Abstract
Background. To date, only mutations in CCR5 have been shown to confer resistance to human immunodeficiency virus type 1 (HIV-1) infection, and these explain only a small fraction of the observed variability in HIV susceptibility.
Methods. We performed a meta-analysis between 2 independent European genomewide association studies, each comparing HIV-1 seropositive cases with normal population controls known to be HIV uninfected, to identify single-nucleotide polymorphisms (SNPs) associated with the HIV-1 acquisition phenotype. SNPs exhibiting P < 10−5 in this first stage underwent second-stage analysis in 2 independent US cohorts of European descent.
Results. After the first stage, a single highly significant association was revealed for the chromosome 8 rs6996198 with HIV-1 acquisition and was replicated in both second-stage cohorts. Across the 4 groups, the rs6996198-T allele was consistently associated with a significant reduced risk of HIV-1 infection, and the global meta-analysis reached genomewide significance: Pcombined = 7.76 × 10−8.
Conclusions. We provide strong evidence of association for a common variant with HIV-1 acquisition in populations of European ancestry. This protective signal against HIV-1 infection is the first identified outside the CCR5 nexus. First clues point to a potential functional role for a nearby candidate gene, CYP7B1, but this locus warrants further investigation.
In the past few years, several genomewide association studies (GWASs) have been conducted to identify host genetic variants involved in control of human immunodeficiency virus type 1 (HIV-1) load and in progression to AIDS [1–10]. Overall, these GWASs emphasized the major role of the HLA chromosome 6 region, particularly the HCP5/HLA-B*57 rs2395029 signal [1–3, 5, 8], and the CXCR6 gene region [7]. These GWASs focused on viral load and disease progression, but genetic correlates of the HIV-1 acquisition phenotype have met limited success: 2 recent GWASs conducted in Malawi reported no significant determinants of HIV infection [11, 12]. Looking for new host factors correlated to HIV susceptibility is critical because the only validated association to date is the 32 base-pair deletion in the CCR5 gene: only 1%–2% of Europeans are homozygous for this mutation and exhibit a near-complete protection against infection by HIV-1 R5 strains [13, 14].
To identify additional genetic factors that might contribute to HIV-1 acquisition, we performed a meta-analysis using GWAS genotypic data from 2 European AIDS progression cohorts, comparing each group of HIV-1–infected patients with uninfected controls of the same ancestry [8, 15]. Next, we replicated the association for the single-nucleotide polymorphism (SNP) showing the smallest P value in the European meta-analysis on 2 independent US cohorts of European ancestry.
MATERIALS AND METHODS
First-Stage Study Subjects
The criteria for subject inclusion in the 2 studies have been described previously [6, 8, 15–17]; demographic characteristics of the study groups are presented in Supplementary Table 1. Each patient provided a written consent for study participation.
French Case and Control Groups
The Genomics of Resistance to Immunodeficiency Virus (GRIV) cohort (n = 360) is composed of French HIV-1 seroprevalent long-term nonprogressors (n = 275) and rapid progressors (n = 85) [6, 8]. The normal population control group used for comparison with GRIV subjects comprised 697 individuals from the DESIR (Data from an Epidemiological Study on Insulin Resistance syndrome) program, which was designed to clarify the development of the insulin resistance syndrome [16]. All subjects were nonobese, normoglycemic, French, and HIV-1 seronegative.
Dutch Case and Control Groups
Four hundred seventeen Dutch HIV-1 seroconverter and seroprevalent subjects were enrolled in the Amsterdam Cohort Study (ACS) [15] and compared with 376 HIV-1 seronegative individuals from the normal Dutch population [17]. The ACS is a longitudinal study established to follow the course of HIV-1 infection in homosexual men and injection drug users.
