Abstract
Background. Natural killer (NK) cells likely contribute to outcome of acute hepatitis C virus (HCV) infection and interferon (IFN)–induced control of chronic HCV infection. We previously observed IFN-αR and NKp30 expression associated with IFN-α–dependent NK cell activity.
Methods. Here, we examined CD16+56−, CD16+56+, and CD16−56+ NK cell subset IFN-αR and NKp30 expression in relation to magnitude of HCV genotype 1 decrease during pegylated IFN-α plus ribavirin therapy.
Results. We observed greater baseline IFN-αR and NKp30 expression on CD16+56+ and CD16−56+ NK subsets in HCV-infected patients than in healthy control subjects. Baseline CD16+56− NK IFN-αR expression was associated with IFN-α–induced pSTAT1, and both were associated with magnitude of HCV decrease during pegylated IFN-α plus ribavirin therapy. Baseline CD16+56− NK IFN-αR expression was associated with race and interleukin 28B genotype, negatively associated with aspartate aminotransferase-to platelet ratio index, and positively associated with increase in NKp30 expression after in vivo IFN-α exposure. Finally, in vitro IFN-α2a–activated NK cytolysis of HCV-infected target cells was in part dependent on NKp30, and CD16+56− NK cell IFN-αR expression correlated with cytolytic activity.
Conclusions. IFN-αR expression on CD16+56− NK cells during chronic HCV infection may in part be genetically determined, and level of expression regulates IFN-α signaling, which in turn may contribute to control of HCV infection.
Acute hepatitis C virus (HCV) infection becomes persistent in a majority of cases [1], and the long-term risk of cirrhosis, liver failure, cancer, and mortality among those with chronic infection highlight the importance of effective therapy [1, 2]. HCV magnitude decrease at 4 and 12 weeks of pegylated interferon (IFN)–α plus ribavirin therapy is predictive of sustained virologic response [3]. Although mechanisms underlying IFN-α responsiveness remain unclear, factors associated with response to IFN-α therapy include HCV genotype, age, race, human immunodeficiency virus (HIV) coinfection, baseline HCV level, and polymorphism near the interleukin 28B (IL-28B) (IFN-λ3) gene locus [3–9]. Despite introduction of protease inhibitors, the need to combine these agents with IFN means that response to newer regimens continues to depend on factors regulating IFN-α responsiveness.
Natural killer (NK) cells provide essential host defense during mouse hepatic viral [10, 11] and human herpesvirus infection [12–15]. They are innate lymphocytes with cytokine-producing, chemokine-producing, and cytotoxic activities regulated by activating and inhibitory receptors [16, 17]. Evidence for NK cells contributing to control of HCV derives from observations that genetically determined NK-KIR (Killer Immunoglobulin like Receptor)/ligand pairing correlates with the course of acute HCV infection [18] and that, during chronic infection, NK KIR2DL3, NKG2C, and NKp30 expression are associated with response to IFN-α–based therapy [19–21]. In addition, TRAIL expression is upregulated on NK cells during IFN-α–based therapy, and this correlates with in vitro cytolysis of HCV JFH-1–infected Huh 7.5 cells [22].
We observed NK IFN-αR and NKp30 expression to associate with IFN-α–dependent killer activity during HIV infection [23]. NK cells of individuals with preserved activity appeared to have enhanced IFN-αR expression. We hypothesized that IFN-αR expression is upregulated during chronic viral infection, in turn determining IFN-α–dependent function. Here, we evaluated NK cell subset IFN-αR and NKp30 expression in HCV genotype 1–infected patients at baseline, longitudinally over the course of IFN-α–based therapy, and in relation to IFN-α signaling capacity and viral decrease.
MATERIALS AND METHODS
Participants
Study participants signed Cleveland Veterans Affairs Medical Center or University Hospitals Case Medical Center institutional review board informed consent. HCV-infected patients (n = 21) were chronically infected (antibody positive for ≥6 months; HCV RNA positive) with HCV genotype 1, naive to HCV therapy, and scheduled to begin pegylated IFN-α2a (180 μg/week) plus weight-based ribavirin (1000–1200 mg/day) therapy. Healthy control subjects (n = 10) were recruited from a comparable age range. Clinical characteristics for study participants are shown in Table 1. HCV-infected and healthy control groups differed by sex and age; thus, analyses comparing groups required consideration of these factors. Three participants were treated with a half-dose of pegylated IFN-α2a because of baseline thrombocytopenia or neutropenia. Analysis was performed with all participant sample data and in the absence of these 3 participant samples. All 21 participants began therapy; 20 continued to receive full-dose therapy at 4 weeks (1 hepatic decompensation–related discontinuation), 19 continued to receive therapy at 8 weeks (1 mental health–related discontinuation), and 15 continued to receive therapy at 12 weeks (4 additional therapy-related discontinuations). Ribavirin dose reduction was required at week 8 (n = 1) and week 12 (n = 2).
Table 1.
Clinical Characteristics
| Longitudinal Therapy Study |
Cross Sectional NK Cytolysis Study |
|||
| Variable | HCV | Healthy Control | HCV | Healthy Control |
| No. | 21 | 10 | 15 | 11 |
| Age, y | 57 (50–65)a | 44 (37–50) | 55 (50–64)a | 31 (26–53) |
| Sex | Male 95%a | Male 50% | Male 93%a | Male 64% |
| Female 5% | Female 50% | Female 7% | Female 36% | |
| Race or ethnic group | Black 48% | Black 30% | Black 60%a | Black 9% |
| White 52% | White 70% | White 33% | White 55% | |
| Hispanic 7% | Hispanic 18% | |||
| Asian 18% | ||||
| Genotype | 1 (100%) | 1 (87%) | ||
| 2 (13%) | ||||
| HCV level (IU/mL) | 996 438 | 1 373 190 | ||
| (171 031–5 384 560) | (36 893–18 800 880) | |||
| AST level (U/mL) | 58 (23–230) | 43 (21–162) | ||
| ALT level (U/mL) | 47 (14–175) | 48 (18–185) | ||
| APRI | 0.72 (0.20–5.10) | 0.44 (0.15–2.59) | ||
| <0.4, 33%; 0.4–1.5, 29%; >1.5, 38% | <0.4, 47%; 0.4–1.5, 40%; >1.5, 13% | |||
| PLT (109 platelets/mL) | 201 (88–262) | 212 (139–370) | ||
| Albumin level (g/dL) | 3.7 (1.8–4.4) | 4.0 (3.6–4.4) | ||
Values are expressed as median (range) for HCV level (by branched chain method or branched DNA), albumin level, PLT count, AST level, ALT level, age, and APRI; calculated as described [24]. Proportions of subjects within each category are given for HCV genotype, sex, APRI, and race.
