Abstract
Mitochondrial genes in animals are especially useful as molecular markers for the reconstruction of phylogenies among closely related taxa, due to the generally high substitution rates. Several insect orders, notably Hymenoptera and Phthiraptera, show exceptionally high rates of mitochondrial molecular evolution, which has been attributed to the parasitic lifestyle of current or ancestral members of these taxa. Parasitism has been hypothesized to entail frequent population bottlenecks that increase rates of molecular evolution by reducing the efficiency of purifying selection. This effect should result in elevated substitution rates of both nuclear and mitochondrial genes, but to date no extensive comparative study has tested this hypothesis in insects. Here we report the mitochondrial genome of a crabronid wasp, the European beewolf (Philanthus triangulum, Hymenoptera, Crabronidae), and we use it to compare evolutionary rates among the four largest holometabolous insect orders (Coleoptera, Diptera, Hymenoptera, Lepidoptera) based on phylogenies reconstructed with whole mitochondrial genomes as well as four single-copy nuclear genes (18S rRNA, arginine kinase, wingless, phosphoenolpyruvate carboxykinase). The mt-genome of P. triangulum is 16,029 bp in size with a mean A+T content of 83.6%, and it encodes the 37 genes typically found in arthropod mt genomes (13 protein-coding, 22 tRNA, and two rRNA genes). Five translocations of tRNA genes were discovered relative to the putative ancestral genome arrangement in insects, and the unusual start codon TTG was predicted for cox2. Phylogenetic analyses revealed significantly longer branches leading to the apocritan Hymenoptera as well as the Orussoidea, to a lesser extent the Cephoidea, and, possibly, the Tenthredinoidea than any of the other holometabolous insect orders for all mitochondrial but none of the four nuclear genes tested. Thus, our results suggest that the ancestral parasitic lifestyle of Apocrita is unlikely to be the major cause for the elevated substitution rates observed in hymenopteran mitochondrial genomes.
Introduction
Mitochondrial genes have been used extensively for phylogenetic studies in insects. Their generally high substitution rates render them especially useful to resolve the relationships among closely related taxa [1]. Deeper phylogenetic splits, however, are usually not well resolved in analyses based on mitochondrial genes, and the high heterogeneity in among-site rate variation may partly be responsible for the poor performance of mitochondrial as compared to nuclear genes [2]. An additional problem with mitochondrial sequences is that differences in mitochondrial evolutionary rates among insect lineages can cause long-branch attraction problems [3] that result in unrelated taxa with high substitution rates erroneously grouping together in phylogenetic trees [4]. A similar effect has been observed as a consequence of occasional reversals in the strand-specific compositional bias that is often pronounced in mitochondrial genomes [5]–[7].
Recently, the availability of an increasing number of complete insect mitochondrial genomes has initiated phylogenomic approaches that have greatly enhanced our understanding of the evolutionary relationships within and among extant hexapod orders [8]–[13]. Despite these efforts, the range of insect taxa for which complete mitochondrial genomes are available remains rudimentary, and many large families are not represented by a single sequence. This is also true for several families within the Hymenoptera, one of the largest insect orders on earth. Notably, no mitochondrial genome sequence is available for the about 8000 species of Crabronidae, although they constitute the sister group to the Apidae, a family of considerable interest due its ecological and economical importance and the wide range of social systems represented in this taxon [14].
Substitution rates of mitochondrial genomes have been found to vary substantially across insect taxa. Notably, Hymenoptera and Phthiraptera exhibit significantly elevated rates of nucleotide substitutions [11], [15]–[17], which has been attributed to the parasitic lifestyle of the extant or ancestral members of these orders [18], [19]. The usually short generation times and small effective population sizes due to frequent founder events typically found in parasitic lineages would be expected to result in elevated nucleotide substitution rates in both mitochondrial and nuclear genes [16], [20]. However, to date no detailed multi-gene study is available that compares evolutionary rates between mitochondrial and nuclear genes in parasitic versus non-parasitic insect taxa (but see [21]).
Here we report on the first complete mitochondrial genome sequence of a crabronid wasp, the European beewolf, Philanthus triangulum (Hymenoptera, Crabronidae). Due to their interesting natural history, Philanthus species have attracted considerable attention among behavioral ecologists, and their biology has been studied in detail [22]–[24]. Recently, Philanthus females have been found to engage in an unusual symbiosis with the actinobacterium ‘Candidatus Streptomyces philanthi’ [25], [26]. These bacteria are cultivated in unique antennal gland reservoirs of female beewolves [27] and transferred to the larval cocoon [28], where they provide protection against pathogenic microorganisms by producing a cocktail of antibiotic substances [29].
Using complete mitochondrial genome sequences as well as four different nuclear gene datasets, we reconstructed the phylogenetic relationships among the four largest holometabolous insect orders (Coleoptera, Hymenoptera, Diptera, and Lepidoptera), and we compared the substitution rates of mitochondrial and nuclear genes among the orders. Based on earlier studies [21], we hypothesized that the ancestral parasitic lifestyle of apocritan Hymenoptera resulted in elevated substitution rates in both mitochondrial and nuclear genomes.
Materials and Methods
Ethics statement
A male European beewolf was obtained from a laboratory culture that had earlier been established with field-collected animals from Würzburg and Erlangen, Germany. No specific permits were required for collecting, as P. triangulum is not an endangered or protected species, and the collecting localities constituted non-protected public areas.
Mitochondrial genome sequencing, assembly and annotation
Whole genomic DNA was extracted from the thorax of the male beewolf by standard phenol-chloroform extraction [30] and stored at 4°C in 100 µl Low-TE (1 mM Tris, 0.1 mM EDTA, pH 8.0). For the PCRs, 2 µl of a 1∶10 dilution were used per 25 µl PCR mix. About half of the mitochondrial genome (7891 bp) could be amplified by using the primers C2LF2 and CBLR5 with the Peqlab MidRange PCR System according to the manufacturer's suggestions (Table S1 and S2). The remaining part was amplified in smaller fragments (Table S2). For fragments spanning the AT-rich control region, the extension temperature had to be reduced to 60°C for successful amplification [31], [32]. The amplicons were sequenced bidirectionally by primer walking on ABI 3700 or ABI 3730 instruments using the Big Dye terminator kit (for sequencing primers see Table S1).
Sequences were edited and assembled using the Phred/Phrap/Consed package [33]–[35]. The whole mitochondrial genome sequence was saved as a FASTA file, and tRNAscan-SE 1.21 was used for tRNA search and secondary structure prediction [36]. Lowering the Cove threshold to 1 yielded all of the expected tRNA genes except for the tRNA-Ser(AGN) gene that has proven difficult to find in earlier studies due to its unusual secondary structure [37]. This gene was detected by aligning other hymenopteran tRNA-Ser(AGN) genes with the P. triangulum mitochondrial genome, and the sequence was manually inspected and the annotation corrected based on the predicted secondary structure. The localization was confirmed by using MOSAS [38]. For annotation of protein-coding genes and rRNAs, the sequence file was imported into DOGMA [39]. All genes were checked and corrected manually on the basis of the predicted amino acid sequences and BLAST searches against available mitochondrial genomes. The rRNA genes were compared to the secondary structure predicted for the honeybee rRNAs [40], and the beginning and end of the genes were assumed to extend to the boundaries of the adjacent tRNA genes [41]. The A+T-rich region was checked for repeat motifs using REPFIND [42]. On the basis of the whole genome sequence and the annotation table exported from DOGMA, a GenBank file was created by using the Sequin 9.00 tool downloaded from GenBank. The file was imported into OrganellarGenomeDRAW for creating a graphical representation of the genome [43]. The complete mitochondrial genome sequence of Philanthus triangulum was deposited in the NCBI database (accession number JN871914).