Genotyping Method and Quality Control
All the HIV-1 infected subjects and the uninfected controls were genotyped using the Illumina Infinium II HumanHap300 BeadChip. In each study, quality control filters (eg, missingness, low minor allele frequency, Hardy-Weinberg equilibrium deviation) were applied to ensure reliable genotyping data as previously described [8, 15]. Potential population stratification was also considered using the Eigenstrat method [18] in a 2-step analysis. First, to confirm continental ancestries, the genotypes of each participant group were combined with the genotypes from the 3 HapMap reference populations [19]. From the ACS group, 13 participants were excluded from further analyses to avoid spurious associations resulting from a non-European ancestry. Then, in each study group of European descent, the top 2 most significant principal components were identified and included as covariates in the regression models described below.
Statistical Analysis
Individual GWAS
For each individual GWAS (French and Dutch), a case-control analysis comparing the HIV-1 seropositive group with the HIV-1 seronegative group was performed to identify SNP association with HIV-1 acquisition. Logistic regressions using a dominant genetic model were computed by including as covariates the 2 principal components identified by the Eigenstrat method.
Meta-analysis
The individual P values obtained in each study were combined to provide a single probability value using the Fisher method [20].
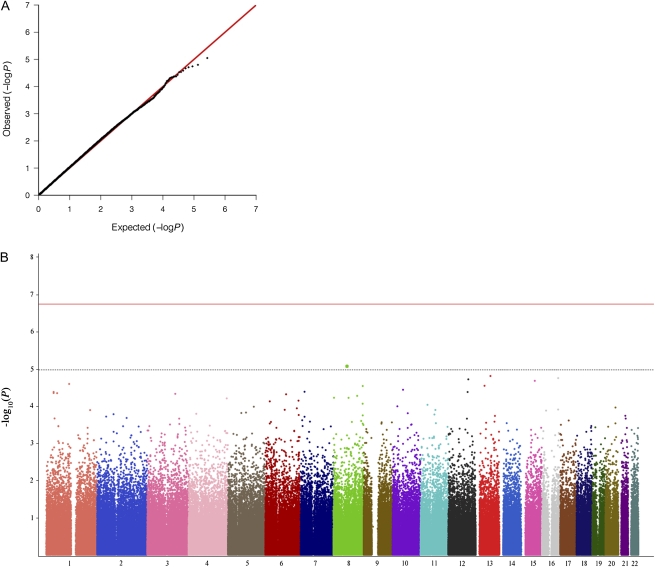
For the meta-analysis results, a quantile-quantile plot and the genomic inflation factor λ [21] were computed in order to test the normality of the P value distribution: neither suggested a significant deviation from the null hypothesis (λ = 1.02), indicating little effect of stratification (Figure 1A).
Figure 1.
A, Quantile-quantile plot for expected (red) vs observed (black) P values from the meta-analysis between the French and Dutch case-control comparisons. X-axis: −log10(expected P values under the null hypothesis); y-axis: −log10(observed P values). The genomic inflation factor was also computed: λ = 1.02. B, Distribution along the human autosomes of −log10(P values) obtained for the meta-analysis between the French and Dutch case-control comparisons. The red line marks the Bonferroni threshold, and the dotted line marks the P < 10−5 cutoff for carrying the single-nucleotide polymorphisms to the second stage.
Multitesting
After quality control steps, a total of 269 962 autosomal SNPs were identified in common between the 2 GWASs. The Bonferroni correction was used to take multiple comparisons into account, and SNPs with P < 1.85 × 10−7 were considered to reach genomewide significance. For all the SNPs meeting the statistical threshold, we checked for potential opposite effects and assigned P = 1 if the odds ratios went in opposite directions.
Second-Stage Analysis
We performed a standard second-stage analysis to explore the polymorphisms exhibiting P < 10−5 in the discovery meta-analysis for replication in 2 European American cohorts by candidate SNP genotyping [22–24]. For each study group, demographic characteristics are presented in Supplementary Table 1.
