Abstract
Background
The use of a multistarter fermentation process with Saccharomyces cerevisiae and non-Saccharomyces wine yeasts has been proposed to simulate natural must fermentation and to confer greater complexity and specificity to wine. In this context, the combined use of S. cerevisiae and immobilized Starmerella bombicola cells (formerly Candida stellata) was assayed to enhance glycerol concentration, reduce ethanol content and to improve the analytical composition of wine. In order to investigate yeast metabolic interaction during controlled mixed fermentation and to evaluate the influence of S. bombicola on S. cerevisiae, the gene expression and enzymatic activity of two key enzymes of the alcoholic fermentation pathway such as pyruvate decarboxylase (Pdc1) and alcohol dehydrogenase (Adh1) were studied.
Results
The presence of S. bombicola immobilized cells in a mixed fermentation trial confirmed an increase in fermentation rate, a combined consumption of glucose and fructose, an increase in glycerol and a reduction in the production of ethanol as well as a modification in the fermentation of by products. The alcoholic fermentation of S. cerevisiae was also influenced by S. bombicola immobilized cells. Indeed, Pdc1 activity in mixed fermentation was lower than that exhibited in pure culture while Adh1 activity showed an opposite behavior. The expression of both PDC1 and ADH1 genes was highly induced at the initial phase of fermentation. The expression level of PDC1 at the end of fermentation was much higher in pure culture while ADH1 level was similar in both pure and mixed fermentations.
Conclusion
In mixed fermentation, S. bombicola immobilized cells greatly affected the fermentation behavior of S. cerevisiae and the analytical composition of wine. The influence of S. bombicola on S. cerevisiae was not limited to a simple additive contribution. Indeed, its presence caused metabolic modifications during S. cerevisiae fermentation causing variation in the gene expression and enzymatic activity of alcohol deydrogenase and pyruvate decarboxilase.
Keywords: Multistarter fermentation, Saccharomyces cerevisiae, Starmerella bombicola, Immobilization, Real-time RT-PCR
Background
Wine fermentation is a complex process in which Saccharomyces and non-Saccharomyces yeasts can coexist and positively interact [1-7]. The control of spontaneous microflora involved during the winemaking process and the use of the inoculum of selected S. cerevisiae strains were considered to be fundamental steps to improve wine quality [8,9]. The use of a multistarter fermentation process with S. cerevisiae and non-Saccharomyces wine yeasts has been proposed to simulate natural must fermentation and to confer greater complexity and specificity to wine. The advantage of this process is to simulate a spontaneous process avoiding the risks of stuck fermentation [2,10-13]. Furthermore, non-Saccharomyces wine strains could have some specific enological characteristics that are absent in S. cerevisiae species, leading to combined, synergic and/or additive effects on the final wine [3,7,14-17].
In general, during a multistarter process, different microbial species live together, and the biotransformation of the nutritional sources is affected not only by the behavior of each microorganism but also by the interactions between different strains. In order to study this interaction, the hardest goal is to monitor the process which tries to understand the involvement of each individual yeast strain or its combined synergistic effect. In typical mixed-culture assays, the routine procedures used for tracking each strain are time-consuming, difficult and expensive [18]. For this reason, in order to monitor the behavior of each yeast during a multistarter experiment, some methods were based on cell separation by means of a porous membrane, while others were based on the cell immobilization technique [19,20].
In this work, we studied these interactions during the mixed fermentation of S. cerevisiae starter strain and a non-Saccharomyces enological yeast species, focusing our attention on S. cerevisiae during the most limiting steps of alcoholic fermentation: the decarboxylation of pyruvate to acetaldehyde and its subsequent reduction to ethanol. These steps are regulated by the expression of ADH and PDC genes. During S. cerevisiae growth on fermentable carbon sources, six PDC genes were identified out of which three structural genes (PDC1, PDC5 and PDC6) were encoded for active Pdc enzymes, independently [21]. These enzymes catalyze an irreversible reaction in which pyruvate is decarboxylated to acetaldehyde. Pdc1 is the predominant isoenzyme form (performing 80-90% of the activity in wild type cells). The regulatory genes PDC2, PDC3 and PDC4 encode proteins that are probably involved in the regulation of PDC1 and PDC5 expression. On the other hand, in S. cerevisiae there are five genes that encode alcohol dehydrogenases involved in ethanol metabolism, from ADH1 to ADH5. Four of these enzymes, Adh1p, Adh3p, Adh4p and Adh5p reduce acetaldehyde to ethanol during glucose fermentation, while Adh2p catalyzes the reverse reaction of oxidizing ethanol to acetaldehyde. The cytosolic ADH1 gene product is the major enzyme that is responsible for converting acetaldehyde to ethanol, and its transcription is repressed when cells are grown on a non-fermentable carbon source such as ethanol or glycerol.
In the present study, we explore the possible influence of a non-Saccharomyces yeast such as Candida stellata strain, recently reclassified as Starmerella bombicola [22], on the fermentation activity of S. cerevisiae during mixed fermentation. Trials were carried out using immobilized S. bombicola cells in order to confine non-Saccharomyces cells and to allow harvesting of S. cerevisiae cells, separately. In previous studies, the combined use of S. cerevisiae and S. bombicola had also been proposed in order to enhance glycerol content and their overall analytical profiles [23-25].
In this context, we evaluated the gene expression and enzymatic activities in S. cerevisiae strain as well as the fermentation products of the resulting wines.
