Abstract
(See the editorial commentary by Stebbing and Bower, on pages 1032–4.)
We studied the presence of Kaposi sarcoma herpesvirus sequences in cell-free DNA (cfDNA) isolated from the blood of patients with AIDS-related Kaposi sarcoma (KS) and primary effusion lymphoma (PEL). The use of paramagnetic beads linked to methyl-CpG binding domain protein allowed separation of virion and cell-derived DNA. Only virion DNA was detected in the blood of KS patients, whereas cell-derived DNA was detected in a patient with AIDS-related PEL. The difference in the origins of cfDNA in these settings may in part reflect very different proliferative indices in KS and PEL tumor tissue.
Kaposi sarcoma (KS) is a tumor characterized by neovascular proliferation [1, 2]. It commonly presents as cutaneous lesions, but lymphadenopathy and gut and lung involvement are not unusual. Physical examination and radiography have been the major tools for assessing tumor. However, hyperpigmentation associated with cutaneous lesions persists for months or years after tumor response so that visual assessment is sometimes misleading. Edema, particularly in the legs, may result from tumor infiltration of the skin, obstruction of lymphatics associated with nodal involvement, or lymphatic scarring resulting from tumor. After chemotherapy, severe and sometimes disabling edema may persist. In some instances, evidence of tumor persistence would lead to further chemotherapy, but distinguishing residual lymphatic scarring from lymphatic obstruction associated with a tumor in edematous legs is not easy. Better tools for assessing tumor persistence or progression might be useful in guiding decision making in this and many other settings.
Tumors are recognized as a source of cell-free DNA (cfDNA) in blood, and it has been suggested that cfDNA may be regarded as a liquid tumor biopsy [3]. Viral DNA may be released into blood from tumor and other cells as other cellular DNA or may be released packaged in virions. Kaposi sarcoma herpesvirus (KSHV; also known as human herpesvirus 8) is associated with tumor cells in all forms of KS [1]. The presence of KSHV virion DNA in blood has been reported by several groups including our own [4, 5]. Evaluation of the relative contribution of virion vs cell-derived viral DNA using deoxyribonuclease (DNase) protection assays in clinical specimens has proven challenging. The degree of protection from DNase afforded by the virion varies as a function of specimen handling. In the investigations reported here, we use the presence of CpG methylation as a marker for cell-derived DNA vs virion DNA.
METHODS
Cell Culture, Control DNA Samples, and DNA Isolation
BC-3 is a primary effusion lymphoma cell line that harbors KSHV episomes [6]. Purified virions were prepared from the supernatant of BC-3 cultures induced with sodium butyrate 0.3 ng/mL (for the initial 24 hours) and 12-O-tetradecanoylphorbol-13-acetate 20 ng/mL (for 5 days). After 5 days the cell suspension was transferred into 50-mL conical tubes and centrifuged at 3500 rpm for 20 minutes at 4°C. Clarified media were centrifuged at 15 000 rpm for 35 minutes at 4°C. DNA was extracted from virus pellets according to the manufacturer’s protocol (QIAamp DNA Blood Mini Kit, Qiagen).
Specimens
Pretreatment plasma specimens from patients with AIDS-related KS enrolled in the AIDS Malignancy Consortium trial 036 [7] were studied, as well as plasma and ascites specimens from patients with AIDS-related primary effusion lymphoma (PEL). Specimens were obtained with written informed consent with approval from the relevant institutional review boards. Patients on the clinical trial underwent physical examination and chest imaging to exclude visceral disease or other malignancy. None of the patients developed lymphoma during the 12 weeks of therapy, again providing reassurance that PELs were not missed at the time of study entry.
Methylated DNA Enrichment
Extracted DNA was added to 10 μL of methyl-CpG binding domain (MBD) bead slurry (MethylMiner DNA Enrichment Kit, Invitrogen) and incubated on a rotating mixer for 1 hour. The DNA in the noncaptured fraction, washes (300 mM and 450 mM sodium chloride), and elution (2000 mM sodium chloride) was ethanol precipitated, resuspended in water, and subjected to real-time polymerase chain reaction (PCR) with Power SYBR Green PCR Master Mix (Applied Biosystems) and the following primers: KSHV ORF 64 (sense: ATGTGGCCATCTTGGATCTC; antisense: CACAGCCTTGAGCATTGTTG), ORF23 (sense: ACACGACACGATGTTTTCCA; antisense: TCATGGAGCGTGCTAACAAC), and K8 (sense: TCCAACTCGCAGATCCAAGAG; antisense: CGACCTGCGCCCTGTTT). KSHV copy numbers were measured by using real-time PCR with primers and a probe that targeted the K8 region, as described previously [5].