US HIV-1 Natural History Study Groups
Three hundred thirty HIV-1–infected participants of European descent were enrolled in 2 US multicenter, natural history HIV/AIDS cohorts (San Francisco City Clinic [SFCC] study and Multicenter Hemophilia Cohort Study [MHCS]) [25]. The SFCC group comprised seroconverter homosexual men originally enrolled in a hepatitis B study (n = 78). The MHCS group was composed of seroconverter and seroprevalent persons with hemophilia (n = 252). Each patient provided a written consent for study participation.
Participants From the Longitudinal Studies of the Ocular Complications of AIDS Cohort
Study subjects were 659 European American individuals enrolled in the Longitudinal Studies of the Ocular Complications of AIDS (LSOCA) cohort [26–28]. The LSOCA is a multicenter, prospective, observational study of persons with incident or prevalent cases of AIDS diagnosed according to the 1993 surveillance case definition of the Centers for Disease Control and Prevention. Eligible patients included those with either incident or prevalent cases of cytomegalovirus (CMV) retinitis, as well as those without CMV retinitis. Each patient provided written consent for study participation.
HIV-1 Seronegative Controls From the United States
Because HIV-1–seronegative controls with known exposure to the virus were not available in the original discovery GWAS, similar controls from the normal population were recruited for the second-stage analysis for consistency in the replication arm of the study. The control genotypes for the US study were obtained from the Illumina Genotyping Control Database for 324 unrelated individuals of European ancestry who were genotyped with HumanHap300 BeadChips (www.illumina.com; downloaded in January 2008).
Candidate SNP Genotyping
For the US study groups, candidate SNP genotyping data was obtained using commercial TaqMan genotype assays (with assay ID C_1929536_1; Applied Biosystems). Conformity to the genotype frequencies expected under Hardy-Weinberg equilibrium was checked.
Control for Population Stratification
Genomewide data were available for the US combined MHCS-SFCC group [10] and for the Illumina HIV seronegative group. Potential population stratification was considered using the Eigenstrat method in a 2-step analysis as described above. Fourteen HIV-seropositive participants were thus excluded, and the top 2 most significant principal components were identified and included as covariates in the statistical analysis described below.
Genomewide data were available only for 379 of 659 LSOCA participants (57%) (data from E. Sezgin, M. L. van Natta, and D. A. Jabs; manuscript in preparation). With the available data, we checked the continental ancestry and confirmed that the LSOCA individuals matched the HapMap group of European descent (Supplementary Figure 1).
Statistical Analysis
Logistic regressions using a dominant genetic model were computed to compare genotypic distribution between each case group and the Illumina control subjects.
RESULTS
A total of 764 HIV-1–seropositive and 1073 HIV-1–seronegative individuals of European descent were genotyped on whole-genome arrays. HIV-1–infected participants were recruited from 2 independent cohorts: the French GRIV cohort (n = 360 HIV positive vs 697 HIV negative) and the Dutch ACS (n = 404 HIV positive vs 376 HIV negative). After quality-control steps, 269 962 autosomal SNPs with reliable genotypes were found in common between both studies. These SNPs were tested for association with HIV-1 acquisition in a meta-analysis combining the individual P values obtained in each study. The global distribution of resulting P values was close to the null hypothesis (λ = 1.02) (Figure 1A), indicating appropriate control of stratification.
This meta-analysis did not reveal any signal respecting the Bonferroni genomewide significance threshold (Pthreshold = 1.85 × 10−7) (Figure 1B). We next did a second-stage analysis in 2 independent cohorts for SNPs exhibiting a P value <10−5 in the discovery analysis: only the chromosome 8 rs6996198 fell below this threshold with P = 8.91 × 10−6 (Figure 1B). This association was independent of age, sex, and CCR5-Δ32 variant (which is not targeted by Illumina arrays but was previously genotyped in both cohorts) when added as covariates in the regression models (data not shown). Similarly, mode of HIV-1 transmission (mucosal or parenteral) does not seem to impact the signal because rs6996198-T frequency was similar in every HIV-infected subgroup (heterosexual contact, homosexual contact, or injecting drug use). Importantly, rs6996198 was not associated with AIDS progression in the ACS seroconverter cohort, and the T allele was not underrepresented in the large rapid progressor subgroup from the GRIV cohort. This observation suggests no survival bias and no influence on rapid AIDS progression.