Results and discussion
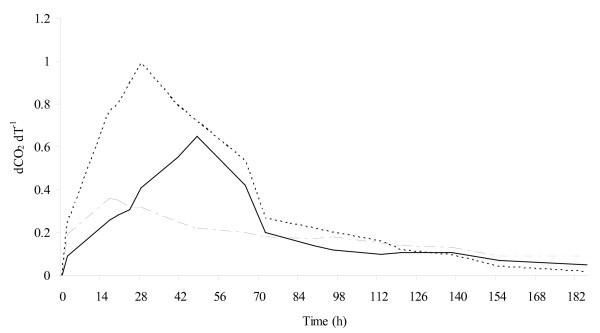
The evolution of the fermentation process is shown in Figure 1. As expected, the maximum fermentation rate (dCO2/dt) of mixed culture was higher than that of S. cerevisiae pure culture and it also reached its maximum level faster than S. cerevisiae pure culture (0.99 g l-1 h-1 at 28th h, 0.65 g l-1 h-1 at 48th h respectively). Indeed, immobilized cells of S. bombicola (10% wet/wt vol-1, corresponding to 1.3 × 108 cell ml-1), despite low fermentation activity, increased fermentation rate because of the high concentration of biomass [23]. Cell release from the beads was very low (< 103 cells ml-1), which indicated that the matrix was stable without any interference with S. cerevisiae cell harvest.
Figure 1.
Fermentation rate. Fermentation kinetics of pure S. cerevisiae culture (continous line), pure S. bombicola culture (mixed line) and mixed culture (dashed line). dCO2/dT, CO2 production rate (grams of CO2 evolved per litre per hour). Results are the mean of two independent biological trials, SD was less than 10%.
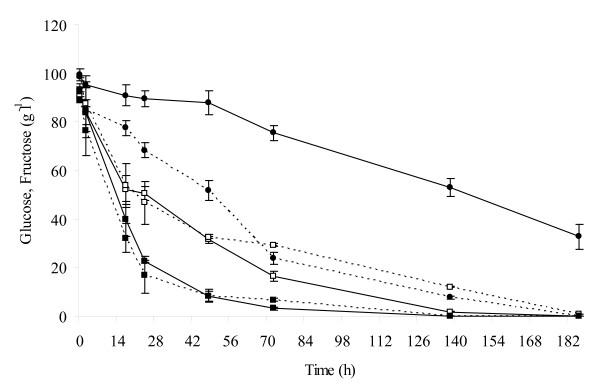
One of the most important features during wine production is the total consumption of sugar. Pure culture of S. cerevisiae consumed glucose and fructose at almost the same rate only during the first 48 hours of fermentation (Figure 2). After that, S. cerevisiae pure culture began to consume glucose faster than fructose determining the total use of sugars at 186th h (glucophilic yeast). Immobilized cells of fructophilic yeast S. bombicola [23], showed opposite behaviour consuming fructose faster than glucose (Figure 2). In fact, fructose was completely consumed at 186th h, while glucose was consumed slowly. Mixed culture fermentation showed that the combined use of S. cerevisiae and immobilized S. bombicola cells leads to contemporary, fast and complete consumption of both sugars (at 138th h).
Figure 2.
Consumption of sugars. Progress of glucose (continuous lines) and fructose (dashed lines) consumption throughout fermentation in mixed culture (■), in S. cerevisiae pure culture (□) and in immobilized S. bombicola pure culture (●).
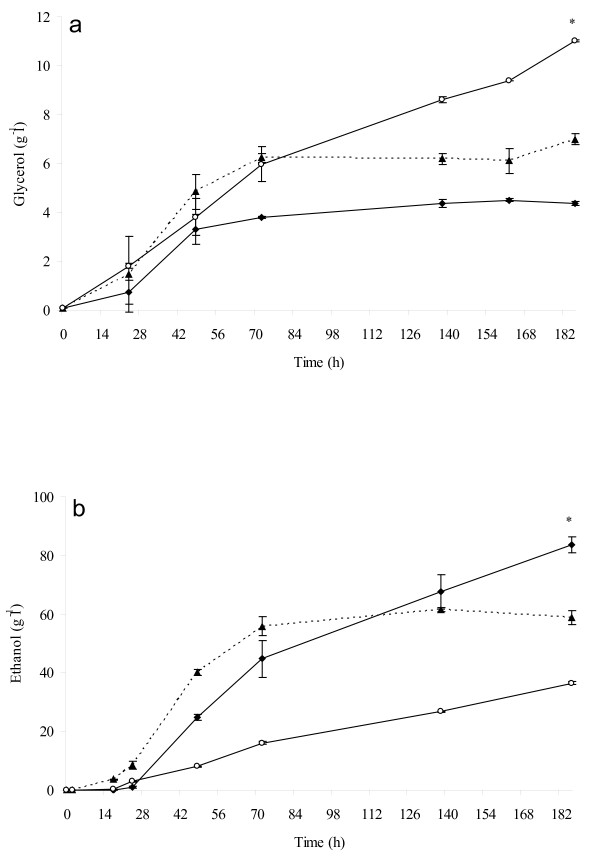
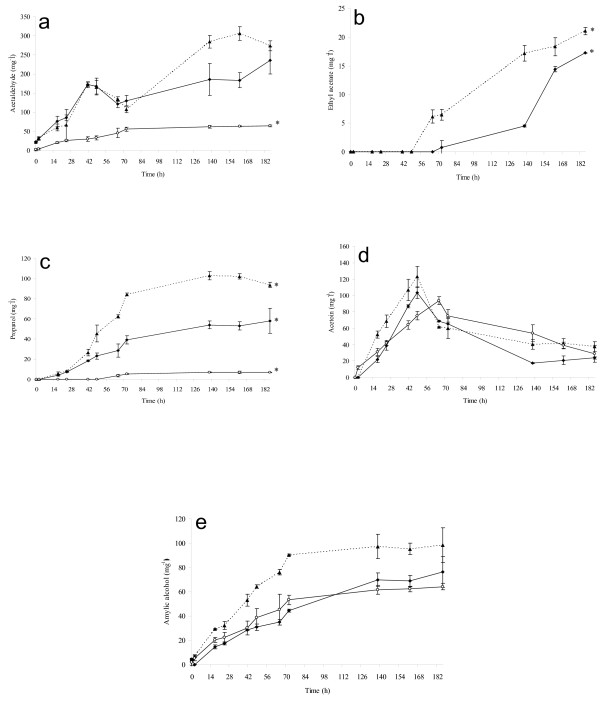
As expected, pure culture of S. bombicola produced high quantity (11 g l-1) of glycerol, confirming previous results [24,25] (Figure 3a). Mixed culture produced glycerol faster and higher than S. cerevisiae pure culture (7.0 g l-1, 4.4 g l-1 respectively) indicating that immobilized S. bombicola cells positively affect the final amounts of glycerol. Evolution of ethanol showed different kinetics in mixed and pure fermentations (Figure 3b). Immobilized cells of S. bombicola in pure culture produced the smallest amount of ethanol (36.5 g l-1). During the first 72 h of fermentation, mixed culture produced ethanol faster and in higher concentration than that exhibited by S. cerevisiae pure culture, remaining stable until the end of fermentation (58.9 g l-1). Pure culture of S. cerevisiae showed a lower trend of ethanol production, but at the end of fermentation, its concentration was higher than that exhibited by mixed culture (83.6 g l-1). This is a very interesting behavior since the reduction of final ethanol concentration in winemaking is one of the most investigated