Abbreviations: ALT, alanine aminotransferase; APRI, AST-to-PLT ratio index; AST, aspartate aminotransferase; HCV, hepatitis C virus; NK, natural killer; PLT, platelet.
P ≤ .05 compared with healthy controls.
For in vitro NK cytolytic function assays, a separate nonoverlapping cohort of chronic HCV-infected patients naive to therapy was recruited (n = 15), along with age range–matched healthy control subjects (n = 11) (Table 1).
Clinical Laboratories
HCV branched-chain polymerase chain reaction (PCR; sensitivity, 615 IU/mL) and HCV transcription-mediated amplification PCR (sensitivity, 15 IU/mL) were performed, and aspartate aminotransferase (AST), alanine aminotransferase (ALT), platelet (PLT), total bilirubin, and albumin levels were measured in a single clinical laboratory. AST-to-PLT ratio index (APRI) was calculated as described elsewhere [24]. IL-28B rs12979860 single-nucleotide polymorphism (SNP) genotype was determined using a strand-specific PCR method (Monogram Biosciences).
NK Cell Subset Frequency, IFN-αR, NKp30, TRAIL, and CD161 Expression
Freshly prepared peripheral blood mononuclear cells (PBMCs) were stained in real time with anti–CD3-PerCP (clone SK7), anti–CD16-APC-Cy7 (clone 3G8), anti–CD56-PE-Cy7 (clone NCAM16.2), anti–CD161-APC (clone DX12), anti–NKp30-PE (clone P30-15; BD Biosciences), and anti–IFN-αR1-FITC (clone 85228; R&D Systems) or isotype controls. IFN-αR1-PE (clone 85228; R&D Systems) was used for bulk NK cytolytic assays. Flow cytometric analysis was performed on a BD LSRII flow cytometer (BD Biosciences) with FACSDiva Software (BD Biosciences).
For NK cell TRAIL expression, cryopreserved PBMCs were stained with anti-CD3PerCP (clone SK7), anti-CD56APC (clone NCAM16.2), anti-CD16FITC (clone 3G8; BD Biosciences), and anti-TRAIL-PE (clone RIK-2) or PE-labeled mouse immunoglobulin G (IgG) 1 (BD Biosciences). Analysis was performed on a BD FACSCalibur flow cytometer (BD Biosciences) with CELLQuest software (BD Biosciences).
IFN-α–Induced pSTAT1
Freshly prepared PBMCs (1 × 106) were analyzed in real time for IFN-α2a–induced pSTAT1 by preincubating cells at 4°C for 30 minutes with anti–CD16-FITC (clone 3G8), anti–CD56-PE (clone NCAM16.2), and anti–CD3-APC (clone SK7; BD Biosciences); washing; resuspending in RPMI 10% fetal calf serum (Hyclone); and culturing for 15 minutes at 37°C with 0, 1000, 3000, or 10 000 U/mL of IFN-α2a (PBL Biomedical Labs). Cells were washed, fixed with BD Cytofix, permeabilized with BD Phosflow Perm Buffer III (BD Biosciences), and stained for 30 minutes at room temperature with anti-human pSTAT1 PerCPCy5.5 (clone 4a) or isotype control (BD Biosciences). Flow cytometric analysis was performed on a BD LSRII flow cytometer with FACSDiva software (Supplementary Figure 1), and specific expression of pSTAT1 was calculated as the mean fluorescence intensity (MFI) above isotype control.
IFN-α–Induced NK Subset IFN-γ by Intracellular Flow Cytometry and IFN-α–Induced PBMC IFN-γ by Enzyme-Linked Immunosorbent Spot Assay
Cryopreserved PBMCs were thawed, plated at 106 cells per well, incubated for 20 hours at 37°C (Brefeldin A [Sigma-Aldrich] was added after 2 hours) in the presence or absence of IFN-α2a (1000 U/mL; PBL Biomedical Labs), fixed, permeabilized, and stained using intracellular cytokine staining protocol with anti–CD3-APC (clone SK7), anti–CD14-PerCP (clone MφP9), anti–CD16 APC-Cy7 (clone 3G8), anti–CD56-PE-Cy7 (clone NCAM16.2), and anti–IFN-γ−FITC (clone 25723.11; BD Biosciences). Thawed cells were also plated at 300 000 and 600 000 cells per well in precoated IFN-γ enzyme-linked immunosorbent spot assay plates (Millipore) and cultured for 20 hours at 37°C in the presence or absence of IFN-α2a (1000 IU/mL), and IFN-γ–secreting cell frequency was determined as previously described [23].
NK Cytolysis of JFH-1–Infected Huh 7.5 Cell Cultures
Huh 7.5 cells were provided by Dr C. M. Rice (Apath LLC). The pJFH1 plasmid was provided by Dr T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan). Infectious JFH1 virus was prepared as previously described [25]; 2000 Huh 7.5 cells were infected by 1 multiplicity of infection of JFH-1 for 2 days in 384-well plates prior to NK cell coculture.
