Abstract
Background
Spot 42 was discovered in Escherichia coli nearly 40 years ago as an abundant, small and unstable RNA. Its biological role has remained obscure until recently, and is today implicated in having broader roles in the central and secondary metabolism. Spot 42 is encoded by the spf gene. The gene is ubiquitous in the Vibrionaceae family of gamma-proteobacteria. One member of this family, Aliivibrio salmonicida, causes cold-water vibriosis in farmed Atlantic salmon. Its genome encodes Spot 42 with 84% identity to E. coli Spot 42.
Results
We generated a A. salmonicida spf deletion mutant. We then used microarray and Northern blot analyses to monitor global effects on the transcriptome in order to provide insights into the biological roles of Spot 42 in this bacterium. In the presence of glucose, we found a surprisingly large number of ≥ 2X differentially expressed genes, and several major cellular processes were affected. A gene encoding a pirin-like protein showed an on/off expression pattern in the presence/absence of Spot 42, which suggests that Spot 42 plays a key regulatory role in the central metabolism by regulating the switch between fermentation and respiration. Interestingly, we discovered an sRNA named VSsrna24, which is encoded immediately downstream of spf. This new sRNA has an expression pattern opposite to that of Spot 42, and its expression is repressed by glucose.
Conclusions
We hypothesize that Spot 42 plays a key role in the central metabolism, in part by regulating the pyruvat dehydrogenase enzyme complex via pirin.
Background
Bacteria contain a class of regulatory non-coding (nc) RNAs that are transcribed in trans from distinct promoters [1]. They are typically between 50 and 200 nt in size [2] and have become known as bacterial small RNAs, or sRNAs. The majority of known sRNA genes are located in intergenic regions, but it is becoming increasingly evident that a relatively large number of RNAs are also being transcribed from within protein coding regions, but from the opposite strand (i.e., anti-sense RNAs). Even though their roles are still mostly unknown, it is likely that anti-sense RNAs also play important roles in gene regulation. cis-encoded anti-sense RNAs share extensive complementarity to their messenger RNA (mRNA) target, whereas trans-encoded RNAs typically show limited complementarity [1]. Although cis-encoded RNA regulators can be encoded from within their mRNA target, they are typically located in front of protein coding regions as part of the mRNA. Here they change the expression of the corresponding protein by binding small metabolites (i.e., riboswitches) [3].
sRNAs typically bind to the 5' end of mRNAs through short imperfect base-paring and induce degradation of itself and the target [1]. Other mechanisms, like direct interaction with proteins to modulate their activities or increase stability of mRNAs, also occur. Finding the function and/or mechanism of sRNAs can however be a daunting task, and the Spot 42 sRNA represents a striking example. It was first described as an unstable RNA species of 109 nt in Escherichia coli that accumulated under growth in the presence of glucose (i.e., when adenosine 3', 5'-cyclic monophosphate (cAMP) is low) [4]. During growth with a non-glucose carbon source (i.e., when cAMP concentrations are high), the Spot 42 concentrations were found to be significantly lower. Later experiments showed that over-expression of Spot 42 (~10 fold increase) resulted in impaired growth and lowered ability to adapt to shifts to richer media or shift from glucose to succinate as the carbon source [5]. Also, deletion of the gene that encodes Spot 42, i.e., spf (spot fourty-two), resulted in viable spf null mutants, which indicated that the Spot 42 RNA is non-essential [6]. The direct responsiveness of Spot 42 levels to glucose and cAMP is due to repression of spf by a cAMP-CRP (cAMP-receptor protein) complex [7,8]. It was unclear for some years if the function of Spot 42 was mediated through the 109 nt RNA itself or through a 14 amino acids long peptide that is hypothetically encoded from within the sRNA. This controversy was settled by Rice et al. [9]. They used a filter binding assay and other methods to show that Spot 42 bound very inefficiently and nonproductive to purified 70S ribosomes, and concluded that Spot 42 function is mediated by the RNA itself. Later, the proximity of spf to polA (encodes DNA polymerase I) lead Dahlberg and co-workers to test whether the products of these genes could influence each other [7,8], and they found that reduction in Spot 42 levels, either by deletion of spf or by manipulating the growth conditions, both resulted in reduction in DNA pol A activity. The underlying mechanism remains however unknown. The first direct Spot 42 target was discovered by Møller et al. [10], who showed that the sRNA can bind specifically by base pairing with a short complementary region of the translation initiation region of galK (encodes a galactokinase), which is the third gene of the galactose operon (galETKM). The Spot 42-binding region overlaps with the galK Shine-Dalgarno region, thereby blocking ribosome binding. Spot 42 is therefore responsible for a discoordinate regulation of the gal operon. Finally, in a recent work Beisel and Storz [11] demonstrated with microarray analysis and reporter fusions that Spot 42 plays a broader role in metabolism by regulating at least fourteen operons dominated by genes involved in uptake and catabolism of non-favored carbon sources. Several of these operons are regulated by both Spot 42 and CRP, and these two regulators can therefore be considered as participating in a feed-forward loop.
In this study, we used our model bacterium Aliivibrio salmonicida, which belongs to the Vibrionaceae family of gamma-proteobacteria, to further address the biological roles of Spot 42. When we started the project, galK was the only known Spot 42 target. Interestingly, we found that A. salmonicida naturally lacks the gal operon, but contains a highly conserved spf gene, and we therefore considered this bacterium as an excellent model for studying Spot 42 roles other than in galK regulation. Even though other Spot 42 targets have since been proposed [11], our study is still valid, and our goal remains the same, i.e., to identify all biological roles of Spot 42. We generated an A. salmonicida spf deletion mutant and used microarray and Northern blot analyses to find more clues to its function. Deletion of spf has a surprisingly large effect on the transcriptome, with the most dramatic effect on expression of a pirin-like protein gene. In the process, we also discovered a neighboring sRNA, named VSsrna24, which is encoded from the same intergenic region (IGR) and 262 nt downstream of spf. Interestingly, this RNA is expressed in a pattern opposite to that of Spot 42, and its expression is repressed by glucose.
Results and Discussion
spf is highly conserved in Vibrionaceae and is located upstream of a previously unrecognized sRNA gene named VSsrna24
We first surveyed available genome sequences from the Vibrionaceae family and found that orthologs of spf are found in virtually all 76 currently available genomes. Available genomes include representatives of the Vibrio, Aliivibrio, Photobacterium, and Grimontia genera. Six V. cholerae genomes lack a recognizable spf gene, but we suspect that this is due to the poor quality of these genomes (i.e., the genome sequences consist of many small unfiltered contigs) or the lack of data. By using the spf sequence of A. salmonicida as the query in a Blastn search, the gene was also identified in fourteen gamma-proteobacteria genera (Pantoea, Xenorhabdus, Salmonella, Citrobacter, Yersinia, Serratia, Edwardsiella, Dickeya, Photorhabdus, Escherichia, Enterobacter, Klebsiella, Rahnella and Shigella) in the Enterobacteriales order.