topics [26-28]. The principal by-products in mixed fermentation were produced mainly due to the metabolic activity of S. cerevisiae strain (Figure 4). Actually, the trend of these compounds is closely related to that showed by S. cerevisiae pure culture, while immobilized S. bombicola cells showed a significantly lower production of acetaldehyde, ethyl acetate and n-propanol (Figures 4a, b and 4c). Mixed fermentation exhibited a significantly higher amount of n-propanol and ethyl acetate (Figures 4b and 4c) even if the final concentration turned out to be far from the sensory threshold level. As regards acetic acid production, we noted that there was a lower level of acetic acid in the mixed culture than that in the pure culture of S. cerevisiae (0.82 g l-1, 0.94 g l-1 respectively), while the pure culture of S. bombicola immobilized cells produced only a small amount of acetic acid (0.23 g l-1).
Figure 3.
Glycerol and ethanol production. Evolution of glycerol (a) and ethanol (b) during fermentation carried out by mixed culture (▲ dashed lines), pure S. cerevisiae culture (♦ continuous line) and immobilized S. bombicola pure culture (○ continuous lines). The data represented are the mean of three technical repetitions for two independent biological samples ± SD. Asterisk represents significantly different values according to the Duncan test (0.05%).
Figure 4.
Fermentation by-products. By products produced during wine fermentation in mixed culture (▲ dashed lines), in S. cerevisiae pure culture (♦continuous lines) and in immobilized S. bombicola pure culture (○ continuous lines). Production of (a) acetaldehyde, (b) ethyl acetate, (c) propanol, (d) acetoin and (e) amylic alcohols. The data represented are the mean of the three technical repetitions for two independent biological samples ± SD. Values displaying asterisk are significantly different according to the Duncan test (0.05%).
The overall results of the metabolic interactions between S. cerevisiae and S. bombicola immobilized cells in mixed culture confirm their strong influence on fermentation rate, glycerol, ethanol production and utilization of sugars. At the same time, a reduced influence on the main by-products was also seen. In this context, we investigated the influence of S. bombicola on the alcoholic fermentation of S. cerevisiae evaluating pyruvate decarboxylase and alcohol deydrogenase gene expression and activity. These are two key enzymes of the alcoholic fermentation process. Pyruvate decarboxylase irreversibly converts pyruvate into acetaldehyde and CO2 , and its gene transcription is higher in cells grown on glucose than that grown on ethanol [29-36], while alcohol deydrogenase 1 is the main cytosolic enzyme involved in the formation of ethanol during glycolysis [37].
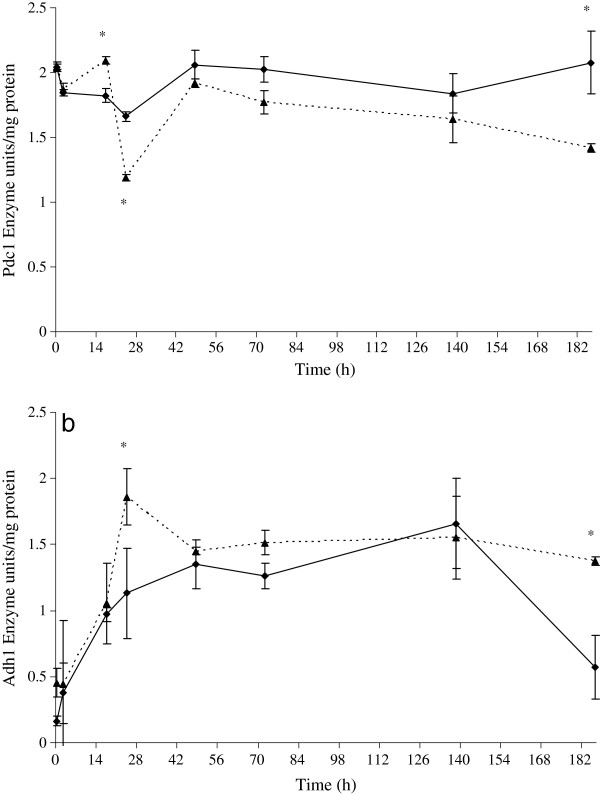
The activity trends of Pdc1 and Adh1 of S. cerevisiae during fermentation in pure and mixed cultures are shown in Figures 5a and 5b. Pdc1 enzyme in mixed fermentation at 17th h showed maximum activity (Figyre. 5a). After this time, the activity of Pdc1 in mixed culture decreased quickly (24th h), then rose again remaining stable and low in comparison with pure culture (Figure 5.5b). On the other hand, the behavior of enzymatic activity in pure culture trial was quite stable during the first stage of fermentation.