Bulk NK cells were prepared by negative bead selection (Stemcell Technologies) depleting CD3/4/14/19/20/36/66b/123/HLA-DR/glycophorin-A–bearing cells (purity, >95%). Bulk NK cells or flow-sorted NK subsets (BD FACSAria; BD Biosciences) were stimulated with 500 U/mL of IFN-α-2a or media alone for 16 hours, washed, added to JFH-1–infected or –uninfected Huh 7.5 target cells at indicated effector: target ratios, and cocultured for 5 hours. A total of 50 μL of supernatant was analyzed for cytolytic activity, measuring glyceraldehyde 3-phosphate dehydrogenase (GAPDH) release with use of aCella-Tox (Cell Technology) as described elsewhere [26, 27] by luminometer (VICTOR3V; PerkinElmer). Mean serum concentration of IFN-α2a during pegylated IFN-α2a (180 μg/week) therapy is 8 ng/mL [28]. IFN-α2a used here is 3–5 pg/U. Therefore, 500–3000 U/mL culture concentration is equivalent to 1.5–15 ng/mL, a range similar to serum IFN-α2a concentrations during therapy.
Statistical Analysis
Statistical analyses were performed using SPSS for Windows, version 19.0 (SPSS). We used the Mann–Whitney U test for 2-way comparisons of continuous variables across and within groups. Associations between continuous variables were evaluated using Spearman rank correlation coefficient. Intragroup variable comparison over time in the same participants was analyzed using Wilcoxon signed-rank test. Linear regression analysis was used to evaluate the joint effects of IFN-αR race and APRI on viral decrease. Jonckheere-Terpstra test for ordered alternatives was used to analyze ascending trends across the 3 IL-28B genotypes. All tests of statistical significance were 2-sided, and P values ≤.05 were considered statistically significant.
RESULTS
IFN-αR Expression Is Higher in Chronic HCV–Infected Patient CD16+56+ and CD16−56+ NK Cells Than in Controls, and HCV-Infected Patient IFN-αR Expression Differs by Race on CD16+56− and CD16+56+ NK Cells
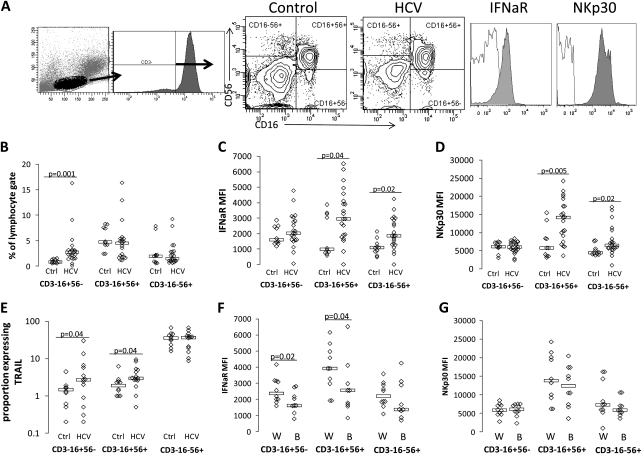
We first evaluated NK subset frequency and IFN-αR and NKp30 expression. Peripheral blood frequencies of CD16+56−, CD16+56+, and CD16−56+ NK cells were quantified as shown (Figure 1A). CD16+56− NK cell frequencies were higher in chronic HCV infection (Figure 1B), as previously described [29]. IFN-αR and NKp30 expression were greater in CD16+56+ and CD16−56+ NK subsets in chronic HCV–infected patients than in controls (P = .04 and P = .02, respectively, for IFN-αR and P = .005 and P = .02, respectively, for NKp30), whereas expression of IFN-αR on CD16+56− NK cells did not differ significantly (Figure 1C and 1D). Because sex differed between groups, we also analyzed data for male patients only. Differences between groups were preserved when analysis was restricted to male participants (P = .002 and P = .008). NK TRAIL expression is associated with in vitro killing in HCV-infected Huh 7.5 cells [22]. TRAIL expression was found to be greater in CD16+56− and CD16+56+ NK cells in chronic HCV–infected patients than in those in controls. Because age differed between HCV-infected and control groups, we evaluated associations between receptor expression (IFN-αR, NKp30, CD161, and TRAIL) and clinical variables (AST, ALT, PLT, total bilirubin, age, race, APRI, and HCV level) in the HCV-infected group. No correlation between IFN-αR or NKp30 and age was observed. We did, however, observe IFN-αR expression, but not NKp30, TRAIL, or CD161expression, to be higher on CD16+56− and CD16+56+ NK cells of white, compared with black, HCV-infected patients. APRI also negatively correlated with IFN-αR expression on CD16+56− NK cells only (r = −0.43; P = .05). Ideally, immune parameters would also be compared as a function of sustained virologic response vs nonresponse to therapy. However, although 20 of the initial 21 participants continued therapy at 4 weeks, only 15 continued therapy at 12 weeks. Four were nonresponders, 3 were partial responders, 3 were responder-relapsers, 2 were sustained virologic responders, and the results for 3 are pending. Because of the number of adverse effect–related dropouts after 4 weeks of therapy, week 4 data were viewed as the most appropriate to focus on here.
Figure 1.
Natural killer (NK) subset interferon (IFN)–αR expression is increased during hepatitis C virus (HCV) infection, and expression differs by race. Peripheral blood mononuclear cells were stained with CD3, CD16, CD56, IFN-αR, and NKp30 or isotype control, and flow cytometric analysis was performed on a BD LSRII. A, Gating strategy used to define NK cell subsets in both healthy control and HCV-infected subjects, followed by example of IFN-αR and NKp30 expression on NK cells of 1 healthy control subject. Isotype shown in nonshaded area, and antibody shown in shaded area. Specific NKp30 and IFN-αR expression were determined as mean fluorescence intensity above isotype control. NK subset frequency (B), IFN-αR (C), NKp30 (D), and TRAIL (E) expression comparison between healthy control subjects (ctrl; n = 10) and HCV-infected patients (n = 21). IFN-αR (F) and NKp30 (G) are shown as a function of race in the HCV-infected group (W = white, B = black). P values ≤.05 are shown.