RESULTS
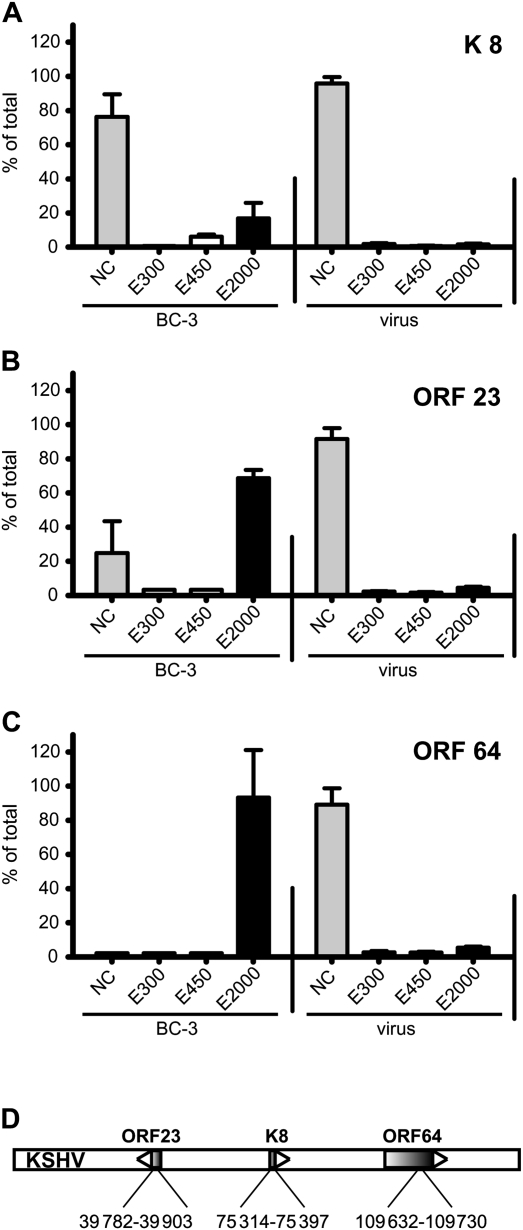
CpG methylation in KSHV DNA from tumor cell lines or from infected endothelial cells has been consistently reported, whereas DNA extracted from virions does not show CpG methylation [8]. Thus it seems likely that the only condition under which KSHV DNA can be CpG methylated is during latent persistence. We applied paramagnetic beads coupled to the MBD of MBD2 to KSHV virion and KSHV cell-derived DNA. Noncapture, wash (E300, E450), and high salt eluate (E2000) fractions were evaluated by real-time PCR with 3 sets of KSHV primers (Figure 1). Virion DNA was never captured by the resin regardless of the region of the viral genome targeted by primers for amplification, consistent with the expectation that virion DNA was not methylated. Cellular viral DNA showed major differences as a function of the region of the viral genome analyzed. DNA from the K8 region was never captured by the resin, but DNA from the ORF23 and ORF64 regions was captured, consistent with a previous analysis using immunoprecipitation with antibodies against methyl-CpG coupled with high-resolution tiling microarray [8]. Whereas only some of the DNA from the ORF23 region was captured, all of the DNA from the ORF64 region was captured. Thus MBD2 paramagnetic beads in combination with appropriate PCR primers can be used to distinguish viral sequences derived from virion vs viral sequences from cellular DNA. The sensitivity of the method was assessed in reconstruction experiments by mixing virion and BC-3 DNA. BC-3 DNA could consistently be distinguished from virion DNA when it constituted >5% of the total viral DNA in the sample (data not shown).
Figure 1.
The methyl-CpG binding domain 2 (MBD2) beads distinguish between unmethylated virion DNA and methylated Kaposi sarcoma herpesvirus (KSHV) episomal DNA. DNA isolated from purified KSHV virions or from latently infected BC-3 cells were subjected to binding on the MBD2 beads. DNA isolated from the noncaptured fraction (NC), washes (300 mM and 450 mM), and elution (2000 mM) was subjected to real-time polymerase chain reaction (PCR) with primers that amplify a region in the K8 ORF (A), ORF23 (B), and ORF64 (C). Each column represents the amount of DNA in the indicated fraction relative to the total DNA detected (100%). The standard deviation of 3 independent real-time PCR reactions is indicated. D, Schematic representation of the KSHV genome. The nucleotide positions within the KSHV genome (human herpesvirus 8 strain GK18, AF148805) of the amplified regions are indicated.