Figure 5.
Enzymatic activity. Enzymatic activity of Pdc1 (a) and Adh1 (b) in S. cerevisiae at different stages of fermentation. Dashed lines represent mixed culture, and continuous lines, pure S. cerevisiae culture. Error bars show standard deviations including three technical repetitions for two independent biological samples. Values displaying asterisk are significantly different according to the Duncan test (0.05%).
Adh1 activity of S. cerevisiae in mixed fermentation was higher than that exhibited by pure culture during the whole fermentation process. After 24 h, it reached its maximum level (72% greater than pure culture) and, after a decrease, remained stable until the end of the fermentation process. In pure culture, there was a progressive increase until the maximum level at 138th h and a notable decrease at the end point (81% lower than in mixed culture). Interestingly, there was an opposite trend of these two enzymes in mixed fermentation, i.e., when Adh1 activity was at its peak, Pdc1 activity was at its minimum.
Since both ADH1 and PDC1 genes encode these two enzymes that catalyze the synthesis of ethanol from pyruvate during alcoholic fermentation, they were expected to be highly expressed under wine fermentation conditions [38,39].
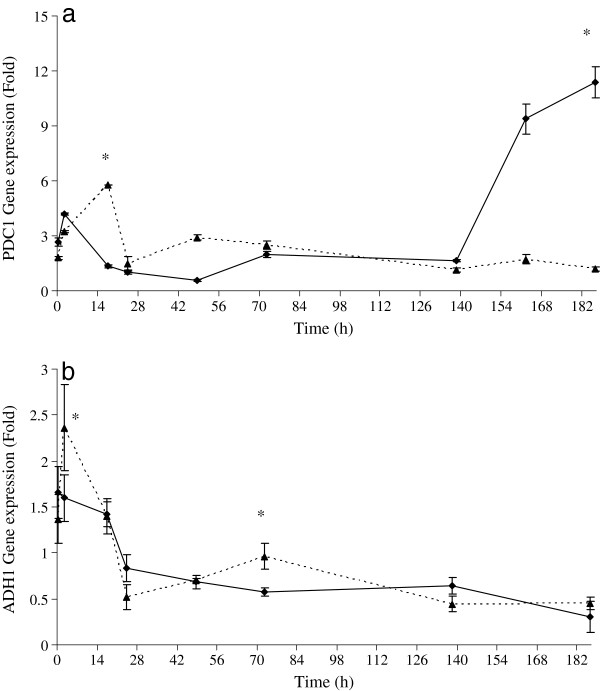
Results of gene expression are showed in Figure 6 where, during the first 72 h, PDC1 expression in mixed culture was higher than that exhibited by pure culture reaching its maximum level at 17th h (5.8 and 1.4 folds, respectively). Subsequently, the expression of PDC1 in mixed culture was stable until the end of fermentation, while in pure culture, a fast enhancement during the final step of fermentation (11.3 folds) was seen. This behavior could be explained by the presence of fructose (ca. 12 g l-1) in this last stage of fermentation (Figure 2) that determined an improvement in gene transcription level. Similar results were reported by Molina and co-workers [40] where, at 60-80% of sugar consumption, a peak of PDC1 expression in synthetic must was noted.
Figure 6.
Gene expression. Expression levels of PDC1 (a) and ADH1 (b) in S. cerevisiae at different stages of fermentation. Dashed lines represent mixed culture, and continuous lines, pure S. cerevisiae culture. Error bars show standard deviations, including three technical repetitions for two independent biological samples. Relative normalized fold expression is calculated using TAF10 as reference gene. Values displaying asterisk are significantly different according to the Duncan test (0.05%).
S. cerevisiae ADH1 for both pure and mixed fermentation was highly induced at the initial step. At the 2nd h, the expression level of this gene quickly increased in mixed fermentation, and it was higher than that seen in pure culture (2.36 and 1.52 folds, respectively). This maximum expression level was followed by a rapid down regulation during the following 22 hours (from 2.36 to 0.52 folds). Furthermore, in previous studies, it has been reported that the biosynthesis of Adh1 takes place basically during the adaptation of the yeasts to the musts (first 4 h) [41,42]. Subsequently, this gene in mixed culture slightly increased the expression level until 72th, to remain constant at 0.44 fold. Similarly, ADH1 expression of pure culture decreased in the first 24 hours and after which it remained stable towards the end of fermentation (0.3 fold).
Similar expression profiles observed for PDC1 and ADH1 strongly support the hypothesis that the enzymes Pdc1 and Adh1 might be co-regulated and participate in the same metabolic pathway [40].
Present results indicate that the activity and gene expression of pyruvate decarboxylase and alcohol deydrogenase in S. cerevisiae are influenced in mixed culture by S. bombicola. The metabolic interactions in mixed yeast culture of S. bombicola immobilized cells and S. cerevisiae under winemaking conditions are complex that lead the modification of the kinetics of fermentation and analytical profile of wine, influencing the activity and gene expression of pyruvate decarboxilase and alcohol deydrogenase.
Conclusions
In mixed fermentation, S. bombicola immobilized cells greatly influenced the fermentation behaviour of S. cerevisiae and the analytical composition of wine. The influence of S. bombicola seems to be very complex and not limited to a synergistic or additive effect on the analytical profile of wines. Metabolic modifications in S. cerevisiae alcoholic fermentation were observed since significant modification of alcohol dehydrogenase and pyruvate decarboxylase gene expression and enzymatic activity was exhibited. Metabolic modifications in S. cerevisiae coupled with S. bombicola immobilized cells showed an enhancement of both pyruvate decarboxylase activity and gene expression during the first stage of fermentation (2-17 h). A similar behavior was shown for alcohol dehydrogenase gene expression, and enzymatic activity also exhibited the same enhancement but with a delay of 7 hours (at 24th h).