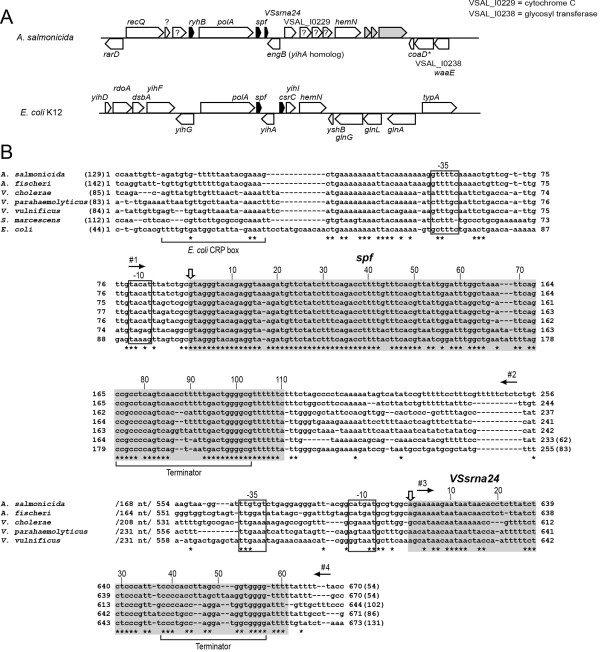
Figure 1A shows the genomic location of spf and its flanking genes in A. salmonicida compared to the corresponding region in E. coli. In both genomes, spf is flanked by polA as the nearest upstream neighbor and yihA (or the engB homolog) as the nearest downstream protein coding gene. The genomic locus is also home to other important sRNA genes. In A. salmonicida, RyhB is encoded from the neighboring and upstream IGR [12]. Interestingly, spf is also neighbor to a putative sRNA gene (VSsrna24), which is located in the same IGR as spf. VSsrna24 was predicted in a global computer-based search for sRNAs that will be reported elsewhere. In E. coli, the IGR downstream of spf encodes the carbon storage regulator C (CsrC) sRNA that interacts directly with and regulates the activity of the carbon storage regulator A protein (CsrA) [13], which functions as a global post transcriptional regulator.
Figure 1.
Synteny comparison and sequence alignment of spf and VSsrna24. (A) The order of genes that flank spf and other sRNA genes (filled arrows) in A. salmonicida and E. coli are shown. Question marks indicate genes with unknown function. (B) Sequence alignment of spf and VSsrna24 from selected members of Vibrionaceae, E. coli and Serratia marcescens. The CRP binding site of E. coli, and -10 and -35 promoter regions in front of spf and VSsrna24 are based on knowledge from E. coli [7,8]. The vertical open arrows denote the 5' end of Spot 42 and VSsrna24 based on 5' RACE data on A. salmonicida presented in this work. The horizontal arrows (#1-4) denote areas deleted in the spf (#1-2) and VSsrna24 (#3-4) deletion strains. Numbers in parenthesizes denote numbers of nt to the nearest CDS. Asterisks indicate invariable positions.
Figure 1B shows an alignment of Spot 42 and VSsrna24 from selected species of Vibrionaceae, and Serratia marcescens and E. coli. Spot 42 is found widespread among gamma-proteobacteria, whereas VSsrna24 is limited to Vibrionaceae, i.e., in A. salmonicida, A. fischeri, V. cholera, V. furnissi, V. vulnificus, V. harveyi, V. parahaemolyticus and V. natriegens. A 5' rapid amplification of cDNA ends (5' RACE) analysis was done to map the 5' end of the A. salmonicida Spot 42 and VSsrna24, and the result showed that the Spot 42 5' end is the same as for E. coli (indicated by vertical arrow in Figure 1B), and that VSsrna24 is approximately 60 nt in length. Ninety-one of the 109 nt in E. coli Spot 42 are invariable when compared to the selected sequences. The highly conserved nature and the wide distribution of spf in Vibrionaceae and other gamma-proteobacteria suggest that, as in E. coli, Spot 42 serves important cellular functions that might be common to this group of bacteria, regardless if the bacterium is a pathogen or not (although conservation is not always a sign of viability). Since 2002 and until recently, galK was the only known target of Spot 42 [10]. A. salmonicida naturally lacks the gal operon (including galK), but at the same time contains a highly conserved spf gene. At the onset of this project we therefore considered the bacterium as an ideal model for studying roles of Spot 42 other than regulation of galK translation. This is still valid even though other targets have been found [11]. In agreement with the lack of a gal operon, the bacterium is unable to utilize galactose when grown in a minimal medium optimized for A. salmonicida, with 44.4 mM galactose as essentially the only carbon source (data not shown). In other words, galactose has little or no effect on growth when compared to a control with no added sugar. In contrast, when 44.4 mM glucose is added as carbon source, the bacterium grows to high densities [i.e, typically to optical density 600 nm (OD600 nm) > 7].
Next, we examined the sequence upstream of spf for potential transcription factor binding sites to gain insights into regulation of spf transcription. Sequences that weakly resemble the E. coli CRP binding site, and -10 and -35 promoter regions were predicted upstream of Vibrionaceae spf sequences.
Expression of Spot 42 and the adjacent VSsrna24 is highly dependent on glucose
It is established that expression of E. coli Spot 42 is repressed by the transcription factor CRP and cAMP [7]. Expression is also highly dependent on concentrations of available glucose since cAMP adenylyl cyclase (i.e., a cAMP-producing enzyme) is inhibited as a side-effect during glucose transport into the cell. As a result, cAMP levels are low when glucose is rich and is being transported into the cell for consumption, and vice versa.
To test if A. salmonicida Spot 42 is expressed similar to that in E. coli, the bacterium was cultured in LB with 2.5% NaCl added (A. salmonicida requires elevated salt concentrations for efficient growth) under standard lab conditions, and samples were collected throughout the growth cycle. At OD600 nm 0.4 the culture was split, and one half was supplemented with 1 mM cAMP (LB medium contains yeast extract and will contain trace amounts of cAMP, e.g. see Polnisch and Hofmann) [14] (Figure 2A). At early growth phase the intracellular levels of cAMP are expected to be low. Similarly, the starting culture was also split at OD600 nm 1.2, and glucose was added to one half to a final concentration of 5 mM (i.e., when glucose is expected to be low/exhausted). Cell samples were collected at various time points after the addition of cAMP or glucose to monitor the levels of Spot 42 by Northern blot analysis. Figure 2B shows that Spot 42 is found at high levels during the early phases of growth, but is gradually lost and is below our level of detection at OD600 nm 1.2. The level of Spot 42 is significantly reduced (3 folds) after addition of cAMP or is significantly increased (16-40 folds) after addition of glucose. These results are in agreement with results from E. coli, which suggest that A. salmonicida Spot 42 serve roles similar to that in E. coli, i.e., in carbohydrate metabolism.
Figure 2.
