Abstract
The conservation of data deficient species is often hampered by inaccurate species delimitation. The galaxiid fishes Aplochiton zebra and Aplochiton taeniatus are endemic to Patagonia (and for A. zebra the Falkland Islands), where they are threatened by invasive salmonids. Conservation of Aplochiton is complicated because species identification is hampered by the presence of resident as well as migratory ecotypes that may confound morphological discrimination. We used DNA barcoding (COI, cytochrome b) and a new developed set of microsatellite markers to investigate the relationships between A. zebra and A. taeniatus and to assess their distributions and relative abundances in Chilean Patagonia and the Falkland Islands. Results from both DNA markers were 100% congruent and revealed that phenotypic misidentification was widespread, size-dependent, and highly asymmetric. While all the genetically classified A. zebra were correctly identified as such, 74% of A. taeniatus were incorrectly identified as A. zebra, the former species being more widespread than previously thought. Our results reveal, for the first time, the presence in sympatry of both species, not only in Chilean Patagonia, but also in the Falkland Islands, where A. taeniatus had not been previously described. We also found evidence of asymmetric hybridisation between female A. taeniatus and male A. zebra in areas where invasive salmonids have become widespread. Given the potential consequences that species misidentification and hybridisation can have for the conservation of these endangered species, we advocate the use of molecular markers in order to reduce epistemic uncertainty.
Introduction
Given the current ratio of species discovery to extinction rates, it is likely that many species will go extinct before they are properly described [1]. Accurate classification is thus an essential first step towards effective conservation of local biodiversity [2]–[3]. This is particularly critical in the Southern Hemisphere, where a high proportion of endemic species are poorly known, and are now threatened by non-native introductions [4]. Biological invasions are a leading cause of animal extinctions [5] and one of the main threats to aquatic biodiversity [6], particularly for fishes [7], which tend to display high rates of endemism.
The diversity of fish species in Patagonia is very low, with only five families represented (Galaxiidae, Trichomycteridae, Diplomystidae, Atherinopsidae and Percichthyidae) [8]. The genus Aplochiton is one of the three genera representing the family Galaxiidae in South America [9] and has two recognised species A. zebra and A. taeniatus (Jenyns 1842) restricted to Patagonia [9], and in the case of A. zebra, also present in the Falkland Islands [10]. The existence of a third species, A. marinus, has been suggested [8]–[9], but its taxonomic status remains unclear [11]. Little is known about the ecology and biogeography of Aplochiton [9], [12], which tend to display a patchy, restricted distribution [9], and whose reproductive ecology has only recently been described [13]. Aplochiton are believed to have an amphidromous life cycle with a marine larval phase followed by juvenile growth and spawning in freshwater [12], [14], although landlocked populations are also known [12]. Apparent declines in the abundance of Aplochiton have been attributed to a number of stressors [15]–[16], most notably predation and/or competition from invasive salmonids, which are widespread and dominate freshwater fish communities throughout much of Aplochiton's range [17]–[20]. A. zebra has been classified as ‘in danger of extinction’ in parts of its range, while the conservation status of A. taeniatus remains unclear due to data deficiency [17].
Fishes from the family Galaxiidae are morphologically very diverse, particularly in relation to buccal arrangements and shape of caudal fin [21]–[23]. Although both A. zebra and A. taeniatus have elongate, fusiform bodies with a slender, deeply forked tail [12], they exhibit morphological differences that appear to be related to their different trophic positions. Thus, A. taeniatus seems to attain a larger size than A. zebra and is considered an specialist that preys mostly on fish, displaying particular adaptations for piscivory such as a large mouth, enlarged teeth and elongated stomach. In contrast, A. zebra is considered a generalist that feeds mainly on aquatic invertebrates [12], and resembles more other galaxiids that are also generalized invertebrate predators [17], [24]–[25]. Differences in body size and trophic ecology probably reflect differences in niche breadth, which could ultimately result in different vulnerability to salmonid invasions through predation and resource competition.
Discrimination between A. zebra and A. taeniatus is currently based on variation in meristic (number of vertebrae, gillrakers and fin rays) and morphological traits (body depth, and relative size of jaw, fins, and head in relation to eye diameter) [11]–[12]. However, phenotypic traits can vary widely among individuals and populations with different life histories [11], particularly in species with diadromous life histories such as Aplochiton, making phenotypic based identification unreliable [9]. In addition, marked changes in allometric relationships between juvenile and adult stages of Aplochiton may cause taxonomical problems [24]. Thus, in order to establish appropriate conservation measures for these species, clarification of the taxonomic status of Aplochiton is urgently needed. To help resolve such conservation challenge, we carried out the molecular analysis of 421 individuals classified as A. zebra and 36 individuals classified as A. taeniatus based on the morphological characteristics described by previous workers [11]–[12]. We used DNA barcoding, a diagnostic technique based on sequence variation at a small segment of the mitochondrial cytochrome c oxidase I gene (COI; [25]), that provides an inexpensive and simple tool for identifying novel species [26], and also for describing cryptic species which are difficult to detect phenotypically [27]–[29]. We sequenced two mitochondrial DNA regions commonly used for fish barcoding (COI and cyt b; [30]–[31]) to discriminate between A. zebra and A. taeniatus, and to clarify their distributions in Chilean Patagonia and the Falkland Islands. In addition, we carried for the first time an analysis of genetic diversity and population differentiation of these endangered species, using a set of microsatellite markers that we have recently developed [32].
