of dietary lipids, spontaneous fat preference, and digestive secretions. J Clin Invest. 2005;115:3177–3184. doi: 10.1172/JCI25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin C, Chevrot M, Poirier H, et al. CD36 as a lipid sensor. Physiol Behav. 2011 doi: 10.1016/j.physbeh.2011.02.029. [DOI] [PubMed] [Google Scholar]
- 31.Sclafani A, Ackroff K, Abumrad NA. CD36 gene deletion reduces fat preference and intake but not post-oral fat conditioning in mice. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1823–1832. doi: 10.1152/ajpregu.00211.2007. [DOI] [PubMed] [Google Scholar]
- 32**.Zhang XJ, Zhou LH, Ban X, et al. Decreased expression of CD36 in circumvallate taste buds of high-fat diet induced obese rats. Acta Histochem. 2011;113:663–667. doi: 10.1016/j.acthis.2010.09.007. Study shows a reduction in the expression of CD36 in circumvallate taste buds from obese high-fat diet induced obese rats. [DOI] [PubMed] [Google Scholar]
- 33.Stewart JE, Feinle-Bisset C, Golding M, et al. Oral sensitivity to fatty acids, food consumption and BMI in human subjects. Br J Nutr. 2010;104:145–152. doi: 10.1017/S0007114510000267. [DOI] [PubMed] [Google Scholar]
- 34.Stewart JE, Seimon RV, Otto B, et al. Marked differences in gustatory and gastrointestinal sensitivity to oleic acid between lean and obese men. Am J Clin Nutr. 93:703–711. doi: 10.3945/ajcn.110.007583. [DOI] [PubMed] [Google Scholar]
- 35.Simons PJ, Kummer JA, Luiken JJ, Boon L. Apical CD36 immunolocalization in human and porcine taste buds from circumvallate and foliate papillae. Acta Histochem. 2010 doi: 10.1016/j.acthis.2010.08.006. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 36.Newberry EP, Davidson NO. Intestinal lipid absorption, GLP-2, and CD36: still more mysteries to moving fat. Gastroenterology. 2009;137:775–778. doi: 10.1053/j.gastro.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz GJ. Gut fat sensing in the negative feedback control of energy balance - Recent advances. Physiology & behavior. 2011;104(4):621–623. doi: 10.1016/j.physbeh.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iqbal J, Hussain MM. Intestinal lipid absorption. Am J Physiol Endocrinol Metab. 2009;296:E1183–1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran TT, Poirier H, Clement L, et al. Luminal lipid regulates CD36 levels and downstream signaling to stimulate chylomicron synthesis. J Biol Chem. 2011;286:25201–25210. doi: 10.1074/jbc.M111.233551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siddiqi S, Saleem U, Abumrad NA, et al. A novel multiprotein complex is required to generate the prechylomicron transport vesicle from intestinal ER. J Lipid Res. 2010;51:1918–1928. doi: 10.1194/jlr.M005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drover VA, Ajmal M, Nassir F, et al. CD36 deficiency impairs intestinal lipid secretion and clearance of chylomicrons from the blood. J Clin Invest. 2005;115:1290–1297. doi: 10.1172/JCI21514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masuda D, Hirano K, Oku H, et al. Chylomicron remnants are increased in the postprandial state in CD36 deficiency. J Lipid Res. 2009;50:999–1011. doi: 10.1194/jlr.P700032-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall D, Mayosi BM, Rahman TJ, et al. Common variation in the CD36 (fatty acid translocase) gene is associated with left-ventricular mass. J Hypertens. 2011;29:690–695. doi: 10.1097/HJH.0b013e3283440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madden J, Carrero JJ, Brunner A, et al. Polymorphisms in the CD36 gene modulate the ability of fish oil supplements to lower fasting plasma triacyl glycerol and raise HDL cholesterol concentrations in healthy middle-aged men. Prostaglandins Leukot Essent Fatty Acids. 2008;78:327–335. doi: 10.1016/j.plefa.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Ghosh A, Murugesan G, Chen K, et al. Platelet CD36 surface expression levels affect functional responses to oxidized LDL and are associated with inheritance of specific genetic polymorphisms. Blood. 2011;117:6355–6366. doi: 10.1182/blood-2011-02-338582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lecompte S, Szabo de Edelenyi F, Goumidi L, et al. Polymorphisms in the CD36/FAT gene are associated with plasma vitamin E concentrations in humans. Am J Clin Nutr. 2011;93:644–651. doi: 10.3945/ajcn.110.004176. [DOI] [PubMed] [Google Scholar]