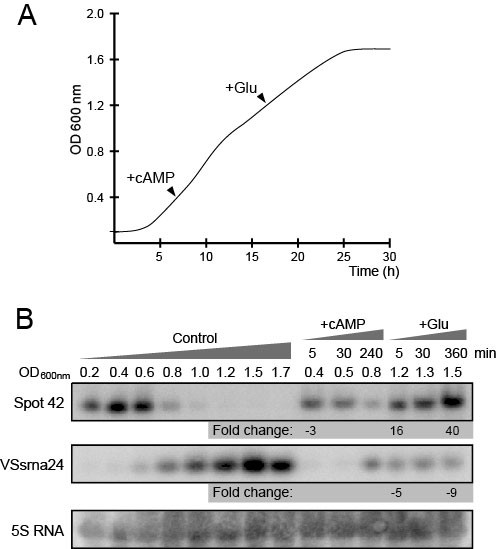
Monitoring Spot 42 and VSsrna24 expression under different treatments. (A) A. salmonicida was cultured for approximately 30 hours (h) starting at OD600 nm 0.1, and ending at 1.8 (stationary phase). A typical growth trajectory of A. salmonicida in LB is illustrated in the figure. Samples were collected throughout the growth cycle. The culture was split and 1 mM cAMP was added at OD600 nm 0.4 to one half. Similarly, a second culture was split and 5 mM glucose was added at OD600 nm 1.2. Untreated cells were used as control. (B) Samples collected in A were subjected to Northern blot analysis. From the control culture samples were collected at OD600 nm 0.2-1.7, and after the additions of cAMP or glucose samples were collected after 5-240 min (OD600 nm 0.4-0.8) or 5-360 (OD600 nm 1.2-1.5) min, respectively. Radio-labeled double-stranded DNA probes were used to monitor the levels of Spot 42 and VSsrna24, and 5S rRNA was used to normalize the result. Fold change values ≥ 2 are shown below samples, and are always comparisons between treated and untreated control samples, at same ODs.
In the same experiment we also monitored the levels of the putative sRNA named VSsrna24, which is encoded from the same IGR as Spot 42. This sRNA produced a signal corresponding to an RNA of approximately 60 nt in length, with an expression pattern opposite to that of Spot 42. Also, expression of VSsrna24 is repressed by glucose (5-9 folds), but appears independent of cAMP. These observations and the close proximity of VSsrna24 to Spot 42 suggested to us that VSsrna24 might also play roles in carbohydrate metabolism, perhaps in concert with Spot 42.
Finally, in a simple experiment we subjected A. salmonicida to different stress conditions (i.e., low iron conditions, oxidative stress, low/high temperatures, and alcohol) or to one of two quorum sensing signal molecules (hexanoyl-L-homoserine lactone or N-3-oxo-hexanoyl-L-homoserine lactone), and monitored levels of Spot 42 and VSsrna24 by Northern blot analysis (provided as Additional File 1). This was done to test if expression of Spot 42 and/or VSsrna24 is dependent on external stress factors or communication molecules, which could indicate potential roles in stress response or quorum sensing, respectively. A. salmonicida was grown to OD600 nm 0.5 and subjected to the different treatments. Samples were then collected at various time points after treatment, but none of the stress conditions dramatically changed the expression pattern, maybe except for low iron conditions (addition of 50 uM 2,2'-dipyridyl) that resulted in reduction in Spot 42 levels (2-5 fold).
To summarize, we found that expression of A. salmonicida Spot 42 is repressed by cAMP and activated by glucose, whereas expression of a new sRNA, named VSsrna24, is repressed by glucose and is independent of cAMP. We also found that expression of both Spot 42 and VSsrna24 is, in general, not affected by external stress factors or by the addition of quorum sensing signals.
Microarray analysis of a A. salmonicida spf deletion mutant identifies potential roles and targets
We constructed a spf deletion mutant (see Methods for details) to identify potential biological roles and possible mRNA targets for A. salmonicida Spot 42. The transcriptome of the spf deletion mutant was compared on a global scale to that of the wild-type strain by using microarray analysis. To create growth conditions in which Spot 42 was highly expressed in the wild-type strain, the cells were cultured in ASMM minimal medium and then stimulated by adding glucose as the main carbon sources. Specifically, in three independent experiments (biological replicates), cells were grown to OD600 nm 0.4, and 44.4 mM glucose was added. Samples were collected 15 min after addition of glucose, total RNA was extracted, and the three independent samples were pooled before cDNA synthesis and hybridization to three A. salmonicida whole genome custom DNA chips (i.e., Vibrio salmonicida V1.0.1 AROS). RNA from glucose treated wild-type and spf deletion cells were labeled with fluorescent dyes and run on same DNA chips.
Using the LIMMA framework in Bioconductor [15] we considered genes with ≥ 2 fold differential expression and adjusted p-values ≤ 0.05. Figure 3 shows a graphical presentation of the functional classes and the number of differentially up- or downregulated genes in the spf deletion mutant 15 min after addition of glucose. Details on differentially regulated genes are shown in Tables 1 and 2 (Additional file 2 contains complete lists of differentially expressed genes). From these data it is clear that the spf deletion is affecting the expression of a relatively large number of genes (e.g., 385 genes are ≥ 2 fold differentially expressed after addition of glucose) that belong to a wide range of categories, such as "foreign DNA", "carbohydrate metabolism", "cell envelope", "transport proteins", "motility", "iron homeostasis", and "quorum sensing". Spot 42 therefore has a broad effect on multiple cellular functions.
Figure 3.
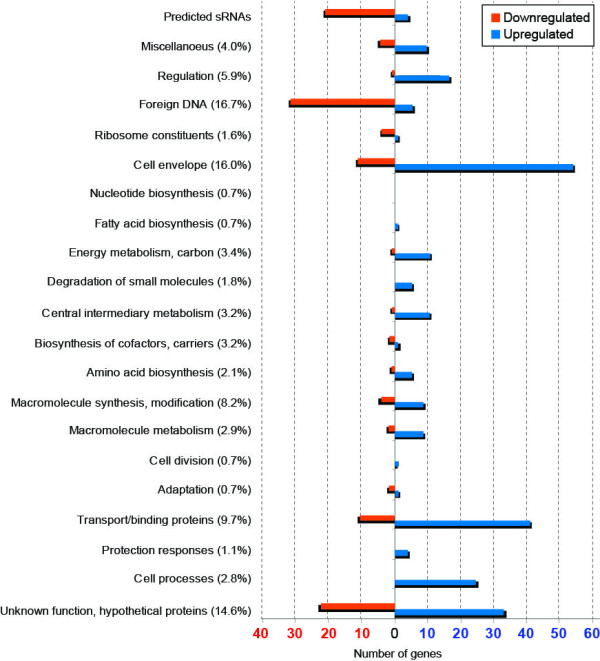
Overview of microarray results with spf deletion mutant compared to the wild-type strain. Bacteria were grown to OD600 nm 0.4, then the culture was split and 44.4 mM glucose was added to one half. Cells were collected after 15 min. CDSs with differential gene expression corresponding to up- or downregulation above or below 1.5 fold are divided into functional categories as defined by the Sanger Institute Pathogen Sequencing Unit http://genprotec.mbl.edu/files/MultiFun.html. Numbers in parenthesises indicate in percentage the share of the total number of genes in the genome that each class represents.
Table 1.