Magnitude of HCV Decrease at 4 Weeks of Pegylated IFN-α Plus Ribavirin Therapy Is Associated With Baseline CD16+56− NK Cell IFN-αR Expression and IL-28B Genotype
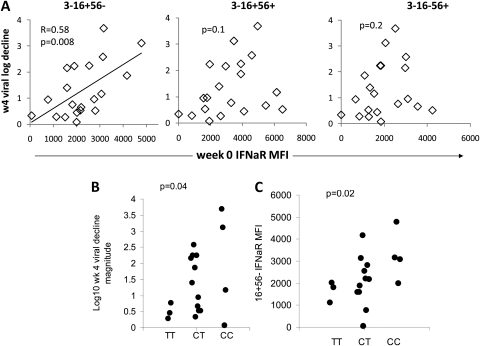
We observed an expected degree of variability in magnitude of viral decrease during the early phase of pegylated IFN-α plus ribavirin therapy, in part associated with race (week 4, 2.2 vs 0.52 log10 decrease in white and black participants, respectively; P = .01) and APRI (r = -0.70; P = .001), as expected. Race and APRI were not associated (P = .2). IL-28B rs12979860 genotype was available for 18 participants (3 [TT], 11 [CT], 4 [CC]). In these genotype groups, 66%, 45%, and 0% of participants, respectively, were black. As expected, IL-28B genotype was also associated with 1-month viral decrease (median, 0.45, 1.39, and 2.14 log decrease in the TT, CT, and CC groups, respectively; P = .04, Figure 2).
Figure 2.
Magnitude of hepatitis C virus (HCV) level decrease during pegylated interferon (IFN)–α plus ribavirin therapy is associated with baseline CD16+56− natural killer (NK) cell interferon (IFN)–αR expression. A, Correlation between log viral decrease at 4 weeks of treatment and baseline IFN-αR in HCV-infected patients (n = 20). B, Interleukin 28B (IL-28B) genotype vs week 4 log HCV decrease (n = 18); C, IL-28B genotype vs baseline CD3–16+56– NK IFN-αR mean fluorescence intensity (n = 18).
When evaluating whether NK IFN-αR, NKp30, CD161, or TRAIL expression was predictive of HCV level decrease, baseline NK IFN-αR expression correlated with magnitude of HCV level decrease at 4 weeks, although only in the CD16+56− NK-cell subset (Figure 2). When 3 participants receiving submaximal IFN-α doses were removed from the analysis, the relationship was preserved (r = 0.59; P = .01). In addition, this relation held for HCV level decrease at week 12, again selectively in the CD16+56− NK-cell subset (r = 0.73; P = .02). Linear regression analysis indicated that the relationship between CD16+56− NK IFN-αR expression and magnitude of viral decrease was not significantly modified by race or APRI (P = .3 and P = .5 for interaction of race or APRI with IFN-αR expression). Furthermore, analysis of this relation in APRI subgroups (above and below median APRI) indicated that the same correlation tended to hold (r = 0.6, P = .09; r = 0.58, P = .06). Baseline NKp30 expression did not significantly correlate with magnitude of viral decrease, nor did baseline CD161 or TRAIL expression. IL-28B genotype was associated with CD16+56− NK IFN-αR expression (median, 1815, 2165, and 3117 MFI in the TT, CT, and CC groups, respectively; P = .02) (Figure 2).
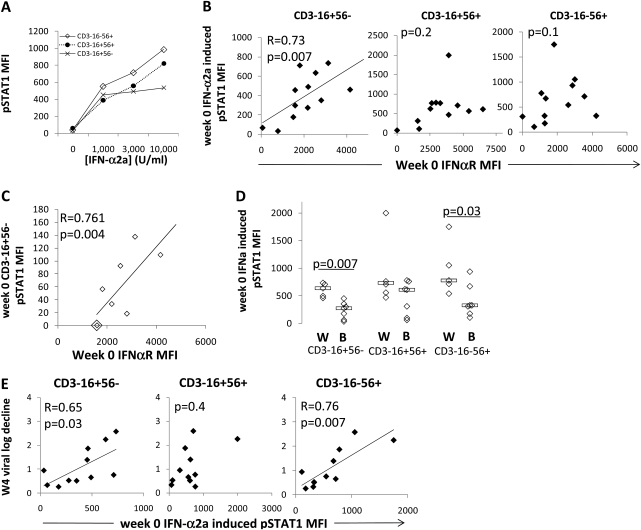
Baseline CD16+56− IFN-αR Expression Is Associated With IFN-α2a-Induced pSTAT1 and With In Vivo IFN-α Plus Ribavirin–Induced NKp30, Both Associated With HCV Level Decrease
To determine whether IFN-αR expression affects IFN-α signaling, we measured IFN-α2a–induced NK subset pSTAT1 by flow cytometry when possible. As shown in Figure 3A, level of IFN-αR expression correlated with IFN-α2a–induced CD16+56− NK cell pSTAT1 in chronic HCV–infected patient samples (Figure 3B). This relationship also held for 1000 and 10 000 U/mL IFN-α2a conditions in the CD16+56− NK subset (r = 0.72, P = .005; r = 0.55, P = .05). In addition, pSTAT1 level correlated with IFN-αR expression on media-treated CD16+56− NK cells (r = 0.80; P = .001), suggesting reflection of in vivo IFN-α signaling. The relationship was not observed for the other NK subsets (Figure 3B), although for the CD16−56+ subset, an association was observed between IFN-αR and the delta pSTAT1 (change from unstimulated condition) induced by 1 concentration of IFN-α-2a (3000 U/mL; r = 0.57; P = .03). When evaluating IFN-α-2a–induced pSTAT1 by race, CD16+56− and CD16−56+ NK cells from white participants were observed to have more IFN-α-2a–induced pSTAT1 than cells from black participants (Figure 3D). When evaluating IFN-α-2a–induced pSTAT1 in relation to therapy-induced viral decrease, IFN-α-2a–induced CD16+56− NK cell pSTAT1 at baseline correlated with IFN-α therapy–induced HCV level decrease at week 4 (Figure 3E). This relationship also held for 0, 1000, and 10 000 U/mL IFN-α2a culture conditions (r = 0.76, P = .004; r = 0.73, P = .007; r = 0.59, P = .04). This relationship also existed for the CD16−56+ NK subset at 1 IFN-α2a concentration (Figure 3E).