By comparing the HIV-1 infected European Americans from the US HIV-1 combined study (n = 316) and the LSOCA (n = 659) cohort with the Illumina European American control group (n = 324), the signal was replicated in both cohorts: P = 1.77 × 10−2 and P = 2.80 × 10−3, respectively (Figure 2). Overall, the combined P value computed by the Fisher method between the 4 cohorts (GRIV, ACS, US HIV-1, and LSOCA) reached the Bonferroni genomewide significance threshold: Pcombined = 7.76 × 10−8 (Table 1 and Figure 2).
Figure 2.
Overall study design and results from each stage. In the first stage, a meta-analysis was performed between the French Genomics of Resistance to Immunodeficiency Virus (white) and the Dutch Amsterdam Cohort Study (light gray) case-control studies; this analysis identified a single single-nucleotide polymorphism (SNP), rs6996198, below the P < 10−5 cutoff for carrying SNPs to the next stage. In the second stage, the rs6996198 association was replicated in 2 independent cohorts of European Americans (dark gray): the US HIV-1 combined study (*) and the Longitudinal Studies of the Ocular Complications of AIDS (#) cohort. Both case groups were compared with a control group from the Illumina Genotyping Control Database.
Table 1.
Associationa of rs6996198T Genotypes (TT and TC) With HIV Acquisition
| Study | HIV-Positive Subjects | HIV-Negative Controls | ||||
| No. (%) | No. (%) | P Value | OR | 95% CI | ||
| Stage 1 | ||||||
| France | GRIV | 360 (25.99) | 697 (33.29) | 7.59 × 10−3 | 0.70 | .52–.94 |
| Netherlands | ACS | 404 (24.26) | 376 (36.97) | 7.65 × 10−5 | 0.55 | .40–.75 |
| 1st meta-analysis | 764 | 1073 | 8.91 × 10−6 | |||
| Stage 2 | ||||||
| United States | Combinedb | 316 (24.68) | 324 (31.79) | 1.77 × 10−2 | 0.68 | .47–.98 |
| LSOCA | 659 (22.91) | 324 (31.79) | 2.80 × 10−3 | 0.64 | .47–.87 | |
| 2nd meta-analysis | 1739 | 1397 | 7.76 × 10−8 |
Abbreviations: ACS, Amsterdam Cohort Study; CI, confidence interval; GRIV, Genomics of Resistance to Immunodeficiency Virus; HIV, human immunodeficiency virus; LSOCA, Longitudinal Studies of the Ocular Complications of AIDS; OR, odds ratio.
Frequency (%), P value, and OR for the dominant genetic model; meta-analysis P value using Fisher’s method.
Participants enrolled in the Multicenter Hemophilia Cohort Study and the San Francisco City Clinic study.
The rs6996198 TT+TC (T dominant) frequency is consistently lower in the HIV-1 positive groups compared with the normal control groups for each study: French GRIV HIV positive (25.99%) vs French HIV-1 seronegative controls (33.29%); Dutch ACS HIV positive (24.26%) vs Dutch HIV-1 seronegative controls (36.97%); US combined cohorts HIV positive (24.68%) and LSOCA HIV positive (22.91%) vs Illumina controls (31.79%), respectively (Figure 2). By examination of the genotype distribution, most of the rs6996198 protective effect appears to be driven by a depletion of heterozygotes in the HIV-1–infected case groups, indicating that the carriers of the T allele are significantly less likely to acquire HIV infection (Supplementary Table 2); dominant is the best genetic model to fit the data. As a confirmation of the frequencies observed in our control groups, we checked the frequency for European descent populations in the following public databases: HapMap (27.70%, n = 65 and 30.69%, n = 101 for the European American CEU and the Italian TSI, respectively), the 1000 Genomes Project (31.80%, n = 283), and the Human Genome Diversity Project (31.71%, n = 246).