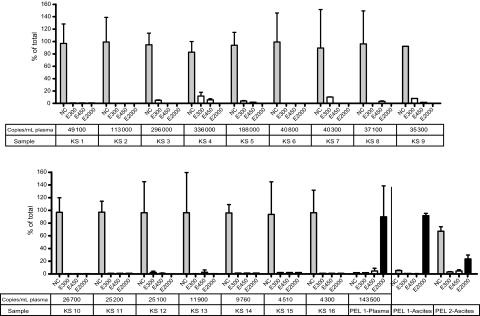
We then applied these methods to DNA isolated from plasma from 16 patients with AIDS-related KS and from plasma from a patient with AIDS-related PEL and DNA extracted from malignant ascites from 2 patients with PEL. As seen in Figure 2, using the MBD2 beads, no CpG methylation was detected in plasma of any KS patient, but CpG methylation was detected in DNA from the plasma of a patient with PEL (Figure 2). CpG methylated DNA was also detected in the ascites from both patients with PEL.
Figure 2.
Analysis of the methylation status of Kaposi sarcoma herpesvirus (KSHV) DNA in patients with Kaposi sarcoma (KS) and primary effusion lymphoma (PEL). DNA isolated from the blood from KS patients and from the blood or ascites fluid from PEL patients was subjected to binding on the methyl-CpG binding domain 2 (MBD2) beads. DNA isolated from the noncaptured fraction (NC), washes (300 mM and 450 mM), and elution (2000 mM) was subjected to real-time polymerase chain reaction with primers that amplify a region in ORF64.
DISCUSSION
In cfDNA from blood of patients with AIDS-related KS, the only KSHV DNA detected failed to bind to the MBD2 beads, consistent with absence of CpG methylation. These results suggest that the viral DNA sequences detected in plasma are virion DNA. In contrast, in blood from a patient with PEL and in ascites from 2 patients with PEL, we detected CpG methylation of viral sequences. Thus it appears that KSHV derived from tumor cells can be detected in cfDNA in the blood of a patient with PEL. However, tumor cell DNA is not readily detected in the cfDNA of patients with AIDS-related KS in the absence of lymphoma. The determinants of the release of nonvirion DNA from cells into blood remains poorly understood. Various investigators have invoked cell death associated with apoptosis or necrosis or a process of DNA secretion from cells as the source of DNA. The amount of tumor DNA detected in cfDNA in blood is believed to reflect tumor bulk, proliferative rate, ongoing rates of apoptosis, necrosis DNA secretion, and nuclease activity [9]. We and others have previously reported that in lymphoma patients, tumor DNA is readily detected in cfDNA in blood [10], so it is not surprising that in a patient with PEL, a rapidly growing aggressive tumor, viral cfDNA with CpG methylation characteristic of cell-derived DNA was detected. We anticipated that viral cfDNA might be a mix of virion and tumor-derived DNA as in the DNA extract from the ascites of a patient with PEL (Figure 2). However, the sensitivity of the method described here only allowed the detection of virion DNA. It should be noted that the patients whose plasma specimens were evaluated were enrolled in a trial for patients with KS but no visceral disease. It might be that in specimens from patients with more extensive or rapidly progressive KS, cfDNA in blood might be more readily detected. It seems likely that methyl-CpG KSHV DNA is not detected in KS patients because few KS tumor cells release cell DNA (through spontaneous apoptosis or other processes). In patients with KS, virions may derive from infected B lymphocytes and other cells that are not tumor cells. This would be consistent with the observation that KSHV-infected B lymphocytes are detected in peripheral blood and in lymph nodes in subjects without KS [5, 11]. It is even possible that KS tumor cells release virions without releasing cell DNA.
It is perhaps worth contrasting these findings with the more extensively studied viral cfDNA detected in the blood of patients with nasopharyngeal carcinoma (NPC) [12]. This is a tumor type that is consistently associated with Epstein–Barr virus (EBV). In contrast to some other EBV-associated malignancies such as posttransplant lymphoproliferative disease, there is little evidence to suggest that NPC is associated with global immunocompromise, and NPC patients do not typically have histories of thrush, Pneumocystis jirovecii pneumonia, or other hallmarks of immune deficiency. Studies of the EBV cfDNA detected in blood from patients with NPC indicate that tumor DNA, rather than virion DNA, predominates. Monitoring EBV cfDNA has become standard for the assessment of NPC progression in high-incidence regions and is arguably the best-established cfDNA tumor marker in blood.
The predominance of virion DNA in cfDNA in patients with AIDS-related KS may reflect 3 factors: (1) These patients are immunocompromised and perhaps less able to limit KSHV replication; (2) virion DNA in blood has a longer half-life than other cfDNA, reflecting the protection provided by the viral capsid; and (3) the proliferative index as measured by Ki67 immunohistochemistry is consistently ≤20% whether in macules-plaques, tumors, or visceral disease [13]. This low Ki-67 percentage contrasts with PEL, in which most cases show a proliferative index ≥90% [14].