This study is the first attempt to investigate yeast metabolic interaction which monitors gene expression and enzymatic activity during controlled mixed fermentation in winemaking.
Methods
Microorganisms
Yeast strains used in this work were commercial strain Lalvin EC1118 Saccharomyces cerevisiae and Starmerella bombicola (formerly Candida stellata) DBVPG 3827 strain coming from the Industrial Yeasts Collection of the Dipartimento di Biologia Vegetale, University of Perugia (DBVPG).
Media
Synthetic grape juice (SGJ) was used in fermentation tests. Each litre of SGJ was composed of three different solutions: solution A (500 ml), solution B (250 ml), and solution C (250 ml). The composition of SGJ was as follows (per litre): solution A: D-glucose, 100 g; D-fructose, 100 g; ergosterol, 10 mg; Tween80, 1 ml; solution B: L-(+)-tartaric acid, 6.0 g; L-(-)-malic acid, 2.0 g; citric acid, 0.5 g; solution C: YNB (yeast nitrogen base without amino acids and ammonium sulfate) (Difco), 1.7 g; CAA (vitamin-free Casamino Acids) (Difco), 2.0 g; CaCl2, 0.2 g; arginine-HCl, 0.8 g; L-(2)-proline, 1.0 g; L-(2)-tryptophan, 0.1 g. Solutions B and C were buffered at pH 3.5 with NH4OH and H3PO4, respectively. Four millilitres of Ergosterol stock solution (Tween 80, 6.25 ml; Ergosterol, 62.5 mg in ethanol to make 25 ml) was added to the glucose-fructose solution to complete solution A. All three solutions were sterilized at 121°C for 20 min separately and then combined aseptically. YPD medium (2% glucose, 2% peptone and 1% yeast extract; all w/v) was used to produce biomass for the immobilization system.
Fermentation conditions and sampling
In order to investigate the influence of immobilized S. bombicola on S. cerevisiae fermentation activity, fermentations in mixed cultures were set up together with pure culture fermentations of both strains as control. The basic principle of immobilization consists in keeping S. bombicola cells separate from free S. cerevisiae cells which can be sampled and analyzed separately. Metabolites from both microorganisms are allowed to pass through the substrate even if two different yeasts grow separately. Duplicate fermentations were carried out in 1-litre glass minifermentors (containing 500 ml of SGJ under static conditions at 25°C) with two ports, one for gas flow and the other for an inoculum of beads, and a septum of frit glass in order to maintain the beads in the medium and to allow carbon dioxide to come out. Cells for immobilization were grown in YPD at 25°C in a rotary shaker (150 rpm) for 72 h (S. bombicola), harvested by centrifugation, washed three times with sterile distilled water and added to 2.5% Na-alginate (Carlo Erba, Milan, Italy) at a ratio of 5% (wet weight vol-1) (biomass moisture, 70%; final concentration, 1.3 × 109 cells per g of beads). By means of a peristaltic pump, this mixture was then dripped into CaCl2 (0.1 M) to induce gelation. After 1 h, the beads were washed several times with sterile distilled water and used immediately. The inoculum for immobilized cells of S. bombicola was 10% (wet weight/vol) of the amount of beads in the medium (corresponding to 1.3 × 108 cells per ml). Yeast culture of S. cerevisiae was pre-incubated in SGJ at 25°C in a rotary shaker (150 rpm) for 48 h, harvested by centrifugation, washed with sterile distilled water, and the procedure was standardized to provide an inoculation level of 106 cells/ml. Before the inoculum and, at the end of fermentation in mixed and pure culture, 10 g of beads were maintained under agitation in 100 ml 1% Na-citrate solution (w/v) for 1 h to release the cells, and cell viability was evaluated by the standard plate count techniques in Lysine medium. The evolution of fermentations was evaluated gravimetrically by weight loss due to the carbon dioxide evolved. Samples of the culture medium were taken at different stages of fermentation from each minifermentor. One part of all the samples was used to determine cell number by light microscopy using a Thoma-Zeiss counting chamber, and optical density was measured at 600 nm (OD600). The other part of each sample was centrifuged for 5 min at 2000 g. Supernatants were filtered through a 15 mm syringe filter Phenex (0.2 μm pore diameter, Phenomenex, Torrence CA, USA), stored at -20°C and analyzed later to determine residual sugars, ethanol, glycerol, acetaldehyde, ethyl acetate, propanol, acetoin and amylic and iso-amylic alcohols concentration. The cell pellet for RNA and protein extraction was mixed with glycerol and conserved at -80°C until use.
Analytical determinations
Ethanol was measured by a gas-liquid chromatography (GLC) analysis [43]. Acetaldehyde, ethyl acetate, acetoin and higher alcohols were determined by direct injection into the GLC system. The samples were injected into a 30 m by 0.32 mm, 0.25 μm film thickness column Zebron ZB-WAXPlus (Phenomenex, Torrance, California, USA) with an internal standard of 1-penthanol (162 mg l-1). Nitrogen was used as the carried gas. A Shimadzu gas chromatograph (Japan), equipped with a flame ionization detector, was used. The oven temperature ranged from 40°C to 200°C. The temperature of the injector and the detector was 150°C.
Glucose, fructose (kit no. 139106), and glycerol concentration (kit no. 148270) were determined by using specific enzymatic kits (Boehringer, Mannheim, Germany). Volatile acidity (expressed as grams of acetic acid per litre) was quantified by steam distillation according to the official analytical methods [44].