Phylogenetic analysis
All completed mitochondrial genomes of holometabolous insects available as of October 2010 (but only one species per genus) as well as the genomes of two hemimetabolous outgroup species (Hemiptera) were downloaded from the NCBI database (Table S3). The hymenopteran dataset was complemented with some unfinished genomes (Primeuchroeus sp.: nad2, nad3, nad5 missing; Perga condei: nad2 missing; Nasonia vitripennis: small part of nad2 missing).
Four nuclear genes were used for the comparative analysis of mitochondrial and nuclear substitution rates: the 18rRNA gene (18S), wingless (Wg; 336 bp), arginine kinase (ArgK; 1029 bp), and phosphoenolpyruvate carboxykinase (PEPCK; 732 bp). For 18S, an alignment based on the predicted secondary structure of the rRNA was obtained from the study of Whiting [44]. Four Apidae sequences were added manually to the alignment (Anthophora montana [AY995678], Apis mellifera [AB126807], Centris rhodopus [AY995680], and Thyreus delumbatus [AY995687]). For the three protein-coding genes, representative sequences from the major holometabolous insect orders Lepidoptera, Diptera, Hymenoptera and Coleoptera as well as outgroup specimens from the hemimetabolous Hemiptera were downloaded from the NCBI database (for accession numbers see Table S4). The ArgK sequence for Philanthus triangulum (JQ083477) was obtained by sequencing of a fragment that had been amplified by PCR with primers ArgK_fwd2 (5′-GACAGCAARTCTCTGCTGAAGAA-3′) and ArgK_KLTrev2 (5′-GATKCCATCRTDCATYTCCTTSACRGC-3′) [45].
For phylogenetic analyses, we aligned protein-coding sequences with ClustalX [46] and with T-Coffee [47] using the respective default parameters. To account for secondary structure of the rRNAs we used R-Coffee [48]. We checked the alignments by eye and corrected small portions of obvious homology missed by the alignment software (MacClade4 [49]) and selected final alignments on the basis of the fits of the alternatives as measured by the color-coded scoring system of T-Coffee and R-Coffee. All nuclear and mitochondrial gene alignments were submitted to TreeBASE.
For the combined analysis of all mitochondrial genes, the sequences were concatenated with MacClade4 [49]. We used likelihood tests in Phyml [50] to determine optimal models of sequence evolution from HKY, GTR, TN93 and their variants with invariant site (I) and rate heterogeneity (G). For the concatenated protein-coding genes, the Likelihood Ratio Test and the Bayesian Information Criterion (BIC) as implemented in Metapiga [51] were used to determine the optimal model. Based on these results, trees and parameters were optimized using MrBayes [52] assuming GTR+I+G maximum likelihood models partitioned by codon position for the protein-coding gene sequences. For the final analysis, the third codon position was excluded. Each analysis comprised two simultaneous runs with four chains each. The chains ran from 500,000 to 1,000,000 generations depending on when the average standard deviation of the split frequencies was consistently less than 0.01. Plots of the number of generations against the maximum likelihood scores indicated stabilization of likelihood scores. Further diagnostics included the potential scale reduction factor (PSRF) that measures the fit of branch length and all parameters. Trees and parameters from the first 25% of the generations were discarded (the burn in) after completion of the MCMC (Markov Chain Monte Carlo) search.
For the maximum likelihood analyses of the individual genes, we estimated parameters for GTR+I+G models of sequence evolution and optimized the tree using PHYML [50]. Our analysis of 1000 bootstrap replicates provided confidence limits on the maximum likelihood tree clades. For the concatenated sequences of several genes we used RAxML [53] to optimize the maximum likelihood tree and to calculate bootstrap proportions under GTR+I+G models partitioned by codon position.
Finally, we tested various alternative topologies (e.g. monophyly of the Hymenoptera and of the Diptera) using Shimodaira-Hasegawa [54] tests with GTR+I+G optimized parameters in PAUP*4b10 [55] to determine whether any differed significantly from the optimal tree. We inferred trees using the log-determinant transformation [56] to correct for base composition bias. We explicitly tested for base composition homogeneity across insect orders with an analysis of variance (ANOVA, JMP Pro 9.0.0) of the first two eigenvectors from a principal components analysis (PCA) of the base frequencies. Unlike the raw base frequencies, the PCA transformed variables (eigenvectors) are uncorrelated and approximately normally distributed and so amenable to ANOVA analysis.
We tested the sequences for the presence of a global molecular clock using the Rambaut multidimensional clock option in PAUP4b10 [55]. For computational facility, calculations of local clock parameters were calculated using maximum likelihood methods of Hyphy2.0 [57] on a smaller (50 taxa) data set rather than the full set. To test for significant differences in substitution rates across taxa for the various phylogenies (concatenated mitochondrial protein-coding genes, mitochondrial SSU and LSU rRNA, respectively, and the nuclear genes ArgK, PEPCK, wingless, and 18S rRNA), we used Hyphy's relative rates test. The Kishino80+G model was used for all datasets except for wingless, because relative rate parameters for this gene could not be resolved using the optimal model. Thus, we excluded the gamma parameter and used the suboptimal K80 test for the wingless dataset. For all analyses, substitution rates were compared for all possible pairwise combinations, using a hemipteran specimen as outgroup (except for the 18S rRNA dataset, for which Libellula [Odonata] was used as outgroup). Additional outgroup specimens were discarded from the analysis. Probability values were corrected for multiple comparisons with the Bonferroni correction.
Results and Discussion
Architecture of the beewolf mitochondrial genome
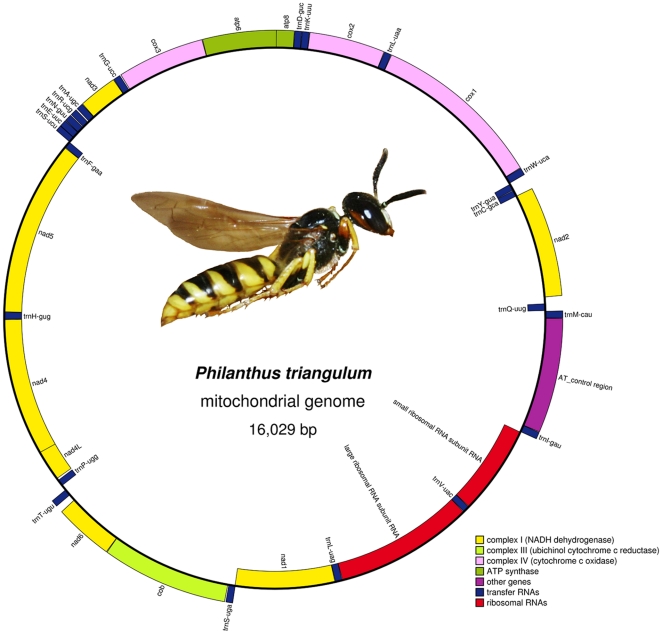
The mitochondrial genome of P. triangulum consists of 16,029 bp, with a mean A+T content of 83.6% (Fig. 1). The 37 genes typical for arthropod mitochondrial genomes were found (22 tRNA genes, two rRNA genes, and 13 protein-coding genes; Table S5). While the protein-coding and rRNA genes were conserved in positions and orientations relative to the inferred ancestral arrangement in insect mitochondrial genomes [58], five translocations of tRNAs were detected (tRNA-Met/-Gln; tRNA-Trp; tRNA-Glu/-Ser(AGN); tRNA-Pro/-Thr; tRNA-Ile). The organization of the highly variable nad3-nad5 junction was identical to that of Vespula germanica and differed only in the relative order of tRNA-Glu/-Ser(AGN) from the inferred ancestral type in insects [59].
Figure 1. Mitochondrial genome sequence of Philanthus triangulum.