Methods
Study populations
Aplochiton spp. were collected by electrofishing at 20 different sites in Chilean Patagonia (n = 376) and 15 sites in the Falkland Islands (n = 80; Table 1; Figure 1). Samples from Chile were collected under permit No. 958, 17 April 2008 from the Chilean Subsecretary of Fishing; samples from the Falkland Islands were collected under licence No. R0221, issued by The Falkland Islands Government, Environmental Planning Department. Individuals were identified in situ as A. zebra or A. taeniatus based on body depth, relative size of the head and the caudal peduncle in relation to body length, and body coloration/pigmentation [11]–[12]. Fin clips were preserved in 95% ethanol and stored at 4°C for genetic analysis. We recorded wet weight (Wt, 0.1 g) and either total length (TL, mm) - measured from the tip of the snout to the tip of the tail, or fork length (FL, mm) – measure from the tip of the snout to the fork of the tail, depending on country and field crew. To standardise body size measurements obtained by different field crews, we converted total length (TL) to fork length (FL) based on the following empirical expression derived from 30 matched samples: FL = −3.076+0.945 TL (R 2 = 0.993, n = 30, P<0.001), and used Fulton's condition factor K = (WT/FL3)×10,000 as a measure of body shape [33]. Size comparisons between locations and species were carried out in SYSTAT v.11.
Table 1. Distribution of Aplochiton zebra, A. taeniatus and hybrids (Hyb), amongst samples collected in Chilean Patagonia and the Falkland Islands.
| River | Area | Latitude | Longitude | Date | A. zebra | A. taeniatus | Hyb | Total |
| Chilean Patagonia | ||||||||
| Blanco-Enco | Mainland | −39.574 | −72.149 | 24-03-09 | 29 | 0 | 0 | 29 |
| Punahue | Mainland | −39.831 | −72.037 | 24-03-09 | 21 | 0 | 0 | 21 |
| Quimán | Mainland | −40.113 | −72.343 | 26-03-09 | 27 | 0 | 0 | 27 |
| Iculpe | Mainland | −40.314 | −72.439 | 31-03-09 | 30 | 0 | 0 | 30 |
| Pitreño | Mainland | −40.326 | −72.319 | 26-03-09 | 30 | 1 | 0 | 31 |
| Futangue | Mainland | −40.331 | −72.266 | 30-03-09 | 30 | 0 | 0 | 30 |
| Lenca | Mainland | −41.605 | −72.682 | 14-04-09 | 17 | 0 | 0 | 17 |
| U24 | Chiloé | −41.811 | −74.031 | 29-11-07 | 0 | 1 | 0 | 1 |
| U25 | Chiloé | −41.814 | −73.971 | 29-11-07 | 0 | 1 | 0 | 1 |
| Huincha | Chiloé | −41.879 | −73.652 | 18-03-09 | 1 | 28 | 1 | 30 |
| U26 | Chiloé | −41.886 | −73.962 | 30-11-07 | 0 | 3 | 0 | 3 |
| U27 | Chiloé | −41.893 | −73.959 | 30-11-07 | 0 | 5 | 0 | 5 |
| Punihuil | Chiloé | −41.931 | −74.023 | 01-12-07 | 0 | 2 | 0 | 2 |
| U28 | Chiloé | −41.946 | −74.024 | 02-12-07 | 22 | 6 | 1 | 29 |
| U29 | Chiloé | −41.959 | −74.040 | 03-12-07 | 21 | 0 | 0 | 21 |
| U30 | Chiloé | −41.984 | −74.012 | 04-12-07 | 0 | 12 | 0 | 12 |
| U34 | Chiloé | −42.110 | −73.484 | 08-12-07 | 16 | 0 | 0 | 16 |
| U17 | Chiloé | −42.115 | −73.484 | 04-11-07 | 20 | 0 | 0 | 20 |
| U33 | Chiloé | −42.168 | −73.479 | 07-12-07 | 25 | 0 | 0 | 25 |
| U20 | Chiloé | −42.208 | −73.401 | 09-11-07 | 26 | 0 | 0 | 26 |
| Total | 315 | 59 | 2 | 376 | ||||
| Falkland Islands | ||||||||
| North Arm | E. Falkland | NA | NA | 2007/08 | 1 | 30 | 0 | 31 |
| Half-way House | E. Falkland | −51.997 | −59.283 | 2008/09 | 4 | 2 | 0 | 6 |
| Findlay Creek | E. Falkland | −51.888 | −59.025 | 2008/09 | 0 | 2 | 0 | 2 |
| N.W. Arm House | E. Falkland | −52.167 | −59.487 | 26-01-09 | 0 | 3 | 0 | 3 |
| Deep Arroyo | E. Falkland | −51.955 | −59.208 | 28-01-09 | 0 | 2 | 0 | 2 |
| Bull Pass | E. Falkland | −51.890 | −59.007 | 27-01-09 | 1 | 2 | 0 | 3 |
| Spots Arroyo | E. Falkland | −52.025 | −59.343 | 27-01-09 | 4 | 3 | 1 | 8 |
| Fish Creek | W. Falkland | −51.891 | −60.368 | 10-12-08 | 3 | 1 | 0 | 4 |
| Stewarts Brook | W. Falkland | −52.048 | −60.682 | 01-12-08 | 0 | 2 | 0 | 2 |