Top 30 list of ≥ 2 fold upregulated genes in spf deletion mutant after addition of 44.4 mM glucose.
| CDS | Gene | Gene product | Log2 ratio | Adjusted P-value |
|---|---|---|---|---|
| VSAL_I1200 | putative pirin | 3.97 | 2.49E-04 | |
| VSAL_I2193 | methyl-accepting chemotaxis protein | 2.92 | 2.97E-04 | |
| VSAL_I4139s | VSsrna140 undefined small RNA | 2.53 | 4.98E-04 | |
| VSAL_I0799 | methyl-accepting chemotaxis protein | 2.53 | 4.02E-04 | |
| VSAL_I2318 | flaD | flagellin subunit D | 2.48 | 4.35E-04 |
| VSAL_II0716 | putative exported protein | 2.39 | 3.54E-03 | |
| VSAL_I2317 | flaA | flagellin subunit E | 2.33 | 4.98E-04 |
| VSAL_II0613 | putative membrane protein | 2.17 | 8.69E-04 | |
| VSAL_I1201 | putative IMP dehydrogenase/GMP reductase | 2.13 | 4.44E-04 | |
| VSAL_I2319 | flaA | flagellin subunit C | 2.09 | 5.58E-04 |
| VSAL_I2517 | flaF | flagellin subunit F | 2.07 | 6.11E-04 |
| VSAL_I2329 | flgM | flagellar hook-associated protein type 3 FlgM | 2.03 | 4.98E-04 |
| VSAL_I2022 | vcmH | multidrug efflux pump | 1.87 | 1.20E-03 |
| VSAL_II0587 | outer membrane protein, OmpA family | 1.87 | 6.40E-04 | |
| VSAL_I2593 | gluconate permease | 1.81 | 6.74E-04 | |
| VSAL_II0091 | fruK | 1-phosphofructokinase | 1.81 | 9.06E-04 |
| VSAL_II0090 | fruB | PTS system, fructose-specific IIA/FPR component | 1.80 | 6.11E-04 |
| VSAL_I2578 | ABC-type [(GlcNAc)2] transporter, permease protein | 1.79 | 6.11E-04 | |
| VSAL_I1857 | hypothetical protein | 1.78 | 6.74E-04 | |
| VSAL_II0715 | cusB | putative cation efflux system protein | 1.77 | 8.23E-04 |
| VSAL_I2338 | flgB | flagellar basal-body rod protein FlgB | 1.75 | 5.13E-04 |
| VSAL_II0231 | cheV | chemotaxis protein CheV | 1.74 | 8.03E-04 |
| VSAL_I2771 | motX | sodium-type polar flagellar protein MotX | 1.71 | 7.15E-04 |
| VSAL_II0331 | putative exported protein | 1.70 | 6.14E-04 | |
| VSAL_I2334 | flgF | flagellar basal-body rod protein FlgF | 1.70 | 8.55E-04 |
| VSAL_II1080 | membrane protein | 1.70 | 7.00E-04 | |
| VSAL_I0474 | mshF | Type IV pilus, mannose-sensitive hemagglutinin protein MshF | 1.68 | 6.11E-04 |
| VSAL_I1401 | tupA | extracellular tungstate binding protein precursor | 1.66 | 6.86E-04 |
| VSAL_I2327 | fliC | flagellin subunit A | 1.62 | 7.00E-04 |
| VSAL_II0785 | putative exported protein | 1.59 | 6.11E-04 | |
| VSAL_I2061 | hypothetical protein | 1.59 | 6.11E-04 |
Table 2.
Top 30 list of ≥ 2 fold downregulated genes in spf deletion mutant after addition of 44.4 mM glucose
| CDS | Gene | Gene product | Log2 ratio | Adjusted P-value |
|---|---|---|---|---|
| VSAL_II0612 | HTH-type transcriptional regulator, LysR family (pseudogene) | -5.71 | 1.24E-04 | |
| VSAL_I0768 | hypothetical protein, putative phage gene | -5.62 | 1.24E-04 | |
| VSAL_I3103s | VSsrna23 small RNA Spot 42 | -5.60 | 1.24E-03 | |
| VSAL_I3178s | VSsrna149 tmRNA | -5.19 | 1.24E-04 | |
| VSAL_I1028 | gpN major capsid protein | -5.17 | 1.46E-04 | |
| VSAL_p320_13 | putative peptidase, S24-like | -5.16 | 2.49E-04 | |
| VSAL_II2002s | VSAsrna3 undefined small RNA | -5.12 | 1.46E-04 | |
| VSAL_I0985 | MrdA penicillin-binding protein 2 | -5.06 | 2.49E-04 | |
| VSAL_I1029 | gpM phage terminase, endonuclease subunit | -5.02 | 1.24E-04 | |
| VSAL_I4069s | VSsrna70 undefined small RNA | -4.72 | 4.98E-04 | |
| VSAL_I0136 | IucC siderophore biosynthesis protein | -4.42 | 1.97E-04 | |
| VSAL_I4155s | VSsrna156 undefined small RNA | -4.20 | 6.14E-04 | |
| VSAL_I1027 | gpO phage capsid scaffolding protein | -4.10 | 3.71E-04 | |
| VSAL_I3104s | VSsrna24 small RNA | -4.10 | 2.97E-04 | |
| VSAL_I0135 | AlcA siderophore biosynthetis protein | -4.04 | 3.71E-04 | |
| VSAL_p54_02 | putative mobilization protein | -3.93 | 3.71E-04 | |
| VSAL_p43_01 | replication initiation protein | -3.92 | 1.24E-04 | |
| VSAL_I0134 | L-2,4-diaminobutyrate decarboxylase | -3.84 | 2.49E-04 | |
| VSAL_I3073r | 5S rRNA 5S rRNA undefined product 93740:93859 forward | -3.82 | 7.53E-04 | |
| VSAL_I1039 | probable exported protein, putative phage gene | -3.72 | 2.49E-04 | |
| VSAL_I3157t | tRNA transfer RNA-Ser | -3.64 | 3.13E-04 | |
| VSAL_I0137 | TonB-dependent iron-siderophore receptor precursor | -3.58 | 3.08E-04 | |
| VSAL_p54_01 | acyltransferase | -3.58 | 4.98E-04 | |
| VSAL_I1751 | TonB1 TonB protein (pseudogene) | -3.56 | 3.71E-04 | |
| VSAL_I3144t | tRNA tRNA transfer RNA-Leu 842679:842760 reverse | -3.46 | 8.25E-04 | |
| VSAL_I3072r | 23S rRNA 23S rRNA undefined product 90756:93646 forward | -3.37 | 1.04E-03 | |
| VSAL_I1040 | hypothetical protein, putative phage gene | -3.33 | 3.13E-04 | |
| VSAL_p320_31 | putative phage intergrase | -3.32 | 3.71E-04 | |
| VSAL_p43_02 | acetyltransferase | -3.25 | 1.97E-04 | |
| VSAL_I3102s | VSsrna22 small RNA RyhB | -3.23 | 3.08E-04 |
Major findings are, first, that a gene encoding a pirin protein is by far the most upregulated gene. The log2 ratio between spf deletion mutant and wild-type is 3.97 (equivalent to ~16 fold increase), whereas the same value for the second most upregulated gene, i.e., VSAL_I2193 that encodes a methyl-accepting chemotaxis protein, is 2.92 (equivalent to ~8 fold increase). The pirin protein (theoretically 283 aa) has 21 and 24% identity, and 45 and 49% similarity to the E. coli (231 aa) and S. marcescens (313 aa) pirin proteins, respectively, when all positions in the protein sequences are considered (approx. 45% identity in conserved regions). Second, the top 30 list of most upregulated genes is highly enriched with genes involved in motility and chemotaxis [cheV, flaA (subunit E), flaA (subunit C), flaD, flaF, flgB, flgF, flgM, fliC, motX, VSAL_I0799 and VSAL_I2193]. Upregulation of genes with functions in motility and chemotaxis is potentially a result of upregulation of the flagellar biosynthesis sigma factor (sigma F or 28), encoded by fliA (VSAL_I2290), the spoIIAA anti-sigma F factor antagonist (VSAL_II0328), and finally the anti-sigma 28 factor (VSAL_I2342). These are the most upregulated genes (i.e., 1.7, 1.6 and 1.6 fold, respectively) of all sigma factor-related genes in the complete dataset. Flagella-related genes are known from other bacteria to be regulated by the sigma 28 factor (or its orthologs, e.g., sigma F). Almost all upregulated motility and chemotaxis genes are located in the same gene cluster (approx. VSAL_I2283 to VSAL_I2343). Third, fruBK are both on the top 30 list of upregulated genes, whereas fruA is also significantly upregulated (2.3 fold upregulated). FruAB constitute the fructose-specific EII-component of the phosphotransferase system (PTS). Therefore, the upregulation of fruABK supports a similar role for A. salmonicida Spot 42 as in E. coli where uptake of less favorable sugars is suppressed by Spot 42 (carbon catabolite repression). Finally, the top 30 list of most downregulated genes is dominated by foreign DNA (phage and plasmid DNA) and genes encoding non-coding RNAs (ncRNAs), including a number of predicted sRNA genes.