Genes on the inside of the circle are located on the complementary strand (−). Genes are color-coded according to gene function: tRNA genes in blue, rRNA genes in red, genes of the NADH dehydrogenase complex in yellow, ATP synthase genes in dark green, cytochrome oxidase genes in light pink, cytochrome b in light green, and the AT-rich control region in magenta.
Coding density
As is characteristic for mitochondrial genomes in general, the mt-genome of P. triangulum exhibits an extremely high coding density. Overlaps between protein-coding regions and/or tRNA genes were found in six locations, with a total of 26 bases shared by two genes (Table S5). tRNA genes in animal mitochondrial genomes have commonly been found to overlap at the discriminator nucleotide [60], and shared nucleotides between protein coding genes and tRNA genes have also been reported for many insect species [11], [41], [61]–[64]. If the A+T-rich region and potential non-coding regions between the rRNA genes and adjacent tRNA genes are not taken into account, a total of 217 bp of non-coding intergenic spacers was found (size range of non-coding regions: 1–61 bp). Thus, coding density in the mitochondrial genome of P. triangulum tends to be even higher than in other hymenopteran mt-genomes (e.g. 811 bp non-coding sequences in Apis mellifera [61]; 486 bp in Melipona bicolor [41]; 880 bp in Bombus ignitus [64]), but slightly lower than in Drosophila yakuba (183 bp [62]).
Protein-coding genes
The 13 protein-coding genes typical for animal mitochondrial genomes were found in the mt-genome of P. triangulum (Fig. 1, Table S5). Conventional start codons ATG or ATT could be assigned to all protein-coding genes except for nad3 and cox2 (Table S5). The nad3 gene apparently uses ATC (codes for Ile) as a start codon, while the start codon for cox2 seems to be TTG (codes for Leu). The use of irregular start codons has been found repeatedly for animal mitochondrial genomes (notably for cox1, e.g. [60]–[63], [65]), but to our knowledge this is the first report of a TTG start codon in cox2 (but GTG has recently been found as the start codon for cox2 in Eriogyna pyretorum, Lepidoptera: Saturniidae, see [66]). Six of the protein-coding genes end in complete stop codons (TAA). The remaining seven protein-coding genes appear to use abbreviated stop codons (TA or T) that are presumably completed by post-transcriptional polyadenylation as reported for other animal mitochondrial genomes [67]–[69]. The nucleotide composition of the P. triangulum mt-genome was strongly biased towards A+T, with a mean A+T content of 83.6%, which lies in the range found for other hymenopteran mt-genomes (Table S6) [9], [41], [61], [63], [64], [70]. Within protein-coding genes, the nucleotide bias differed strongly among codon positions. The highest A+T bias was found for the third positions (94.7%), while the first and second positions had A+T contents lower than the average of the complete genome (79.0% and 74.4%, respectively) (Table S6).
Ribosomal and transfer RNAs
The anticodons of the tRNAs in the European beewolf mitochondrial genome were identical to those of Bombus ignitus [64] and Apis mellifera [61], but differed from those found for other insects: in P. triangulum, GAT is the anticodon of tRNA-Ile, TTT of tRNA-Lys, and TCT of tRNA-Ser(AGN), while these are CCT, CTT, and GCT in most other insects, respectively [64]. Of the 22 tRNAs, 21 showed the typical cloverleaf secondary structure. As in several other insect species, the tRNA-Ser(AGN) showed an aberrant structure, with the D-arm forming a simple loop [64], [71].
Two rRNAs were found in the mitochondrial genome of P. triangulum: rrnL is located between tRNA-Leu(CTN) and tRNA-Val, and rrnS between tRNA-Val and tRNA-Ile. Their lengths (1328 bp for rrnL and 863 bp for rrnS) are similar to those of other hymenopteran mt-rRNAs [61], [64].
A+T-rich region
Both length (1039 bp) and AT-content (85.7%) of the A+T-rich region in the beewolf mt-genome are within the range of other insect mitochondrial genomes (Table S6) [64], [72]. This region is by far the longest non-coding sequence in the beewolf mitochondrial genome, and it is likely to play a role in the initiation of replication as well as the regulation of transcription, as has been shown for Drosophila yakuba [62], [73]. In the A+T-rich region of the beewolf mitochondrial genome, a 43 bp-tandem repeat (two repeat copies) as well as several (AT)n microsatellite-like tandem repeats of up to 13 copies were discovered. Such repeats are typical for insect mitochondrial A+T-rich regions and may cause heteroplasmy [72].
Phylogeny of holometabolous insects based on mitochondrial genomes
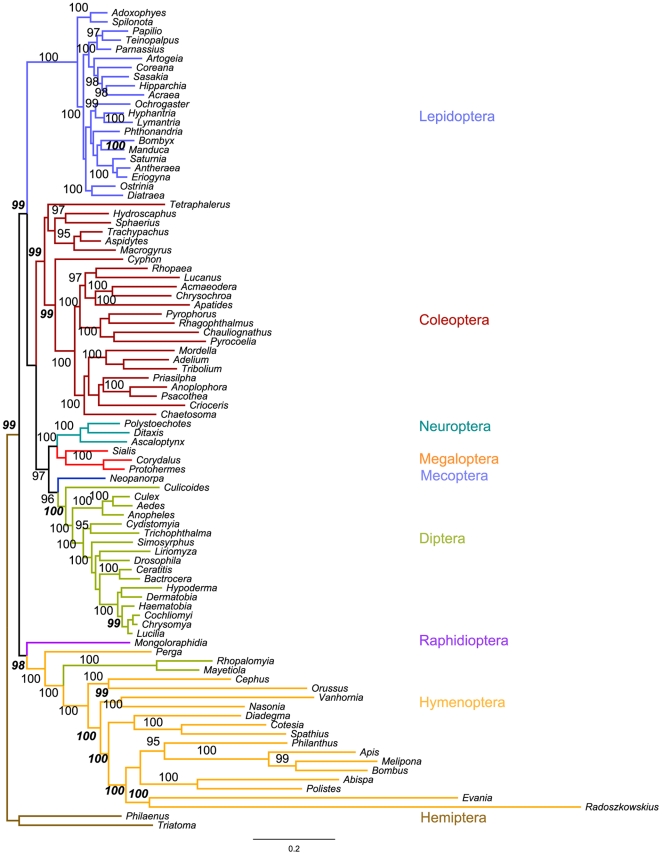
Bayesian and maximum likelihood analyses were used to reconstruct the phylogeny of holometabolous insects based on mitochondrial protein-coding and rRNA genes. Analyses including the 1st and 2nd positions of all protein-coding genes recovered most of the expected relationships on the order, suborder, superfamily and family level, with three notable exceptions: The gall midge genera Rhopalomyia and Mayetiola (Diptera, Cecidomyiidae) were consistently misplaced in the Hymenoptera; the only Raphidiopteran was placed at the base of the Hymenoptera instead of within the Neuropterida (Neuroptera+Mecoptera); and the non-aculeate genus Evania erroneously grouped as a sister group of Radoszkowskius within the Aculeata (Fig. 2) [74], [75]. As in previous phylogenetic studies, the Hymenoptera were recovered as the most basal of the holometabolous insect orders [75], [76], Diptera and Mecoptera were sister groups and together represented the sister group of the Neuropterida [77], which disagrees with a recent study based on single-copy nuclear genes that revealed the Coleoptera and Strepsiptera to be the closest relatives to the Neuropterida [75]. Including the 3rd positions in the phylogenetic analysis led to the loss of monophyly in the Coleoptera, and an analysis with only the 3rd positions did not recover the Coleoptera or the Lepidoptera as monophyletic clades, and several of the expected relationships below order level were not recovered as monophyletic clades (Fig. S1). A maximum likelihood phylogeny of the two mitochondrial rRNA genes with a subset of 50 taxa recovered the Lepidoptera, Diptera, and Hymenoptera as monophyletic, but not the Coleoptera or many of the relationships on lower taxonomic levels (Fig. 3). Our results agree with an earlier study indicating that Bayesian analyses of 1st and 2nd protein-coding positions are most useful to recover uncontroversial relationships within the Hymenoptera when using mitochondrial genomes [8].