The rs6996198 T>C polymorphism is located 29.4 kb upstream from the BHLHE22 gene and 45.2 kb downstream from the CYP7B1 gene. We could not find any other SNP in strong linkage disequilibrium (r2 ≥0.8) in HapMap for the European population. Interestingly, according to the Genevar messenger RNA expression database [29], the rs6996198-T allele is associated with higher CYP7B1 gene expression (P = 5 × 10−3) in the HapMap European adult child population.
As was done by Petrovski et al [12], we performed an additional subset analysis on the genomewide meta-analysis data by focusing on 22 candidate genes previously reported as associated with HIV-1 susceptibility. For each gene, we reported the candidate SNP, when possible, or otherwise the best available proxy in the European population (based on the CEU HapMap population with r2 > 0.8). We also provided the lowest P value observed within each candidate gene or the 2 kb flanking regions. Finally, we corrected the P value for the number of SNPs tested in the corresponding gene and for all SNPs tested in this subset analysis (Supplementary Table 3). We could not examine any single variant within 4 genes (APOBEC3G, CCR5, CD209, and PPIA). For the 18 remaining genes, we were able to directly test 6 previously reported SNPs and found a good proxy for 7 additional SNPs. Of these 13 SNPs, only ABCB1-rs1045642 and IL10-rs1800896 exhibited a P value <.05 in our discovery meta-analysis (P = 1.44 × 10−2 and P = 3.40 × 10−2, respectively), but both signals were tracked only by the GRIV cohort; the P value remained nonsignificant in ACS. In addition to these 2 genes, CCL7 also displayed an SNP with P < .05 (P = 2.53 × 10−3), but the signal was only allocated to ACS. After correcting for the number of SNPs per gene, only the CCL7 variant remains significant (corrected for only 1 SNP), but this signal disappeared (P = .18) when correcting for all 73 SNPs tested across the 22 candidate genes. For the genes in which the originally reported variant was not covered by our genotyping arrays, failure to find a significant signal does not mean there is no association. For the others, failure to replicate the initial signal could stem from a power issue (either in the original study or in ours), from differences in population ancestry, or from a false-positive or false-negative signal.
DISCUSSION
In this study, we focused on the HIV-1 acquisition phenotype, a phenotype not yet investigated for Europeans by GWAS. The comparison of HIV-1 seropositive cases with HIV-1 seronegative controls in a 2-stage meta-analysis between 4 independent cohorts of European ancestry revealed a genomewide significant signal for the chromosome 8 rs6996198 (Pcombined = 7.76 × 10−8). Each independent study group steadily showed a nominally statistical significance and a similar pattern in direction of effect: The rs6996198-T allele was associated with reduced risk of HIV-1 infection.
A major limitation of GWAS is that in an effort to avoid false-positive associations, false-negative associations may be inflated; the extremely low P values needed to achieve genomewide significance make it unlikely that small or modest effect size associations will be detected without very large sample numbers. In this study, we applied a meta-analysis approach to genotypic data from 2 independent European AIDS GWASs to identify a single noteworthy SNP that approached but did not reach genomewide significance; this SNP was moved forward for replication and found to be statistically significant in both US studies. A second meta-analysis on the 4 groups reached genomewide statistical significance. Importantly, this study is the first to use a 2-step meta-analysis for gene discovery study and to identify a common variant associated with HIV-1 infection that is not in a gene encoding a chemokine receptor or ligand. The consistency in direction and effect sizes (odds ratio between 0.55 and 0.70) among the 4 independent case-control groups limits the risk of propagating a false-positive association and strongly implicates the region on chromosome 8 as having a role in HIV-1 acquisition.