In summary, the microarray analysis shows that deletion of spf has a broad effect on many genes and cellular functions. Interestingly, the most upregulated gene encodes a putative pirin protein. In S. marcescens, pirin has been found to regulate the activity of pyruvat dehydrogenase E1 [16]. Pirin is therefore a key regulator in the central metabolism by selecting the destiny of pyruvat, either through fermentation or respiration through the tricarboxylic acid (TCA) cycle and electron transport. Because A. salmonicida Spot 42 directly or indirectly regulates pirin, Spot 42 likely plays a critical regulatory role in the central metabolism.
Upregulation of pirin and repression of multiple sRNAs are supported by Northern blot analysis
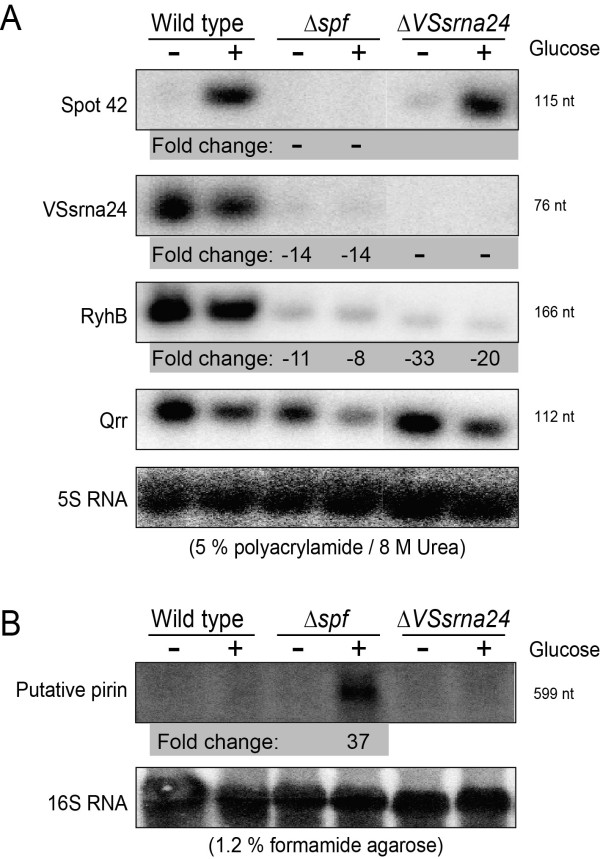
Inspired by findings from the microarray analysis we next used Northern blot analysis to evaluate results of particular interest. First, we wanted to validate levels of Spot 42, and therefore generated a corresponding [α-32P]-labeled DNA probe. Figure 4A shows the resulting autoradiogram which confirms that Spot 42 is rich in wild-type cells in the presence of glucose, and is absent from spf deletion cells. As expected, Spot 42 is virtually absent in wild-type cells grown without any added glucose.
Figure 4.
Validation of selected microarray results with Northern blot analysis. (A) RNAs from wild-type and spf and VSsrna24 deletion mutants (Δspf and ΔVSsrna24, respectively) were separated on 5% denaturating polyacrylamide gels, transferred to membranes and tested for presence of the sRNAs Spot 42, RyhB, Qrr and VSsrna24. 5S rRNA was used as control and to normalize the results. Plus (+) indicates that glucose was added to the culture 15 min prior to sampling, whereas minus indicates that no glucose was added. Numbers to the right of gel pictures indicate the length of RNAs as measured from the gel. Fold change values ≥ 2 are shown below samples, and are always comparisons between deletion mutants and wild-type control samples, at same conditions. (B) Same samples as described above separated on a 1.2% denaturating formamide gel.
We were intrigued by the dramatic effect of spf deletion on levels of a putative pirin-encoding mRNA. In agreement with the microarray result a pirin probe produced no visible band when using total RNA from wild-type cells, whereas total RNA from spf deletion mutant cells treated with glucose produced a distinct and readily visible band of the expected size (Figure 4B). The increase in intensity corresponds to a fold change value of approx. 37 folds (approx. 16 folds in microarray analysis). We next used the RNAhybrid software [17] to test for potential interactions between Spot 42 and the 5' region of the pirin mRNA. This software calculates the most energetically favorable base pairing between a small RNA and a larger target RNA [17]. Additional file 3 shows that when using A. salmonicida Spot 42 (excluding the terminator) and a sequence extending from -100 nt to + 50 relative to the pirin start codon as input sequences to the program, significant potential for base-pairing between Spot 42 and the 5' untranslated region (UTR) of the pirin mRNA is found. Given that Spot 42 binds directly to this region then this would be similar to the most common form of sRNA-mRNA interaction, i.e., base pairing within the 5' UTR region [1]. Further analysis to address potential interactions between Spot 42 and pirin mRNA will come from biochemical methods, such as gel mobility shift assay, fusion-reporter assays, structure probing and site directed mutagenesis.