Figure 3.
Baseline CD16+56− interferon (IFN)–αR expression is associated with IFN-α–induced CD16+56− pSTAT1 and magnitude of hepatitis C virus (HCV) level decrease. A, Representative example of IFN-α2a–induced pSTAT1 expression on each of the 3 natural killer (NK) cell subsets over IFN-α2a concentration range. B, Correlation between baseline IFN-α2a (3000 U/mL) induced pSTAT1 and baseline IFN-αR mean fluorescence intensity (MFI) on each of the 3 NK subsets (12 hepatitis C virus [HCV]–infected patients). C, Correlation between baseline spontaneous (0 U/mL IFN-α) induced pSTAT1 and baseline IFN-αR MFI for CD3–16+56– NK cells of HCV-infected patients (n = 8). D, Racial comparison of IFN-α2a–induced pSTAT1 on each of the 3 NK subsets at 3000 U/mL IFN-α2a. E, Correlation between log of viral decrease at 4 weeks of treatment and baseline IFN-α2a–induced pSTAT1 on each of the 3 NK subsets at 3000 U/mL IFN-α2a (11 HCV-infected patients).
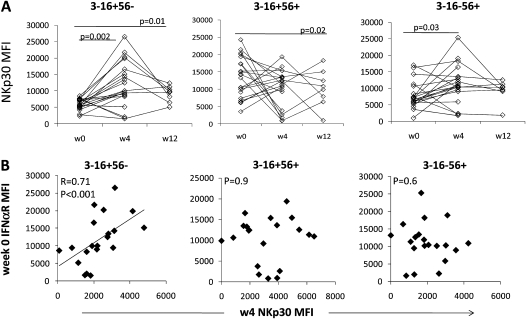
We next focused on downstream consequences of IFN-α signaling. Baseline IFN-γ–producing PBMC frequency and NK subset IFN-γ in response to IFN-α2a were not correlated with the magnitude of HCV level decrease. IFN-α has been shown to induce NK cell NKp30 expression [30]. We therefore evaluated IFN-αR and NKp30 expression longitudinally over IFN-α therapy. Expression of NKp30 was greater in CD16+56− NK cells at week 4 of therapy (Figure 4A), and there was a trend toward week 4 CD16+56− NK cell subset NKp30 expression to correlate with week 4 viral decrease, although only when considering participants who received full-dose pegylated IFN-α2a (r = 0.44; P = .07). In addition, baseline CD16+56− NK cell subset IFN-αR expression correlated with week 4 NKp30 expression, consistent with IFN-αR–mediated regulation of NKp30 expression (Figure 4B). CD16−56+ NKp30 modestly increased at week 4 (Figure 4A), although this was not correlated with baseline IFN-αR expression. At the same time, CD16+56− NK subset frequency decreased in the peripheral blood at week 4 (week 0 vs 4: median, 2.73% [interquartile range {IQR}, 1.59%–3.20%] vs 0.81% [IQR, 0.44–1.70]; P < .01) as previously described [29]. IFN-αR expression was somewhat increased at week 4, compared with baseline, in CD16+56− NK cells (week 0 vs 4: median MFI, 2020 [IQR, 1596–2802] vs 2979 [IQR, 2479–5129]; P < .05).
Figure 4.
Baseline CD16+56− interferon (IFN)–αR expression is associated with pegylated IFN-α plus ribavirin–induced CD16+56− natural killer (NK) p30 expression. A, NKp30 expression over course of treatment on each of the 3 NK subsets (20 hepatitis C virus [HCV]–infected patients). B, Correlation between baseline IFN-αR expression and week 4 NKp30 expression on each of the 3 NK subsets.
NKp30 Contributes to Cytolysis of HCV JFH1-Infected Huh 7.5 Cells, CD16+56− NK Cells Contribute to Activity, and CD16+56– NK IFN-αR Expression Is Associated With Cytolytic Activity
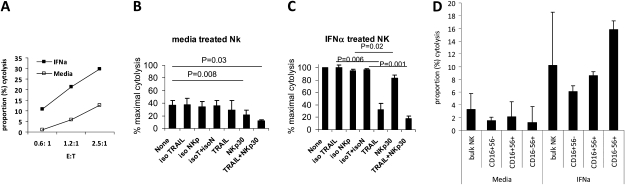
To analyze the role of NKp30 in HCV-infected target cell cytolysis, we performed 5-hour cocultures of negatively selected bulk NK cells and HCV JFH1–infected Huh 7.5 cells. NK-dependent cytolysis of Huh 7.5 targets was enhanced by the presence of HCV infection and IFN-α2a pretreatment, as previously described [22]. Activity was partially dependent on TRAIL (Figure 5), as previously described [22], mainly in the presence of IFN-α2a pretreatment (Figure 5C). Blockade of NKp30 inhibited killing efficiency in both the absence and presence (Figure 5C) of IFN-α2a pretreatment, although the effect was more robust in the absence of IFN-α2a. These data indicate a role for NKp30 in HCV-infected target killing.
Figure 5.