A limitation of our study is the absence of data on HIV-1 exposure risk in our control groups. By including individuals who have probably never been exposed to the virus in our control groups, our design reduces power for discovering genetic factors associated with HIV-1 acquisition and possibly inflates false-negative signals. The constancy in replication and the observation of similar allele frequencies among subgroups infected by a different transmission mode (heterosexual contact, homosexual contact, injection drug use, blood transfusion) reduce the risk of propagating a false positive signal. Nevertheless, additional genetic studies will be necessary to securely confirm the association with rs6996198—especially by using control groups with known data on exposure risk—and to identify functional, causal sequence variation.
Although the rs6996198 SNP was targeted by genotyping arrays used in the 2 recent GWASs focusing on HIV-1 acquisition [11, 12], none of them identified a strong signal for this variant. Several differences between these studies and ours could explain why the rs6996198 association was not previously emphasized: (1) study design (Joubert et al and Petrovski et al focused on mother-to-child transmission and sexual transmission, respectively [11, 12]); (2) sample size and power—both studies were smaller than our discovery meta-analysis and may not have been powered to detect a modest association with odds ratios between 0.55 and 0.70 (100 HIV positive vs 126 HIV negative, and 531 HIV positive vs 848 HIV negative); (3) continental ancestry (both previous studies were conducted on Africans from Malawi whereas ours comprised Europeans). The differences in SNP frequency across populations from different ancestries exemplify this last point: approximately 47% for African populations vs approximately 31% for European populations according to the public databases (HapMap, 1000 Genomes Project, and Human Genome Diversity Project). The difficulty in identifying associations with HIV-1 susceptibility could stem from the involvement of complex interactions between genetic and/or environmental factors, weak signals undetectable without a highly powered study, or rare or common polymorphisms not targeted by the arrays.
Neither of the adjacent genes to rs6996198 was identified by the previous whole-genome RNA interference screens designed to identify intracellular HIV-dependency factors essential for viral replication [30–33]. Although all nearby genes are viable candidates for harboring a causal allele, rs6996198 is in strong linkage disequilibrium with no known SNP. The rs6996198-T allele was previously associated with higher CYP7B1 transcription levels. CYP7B1 encodes a cytochrome P450 enzyme catalyzing hydroxylation of several steroids and oxysterols, especially in the liver, brain, and reproductive tract. CYP7B1 variants have been associated with liver failure in infants, with prostate and ovarian abnormalities, and with spastic paraplegia type 5, a neurodegenerative disease (for a review, see [34]). Interestingly, CYP7B1 is also involved in immune control and regulation. Stimulation of macrophage Toll-like receptors induces secretion of 25-hydroxycholesterol, which in turn suppresses immunoglobulin A (IgA) production [35, 36]. Mice knocked-out for Cyp7b1, which metabolizes 25-hydroxycholesterol in mice and humans, display increased levels of 25-hydroxycholesterol and low IgA levels in serum, lung, and mucosa [35]. A role in the induction of programmed cell death has also been attributed to 25-hydroxycholesterol [37–39]. Proinflammatory cytokines (eg, tumor necrosis factor α, interleukin 1β) released by activated immune cells induce CYP7B1 expression and activity in mice lungs [40] and human joints [41], which could contribute to maintaining inflammation. The rs6996198-T allele, which is correlated to upregulation of CYP7B1 expression, is more frequent in controls than in seropositive subjects and may potentially lead to higher levels of IgA, to a bypass of programmed cell death, and to support of the inflammatory state. How decreased levels of CYP7B1 may prevent HIV infection is unknown. Before any definitive conclusions, this newly identified SNP and surrounding region on chromosome 8 will require further analyses in other HIV cohort studies to securely confirm the association and to investigate the potential functional role for CYP7B1 or a neighboring gene in HIV acquisition.