Our microarray data suggest that many predicted sRNAs are significantly downregulated in the spf deletion mutant. This can potentially be explain by reduction in Hfq (encoded by hfq) and/or upregulation of RNaseE (encoded by rne), which can destabilize sRNAs in general. However, the microarray data indicate that mRNA levels of the corresponding mRNAs are virtually unchanged. Next, we designed probes against the new VSsrna24, and the RyhB and Qrr sRNAs that are critical components in regulation of iron homeostasis and quorum sensing, respectively. These sRNAs were found to be downregulated approx. 14, 10 and 2 folds, which is in good agreement with the corresponding numbers (approx. 17, 10 and 4 folds, respectively) from the microarray analysis. The underlying mechanism(s) for the apparent reduction in sRNA levels remains however unclear. It should however be noted that VSsrna24 is encoded 219 nt downstream of the deleted region in the spf mutant strain, and it is therefore possible that the deletion has affected the promoter and therefore expression of VSsrna24. In the same experiment we also included a VSsrna24 deletion mutant. Here, RyhB is also significantly downregulated (19/35 folds with/without glucose), whereas levels of Spot 42 and Qrr appear unchanged.
Conclusions
We have studied potential biological roles of a A. salmonicida Spot 42 homolog and discovered a neighboring sRNA, named VSsrna24. The expression of Spot 42 is similar to that in E. coli, i.e., expression is repressed by cAMP and activated by glucose. Expression of VSsrna24 is, in contrast, repressed by glucose but unaffected by cAMP. A microarray analysis revealed that deletion of spf affects a wide range of cellular processes and the expression of a surprisingly large number of genes. After addition of glucose at mid exponential growth phase (i.e., when Spot 42 is highly expressed in the wild-type strain), the most differentially upregulated gene in the spf deletion mutant was VSAL_I1200, which encodes a pirin protein with 21% identity and 45% similarity to the E. coli pirin. Other notable results are upregulation of many genes involved in motility and chemotaxis, and downregulation of predicted sRNA genes and foreign DNAs. Expression of Spot 42, VSsrna24, Qrr, RyhB and pirin were validated by Northern blot analysis. Figure 5 shows a hypothesis of the main findings. It should be stressed that it should be regarded as a hypothesis that must be tested further, and solid evidence can come from e.g., gel mobility shift assay or fusion-reporter assays to address potential interactions between Spot 42 and potential mRNA targets, or complementation assay to restore Spot 42 functions.
Figure 5.
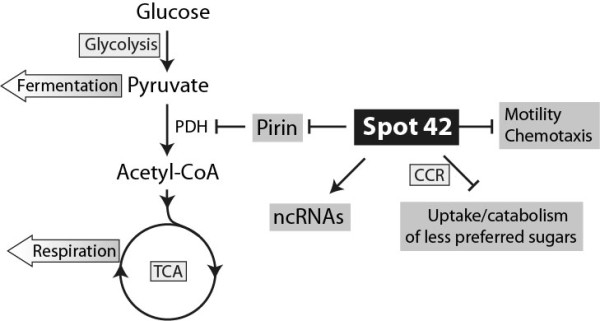
A model for Spot 42 regulation in A. salmonicida. The model is based on microarray and Northern blot analyses from this study, which suggest that Spot 42 downregulates Pirin (key regulator of puruvat dehydrogenase complex in central metabolism), genes for uptake/catabolism of less preferred sugars in a carbon catabolite repression (CRR)-like manner, and genes involved in motility and chemotaxis. The results also suggest that Spot 42 activates expression of other ncRNAs (e.g., the sRNA RyhB).
The majority of bacterial regulatory RNAs, like sRNAs, probably remain to be identified. Eighty-seven sRNAs are known in E. coli [2], whereas the corresponding number is much lower for other bacteria. Once identified, finding the function and targets of each sRNA represents a formidable task. In general, the current knowledge on sRNA function is therefore limited. Although one or a few targets have been identified for a given sRNA, it is likely that many additional targets remain to be found. For example, in E. coli, the transcript of the multi-cistronic gal operon was for many years the only known Spot 42 target [10], but a recent microarray analysis in which over-expression of spf with an isopropyl β-D-thiogalactopyranoside (IPTG)-inducible promoter was used, suggests many additional targets [11]. The list of targets is enriched in genes involved in sugar transport (dppB, IldP, nanC, nanT, srlA and xylF) or sugar catabolism (ebgC, fucI, fucK and galK). We have used a knock-out approach to find overexpressed genes that could be potential Spot 42 targets in A. salmonicida. Interestingly, the majority of genes that corresponds to mRNA targets in E. coli are apparently not present (i.e., they produce no significant hits in BLASTP searches) in the A. salmonicida genome (i.e., IldP, nanC, srlA, ebgC, fucI, galK and gsp). Also, apparent homologs of the remaining E. coli targets were in general not significantly differentially expressed in our dataset. These results suggest that there are significant differences in Spot 42 regulation between E. coli and A. salmonicida.
Perhaps our most intriguing finding is that A. salmonicida Spot 42 appears to directly or indirectly regulate the levels of pirin mRNA. This is in agreement with its currently known role in carbohydrate metabolism, and that the level of Spot 42 is repressed by cAMP and activated by glucose. We are also pursuing potential roles of the neighboring sRNA, named VSsrna24. Its expression is repressed by glucose, and we suspect that this sRNA also could have important roles in carbohydrate metabolism.
Methods
Bacterial strains and growth
A. salmonicida LFI1238 and the corresponding spf deletion mutant were cultured in LB medium with 2.5% NaCl added, or in the A. salmonicida-specific minimal media ASMM, at 12-16°C in a shaking incubator at 200-230 rpm. ASMM contains the following compounds in mM: NaCl, 256.7; KCl, 9.4; MgCl2 · 6H2O, 50.075; FeCl3 · 6H2O, 0.0126; NH4Cl, 18.7, K2HPO4, 0.58; Trizma base, 41.3; CaCl2 · 2H2O, 0.225; ZnCl2, 0.0085, CoCl2 · 6H2O, 0.0055; CuSO4 · 5 H2O, 0.005; Na2MoO4, 0.007; MnCl4 · 2H2O, 0.011; cycteine, 0.5; isoleucine, 0.5; methionine, 0.5; valine, 2.0; glutamic acid, 40; glucose or galactose (added when indicated), 44.4 mM.
A. salmonicida was subjected to multiple treatments by first growing the bacterium in LB medium with 2.5% NaCl at 16°C to mid-exponential growth phase (OD600 nm ~0.6). Cultures were then divided into nine equally sized cultures, one for each stimulation, i.e., 50 μM 2,2'-dipyridyl iron chelator, 1 mM hydrogen peroxide, 500 μM paraquat (N, N'-dimethyl-4,4'-bipyridinium dichloride), 0.5% ethanol, 2 μg/ml hexanoyl-L-homoserine lactone (HHL), 2 μg/ml N-3-oxo-hexanoyl-L-homoserine lactone (OHHL), 4°C or 20°C growth temperatures, and one non-stimulated control culture. Ten ml samples were collected 0, 2, 5, 10 and 30 min after stimulation. Cells were harvested by centrifugation at 3,500 × g for 10 min. Pelleted cells were then snap-frozen in a combination of dry-ice and ethanol, and kept at -70°C prior to RNA isolation.