- 47.Borel P, de Edelenyi FS, Vincent-Baudry S, et al. Genetic variants in BCMO1 and CD36 are associated with plasma lutein concentrations and macular pigment optical density in humans. Ann Med. 2011;43:47–59. doi: 10.3109/07853890.2010.531757. [DOI] [PubMed] [Google Scholar]
- 48.Goyenechea E, Collins LJ, Parra D, et al. CD36 gene promoter polymorphisms are associated with low density lipoprotein-cholesterol in normal twins and after a low-calorie diet in obese subjects. Twin Res Hum Genet. 2008;11:621–628. doi: 10.1375/twin.11.6.621. [DOI] [PubMed] [Google Scholar]
- 49.Heni M, Mussig K, Machicao F, et al. Variants in the CD36 gene locus determine whole-body adiposity, but have no independent effect on insulin sensitivity. Obesity (Silver Spring) 2011;19:1004–1009. doi: 10.1038/oby.2010.251. [DOI] [PubMed] [Google Scholar]
- 50*.Bokor S, Legry V, Meirhaeghe A, et al. Single-nucleotide polymorphism of CD36 locus and obesity in European adolescents. Obesity (Silver Spring) 2010;18:1398–1403. doi: 10.1038/oby.2009.412. This study suggested CD36 SNPs associate with measures of obesity in a European population. [DOI] [PubMed] [Google Scholar]
- 51**.Kondo N, Honda S, Kuno S, Negi A. Positive association of common variants in CD36 with neovascular age-related macular degeneration. Aging (Albany NY) 2009;1:266–274. doi: 10.18632/aging.100006. This study refutes the Bokor et. al. (reference 50) study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love-Gregory L, Sherva R, Sun L, et al. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum Mol Genet. 2008;17:1695–1704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arnett DK, Li N, Tang W, et al. Genome-wide association study identifies single-nucleotide polymorphism in KCNB1 associated with left ventricular mass in humans: the HyperGEN Study. BMC Med Genet. 2009;10:43. doi: 10.1186/1471-2350-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corpeleijn E, van der Kallen CJ, Kruijshoop M, et al. Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with Type 2 diabetes mellitus and insulin resistance. Diabet Med. 2006;23:907–911. doi: 10.1111/j.1464-5491.2006.01888.x. [DOI] [PubMed] [Google Scholar]
- 55.Taguchi K, Yamagata HD, Zhong W, et al. Identification of hippocampus-related candidate genes for Alzheimer’s disease. Ann Neurol. 2005;57:585–588. doi: 10.1002/ana.20433. [DOI] [PubMed] [Google Scholar]
- 56**.Ikram MA, Seshadri S, Bis JC, et al. Genomewide association studies of stroke. N Engl J Med. 2009;360:1718–1728. doi: 10.1056/NEJMoa0900094. This study identifies strong associations be CD36 SNPs and stroke phenoypes in a genome wide association study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knowles JW, Wang H, Itakura H, et al. Association of polymorphisms in platelet and hemostasis system genes with acute myocardial infarction. Am Heart J. 2007;154:1052–1058. doi: 10.1016/j.ahj.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morii T, Ohno Y, Kato N, et al. CD36 single nucleotide polymorphism is associated with variation in low-density lipoprotein-cholesterol in young Japanese men. Biomarkers. 2009;14:207–212. doi: 10.1080/13547500902811274. [DOI] [PubMed] [Google Scholar]