RNA isolation and cDNA synthesis
Total RNA was extracted using the NucleoSpin RNAII kit (Macherey-Nagel, Düren, Germany) from samples containing approximately 107 cells following the protocol provided by the manufacturer. RNA concentration was determined using Nanodrop ND 1000 (Thermo Fisher scientific, Wilmington, DE, USA), and RNA quality was tested by electrophoresis on 1.5% agarose gel.
cDNA was synthesised from the isolated RNA by the Maxima First Strand cDNA Synthesis kit for RT-qPCR (Fermentas) as recommended by the manufacturer using oligo (dT)18 and random hexamer primers to prime synthesis of first strand cDNA.
Primer design
In order to identify the housekeeping and target genes of S. cerevisiae coding for selected proteins, we used the Genome database available at NCBI (http://www.ncbi.nlm.nih.gov). TAF10 gene (RNA Pol II transcription factor activity/transcription initiation and chromatin modification) was used as housekeeping reference because it turned out to be one of the genes whose expression remained stable, independent of the growth conditions as highlighted by Teste and co-workers [45].
Real-time PCR primers were designed using the OligoAnalyzer 3.0 software (available at http://eu.idtdna.com/analyzer/) and synthesised by MWG Biotech (MWG Biotech, Germany). Annealing temperature of all primer pairs was 60°C. Amplification efficiency was determined by the serial dilution method beginning from a cDNA pool [46], and PCR efficiency was calculated by the equation E = 10[-1/slope]. Each primer was tested on agarose gel to verify its specificity. Table 1 shows the primer sequences used in this study along with their amplicon sizes, and their resulting efficiencies. Primers have been checked for absence of cross-amplification using S. bombicola cDNA as template.
Table 1.
PCR primers used in this study
| Gene | Sequence (5'-3') | Amplicon length (bp) |
Primer efficiency |
|---|---|---|---|
| ADH1 | ATCCAACTGTCCTCACGCTGACTT | 104 | 1.01 |
| TACCTTGAGGAATGTGAGCGGCTT | |||
| PDC1 | TGTCGAATTCCACTCCGACCACAT | 114 | 1.00 |
| TAACCCTTAGCGGCGTCAGCAATA | |||
| TAF10 | GCAGCTATTGCAAGGACAGCAACA | 142 | 1.08 |
| ATTGAGCCCGTATTCAGCAACAGC | |||
Gene expression analysis by real time PCR
The expression levels were determined using real time PCR. All real-time reactions were performed using Mastercycler® ep realplex (Eppendorf, Hamburg, Germany). A ready-to-use RealMasterMix SYBR ROX 2.5X (5 Prime, Hamburg, Germany) was used according to the manufacturer's instructions. Reactions were performed in 150 μl twin.tec real time PCR plates 96 (Eppendorf), and each 10 μl reaction mixture contained: 200 nM of each primer, 4.5 μl of 2.5X RealMasterMix SYBR ROX, 4 μl of cDNA and H2O to reach a final volume. All real time PCR experiments were carried out using two biological repetitions, and the samples were considered in triplicate. A negative control without a cDNA template was included to ensure that the samples did not have any unspecific SYBR Green fluorescence. The program used was the follow: 95°C for 2 min; 40 cycles of 95°C for 20 s, 20 s at 55°C, 68°C for 30 s; 1 cycle of 68°C for 1 min. After this, melt curve data were then collected. Gene expression levels are shown as the concentration of the studied gene normalized with the concentration of the housekeeping TAF10 gene.
Enzyme assay
Whole-cell homogenates were prepared following the method by Blumer and co-workers [47]. Protein concentrations of cell extracts were determined by the Lowry method with a bovine serum albumin (Sigma) as the standard. Enzyme assays were performed at 25°C with a Shimadzu UV1800 spectrophotometer at 340 nm. Reaction rates were linearly proportional to the amount of cell extract added. One unit (U) is defined as the amount of enzyme catalyzing the conversion of one micromole of substrate min-1. Specific activity is expressed as U per mg of protein. The assay mixture for each enzyme is described below.
Alcohol dehydrogenase (NAD-dependent, ADH1 EC.1.1.1.1). The assay mixture contained 60 mM sodium pyrophosphate buffer (pH 8.5), 100 mM ethanol and 50 mM NAD+. The reaction was started with a cell extract.
Pyruvate decaroxylase (PDC1 EC.4.1.1.1). The reaction mixture contained 40 mM imidazole HCl buffer pH (6.5), 5 mM MgCl2, 0.2 mM thiamine pyrophosphate (TPP), 0.15 mM NADH, 88 U of alcohol dehydrogenase and a cell extract. The reaction was started with 50 mM pyruvate.
Statistical analysis
Analysis of variance (ANOVA) was applied to the experimental data for the by-products evaluated during pure and mixed fermentations. The means were analyzed using the SuperANOVA software, version 1.1, for Mac OS 9.1. The significant differences were determined by means of the Duncan test, and the results were considered significant if the associated P values were below 0.05.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
VM, LO, MC and FC contributed equally to this manuscript. All authors participated in the design and discussion of the research. VM and LO carried out the experimental part of the work. VM, LO, MC and FC carried out the analysis of the data and wrote the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Vesna Milanovic, Email: v.milanovic@univpm.it.
Maurizio Ciani, Email: m.ciani@univpm.it.
Lucia Oro, Email: l.oro@univpm.it.
Francesca Comitini, Email: f.comitini@univpm.it.