Figure 2. Bayesian phylogeny inferred from the 1st and 2nd codon positions of the 13 mitochondrial protein-coding genes.
Because most of the maximum likelihood values differed very little from the posterior probabilities, branches are labeled with a single number for reading clarity. Most are maximum likelihood bootstrap proportions (regular text) but are replaced by posterior probabilities (bold italics) for branches with a difference of more than five between the two values. Branches are color-coded based on order-level taxonomic affiliations.
Figure 3. Maximum likelihood tree inferred from the mitochondrial 12S and 16SrRNA gene sequences of 50 representative taxa from the four major holometabolous insect orders.
Branches are color-coded based on order-level taxonomic affiliations (see Fig. 2).
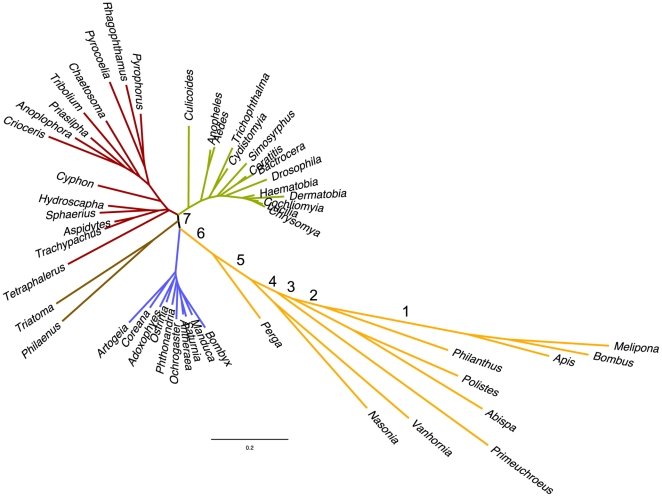
All phylogenetic analyses showed markedly longer branches for the apocritan Hymenoptera as well as for Orussus and, to a lesser extent, Cephus, than for any other taxa (with the exception of the two dipteran genera Rhopalomyia and Mayetiola, which will be discussed below), indicating unusually high mitochondrial substitution rates in the Hymenoptera. High substitution rates in hymenopteran mt-genes have been reported repeatedly and implicated in the problems of reconstructing holometabolous insect phylogenies based on mitochondrial genes due to long-branch attraction effects [8], [11], [18]. Although tree topologies varied markedly among single genes, we observed long branches leading to hymenopteran taxa for all protein-coding genes with the possible exception of Atp8 (see Fig. S2). Molecular clock analyses including all protein-coding genes showed that the substitution rates within Hymenoptera deviated significantly from molecular clock assumptions (Fig. 4). Relative rate tests confirmed the significant differences between Hymenoptera and the other major holometabolous insect orders for the concatenated protein-coding gene dataset and to a lesser extent for the SSU rRNA, but not for the LSU rRNA (Table 1).
Figure 4. Molecular clock analysis of 50 representative taxa from the four major holometabolous insect orders (global molecular clock lnL = −311050, no clock lnL = −309424).
Numbers correspond to significantly different local clock log likelihood values as follows: ΔlnL = 24 df = 2 (branch 1), ΔlnL = 125 df = 3(branch 2), ΔlnL = 170 df = 5 (branch 3), ΔlnL = 172 df = 6 (branch 4), ΔlnL = 263 df = 8 (branch 5), ΔlnL = 817 df = 9 (branch 6), ΔlnL = 2363 df = 19 (branch 7). Branches are color-coded based on order-level taxonomic affiliations (see Fig. 2).
Table 1. Comparative analysis of relative substitution rates in hymenopteran and non-hymenopteran taxa for three mitochondrial and four nuclear phylogenies.
| Number of tests | ||||||
| Genome | Genes | Pairwise comparisons | signif.(1) | non-signif. | total | % signif. |
| Mito | all protein- | Apocrita vs. non-Hymenoptera | 930 | 6 (2) | 936 | 99 |
| coding | Apocrita vs. Tenthredinoidea ( Perga ) | 13 | 13 | 100 | ||
| genes | Apocrita vs. Cephoidea ( Cephus ) | 12 | 1 | 13 | 92 | |
| combined | Apocrita vs. Orussoidea ( Orussus ) | 7 | 6 | 13 | 54 | |
| Apocrita vs. Apocrita | 29 | 49 | 78 | 37 | ||
| Orussoidea ( Orussus ) vs. non-Hymenoptera | 70 | 2 (3) | 72 | 97 | ||
| Orussoidea ( Orussus ) vs. Tenthred. ( Perga ) | 1 | 1 | 100 | |||
| Orussoidea (Orussus) vs. Cephoidea (Cephus) | 1 | 1 | 0 | |||
| Cephoidea ( Cephus ) vs. non-Hymenoptera | 69 | 3 (4) | 72 | 96 | ||
| Cephoidea (Cephus) vs. Tenthred. (Perga) | 1 | 1 | 0 | |||
| Tenthredinoidea (Perga) vs. non-Hymenoptera | 22 | 50 | 72 | 31 | ||
| non-Hymenoptera vs. non-Hymenoptera | 239(5) | 2317 | 2556 | 9 | ||
| Total | 1392 | 2436 | 3828 | 36 | ||
| SSU rRNA | Apocrita vs. non-Hymenoptera | 177 | 165 | 342 | 52 | |
| Apocrita vs. Symphyta (Perga) | 9 | 9 | 0 | |||
| Apocrita vs. Apocrita | 36 | 36 | 0 | |||
| Symphyta (Perga) vs. non-Hymenoptera | 2 | 36 | 38 | 5 | ||
| non-Hymenoptera vs. non-Hymenoptera | 3 | 700 | 703 | 0 | ||
| Total | 182 | 946 | 1128 | 16 | ||
| LSU rRNA | Apocrita vs. non-Hymenoptera | 17 | 325 | 342 | 5 | |
| Apocrita vs. Symphyta (Perga) | 9 | 9 | 0 | |||
| Apocrita vs. Apocrita | 36 | 36 | 0 | |||
| Symphyta (Perga) vs. non-Hymenoptera | 38 | 38 | 0 | |||
| non-Hymenoptera vs. non-Hymenoptera | 4 | 699 | 703 | 1 | ||
| Total | 21 | 1107 | 1128 | 2 | ||
| Nuclear | ArgK | Hymenoptera vs. non-Hymenoptera | 98 | 98 | 0 | |
| Hymenoptera vs. Hymenoptera | 21 | 21 | 0 | |||
| non-Hymenoptera vs. non-Hymenoptera | 91 | 91 | 0 | |||
| Total | 0 | 210 | 210 | 0 | ||
| PEPCK | Hymenoptera vs. non-Hymenoptera | 350 | 350 | 0 | ||
| Hymenoptera vs. Hymenoptera | 1 | 90 | 91 | 1 | ||
| non-Hymenoptera vs. non-Hymenoptera | 300 | 300 | 0 | |||
| Total | 1 | 740 | 741 | 0 | ||
| wingless | Hymenoptera vs. non-Hymenoptera | 42 | 322 | 364 | 12 | |
| Hymenoptera vs. Hymenoptera | 9 | 69 | 78 | 12 | ||
| non-Hymenoptera vs. non-Hymenoptera | 378 | 378 | 0 | |||
| Total | 51 | 769 | 820 | 6 | ||
| 18S rRNA | Hymenoptera vs. non-Hymenoptera | 506 | 454 | 960 | 53 | |
| Hymenoptera vs. Hymenoptera | 190 | 190 | 0 | |||
| non-Hymenoptera vs. non-Hymenoptera | 564 | 564 | 1128 | 50 | ||
| Total | 1070 | 1208 | 2278 | 47 | ||
significant at P<0.01 after Bonferroni correction.
all involving Rhopalomyia or Mayetiola.