It is worth noting that methyl-CpG–free KSHV DNA identifies the same molecules that we previously identified by DNase protection assays (ie, those DNA molecules protected by virions). However, we have found that DNase protection assays are difficult to standardize and replicate. The DNase protection approach must be optimized for each DNase preparation and is highly sensitive to specimen handling. It was in part our frustration with implementing the quantitation of DNase sensitivity/resistance for large numbers of clinical specimens that led us to the present approach. The methyl-DNA–binding domain matrix was first developed for enrichment of methyl-CpG DNA from large numbers of prostate cancer biopsy specimens [15].
Monitoring KSHV copy number in whole blood, blood mononuclear cells or blood cfDNA may ultimately play a role in the diagnosis and management of patients with KSHV-associated disorders. However, this study shows that the cfDNA that predominates in patients with AIDS-related KS in the United States without visceral KS lesions is almost exclusively virion rather than tumor DNA derived. Extrapolation with regards to the utility of measuring viral DNA based on the experiences in other settings must be undertaken cautiously because the biologic determinants of virion DNA and tumor-derived viral DNA in blood are different. That said, application of the approach described here to the monitoring of tumor DNA in patients with PEL or rapidly progressive KS should be straightforward and, we believe, warrants further assessment.
Notes
Financial support.
This work was supported by grants from the National Institutes of Health (P50CA96888, U01CA121947, P30CA006973, P01CA113239).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Moore PS, Chang Y. KSHV: forgotten but not gone. Blood. 2011;117:6973–4. doi: 10.1182/blood-2011-05-350306. [DOI] [PubMed] [Google Scholar]
- 2.Uldrick TS, Whitby D. Update on KSHV epidemiology, Kaposi sarcoma pathogenesis, and treatment of Kaposi sarcoma. Cancer Lett. 2011;305:150–62. doi: 10.1016/j.canlet.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 4.Campbell TB, Borok M, White IE, et al. Relationship of Kaposi sarcoma (KS)–associated herpesvirus viremia and KS disease in Zimbabwe. Clin Infect Dis. 2003;36:1144–51. doi: 10.1086/374599. [DOI] [PubMed] [Google Scholar]
- 5.Lin L, Lee JY, Kaplan LD, et al. Effects of chemotherapy in AIDS-associated non-Hodgkin’s lymphoma on Kaposi’s sarcoma herpesvirus DNA in blood. J Clin Oncol. 2009;27:2496–502. doi: 10.1200/JCO.2008.20.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvanitakis L, Mesri EA, Nador RG, et al. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–54. [PubMed] [Google Scholar]
- 7.Koon HB, Fingleton B, Lee JY, et al. Phase II AIDS Malignancy Consortium trial of topical halofuginone in AIDS-related Kaposi sarcoma. J Acquir Immune Defic Syndr. 2011;56:64–8. doi: 10.1097/QAI.0b013e3181fc0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Günther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma–associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva JM, Dominguez G, Garcia JM, et al. Presence of tumor DNA in plasma of breast cancer patients: clinicopathological correlations. Cancer Res. 1999;59:3251–6. [PubMed] [Google Scholar]
- 10.Wagner-Johnston ND, Gellert L, Gocke CD, et al. Clonal immunoglobulin DNA in the plasma of patients with AIDS lymphoma. Blood. 2011;117:4860–2. doi: 10.1182/blood-2010-12-324657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell TB, Staskus KA, Folkvord J, et al. Persistence of Kaposi sarcoma–associated herpesvirus (KSHV)–infected cells in KSHV/HIV-1–coinfected subjects without KSHV-associated diseases. J Infect Dis. 2005;191:367–71. doi: 10.1086/427194. [DOI] [PubMed] [Google Scholar]
- 12.Leung SF, Zee B, Ma BB, et al. Plasma Epstein–Barr viral deoxyribonucleic acid quantitation complements tumor-node-metastasis staging prognostication in nasopharyngeal carcinoma. J Clin Oncol. 2006;24:5414–8. doi: 10.1200/JCO.2006.07.7982. [DOI] [PubMed] [Google Scholar]
- 13.Penin RM, Fernandez-Figueras MT, Puig L, Rex J, Ferrandiz C, Ariza A. Over-expression of p45(SKP2) in Kaposi’s sarcoma correlates with higher tumor stage and extracutaneous involvement but is not directly related to p27(KIP1) down-regulation. Mod Pathol. 2002;15:1227–35. doi: 10.1097/01.MP.0000036589.99516.D6. [DOI] [PubMed] [Google Scholar]
- 14.Carbone A, Gloghini A, Bontempo D, et al. Proliferation in HHV-8–positive primary effusion lymphomas is associated with expression of HHV-8 cyclin but independent of p27(kip1) Am J Pathol. 2000;156:1209–15. doi: 10.1016/S0002-9440(10)64991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res. 2006;34:e19. doi: 10.1093/nar/gnj022. [DOI] [PMC free article] [PubMed] [Google Scholar]