In summary, this work confirms the power of meta-analysis to detect new associations of small to medium effect size with complex diseases and underlines the utility of international collaborative efforts to exploit existing GWAS and phenotypic data for genes associated with HIV susceptibility beyond the CCR5 nexus. These efforts are critical to improve the understanding of AIDS pathogenesis and identify genes and pathways for therapeutic or preventive interventions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
The authors thank all the patients and the medical staff who kindly collaborated with the genetic association studies. We also thank Bailey Kessing, Elizabeth Binns-Roemer, and Yu-Chen Zhou for excellent technical assistance and Dr James J. Goedert for providing samples from the MHCS.
Financial support.
This work was supported by Agence Nationale de Recherche sur le SIDA: Sidaction: the Conservatoire National des Arts et Metiers: Neovacs SA: and Vaxconsulting.
The authors acknowledge funding from the Netherlands Organization for Scientific Research (TOP; registration number 9120.6046). The Amsterdam Cohort Studies on HIVI infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Research, and the University Medical Center Utrecht, are part of the Netherlands HIV Monitoring Foundation and are financially supported by the Netherlands National Institute for Public Health and the Environment.
This project has been funded in part by federal funds from the National Cancer Institute (NCI), National Institutes of Health (NIH) (contract HHSN26120080001E) and by the Intramural Research Program of the NIH, NCI, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
The LSOCA is supported by cooperative agreements from the National Eye Institute, NIH, to The Mount Sinai School of Medicine (U10 EY 08052), The Johns Hopkins University Bloomberg School of Public Health (U10 EY 08057), and the University of Wisconsin, Madison School of Medicine (U10 EY 08067).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Dalmasso C, Carpentier W, Meyer L, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PloS One. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–7. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbeck JT, Gottlieb GS, Winkler CA, et al. Multistage genomewide association study identifies a locus at 1q41 associated with rate of HIV-1 disease progression to clinical AIDS. J Infect Dis. 2010;201:618–26. doi: 10.1086/649842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereyra F, Jia X, et al. International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–7. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Clerc S, Limou S, Coulonges C, et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03) J Infect Dis. 2009;200:1194–201. doi: 10.1086/605892. [DOI] [PubMed] [Google Scholar]
- 7.Limou S, Coulonges C, Herbeck JT, et al. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. J Infect Dis. 2010;202:908–15. doi: 10.1086/655782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limou S, Le Clerc S, Coulonges C, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 9.Pelak K, Goldstein DB, Walley NM, et al. Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010;201:1141–9. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troyer JL, Nelson GW, Lautenberger JA, et al. Genome-wide association study implicates PARD3B-based AIDS restriction. J Infect Dis. 2011;203:1491–502. doi: 10.1093/infdis/jir046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert BR, Lange EM, Franceschini N, Mwapasa V, North KE, Meshnick SR. A whole genome association study of mother-to-child transmission of HIV in Malawi. Genome Med. 2010;2:17. doi: 10.1186/gm138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrovski S, Fellay J, Shianna KV, et al. Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS. 2011;25:513–8. doi: 10.1097/QAD.0b013e328343817b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 14.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 15.van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MA, van't Wout AB, Schuitemaker H. Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS. 2009;23:19–28. doi: 10.1097/QAD.0b013e32831db247. [DOI] [PubMed] [Google Scholar]