- 59.Noel SE, Lai CQ, Mattei J, et al. Variants of the CD36 gene and metabolic syndrome in Boston Puerto Rican adults. Atherosclerosis. 2010;211:210–215. doi: 10.1016/j.atherosclerosis.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsieh J, Longuet C, Maida A, et al. Glucagon-like peptide-2 increases intestinal lipid absorption and chylomicron production via CD36. Gastroenterology. 2009;137:997–1005. 1005, e1–4. doi: 10.1053/j.gastro.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 61.Schwartz GJ, Fu J, Astarita G, et al. The lipid messenger OEA links dietary fat intake to satiety. Cell Metab. 2008;8:281–288. doi: 10.1016/j.cmet.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thabuis C, Destaillats F, Lambert DM, et al. Lipid transport function is the main target of oral oleoylethanolamide to reduce adiposity in high-fat-fed mice. J Lipid Res. 2011;52:1373–1382. doi: 10.1194/jlr.M013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McQuaid SE, Hodson L, Neville MJ, et al. Downregulation of adipose tissue fatty acid trafficking in obesity: a driver for ectopic fat deposition? Diabetes. 2011;60:47–55. doi: 10.2337/db10-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mittendorfer B, Magkos F, Fabbrini E, et al. Relationship between body fat mass and free fatty acid kinetics in men and women. Obesity (Silver Spring) 2009;17:1872–187. doi: 10.1038/oby.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr Diab Rep. 2010;10:306–315. doi: 10.1007/s11892-010-0122-6. [DOI] [PubMed] [Google Scholar]
- 66.Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogenesis of insulin resistance and type 2 diabetes. Endocr Rev. 2002;23:201–229. doi: 10.1210/edrv.23.2.0461. [DOI] [PubMed] [Google Scholar]
- 67.Goldberg IJ, Eckel RH, Abumrad NA. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J Lipid Res. 2009;50 (Suppl):S86–90. doi: 10.1194/jlr.R800085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68**.Koutsari C, Ali AH, Mundi MS, Jensen MD. Storage of Circulating Free Fatty Acid in Adipose Tissue of Postabsorptive Humans: Quantitative Measures and Implications for Body Fat Distribution. Diabetes. 2011 Aug;60(8):2032–40. doi: 10.2337/db11-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poirier P, Giles TD, Bray GA, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss. Arterioscler Thromb Vasc Biol. 2006;26:968–76. doi: 10.1161/01.ATV.0000216787.85457.f3. [DOI] [PubMed] [Google Scholar]
- 70.Bays HE. Adiposopathy is “sick fat” a cardiovascular disease? J Am Coll Cardiol. 2011;57:2461–2473. doi: 10.1016/j.jacc.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 71.Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 72*.Kennedy DJ, Kuchibhotla S, Westfall KM, et al. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc Res. 2011;89:604–613. doi: 10.1093/cvr/cvq360. Interesting study using CD36 −/− mice fed a high-fat diet suggests that there is a CD36-dependent inflammatory pathway connecting macrophages and adipocytes in insulin resistance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallerie SN, Hotamisligil GS. The role of JNK proteins in metabolism. Sci Transl Med. 2010;2:60rv5. doi: 10.1126/scitranslmed.3001007. [DOI] [PubMed] [Google Scholar]
- 74.Choquet H, Labrune Y, De Graeve F, et al. Lack of association of CD36 SNPs with early onset obesity: a meta-analysis in 9,973 European subjects. Obesity (Silver Spring) 2011;19:833–839. doi: 10.1038/oby.2010.226. [DOI] [PubMed] [Google Scholar]
- 75.Yun YM, Song EY, Song SH, et al. CD36 polymorphism and its relationship with body mass index and coronary artery disease in a Korean population. Clin Chem Lab Med. 2007;45:1277–1282. doi: 10.1515/CCLM.2007.270. [DOI] [PubMed] [Google Scholar]
- 76.Kuda O, Jenkins CM, Skinner JR, et al. CD36 protein is involved in store-operated calcium flux, phospholipase A2 activation, and production of prostaglandin E2. J Biol Chem. 2011;286:17785–17795. doi: 10.1074/jbc.M111.232975. [DOI] [PMC free article] [PubMed] [Google Scholar]