For cAMP and glucose stimulations A. salmonicida was cultured in LB medium at 12°C and split two times during growth. At OD600 nm 0.4, the culture was split and 1 mM cAMP was added to one half and grown under the same conditions as the untreated control for 240 min. Ten ml samples were collected 5, 30 and 240 min after stimulation. Similarly, at OD600 nm 1.2, the untreated culture was again split and one half was stimulated with 5 mM D(+)-glucose and grown under the same conditions as the untreated control culture for 360 min. Ten ml samples were collected 5, 30 and 360 min after stimulation.
Samples used for Northern blot and microarray analyses were prepared by culturing A. salmonicida wild-type and a spf deletion mutant strains at 12°C in ASMM without any added carbon source to OD600 nm 0.4. At this density the culture was split into two halves. Glucose to 44.4 mM was added to one half and the other was used as the untreated control. A 15 ml sample was collected 15 min after stimulation and harvested as described above.
Construction of Spf and VSsrna24 deletion strains
The construction of Spf and VSsrna24 deletion strains was performed essentially as previously described [18]. A suicide vector (pDM4) containing regions flanking the spf or VSsrna24 genes were used. The flanking regions were joined by overlap PCR, i.e., two PCR products containing the flanking regions with overlapping ends are extended and then used as template with the outermost primers from the first two PCRs. The final PCR product was cloned into the pDM4 plasmid and introduced into A. salmonicida by conjugal mating with the E. coli S17-1 λ pir donor strain. Transconjugants were selected by plating the conjugation mix on LB plates containing 2 μg/ml chloramphenicol at 12°C. E. coli grows very poorly at 12°C and was therefore selected against. Colonies were next transferred to LB plates with 5% sucrose to induce expression of the sacB suicide gene. The product is lethal to Gram-negative bacteria [19]. This step selects against the plasmid, and integrated plasmids (they are most likely to have integrated into chromosomal locations similar to those that were introduced into the plasmid) must therefore be removed from chromosomal positions in a recombination event in order for the bacterium to survive. Resulting chloramphenicol-sensitive colonies were analyzed by PCR to screen for bacteria with the desired chromosomal deletion. PCR products of expected sizes were sequenced to confirm the deletion.
5' rapid amplification of cDNA ends (RACE) for mapping of the 5' end
5' RACE assays were carried out essentially as previously described [20] using the GeneRacer kit (Invitrogen), 6 μg of total RNA, and a Spot 42 or VSsrna24-specific oligonucleotide (5'-GCCAAATCCAATAACGTGAAAC-3' or 5'-GCTAAGGTGGGGAAATGG-3', respectively).
Microarray analyses
Total RNA was extracted from cells using the RNAisol reagent (5 PRIME) followed by a subsequent DNA removing step with DNAfree kit (Applied Biosystems). Salts and leftover traces of DNA were removed by RNeasy Minelute Cleanup kit (Qiagen). Quality of extracted RNA was examined by NanoDrop, and presence of RNase or DNase activity was checked by incubating the RNA with 500 ng plasmid DNA for 1 h at 37°C, followed by inspection of degradation on a EtBr-stained 1% agarose gel. cDNA was constructed from 15 μg purified RNA using Aminoallyl cDNA labeling kit (Applied Biosystems) and CyDye™ Post-Labeling Reactive Dye Pack (GE Healthcare). The labelled samples were hybridized to "Vibrio salmonicida V1.0.1 AROS" slides at 42°C on a TECAN HS4800 hybridisation station. Following the hybridizations, the slides were washed once in 0.1 × SSC/0.1% SDS for 5 min at 42°C, once in 0.1 × SSC/0.1% SDS for 10 min at room temperature, and finally four times in 0.1 × SSC for 1 min at room temperature. The slides were run in triplets, including one dye swap per triplet. Slides were scanned using a GenePix 4000B scanner (Axon Instruments Inc.) and GenePix Pro v6.1 software. The expression data were analysed using the LIMMA framework in Bioconductor http://www.bioconductor.org. Microarray data is available at the NCBI Gene Expression Omnibus (GEO) database with the GSE28087 as accession number.
Northern blot analyses
Total RNA was isolated from bacterial cultures using the TRIzol RNA isolation procedure (Invitrogen) and quantified by spectrophotometric methods. Northern blot analysis was done as previously described [12]. Briefly, 10 μg total RNA was separated on 1.2% formamide agarose gels or 8 M urea/5% polyacrylamide gels, and then transferred to a nylon membrane by upward capillary transfer or electroblotting, respectively. RNA species were detected on membranes using [α-32P]dCTP-labeled double-stranded DNA probes (PCR products), and signals were collected on phosphoimaging screens (Fujifilm) and scanned on a BAS-5000 phosphoimager (Fujifilm). ImageGauge v4.0 (Fujifilm) was used to measure the strength of signals, and 5S ribosomal RNA was used to normalize the resulting values.
Promoter and TFBS prediction
The region immediately upstream of spf was screened for CRP binding sites. A consensus sequence, made from 211 CRP binding sites (5'-WWWTGTGATCTRBRTCACAWWT-3') from RegulonDB [21], was used along with the pattern search algorithm Fuzznuc (part of EMBOSS software analysis package) [22]. A maximum of 6-9 mismatches of the 22 bp consensus were allowed. This predicted four overlapping sites stretching 44 nt in length. The CRP binding site (5'-TTTTGTGATGGCTATTAGAAAT-3') upstream of the E. coli spf gene [7,8] has 5 mismatches to this consensus. E.coli and A. salmonicida CRP binding sites share 12 conserved nt. In addition, the MEME [23] and Consensus [24] software tools were also used to search for any additional conserved sequence motifs in the promoter region of the vibrio Spot42 sequences. Both methods predicted a 42 nt conserved site overlapping the CRP site predicted from the known consensus. However, there is weak conservation among the predicted CRP sites in vibrios. The BPROM program (from http://www.softberry.com) was used to predict -10 and -35 promoter sequences.
Computational prediction of Spot42-pirin mRNA base-pairing
The program RNAhybrid [17] was used to identify potential base-pair interactions between Spot 42 and the 5' UTR region of the pirin mRNA (VSAL_I1200). The search was restricted to mRNA sequences located 100 nt upstream and 50 nt downstream of the start codon, with default program settings.
Authors' contributions
GÅH participated in the design of experiments, performed experiments and participated in drafting the manuscript. RA participated in design of experiments and helped draft the manuscript. Bioinformatics analyses of microarray data were done by CGF and EH. NPW and PH conceived the study and cosupervised the work. All authors read and approved the final manuscript.