Orussus vs. Rhopalomyia and Mayetiola.
Cephus vs. Rhopalomyia, Mayetiola, and Pyrocoelia.
of those 140 involving Rhopalomyia or Mayetiola.
Given are the numbers of significant (after Bonferroni-correction) and non-significant pairwise comparisons of relative rates, as well as the proportion of significant tests (in percent). Comparisons with >50% significant tests are highlighted in bold. To elucidate the origin of elevated substitution rates in Hymenoptera, the results for the basal symphytan taxa are listed individually for the genes for which sequences of these taxa were available. A hemimetabolous species was used as the outgroup for each analysis.
Surprisingly, more detailed analyses of the basal Hymenoptera (“Symphyta”) indicate that elevated substitution rates are not confined to the Apocrita, as had been suggested earlier [18], but also occur in Orussus (Orussoidea) and Cephus (Cephoidea), both of which show significantly higher rates than >95% of all investigated non-hymenopteran taxa (with 2/2 and 2/3 non-significant pairwise comparisons involving the dipteran taxa Rhopalomyia and Mayetiola, respectively, which also exhibit unusually high substitution rates [see below]). Even in Perga condei (Tenthredinoidea), more than 30% of the pairwise comparisons suggest elevated substitution rates as compared to non-hymenopteran taxa, which is significantly more than the proportion of significant paiwise tests among non-hymenopterans (9.35%, Chi2 = 35.2, P<0.001). It has to be noted, however, that all three symphytan taxa show significantly lower substitution rates than most Apocrita (Table 1), so their substitution rates should be considered intermediate between Apocrita and the non-hymenopteran taxa.
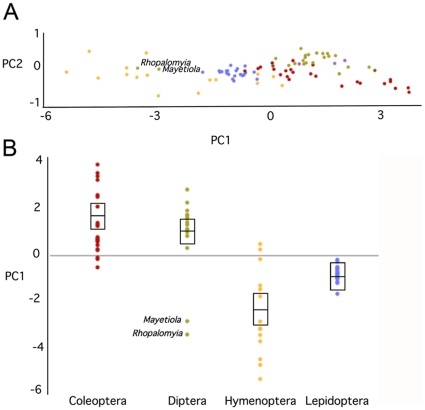
Our analyses consistently misplaced the gall midge genera Rhopalomyia and Mayetiola within the Hymenoptera rather than the Diptera (Fig. 2), and a Shimodaira-Hasegawa test significantly rejected a tree with a monophyletic dipteran clade (ΔlnL = 733.3, P<0.0001). To explain the presumably false attraction of these dipteran sequences to the hymenopteran clade, we investigated sources of bias uncorrected by the optimal GTR+I+G model. First, we used a logDet transform [56] that mitigates the effect of base composition bias among the sequences. This tree, however, also included the two dipteran sequences in the hymenopteran clade (data not shown). In order to statistically test for homogeneity of base composition across the sequences, we transformed the four correlated base frequency variables to uncorrelated, normally distributed variables with principal component analysis (Table S7). The resulting first two principal components (PCs) explain 93% and 5% of the variance, respectively, and the sign and magnitude of the eigenvectors indicates that the first PC contrasts AT against CG frequencies across the sequences. A scatterplot of the two PCs shows that many of the Hymenoptera and the two Diptera (Rhopalomyia and Mayetiola) cluster in a group separated from most of the other sequences (Fig. 5A). A comparison of PC1 across insect orders confirmed that the Hymenoptera differ significantly in base composition from all of the other three major holometabolous insect orders (Fig. 5B; ANOVA, P<0.001; Tukey HSD post-hoc tests: P<0.01 for all pairwise comparisons including Hymenoptera). However, both Rhopalomyia and Mayetiola exhibit base composition biases that are in the range of Hymenoptera rather than other Diptera. Thus, the misplacement of these gall midge genera within the Hymenoptera is likely due to their bias in base composition (especially the extreme AT bias) that is more similar to the Hymenoptera than to the Diptera. Although the authors of the original description of the Rhopalomyia and Mayetiola mitochondrial genomes did not report on elevated substitution rates in gall midges as compared to other Diptera, they commented on several unusual features like the very small size, the rearrangements and truncation of tRNA genes, and, notably, an unusually high AT content in the coding regions [78].
Figure 5. Principal components analysis of base frequencies among mitochondrial genome sequences.
A Plot of the first principal component against the second principal component (explaining 93% and 5% of the variability, respectively). B Boxplot of PC1 according to insect orders. An ANOVA of the first principal component shows that the base composition in Hymenoptera is significantly biased relative to the other orders (P<0.01 for all pairwise comparisons involving Hymenoptera), due to very high AT frequencies. Samples are color-coded based on order-level taxonomic affiliations (see Fig. 2).
Nuclear phylogeny of holometabolous insects
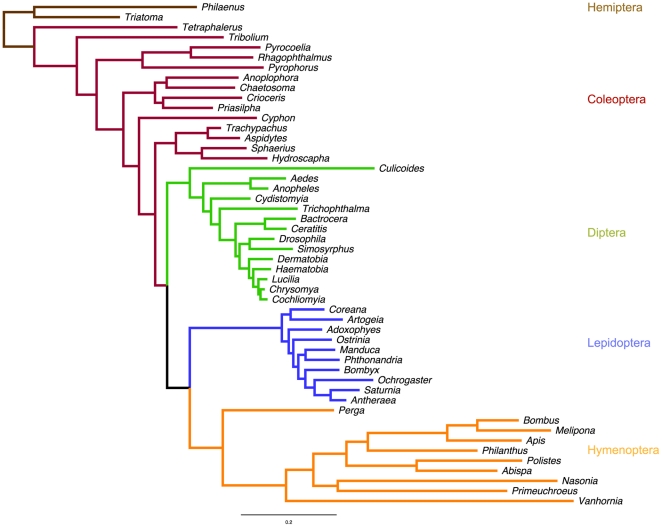
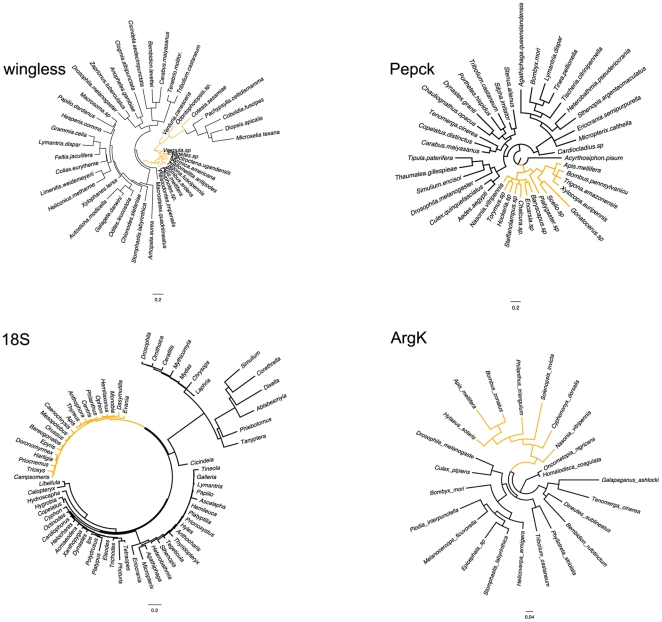
Based on four nuclear gene datasets, we reconstructed the phylogenetic relationships among holometabolous insect orders (Fig. 6). While PEPCK and ArgK recovered the four major holometabolous insect orders as monophyletic, wingless failed to group the hymenopteran taxa in a monophyletic clade, and the Coleoptera were paraphyletic in the 18S phylogeny, as had been reported earlier [44]. Relative rate tests yielded no evidence for higher substitution rates in Hymenoptera for the any of the four nuclear genes (Table 1). For both wingless and 18S rRNA, the observed differences in relative rates between hymenopteran and non-hymenopteran taxa were due to lower rather than higher rates in Hymenoptera (all 42 and 506 significant differences for wingless and 18S rRNA, respectively). These results are in contrast to an earlier study suggesting that both mitochondrial and nuclear genes exhibit elevated substitution rates [21]. However, other phylogenetic analyses of the extant hexapod orders based on nuclear genes did not comment on conspicuously elevated substitution rates in Hymenoptera as compared to other hexapod lineages (see [44], [79], but Whiting et al. [80] comment briefly on long branches in Hymenoptera) or found even shorter phylogenetic branches leading to Hymenoptera than other insect orders [76], which agrees with our results for wingless and 18S rRNA.