References
- Nissen P, Nielsen D, Arneborg N. Viable Saccharomyces cerevisia cells at high concentrations cause early growth arrest of non-Saccharomyce yeasts in mixed cultures by a cell-cell contact mediated mechanism. Yeast. 2003;20:331–341. doi: 10.1002/yea.965. [DOI] [PubMed] [Google Scholar]
- Ciani M, Beco L, Comitini F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int J Food Microbiol. 2006;108:239–245. doi: 10.1016/j.ijfoodmicro.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Bely M, Stoeckle P, Masnuef-Pomarède I, Dubourdieu D. Impact of mixed Torulaspora delbruecki-Saccharomyces cerevisia culture on high-sugar fermentation. Int J Food Microbiol. 2008;122:312–320. doi: 10.1016/j.ijfoodmicro.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Toro ME, Vazquez F. Fermentation behaviour of controlled mixed and sequential cultures of Candida cantarelli and Saccharomyces cerevisia wine yeasts. World J Microb Biot. 2002;18:347–354. [Google Scholar]
- Moreira N, Mendes F, Hogg T, Vasconcelos I. Alcohols, esters and heavy sulphur compound production by pure and mixed cultures of apiculate wine yeasts. Int J Food Microbiol. 2005;103:285–294. doi: 10.1016/j.ijfoodmicro.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Garcia A, Carcel C, Dalau L, Samson A, Aguera E, Agosin E, Gunata Z. Influence of a mixed culture with Debaromyces vanrij and Saccharomyces cerevisia on the volatiles in a Muscat wine. J Food Sci. 2002;67:1138–1143. doi: 10.1111/j.1365-2621.2002.tb09466.x. [DOI] [Google Scholar]
- Comitini F, Gobbi M, Domizio P, Romani C, Lencioni L, Mannazzu I, Ciani M. Selected non-Saccharomyce wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisia. Food Microbiol. 2011;28:873–882. doi: 10.1016/j.fm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Jolly NP, Augustyn OPH, Pretorius IS. The role and use of non-Saccharomyce yeasts in wine production. S Afr J Enol Vitic. 2006;27:15–38. [Google Scholar]
- Soden A, Francis IL, Oakey H, Henschke PA. Effect of co-fermentation with Candida stellat and Saccharomyces cerevisia on the aroma and composition of Chardonnay wine. Aust J Grape Wine Res. 2000;6:21–30. doi: 10.1111/j.1755-0238.2000.tb00158.x. [DOI] [Google Scholar]
- Ciani M, Comitini F, Mannazzu I, Domizio P. Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyce yeasts in winemaking. FEMS Yeast Res. 2010;10:123–133. doi: 10.1111/j.1567-1364.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- Mendoza LM, Manca de Nadra MC, Farìas ME. Kinetics and metabolic behavior of a composite culture of Kloeckera apiculat and Saccharomyces cerevisia wine related strains. Biotechnol Lett. 2007;29:1057–1063. doi: 10.1007/s10529-007-9355-0. [DOI] [PubMed] [Google Scholar]
- Anfang N, Brajkovich M, Goddard MR. Co-fermentation with Pichia kluyver increases varietal thiol concentrations in Savignon Blanc. Aust J Grape Wine Res. 2009;15:1–8. doi: 10.1111/j.1755-0238.2008.00031.x. [DOI] [Google Scholar]
- Languet P, Aguera E, Samson A, Ortiz-Julien A, Salmon JM. Valorization aromatique des mouts par l'utilisation séquentielle des levures d'espécies non-Saccharomyce et Saccharomyce. Rev Oenolog. 2005;117:31–33. [Google Scholar]
- Strauss MLA, Jolly NP, Lambrechts MG, van Rensburg P. Screening for the production of extracellular hydrolytic enzymes by non-Saccharomyce wine yeasts. J Appl Microbiol. 2001;91:182–190. doi: 10.1046/j.1365-2672.2001.01379.x. [DOI] [PubMed] [Google Scholar]
- Viana F, Gil JV, Genovés S, Vallés S, Manzanares P. Rational selection of non-Saccharomyce wine yeasts for mixed starters based on ester formation and enological traits. Food Microbiol. 2008;25:778–785. doi: 10.1016/j.fm.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Domizio P, Romani C, Lencioni L, Comitini F, Gobbi M, Mannazzu I, Ciani M. Outlining a future for non-Saccharomyce yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisia for grape juice fermentation. Int J Food Microbiol. 2011;147:170–180. doi: 10.1016/j.ijfoodmicro.2011.03.020. [DOI] [PubMed] [Google Scholar]
- Salgado Manjarrez E, Albasi CY, Riba JP. A two-reservoir, hollow fiber bioreactor for the study of mixed population dynamics: Design aspects and validation of the approach. Biotechnol Bioeng. 2000;69(Suppl 4):401–408. doi: 10.1002/1097-0290(20000820)69:4<401::aid-bit6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Pestchanker LJ, Ercoli EC. A novel membrane reactor design for the controlled studies of interacting populations (Simulation of the interaction between microorganism and plant suspension cultures) Biotechnol Bioeng. 1997;55:609–615. doi: 10.1002/(SICI)1097-0290(19970820)55:4<609::AID-BIT3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Verbelen PJ, De Schutter DP, Delvaux F, Verstrepen KJ, Delvaux FR. Immobilized yeast cell systems for continuous fermentation. Biotechnol Lett. 2006;28:1515–1525. doi: 10.1007/s10529-006-9132-5. [DOI] [PubMed] [Google Scholar]
- Hohmann S, Cederberg H. Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC and PDC. Eur J Biochem. 1990;188(Suppl 3):615–621. doi: 10.1111/j.1432-1033.1990.tb15442.x. [DOI] [PubMed] [Google Scholar]