- 16.Balkau B. An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome. Rev Epidemiol Sante Publique. 1996;44:373–5. [PubMed] [Google Scholar]
- 17.van Es MA, Veldink JH, Saris CG, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–7. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 20.Fisher R. Statistical methods for research workers. Edinburgh, Scotland Oliver and Boyd; 1932. [Google Scholar]
- 21.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 22.Lessard CJ, Adrianto I, Kelly JA, et al. Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am J Hum Genet. 2011;88:83–91. doi: 10.1016/j.ajhg.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YJ, Go MJ, Hu C, et al. Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet. 2011;43:990–5. doi: 10.1038/ng.939. [DOI] [PubMed] [Google Scholar]
- 24.van Heel DA, Franke L, Hunt KA, et al. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–9. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An P, Duggal P, Wang LH, et al. Polymorphisms of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jabs DA, Ahuja A, Van Natta M, Lyon A, Srivastava S, Gangaputra S. Course of cytomegalovirus retinitis in the era of highly active antiretroviral therapy: five-year outcomes. Ophthalmology. 2010;117:2152–61. doi: 10.1016/j.ophtha.2010.03.031. e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jabs DA, Van Natta ML, Holbrook JT, Kempen JH, Meinert CL, Davis MD. Longitudinal study of the ocular complications of AIDS: 1. Ocular diagnoses at enrollment. Ophthalmology. 2007;114:780–6. doi: 10.1016/j.ophtha.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Jabs DA, Van Natta ML, Holbrook JT, Kempen JH, Meinert CL, Davis MD. Longitudinal study of the ocular complications of AIDS: 2. Ocular examination results at enrollment. Ophthalmology. 2007;114:787–93. doi: 10.1016/j.ophtha.2006.07.065. [DOI] [PubMed] [Google Scholar]
- 29.Stranger BE, Forrest MS, Clark AG, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brass AL, Dykxhoorn DM, Benita Y, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 31.Konig R, Zhou Y, Elleder D, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–73. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou H, Xu M, Huang Q, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 34.Stiles AR, McDonald JG, Bauman DR, Russell DW. CYP7B1: one cytochrome P450, two human genetic diseases, and multiple physiological functions. J Biol Chem. 2009;284:28485–9. doi: 10.1074/jbc.R109.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauman DR, Bitmansour AD, McDonald JG, Thompson BM, Liang G, Russell DW. 25-Hydroxycholesterol secreted by macrophages in response to Toll-like receptor activation suppresses immunoglobulin A production. Proc Natl Acad Sci U S A. 2009;106:16764–9. doi: 10.1073/pnas.0909142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diczfalusy U, Olofsson KE, Carlsson AM, et al. Marked upregulation of cholesterol 25-hydroxylase expression by lipopolysaccharide. J Lipid Res. 2009;50:2258–64. doi: 10.1194/jlr.M900107-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ayala-Torres S, Moller PC, Johnson BH, Thompson EB. Characteristics of 25-hydroxycholesterol-induced apoptosis in the human leukemic cell line CEM. Exp Cell Res. 1997;235:35–47. doi: 10.1006/excr.1997.3630. [DOI] [PubMed] [Google Scholar]
- 38.Christ M, Luu B, Mejia JE, Moosbrugger I, Bischoff P. Apoptosis induced by oxysterols in murine lymphoma cells and in normal thymocytes. Immunology. 1993;78:455–60. [PMC free article] [PubMed] [Google Scholar]
- 39.Rusinol AE, Thewke D, Liu J, Freeman N, Panini SR, Sinensky MS. AKT/protein kinase B regulation of BCL family members during oxysterol-induced apoptosis. J Biol Chem. 2004;279:1392–9. doi: 10.1074/jbc.M308619200. [DOI] [PubMed] [Google Scholar]
- 40.Stoilov I, Krueger W, Mankowski D, et al. The cytochromes P450 (CYP) response to allergic inflammation of the lung. Arch Biochem Biophys. 2006;456:30–8. doi: 10.1016/j.abb.2006.09.029. [DOI] [PubMed] [Google Scholar]
- 41.Dulos J, van der Vleuten MA, Kavelaars A, Heijnen CJ, Boots AM. CYP7B expression and activity in fibroblast-like synoviocytes from patients with rheumatoid arthritis: regulation by proinflammatory cytokines. Arthritis Rheum. 2005;52:770–8. doi: 10.1002/art.20950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.