Figure 6. Phylogenetic trees from analyses of four nuclear genes for representative taxa of the four major holometabolous insect orders.
Sequences for the analysis of wingless, PEPCK, and ArgK were obtained from the NCBI database, the 18S dataset represented a reduced dataset from the analysis of Whiting (2002) that was supplemented with some additional taxa of Apidae from the NCBI database. Hymenopteran taxa are highlighted by yellow branches.
Comparison of mitochondrial and nuclear phylogenies
Our results indicate that substitution rates in Hymenoptera as compared to other holometabolous insect orders are significantly and consistently elevated for mitochondrial (with the exception of the LSU rRNA) but not for any of the four nuclear genes under investigation (Table 1). Interestingly, the inclusion of several non-apocritan Hymenoptera (Perga, Cephus, Orussus) provides evidence for elevated mitochondrial substitution rates also in Orussus, to a lesser extent in Cephus, and possibly even in Perga. Thus, either substitution rates of mitochondrial genes already started to increase within the paraphyletic suborder Symphyta prior to the origin of the Apocrita, or the elevated rates evolved independently in the Apocrita and in the only ectoparasitic group within the Symphyta, the Orussoidea, as has been suggested previously by Dowton and Austin [18]. However, the intermediate to high branch lengths and the significantly elevated substitution rates found for Cephus in our analyses provide support for the former hypothesis (see Table 1, Fig. 2 and S1).
Mechanistically, substitution rates are determined by the mutation rate and the probability that a given mutation reaches fixation [81]. Although analyses based on sequence comparisons fail to record mutations that never reach fixation and generally do not distinguish between the effects of mutation rate and probability of fixation, it is important to consider these as two distinct evolutionary factors affecting substitution rates [82]. Mutation rates are generally thought to be positively correlated with metabolic rate and mutational pressure (e.g. mutagens, oxidative stress), but negatively with the efficiency of the DNA repair machinery. Fixation rates, on the other hand, depend on the effective population size, cladogenesis rate, and the selective pressures acting upon the gene of interest [83]. In many of these factors, mitochondrial and nuclear genomes differ markedly, which may explain the generally higher substitution rates observed in mitochondrial as compared to nuclear genomes [84], [85]. Additionally, Oliveira et al. [70] proposed the compensation-draft-feedback model to explain the high substitution rates in Nasonia spp. mitochondrial genomes: Due to the lack of recombination in mitochondria, any fixation of an adaptive mitochondrial mutation can drag along mildly deleterious substitutions in the same haplotype. Compensatory mutations in the same or in interacting mitochondrial genes would then be selected for, which can lead to a second selective sweep to fixation. Thus, this mechanism could potentially result in a cascade of adaptive and non-adaptive mutations in mitochondrial but not nuclear genes, because recombination impedes the compensation-draft-feedback mechanism [70]. However, the available mechanistic models fail to explain the exceptionally high substitution rates observed in mitochondrial genomes of Hymenoptera as compared to other holometabolous insect orders.
Previously, an ecological hypothesis has been proposed to account for the high mt-substitution rates in Hymenoptera as well as in lice (Phthiraptera) [15], [16] and mites [19]: In all three taxa, ectoparasitism is deemed to be the current (lice and mites) or ancestral (apocritan Hymenoptera) lifestyle [86], [87], which entails short generation times and frequent founder events with small effective population sizes. Since these factors are known to be associated with increased substitution rates, the parasitic lifestyle has been hypothesized to be responsible for the elevated evolutionary rates in these insect lineages [16], [18], [19], [21]. A priori, however, the hypothesis predicts elevated substitution rates in both mitochondrial and nuclear genes, and we fail to find evidence for the latter in our nuclear gene phylogenies of Hymenoptera. In addition, it has been noted earlier that several parasitic lineages do not show elevated mitochondrial substitution rates or higher numbers of gene rearrangements [21], [88], [89]. Furthermore, our analyses suggest that mitochondrial substitution rates already began to increase in a non-parasitic ancestor before the origin of the Apocrita and continue to do so in the extant apocritan families, despite the fact that several of them are not parasitic anymore (Fig. 2).
In conclusion, our results do not provide support for the hypothesis that the parasitic lifestyle alone can explain the pattern of evolutionary rates observed in insect mitochondrial genomes. Considering the immense ecological diversity of Hymenoptera, it seems possible that elevated mitochondrial mutation rates evolved originally as a pleiotropic or epistatic side-effect of an adaptive (mitochondrial or nuclear) mutation. Subsequent processes like the compensation-draft feedback may have led to an increase in fixation rates and a further accumulation of mutations, thus resulting in the strongly elevated substitution rates in Hymenoptera that we observe today.
Supporting Information
Bayesian phylogeny inferred from the 3rd codon postions of 13 mitochondrial protein-coding genes. Branches are color-coded based on order-level taxonomic affiliations (see Fig. 3).
(TIF)
Bayesian phylogenies inferred from single mitochondrial protein-coding genes.
(TIF)
Primers used for amplification and sequencing of the mitochondrial genome of P. triangulum.
(DOCX)
Primer combinations and PCR conditions used for the amplification of the mitochondrial genome of P. triangulum.
(DOCX)
Taxonomy and GenBank accession numbers of mitochondrial genomes used for the phylogenetic analysis.
(DOCX)
Taxonomy and GenBank accession numbers of nuclear genes used for the phylogenetic analysis.
(DOCX)
Summary of the mitochondrial genome of P. triangulum.
(DOCX)
Nucleotide composition of the protein-coding, ribosomal RNA and transfer RNA genes in the mitochondrial genome of P. triangulum. Nucleotide frequencies for all genes were calculated for the coding strand. The strand coding for the cox1-3 genes (+) was arbitrarily chosen for the calculation of the average nucleotide frequencies of the complete genome (“All sites”).
(DOCX)
Principle Components Analysis (PCA) of base composition in mitochondrial genomes of holometabolous insects. The 1st principle component accounts for most of the variability (93%) among the four nucleotides and contrasts A and T frequencies against C and G frequencies.