- Šipiczki M, Ciani M, Csoma H. Taxonomic reclassification of Candida stellat DBVPG 3827. Folia Microbiol. 2005;50(Suppl 6):494–498. doi: 10.1007/BF02931436. [DOI] [PubMed] [Google Scholar]
- Ciani M, Ferraro L. Enhanced glycerol content in wines made with immobilized Candida stellat cells. Appl Environ Microbiol. 1996;62:128–132. doi: 10.1128/aem.62.1.128-132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M, Ferraro L. Combined use of immobilized Candida stellat cells and Saccharomyces cerevisia to improve the quality of wines. J Appl Microbiol. 1998;85:247–254. doi: 10.1046/j.1365-2672.1998.00485.x. [DOI] [PubMed] [Google Scholar]
- Ferraro L, Fatichenti F, Ciani M. Pilot scale vinification process using immobilized Candida stellat cells and Saccharomyces cerevisia. Process Biochem. 2000;35:1125–1129. doi: 10.1016/S0032-9592(00)00148-5. [DOI] [Google Scholar]
- Pickering GJ. Low and reduced-alcohol wine: A review. J Wine Res. 2010;11(Suppl 2):129–144. [Google Scholar]
- Kutyna DR, Varela C, Henschke PA, Chambers PJ, Stanley GA. Microbiological approaches to lowering ethanol concentration in wine. Trends Food Sci Tech. 2010;21:293–302. doi: 10.1016/j.tifs.2010.03.004. [DOI] [Google Scholar]
- Scott JA, Huxtable SM. Removal of alcohol from beverages. J Appl Bacteriol. 1995;79:19–28. [Google Scholar]
- Schmitt HD, Zimmermann FK. Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Biotechnol. 1982;151:1146–1152. doi: 10.1128/jb.151.3.1146-1152.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt HD, Ciriacy M, Zimmermann FK. The synthesis of yeast pyruvate decarboxylase is regulated by large variations in the messenger RNA level. Mol Gen Genet. 1983;192:247–252. doi: 10.1007/BF00327674. [DOI] [PubMed] [Google Scholar]
- Seeboth PG, Bohnsack K, Hollenberg CP. pd(0) mutants of Saccharomyces cerevisia give evidence for an additional structural PD gene: cloning of PDC5 a gene homologous to PDC. J Bacteriol. 1990;172:678–685. doi: 10.1128/jb.172.2.678-685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. PDC, a weakly expressed pyruvate decarboxylase gene from yeast, is activated when fused spontaneously under the control of the PDC promoter. Curr Genet. 1991;20:373–378. doi: 10.1007/BF00317064. [DOI] [PubMed] [Google Scholar]
- Schellenberger A, Hubner G. Binding of the substrate in yeast pyruvate decarboxylase. Angew Chem Int Edit Eng. 1968;7:68–69. doi: 10.1002/anie.196800682. [DOI] [PubMed] [Google Scholar]
- Hubner G, Weidhase R, Schellenberger A. The mechanism of substrate activation of pyruvate decarboxylase, a first approach. European Journal of Biotechnology. 1978;92:175–181. doi: 10.1111/j.1432-1033.1978.tb12735.x. [DOI] [PubMed] [Google Scholar]
- Lu G, Dobritzch D, Baumann S, Schneider G, Konig S. The structural basis of substrate activation in yeast pyruvate decarboxylase: a crystallographic and kinetic study. European Journal of Biochemistry. 2000;267:861–868. doi: 10.1046/j.1432-1327.2000.01070.x. [DOI] [PubMed] [Google Scholar]
- Sergienko EA, Jordan F. New model for activation of yeast pyruvate decarboxylase by substrate consistent with the alternating sites mechanism. Demonstration of the existence of two active forms of the enzyme. Biochemistry. 2002;41:3952–3967. doi: 10.1021/bi011860a. [DOI] [PubMed] [Google Scholar]
- Bakker B, Bro C, Kotter P, Luttik MAH, van Dijken JP, Pronk JT. The mitochondrial alcohol dehydrogenase Adh3 is involved in a redox shuttle in Saccharomyces cerevisia. J Biotechnol. 2000;182:4730–4737. doi: 10.1128/jb.182.17.4730-4737.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton R, Singleton V, Bisson L, Kunkee R. Principles and practices of winemaking. New York: Chapman and Hall; 1996. [Google Scholar]
- Rossignol T, Dulau L, Julien A, Blondin B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast. 2003;20(Suppl 16):1369–1385. doi: 10.1002/yea.1046. [DOI] [PubMed] [Google Scholar]
- Molina AM, Swiegers JH, Varela C, Pretorius IS, Agosin E. Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl Microbial Biotechnol. 2007;77:675–687. doi: 10.1007/s00253-007-1194-3. [DOI] [PubMed] [Google Scholar]
- Mauricio JC, Moreno JJ, Ortega JM. In vitr specific activities of alcohol and aldehyde dehydrogenases from two flour yeasts during controlled wine aging. J Agric Food Chem. 1997;45(Suppl 5):1967–1971. [Google Scholar]
- Salvadó Z, Chiva R, Rodríguez-Vargas S, Rández-Gil F, Mas A, Guillamón JM. Proteomic evolution of a wine yeast during the first hours of fermentation. FEMS Yeast Res. 2008;8:1137–1146. doi: 10.1111/j.1567-1364.2008.00389.x. [DOI] [PubMed] [Google Scholar]
- AOAC. Official Methods of Analysis of the Association of Official Analytical Chemists. 15. II. The Association: Arlington, VA; 1990. Sec.985.29. [Google Scholar]
- EC, December 19, 2000. Community reference methods for the analysis of spirit drinks. Off J Eur Commun Reg. EC No 2870/00. Brussels.
- Teste MA, Duquenne M, Francois JM, Parrou JL. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisia. BMC Mol Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer KJ, Reneke JE, Thorner J. The STE2 gene product in the ligand-bilding component on the a-factor receptor of Saccharomyces cerevisia. J Biol Chem. 1988;263:10836–10842. [PubMed] [Google Scholar]