Supplementary Material
Monitoring Spot 42 and VSsrna24 expression under different treatments or stress conditions with Northern blot analysis. A. salmonicida was grown to mid exponential phase (OD600nm 0.5) and split into nine smaller cultures. These were subjected to hexanoyl-L-homoserine lactone (HHL; 2 μg/ml), N-3-oxo-hexanoyl-L-homoserine lactone, (OHHL; 2 μg/ml), N, N'-dimethyl-4,4'-bipyridinium dichloride, (paraquat; 500 μM), hydrogen peroxide (H2O2; 1 mM), iron-chelator (2,2-dipyridyl; 50 μM), ethanol (EtOH; 0,5%), 4°C or 20°C, or used as control (16°C). Samples were harvested between 0-30 min, and after 8 and 20 hours (h) for the control culture. RNA samples were run on two gels and transferred to two membranes (Membranes 1 and 2) for practical reasons, and subjected to Northern blot analysis. Radio-labeled double-stranded DNA probes were used to monitor the levels of Spot 42 and VSsrna24, and 5S rRNA was used to normalize the result. None of the conditions resulted in significant changes in the expression pattern, except after addition of 50 uM 2,2'-dipyridyl which produced 2-5 fold reduction in Spot 42 levels.
Complete lists of differentially expressed genes. Microarray data from A. salmonicida spf deletion mutant grown in ASSM minimal medium and stimulated by added glucose.
Potential for base-pairing between Spot 42 and pirin mRNA. The RNAhybrid software was used to calculate the most energetically favorable base pairing between A. salmonicida Spot 42 (excluding the terminator stem) and the 5' region of the pirin mRNA (-100 nt to + 50 relative to the pirin start codon).
Contributor Information
Geir Å Hansen, Email: geir.a.hansen@uit.no.
Rafi Ahmad, Email: rafi.ahmad@uit.no.
Erik Hjerde, Email: erik.hjerde@uit.no.
Christopher G Fenton, Email: christopher.fenton@uit.no.
Nils-Peder Willassen, Email: nils-peder.willassen@uit.no.
Peik Haugen, Email: peik.haugen@uit.no.
Acknowledgements
This work was funded by grants from the Norwegian Research Council and the University of Tromsø. We wish to thank Brian Tjaden (Wellesley College) for help with sRNA target predictions, Henrik Nielsen (University of Copenhagen) for providing RNA protocols, Debra Milton for kindly providing the knock-out system, Sunniva K. Thode for assistance with microarray analysis, The Microarray Resource Centre in Tromsø (MRCT) for offering facilities and equipment to run the microarray experiments, and finally Hilde Hansen for her generous help with knock-out constructions.
References
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HY, Chang HY, Chou CH, Tseng CP, Ho SY, Yang CD, Ju YW, Huang HD. sRNAMap: genomic maps for small non-coding RNAs, their regulators and their targets in microbial genomes. Nucleic Acids Res. 2009. pp. D150–154. [DOI] [PMC free article] [PubMed]
- Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahagan BG, Dahlberg JE. A small, unstable RNA molecule of Escherichia coli: spot 42 RNA. I. Nucleotide sequence analysis. J Mol Biol. 1979;131(3):573–592. doi: 10.1016/0022-2836(79)90008-1. [DOI] [PubMed] [Google Scholar]
- Rice PW, Dahlberg JE. A gene between polA and glnA retards growth of Escherichia coli when present in multiple copies: physiological effects of the gene for spot 42 RNA. J Bacteriol. 1982;152(3):1196–1210. doi: 10.1128/jb.152.3.1196-1210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Joyce CM. Deletion of the spf (spot 42 RNA) gene of Escherichia coli. J Bacteriol. 1986;166(3):746–750. doi: 10.1128/jb.166.3.746-750.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP-cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol. 1988;170(7):3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polayes DA, Rice PW, Dahlberg JE. DNA polymerase I activity in Escherichia coli is influenced by spot 42 RNA. J Bacteriol. 1988;170(5):2083–2088. doi: 10.1128/jb.170.5.2083-2088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice PW, Polayes DA, Dahlberg JE. Spot 42 RNA of Escherichia coli is not an mRNA. J Bacteriol. 1987;169(8):3850–3852. doi: 10.1128/jb.169.8.3850-3852.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16(13):1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell. 2011;41(3):286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad R, Hjerde E, Hansen GA, Haugen P, Willassen NP. Prediction and experimental testing of ferric uptake regulator regulons in vibrios. J Mol Microbiol Biotechnol. 2009;16(3-4):159–168. doi: 10.1159/000128322. [DOI] [PubMed] [Google Scholar]
- Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67(17):2897–2908. doi: 10.1007/s00018-010-0381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polnisch E, Hofmann K. Cyclic AMP, fructose-2,6-bisphosphate and catabolite inactivation of enzymes in the hydrocarbon-assimilating yeast Candida maltosa. Arch Microbiol. 1989;152(3):269–272. doi: 10.1007/BF00409662. [DOI] [PubMed] [Google Scholar]
- Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editor. New York: Springer; 2005. Limma: linear models for microarray data. [Google Scholar]
- Soo PC, Horng YT, Lai MJ, Wei JR, Hsieh SC, Chang YL, Tsai YH, Lai HC. Pirin regulates pyruvate catabolism by interacting with the pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J Bacteriol. 2007;189(1):109–118. doi: 10.1128/JB.00710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006. pp. W451–454. [DOI] [PMC free article] [PubMed]
- Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178(5):1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J Bacteriol. 1985;164(2):918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG, Vogel J. Handbook of RNA Biochemistry. Vol. 1. WILEY-VCH, Verlag Gmbh & Co; 2005. [Google Scholar]
- Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, Garcia-Sotelo JS, Lopez-Fuentes A, RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011. pp. D98–105. [DOI] [PMC free article] [PubMed]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15(7-8):563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Monitoring Spot 42 and VSsrna24 expression under different treatments or stress conditions with Northern blot analysis. A. salmonicida was grown to mid exponential phase (OD600nm 0.5) and split into nine smaller cultures. These were subjected to hexanoyl-L-homoserine lactone (HHL; 2 μg/ml), N-3-oxo-hexanoyl-L-homoserine lactone, (OHHL; 2 μg/ml), N, N'-dimethyl-4,4'-bipyridinium dichloride, (paraquat; 500 μM), hydrogen peroxide (H2O2; 1 mM), iron-chelator (2,2-dipyridyl; 50 μM), ethanol (EtOH; 0,5%), 4°C or 20°C, or used as control (16°C). Samples were harvested between 0-30 min, and after 8 and 20 hours (h) for the control culture. RNA samples were run on two gels and transferred to two membranes (Membranes 1 and 2) for practical reasons, and subjected to Northern blot analysis. Radio-labeled double-stranded DNA probes were used to monitor the levels of Spot 42 and VSsrna24, and 5S rRNA was used to normalize the result. None of the conditions resulted in significant changes in the expression pattern, except after addition of 50 uM 2,2'-dipyridyl which produced 2-5 fold reduction in Spot 42 levels.
Complete lists of differentially expressed genes. Microarray data from A. salmonicida spf deletion mutant grown in ASSM minimal medium and stimulated by added glucose.
Potential for base-pairing between Spot 42 and pirin mRNA. The RNAhybrid software was used to calculate the most energetically favorable base pairing between A. salmonicida Spot 42 (excluding the terminator stem) and the 5' region of the pirin mRNA (-100 nt to + 50 relative to the pirin start codon).