(DOCX)
Acknowledgments
We thank Kerstin Roeser-Mueller for providing the P. triangulum ArgK sequence for phylogenetic analysis, Gudrun Herzner for the beewolf picture, and two anonymous reviewers for excellent comments on the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: We gratefully acknowledge financial support from the Volkswagen foundation through a postdoctoral fellowship to MK (I/82682) and from the Max Planck Society (MK). This study was partly supported by the German Science Foundation (DFG STR-532/2-2 to ES and MK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin CP, Danforth BN. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol Phylogenet Evol. 2004;30:686–702. doi: 10.1016/S1055-7903(03)00241-0. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- 4.Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- 5.Hassanin A, Leger N, Deutsch J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst Biol. 2005;54:277–298. doi: 10.1080/10635150590947843. [DOI] [PubMed] [Google Scholar]
- 6.Foster PG, Hickey DA. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol. 1999;48:284–290. doi: 10.1007/pl00006471. [DOI] [PubMed] [Google Scholar]
- 7.Dowton M, Austin AD. The evolution of strand-specific compositional bias. A case study in the hymenopteran mitochondrial 16S rRNA gene. Mol Biol Evol. 1997;14:109–112. doi: 10.1093/oxfordjournals.molbev.a025696. [DOI] [PubMed] [Google Scholar]
- 8.Dowton M, Cameron SL, Austin AD, Whiting MF. Phylogenetic approaches for the analysis of mitochondrial genome sequence data in the Hymenoptera - a lineage with both rapidly and slowly evolving mitochondrial genomes. Mol Phylogenet Evol. 2009;52:512–519. doi: 10.1016/j.ympev.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Cameron SL, Dowton M, Castro LR, Ruberu K, Whiting MF, et al. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome. 2008;51:800–808. doi: 10.1139/G08-066. [DOI] [PubMed] [Google Scholar]
- 10.Cameron SL, Lambkin CL, Barker SC, Whiting MF. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol. 2007;32:40–59. [Google Scholar]
- 11.Castro LR, Dowton M. The position of the Hymenoptera within the Holometabola as inferred from the mitochondrial genome of Perga condei (Hymenoptera: Symphyta: Pergidae). Mol Phylogenet Evol. 2005;34:469–479. doi: 10.1016/j.ympev.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Castro LR, Dowton M. Mitochondrial genomes in the Hymenoptera and their utility as phylogenetic markers. Syst Entomol. 2007;32:60–69. [Google Scholar]
- 13.Fenn JD, Song H, Cameron SL, Whiting MF. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Mol Phylogenet Evol. 2008;49:59–68. doi: 10.1016/j.ympev.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Michener CD. The bees of the world. Baltimore, MD: The Johns Hopkins University Press; 2000. 913 [Google Scholar]
- 15.Johnson KP, Cruickshank RH, Adams RJ, Smith VS, Page RDM, et al. Dramatically elevated rate of mitochondrial substitution in lice (Insecta: Phthiraptera). Mol Phylogenet Evol. 2003;26:231–242. doi: 10.1016/s1055-7903(02)00342-1. [DOI] [PubMed] [Google Scholar]
- 16.Page RDM, Lee PLM, Becher SA, Griffiths R, Clayton DH. A different tempo of mitochondrial DNA evolution in birds and their parasitic lice. Mol Phylogenet Evol. 1998;9:276–293. doi: 10.1006/mpev.1997.0458. [DOI] [PubMed] [Google Scholar]
- 17.Crozier RH, Crozier YC, Mackinlay AG. The CO-I and CO-II region of honeybee mitochondrial DNA - evidence for variation in insect mitochondrial evolutionary rates. Mol Biol Evol. 1989;6:399–411. doi: 10.1093/oxfordjournals.molbev.a040553. [DOI] [PubMed] [Google Scholar]
- 18.Dowton M, Austin AD. Increased genetic diversity in mitochondrial genes is correlated with the evolution of parasitism in the hymenoptera. J Mol Evol. 1995;41:958–965. doi: 10.1007/BF00173176. [DOI] [PubMed] [Google Scholar]
- 19.Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: Strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol. 2006;38:100–116. doi: 10.1016/j.ympev.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 21.Castro LR, Austin AD, Dowton M. Contrasting rates of mitochondrial molecular evolution in parasitic diptera and hymenoptera. Mol Biol Evol. 2002;19:1100–1113. doi: 10.1093/oxfordjournals.molbev.a004168. [DOI] [PubMed] [Google Scholar]
- 22.Evans HE, O'Neill KM. The natural history and behavior of North American beewolves. Ithaca, NY: Cornell University Press; 1988. 278 [Google Scholar]
- 23.Tinbergen N. Über die Orientierung des Bienenwolfes (Philanthus triangulum Fabr.). Z vergl Physiol. 1932;16:305–335. [Google Scholar]
- 24.Strohm E. Allokation elterlicher Investitionen beim Europäischen Bienenwolf Philanthus triangulum Fabricius (Hymenoptera: Sphecidae) Berlin: Verlag Dr. Köster; 1995. 198 [Google Scholar]
- 25.Kaltenpoth M, Goettler W, Dale C, Stubblefield JW, Herzner G, et al. ‘Candidatus Streptomyces philanthi’, an endosymbiotic streptomycete in the antennae of Philanthus digger wasps. Int J Syst Evol Microbiol. 2006;56:1403–1411. doi: 10.1099/ijs.0.64117-0. [DOI] [PubMed] [Google Scholar]
- 26.Kaltenpoth M, Goettler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15:475–479. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 27.Goettler W, Kaltenpoth M, Herzner G, Strohm E. Morphology and ultrastructure of a bacteria cultivation organ: The antennal glands of female European beewolves, Philanthus triangulum (Hymenoptera, Crabronidae). Arthropod Struct Dev. 2007;36:1–9. doi: 10.1016/j.asd.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Kaltenpoth M, Goettler W, Koehler S, Strohm E. Life cycle and population dynamics of a protective insect symbiont reveal severe bottlenecks during vertical transmission. Evol Ecol. 2010;24:463–477. [Google Scholar]
- 29.Kroiss J, Kaltenpoth M, Schneider B, Schwinger M-G, Hertweck C, et al. Symbiotic streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6:261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. 1659 [Google Scholar]
- 31.Hu M, Chilton NB, Gasser RB. Long PCR-based amplification of the entire mitochondrial genome from single parasitic nematodes. Mol Cell Probe. 2002;16:261–267. doi: 10.1006/mcpr.2002.0422. [DOI] [PubMed] [Google Scholar]
- 32.Lessinger AC, Azeredo-Espin AM. Evolution and structural organisation of mitochondrial DNA control region of myiasis-causing flies. Med Vet Entomol. 2000;14:71–80. doi: 10.1046/j.1365-2915.2000.00209.x. [DOI] [PubMed] [Google Scholar]
- 33.Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 34.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 35.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 36.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheffield NC, Song H, Cameron L, Whiting MF. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 2008;25:2499–2509. doi: 10.1093/molbev/msn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheffield NC, Hiatt KD, Valentine MC, Song H, Whiting MF. Mitochondrial genomics in Orthoptera using MOSAS. Mitochondr DNA. 2010;21:87–104. doi: 10.3109/19401736.2010.500812. [DOI] [PubMed] [Google Scholar]
- 39.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 40.Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): structure, organization, and retrotransposable elements. Insect Mol Biol. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silvestre D, Dowton M, Arias MC. The mitochondrial genome of the stingless bee Melipona bicolor (Hymenoptera, Apidae, Meliponini): Sequence, gene organization and a unique tRNA translocation event conserved across the tribe Meliponini. Genet Mol Biol. 2008;31:451–460. [Google Scholar]
- 42.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 43.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 44.Whiting MF. Phylogeny of the holometabolous insect orders: molecular evidence. Zool Scr. 2002;31:3–15. [Google Scholar]
- 45.Kawakita A, Sota T, Ascher JS, Ito M, Tanaka H, et al. Evolution and phylogenetic utility of alignment gaps within intron sequences of three nuclear genes in bumble bees (Bombus). Mol Biol Evol. 2003;20:87–92. doi: 10.1093/molbev/msg007. [DOI] [PubMed] [Google Scholar]