Interferon (IFN)–α–activated natural killer (NK) cytolysis of hepatitis C virus (HCV)–infected targets is partially dependent on NKp30 and partially mediated by CD16+56− NK cells. A, Negatively selected NK cells from 1 healthy control subject were cultured for 16 h with media or 500 U/mL IFN-α2a, washed, and cocultured with HCV JFH-1–infected target cells for 5 h, and cytolysis was measured (y-axis) for differing E:T ratios (x-axis). Background target cell cytolysis in the absence of added NK cells was <10% in each experiment, and this background was subtracted from the data shown. Ten microliters of lytic reagent was added to target cells as a positive control. Percentage cytotoxicity was calculated as: [(experimental glyceraldehyde 3-phosphate dehydrogenase [GAPDH] release-spontaneous GAPDH release from effector cells alone – spontaneous GAPDH release from target cells alone)/(maximum induced GAPDH release from target cells-spontaneous GAPDH release from target cells)] × 100. B, Negatively selected NK cells from 3 healthy control subjects were cultured for 16 h with media then cocultured with HCV JFH1–infected target cells at 2.5:1 E:T 5 h as in (A). For antibody-blocking experiments, NK cells were treated with 10 μg/mL of goat polyclonal anti-NKp30 IgG and vs isotype control (goat IgG) (R&D Systems); or mouse anti-TRAIL monoclonal antibodies (clone, 2E5) vs isotype control (mouse IgG1) (Enzo Life Sciences) at 37°C for 1 h, then added to target cell cultures. Means and standard deviations are shown. C, Same as (B), except NK cells were treated with IFN-α2a, then washed before coculture with HCV-infected target cells. D, Bulk NK cells from 3 healthy controls were cultured in the presence vs absence of 500 U/mL IFN-α2a for 16 h, washed, and cocultured with HCV JFH-1–infected target cells at E/T 1.2:1, or stained with antibodies to CD3, CD16, and CD56 followed by flow cytometric–based cell sorting of NK subsets that were utilized in killer assays at E:T 1.2:1.
We next evaluated the role of NK subsets in HCV-infected target killer function. CD16+56− cells of healthy controls were found to be capable of cytolytic activity (Figure 5D), although at a somewhat reduced efficiency in comparison with CD16−CD56+ NK cells. Cytolytic activity was enhanced by IFN-α2a treatment of all 3 NK subsets, particularly for CD16−CD56+ NK cells. These data indicate that all 3 NK subsets are capable of contributing with varying degrees to HCV-directed cytolytic activity.
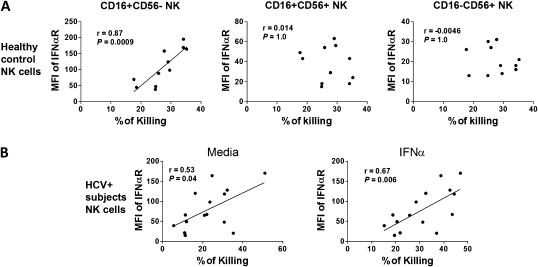
We next evaluated NK subset IFN-αR expression in relation to cytolytic function in groups of persons not overlapping with the therapy cohort (Table 1). Healthy control CD16+56− NK IFN-αR expression at baseline was correlated with IFN-α–activated bulk NK cytolysis of HCV-infected Huh 7.5 target cells, whereas IFN-αR expression on CD16+56+ or CD16−56+ subsets was not (Figure 6A). This relation also held at E:T 1.25:1 (r = 0.8; P = .005). No relationship was observed between IFN-αR expression in any NK subset and cytolysis of uninfected Huh 7.5 target cells for either healthy control subject or HCV-infected participant NK samples or media-treated healthy control NK cell cytolysis of HCV-infected Huh 7.5 targets. In contrast, HCV-infected patient CD16+56− NK cell IFN-αR expression was associated with NK cytolysis of HCV-infected targets when NK cells were treated with either media or IFN-α2a (Figure 6B). This relationship held for all E:T in the presence of IFN-α2a stimulation (E:T 1.25:1, r = 0.56, P = .03; E:T 0.6:1, r = 0.53, P = .04).
Figure 6.
CD16+56− natural killer (NK) interferon (IFN)–αR expression is correlated with in vitro hepatitis C virus (HCV)–targeted NK cytolytic function. Negatively selected NK cells from healthy control subjects or HCV-infected patients were cultured for 16 h with media or IFN-α2a, washed, then cocultured with HCV JFH1–infected target cells at 2.5:1 E:T 5 h as in Figure 5 (E:T 0.6:1 and 1.25:1 also performed, not shown). Background target cell cytolysis in the absence of added NK cells was <10% in each experiment, and this background is subtracted from the data shown. A, Healthy control (n = 11) subject NK subset IFN-αR expression vs IFN-α2a–treated NK cytolytic activity. B, HCV-infected subject (n = 15) CD16+56− NK subset IFN-αR expression vs NK cytolytic activity in absence or presence of IFN-α2a pretreatment.
DISCUSSION
Data here highlight racially associated NK cell IFN-αR expression during chronic HCV infection coinciding with known racial differences in IFN-α therapy efficacy. CD16+56− NK cell IFN-αR expression at baseline correlated positively with IFN-α signaling capacity and magnitude of viral decrease in HCV genotype 1–infected patients treated with pegylated IFN-α plus ribavirin. One consequence of IFN-α signaling is enhanced NKp30 expression, and CD16+56− NK cell IFN-αR expression at baseline correlated both with increased CD16+56− NK cell NKp30 expression at week 4 of therapy and magnitude of viral decrease. NKp30 and CD16+56− NK cells contributed to HCV-infected target killing efficiency in vitro. Furthermore, CD16+56– NK cell IFN-αR expression correlated with HCV-infected cell targeting, suggesting that IFN-α–dependent activity of this subset may be more dependent on IFN-αR expression than that of other NK subsets (in which other factors are likely to be rate limiting). These data suggest that CD16+56− NK cell IFN-αR expression level may contribute to control of HCV infection during IFN-α plus ribavirin therapy and suggest a potential mechanism by which racial differences in IFN-α therapy response are mediated. Because the IL-28B promoter polymorphism is also associated with CD16+56− NK cell IFN-αR expression, one potential mechanism underlying the IL-28B genetic association may be directly or indirectly through IFN-αR expression.