- 46.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 47.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 48.Moretti S, Wilm A, Higgins DG, Xenarios I, Notredame C. R-Coffee: a web server for accurately aligning noncoding RNA sequences. Nucleic Acids Res. 2008;36:W10–W13. doi: 10.1093/nar/gkn278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maddison DR, Maddison WP. MacClade 4.08: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer Associates; 2005. [Google Scholar]
- 50.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 51.Helaers R, Milinkovitch MC. MetaPIGA v2.0: Maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics. 2010;11:379. doi: 10.1186/1471-2105-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 53.Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 54.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 55.Swofford DL. PAUP* 4.10b. Phylogenetic analysis using parsimony (and other methods), Version 4. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 56.Lockhart PJ, Steel MA, Hendy MD, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol Biol Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 57.Kosakovsky Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 58.Shao RF, Barker SC. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta: Thysanoptera): Convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol Biol Evol. 2003;20:362–370. doi: 10.1093/molbev/msg045. [DOI] [PubMed] [Google Scholar]
- 59.Dowton M, Castro LR, Campbell SL, Bargon SD, Austin AD. Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J Mol Evol. 2003;56:517–526. doi: 10.1007/s00239-002-2420-3. [DOI] [PubMed] [Google Scholar]
- 60.Boore JL, Macey JR, Medina M. Sequencing and comparing whole mitochondrial genomes of animals. In: Zimmer EA, Roalson EH, editors. Molecular Evolution: Producing The Biochemical Data, Part B. Burlington, Massachusetts: Elsevier; 2005. pp. 311–348. [DOI] [PubMed] [Google Scholar]
- 61.Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133:97–117. doi: 10.1093/genetics/133.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clary DO, Wolstenholme DR. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 63.Castro LR, Ruberu K, Dowton M. Mitochondrial genomes of Vanhornia eucnemidarum (Apocrita: Vanhorniidae) and Primeuchroeus spp. (Aculeata: Chrysididae): evidence of rearranged mitochondrial genomes within the Apocrita (Insecta: Hymenoptera). Genome. 2006;49:752–766. doi: 10.1139/g06-030. [DOI] [PubMed] [Google Scholar]
- 64.Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, et al. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 2007;392:206–220. doi: 10.1016/j.gene.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 65.Fenn JD, Cameron SL, Whiting MF. The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol Biol. 2007;16:239–252. doi: 10.1111/j.1365-2583.2006.00721.x. [DOI] [PubMed] [Google Scholar]
- 66.Jiang ST, Hong GY, Yu M, Li N, Yang Y, et al. Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int J Biol Sci. 2009;5:351–365. doi: 10.7150/ijbs.5.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson S, Bankier AT, Barrell BG, Debruijn MHL, Coulson AR, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 68.Ojala D, Merkel C, Gelfand R, Attardi G. The transfer-RNA genes puctuate the reading of genetic information in human mitochondrial DNA. Cell. 1980;22:393–403. doi: 10.1016/0092-8674(80)90350-5. [DOI] [PubMed] [Google Scholar]
- 69.Ojala D, Montoya J, Attardi G. Transfer RNA puctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 70.Oliveira D, Raychoudhury R, Lavrov DV, Werren JH. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp Nasonia (Hymenoptera: Pteromalidae). Mol Biol Evol. 2008;25:2167–2180. doi: 10.1093/molbev/msn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolstenholme DR. Animal mitochondrial DNA - structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 72.Zhang DX, Hewitt GM. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997;25:99–120. [Google Scholar]
- 73.Clary DO, Wolstenholme DR. Drosophila mitochondrial DNA - conserved sequences in the A+T-rich region and supporting evidence for a secondary structure model of the small ribosomall RNA. J Mol Evol. 1987;25:116–125. doi: 10.1007/BF02101753. [DOI] [PubMed] [Google Scholar]
- 74.Peters RS, Meyer B, Krogmann L, Borner J, Meusemann K, et al. The taming of an impossible child: a standardized all-in approach to the phylogeny of Hymenoptera using public database sequences. BMC Biol. 2011;9:55. doi: 10.1186/1741-7007-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiegmann BM, Trautwein MD, Kim J-W, Cassel BK, Bertone MA, et al. Single-copy nuclear genes resolve the phylogeny of the holometabolous insects. BMC Biol. 2009;7:1–16. doi: 10.1186/1741-7007-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, et al. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of holometabolous insects. Genome Res. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to holometabolous insects. BMC Genomics. 2010;11:371. doi: 10.1186/1471-2164-11-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beckenbach AT, Joy JB. Evolution of the mitochondrial genomes of gall midges (Diptera: Cecidomyiidae): Rearrangement and severe truncation of tRNA genes. Genome Biol Evol. 2009;1:278–287. doi: 10.1093/gbe/evp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wheeler WC, Whiting M, Wheeler QD, Carpenter JM. The phylogeny of the extant hexapod orders. Cladistics. 2001;17:113–169. doi: 10.1111/j.1096-0031.2001.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 80.Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC. The Strepsiptera problem: Phylogeny of the holometabolous insect orders inferred from 18S and 28S ribosomal DNA sequences and morphology. Syst Biol. 1997;46:1–68. doi: 10.1093/sysbio/46.1.1. [DOI] [PubMed] [Google Scholar]
- 81.Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. 384 [Google Scholar]
- 82.Rand DM. The units of selection on mitochondrial DNA. Annu Rev Ecol Syst. 2001;32:415–448. [Google Scholar]
- 83.Mindell DP, Thacker CE. Rates of molecular evolution: phylogenetic issues and applications. Annu Rev Ecol Syst. 1996;27:279–303. [Google Scholar]
- 84.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 85.Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc R Soc B-Biol Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dowton M, Austin AD. Simultaneous analysis of 16S, 28S, COI and morphology in the Hymenoptera: Apocrita - evolutionary transitions among parasitic wasps. Biol J Linnean Soc. 2001;74:87–111. [Google Scholar]
- 87.Whitfield JB. Phylogenetic insights into the evolution of parasitism in Hymenoptera. Adv Parasit. 2003;54:69–100. doi: 10.1016/s0065-308x(03)54002-7. [DOI] [PubMed] [Google Scholar]
- 88.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 89.Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol Biol Evol. 2009;26:1607–1617. doi: 10.1093/molbev/msp072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian phylogeny inferred from the 3rd codon postions of 13 mitochondrial protein-coding genes. Branches are color-coded based on order-level taxonomic affiliations (see Fig. 3).
(TIF)
Bayesian phylogenies inferred from single mitochondrial protein-coding genes.
(TIF)
Primers used for amplification and sequencing of the mitochondrial genome of P. triangulum.
(DOCX)
Primer combinations and PCR conditions used for the amplification of the mitochondrial genome of P. triangulum.
(DOCX)
Taxonomy and GenBank accession numbers of mitochondrial genomes used for the phylogenetic analysis.
(DOCX)
Taxonomy and GenBank accession numbers of nuclear genes used for the phylogenetic analysis.
(DOCX)
Summary of the mitochondrial genome of P. triangulum.
(DOCX)
Nucleotide composition of the protein-coding, ribosomal RNA and transfer RNA genes in the mitochondrial genome of P. triangulum. Nucleotide frequencies for all genes were calculated for the coding strand. The strand coding for the cox1-3 genes (+) was arbitrarily chosen for the calculation of the average nucleotide frequencies of the complete genome (“All sites”).
(DOCX)
Principle Components Analysis (PCA) of base composition in mitochondrial genomes of holometabolous insects. The 1st principle component accounts for most of the variability (93%) among the four nucleotides and contrasts A and T frequencies against C and G frequencies.
(DOCX)