Peripheral blood NK subset skewing exists during chronic HCV infection, with increased CD16+56− and decreased CD16+56+ NK subset frequencies [29, 31]. CD16+56− NK cells have lower TNF-α– and IFN-γ–secreting activity but similar CD107a and chemokine production in response to K562 targets [29]. Further investigation of IFN-αR–dependent activity of this subset is warranted.
Race is known to associate with response to IFN-α–based therapy [3]. We found that NK subset IFN-αR expression and IFN-α2a–induced pSTAT1 associate with race, providing 1 plausible mechanistic link between race and IFN-α response. Of note, the black population in America is admixed 10%–20% with the population of European ancestry [32]. This means that the differences observed here are minimum estimates. IL-28B SNPs (within the IFN-λ gene region) are thought to account for a substantial portion of racially based variability in pegylated IFN-α plus ribavirin therapy response [8, 9, 33–35]. IL-28B SNPs have been associated with variable levels of IFN-λ messenger RNA expression [34, 35], although the mechanism accounting for the IL-28B SNP link to IFN-α therapy response is not known [36]. We identify an association between IL-28B genotype and CD16+56− NK subset IFN-αR expression, providing 1 possible mechanistic link. Such an association does not necessarily indicate a direct effect of IL-28B on NK cells, although NK cell IL-28B receptor expression has been described without identified function [37].
Change in NK subset frequency and/or phenotype observed in the peripheral blood over the course of therapy may reflect anatomic compartment redistribution, cell differentiation, cell expansion, cell death, or change in receptor expression in the same cells. Peripheral blood CD16+56− NK cells have been previously observed to decrease in frequency over the first 4 weeks of therapy [38]. Results here are in agreement. Whether these cells redistribute, differentiate, or die is unknown.
IFN-αR expression was observed to negatively correlate with APRI, indicating that disease stage may affect NK subset IFN-αR expression. Of note, the relationship between viral decrease and IFN-αR expression tended to exist in both high and low APRI subgroups, indicating that the relationship between IFN-αR expression and viral decrease is likely to be independent of APRI. This was also supported by linear regression analysis. However, linear regression analysis also revealed that the relationship between IFN-α-2a–induced pSTAT1 and viral decrease tended to be attenuated in persons with higher APRI (P = .07). On the surface, these relationships appear to be complex, especially because peripheral blood NK IFN-αR expression may or may not reflect that in the liver. One possibility is that hepatic parenchymal sufficiency may be required to facilitate NK IFN-αR expression. In addition, STAT1 levels have been associated with IFN-α2a–induced NK pSTAT1 activity [39], and NK STAT1 levels may differ as a function of liver disease.
Week 4 NKp30 expression correlates with baseline CD16+56− NK cell IFN-αR expression. In addition, IFN-αR and NKp30 expression level were associated with each other on the same cells at week 4 (data not shown), indicating that NKp30 upregulation likely occurred on the same cell expressing higher levels of IFN-αR. The latter is supported by prior data indicating that IFN-α stimulation results in upregulation of NK NKp30 expression [30]. It is plausible that 1 mechanism underlying variability in IFN-α–mediated control of HCV in vivo is through the level of NK subset IFN-αR expression that determines IFN-α–induced signaling magnitude, in turn leading to enhanced NKp30 (and other modulators of effector function), which in turn contribute to control of HCV infection. Certainly, NKp30 expression during IFN-α therapy has recently been shown to associate with favorable response to therapy [20, 21]. Although this is a small data set, clearly, further investigation of this NK subset is warranted, with specific emphasis on investigation of genetically encoded and environmental factors contributing to enhanced IFN-αR expression and mechanisms underlying downstream associations with magnitude of viral decrease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
We thank the study participants, and Laura Napolitano, Yolanda Lie, and Eoin Coakley at Monogram Biosciences for helping to make IL-28B genotyping possible.
Financial support.
This work was supported by the National Institutes of Health (grant numbers R01 DK068361 and R01 AI069195).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 2.Freeman AJ, Dore GJ, Law MG, et al. Estimating progression to cirrhosis in chronic hepatitis C virus infection. Hepatology. 2001;34:809–16. doi: 10.1053/jhep.2001.27831. [DOI] [PubMed] [Google Scholar]
- 3.Thompson AJ, Muir AJ, Sulkowski MS, et al. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120–9. doi: 10.1053/j.gastro.2010.04.013. e18. [DOI] [PubMed] [Google Scholar]
- 4.Carrat F, Bani-Sadr F, Pol S, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. JAMA. 2004;292:2839–48. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 5.Chung RT, Andersen J, Volberding P, et al. Peginterferon alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004;351:451–9. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–82. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 7.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351:438–50. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 8.O'Brien TR. Interferon-alfa, interferon-lambda and hepatitis C. Nat Genet. 2009;41:1048–50. doi: 10.1038/ng.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 10.Habu S, Akamatsu K, Tamaoki N, Okumura K. In vivo significance of NK cell on resistance against virus (HSV-1) infections in mice. J Immunol. 1984;133:2743–7. [PubMed] [Google Scholar]
- 11.McIntyre KW, Welsh RM. Accumulation of natural killer and cytotoxic T large granular lymphocytes in the liver during virus infection. J Exp Med. 1986;164:1667–81. doi: 10.1084/jem.164.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells [see comments] N Engl J Med. 1989;320:1731–5. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 13.Cauda R, Laghi V, Tumbarello M, Ortona L, Whitley RJ. Immunological alterations associated with recurrent herpes simplex genitalis. Clin Immunol Immunopathol. 1989;51:294–302. doi: 10.1016/0090-1229(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 14.Goodyear HM, McLeish P, Randall S, et al. Immunological studies of herpes simplex virus infection in children with atopic eczema. Br J Dermatol. 1996;134:85–93. [PubMed] [Google Scholar]
- 15.Joncas J, Monczak Y, Ghibu F, et al. Brief report: killer cell defect and persistent immunological abnormalities in two patients with chronic active Epstein-Barr virus infection. J Med Virol. 1989;28:110–7. doi: 10.1002/jmv.1890280211. [DOI] [PubMed] [Google Scholar]
- 16.Lanier LL, Spits H, Phillips JH. The developmental relationship between NK cells and T cells. Immunol Today. 1992;13:392–5. doi: 10.1016/0167-5699(92)90087-N. [DOI] [PubMed] [Google Scholar]
- 17.Moretta L, Ciccone E, Poggi A, Mingari MC, Moretta A. Ontogeny, specific functions and receptors of human natural killer cells. Immunol Lett. 1994;40:83–8. doi: 10.1016/0165-2478(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 18.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–4. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 19.Vidal-Castineira JR, Lopez-Vazquez A, Diaz-Pena R, et al. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol. 2010;84:475–81. doi: 10.1128/JVI.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahlenstiel G, Edlich B, Hogdal LJ, et al. Early changes in natural killer cell function indicate virologic response to interferon therapy for hepatitis C. Gastroenterology. 2011;141:1231–9. doi: 10.1053/j.gastro.2011.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bozzano F, Picciotto A, Costa P, et al. Activating NK cell receptor expression/function (NKp30, NKp46, DNAM-1) during chronic viraemic HCV infection is associated with the outcome of combined treatment. Eur J Immunol. 2011;41:2905–14. doi: 10.1002/eji.201041361. [DOI] [PubMed] [Google Scholar]
- 22.Stegmann KA, Bjorkstrom NK, Veber H, et al. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology. 2010;138:1885–97. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 23.Conry SJ, Milkovich KA, Yonkers NL, et al. Impaired plasmacytoid dendritic cell (PDC)-NK cell activity in viremic human immunodeficiency virus infection attributable to impairments in both PDC and NK cell function. J Virol. 2009;83:11175–87. doi: 10.1128/JVI.00753-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder N, Gajula L, Xiao SY, et al. APRI: an easy and validated predictor of hepatic fibrosis in chronic hepatitis C. J Clin Gastroenterol. 2006;40:535–42. doi: 10.1097/00004836-200607000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Russell RS, Yonkers NL, et al. Differential effects of hepatitis C virus JFH1 on human myeloid and plasmacytoid dendritic cells. J Virol. 2009;83:15. doi: 10.1128/JVI.02671-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogbomo H, Hahn A, Geiler J, Michaelis M, Doerr HW, Cinatl J., Jr NK sensitivity of neuroblastoma cells determined by a highly sensitive coupled luminescent method. Biochem Biophys Res Commun. 2006;339:375–9. doi: 10.1016/j.bbrc.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 27.Ogbomo H, Michaelis M, Kreuter J, Doerr HW, Cinatl J., Jr Histone deacetylase inhibitors suppress natural killer cell cytolytic activity. FEBS Lett. 2007;581:1317–22. doi: 10.1016/j.febslet.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 28.Dahari H, Affonso de Araujo ES, Haagmans BL, et al. Pharmacodynamics of PEG-IFN-alpha-2a in HIV/HCV co-infected patients: implications for treatment outcomes. J Hepatol. 2010;53:460–7. doi: 10.1016/j.jhep.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez VD, Falconer K, Bjorkstrom NK, et al. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J Immunol. 2009;183:6612–8. doi: 10.4049/jimmunol.0901437. [DOI] [PubMed] [Google Scholar]
- 30.Bozzano F, Costa P, Passalacqua G, et al. Functionally relevant decreases in activatory receptor expression on NK cells are associated with pulmonary tuberculosis in vivo and persist after successful treatment. Int Immunol. 2009;21:779–91. doi: 10.1093/intimm/dxp046. [DOI] [PubMed] [Google Scholar]
- 31.Morishima C, Paschal DM, Wang CC, et al. Decreased NK cell frequency in chronic hepatitis C does not affect ex vivo cytolytic killing. Hepatology. 2006;43:573–80. doi: 10.1002/hep.21073. [DOI] [PubMed] [Google Scholar]
- 32.Parra EJ, Marcini A, Akey J, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balagopal A, Thomas DL, Thio CL. IL28B and the control of hepatitis C virus infection. Gastroenterology. 2010;139:1865–76. doi: 10.1053/j.gastro.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suppiah V, Moldovan M, Ahlenstiel G, et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–4. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka Y, Nishida N, Sugiyama M, et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–9. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 36.Wapner J. Pharmacogenomics. Gene variants affect hepatitis C treatment, but link is elusive. Science. 2010;330:579. doi: 10.1126/science.330.6004.579. [DOI] [PubMed] [Google Scholar]
- 37.Witte K, Gruetz G, Volk HD, et al. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009;10:702–14. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez VD, Falconer K, Michaelsson J, et al. Expansion of CD56- NK cells in chronic HCV/HIV-1 co-infection: reversion by antiviral treatment with pegylated IFNalpha and ribavirin. Clin Immunol. 2008;128:46–56. doi: 10.1016/j.clim.2008.03.521. [DOI] [PubMed] [Google Scholar]
- 39.Miyagi T, Takehara T, Nishio K, et al. Altered interferon-alpha-signaling in natural killer cells from patients with chronic hepatitis C virus infection. J Hepatol. 2010;53:424–30. doi: 10.1016/j.jhep.2010.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.