| Gibraltar Stream | W. Falkland | −52.091 | −60.331 | 05-12-08 | 1 | 3 | 0 | 4 |
| First Arroyo | W. Falkland | −52.083 | −60.534 | 2008/09 | 7 | 0 | 0 | 7 |
| Outflow L. Sullivan | W. Falkland | −51.792 | −60.211 | 11-12-08 | 0 | 2 | 0 | 2 |
| Poncho Valley | W. Falkland | −51.973 | −60.435 | 08-12-08 | 1 | 1 | 0 | 2 |
| Mt Rosalie House | W. Falkland | −51.485 | −59.368 | 21-01-09 | 0 | 1 | 0 | 1 |
| Red Pond | W. Falkland | −51.557 | −59.612 | 17-12-08 | 0 | 3 | 0 | 3 |
| Total | 22 | 57 | 1 | 80 | ||||
| Grand total | 337 | 116 | 3 | 456 |
Figure 1. Accepted distribution of the genus Aplochiton in Chilean Patagonia and the Falkland Islands, based on data published in [16], [17] (represented by circles) and samples collected during the present study (represented by squares).
mtDNA analysis
DNA was extracted using the Wizard® SV 96 DNA Purification Kit following manufacturer's instructions. For mtDNA analysis, regions of the COI (cytochrome c oxidase subunit I) and cyt b (cytochrome b) genes were amplified. A region of 515 bp of the 5′ region of the mitochondrial COI gene was amplified for 367 fish using primers FishF1 and FishR1 [30]. In addition, the primers L14724 [34] and H15149 [35] were used to amplify a region of 354 bp of the cyt b gene for 105 fish. PCR was conducted using an initial denaturation step at 95°C for 5 minutes, followed by 35 cycles at 94°C for 30 seconds, 55°C for 1 minute, 72°C for 2 minutes and one cycle for a final extension of 10 minutes at 72°C. Double stranded DNA was purified from the PCR using the High-Throughput Wizard® SV 96 PCR Clean-Up System, quantified using the NanoDrop1000 v.3.7 Spectrophotometer (Thermo Fisher Scientific), and both strands were sequenced on an ABI 3100 DNA analyser (Applied Biosystems CA, USA). Sequences were aligned using BioEdit v. 7.0.9 [36] and corrected by eye. Cyt b sequences were aligned against the Aplochiton cyt b sequence deposited in Genbank [37].
Intraspecific diversity was estimated by the number of haplotypes (H) and nucleotide diversity (π) [38] using DnaSP v.5 [39]. Intra- and interspecific divergence were calculated using the Kimura-2- parameter (K2P) distance [40] in MEGA 4.0 [41].
Microsatellite analysis
Amplifications were performed for thirteen microsatellite markers (Aze1-Aze13) originally designed for A. zebra [32], in three separate multiplex PCR reactions (multiplex1: Aze1, Aze2, Aze3, Aze4, Aze5, Aze6; multiplex 2: Aze8, Aze9 and Aze10; multiplex 3: Aze11, Aze12, Aze13 and Aze14) using the QIAGEN Multiplex PCR kit (QIAGEN, Sussex, UK). Touchdown PCR was performed using an initial denaturing step of 15 min at 95°C followed by 8 cycles of 95°C for 45 s, 64°C −56°C annealing for 90 s and extension at 72°C for 1 min. 25 additional cycles were then performed using an annealing temperature of 56°C and a final extension at 72°C for 10 min. PCR products were resolved on an ABI3130×l sequencer and analyzed using GeneMapper v 4.0 (Applied Biosystems, USA).
Microsatellite loci were examined for evidence of gametic disequilibrium using GENEPOP [42]. FSTAT [43] was used to estimate Hardy Weinberg proportions (HWE), number of alleles (A), observed and expected heterozygosities (Ho and He) and genetic distance among populations (FST). Levels of significance were adjusted by sequential Bonferroni correction for multiple tests [44]. Analysis of Molecular Variance (AMOVA) was performed in Arlequin 3.1 [45] in order to estimate the level of genetic variance owed to species differentiation. An UPGMA tree based on Nei's distance [46] was built in TFPGA [47] to provide a graphical representation of the divergence between and within Aplochiton species. Statistical confidence on the UPGMA tree nodes was computed by 10,000 bootstrap permutations.
Hybrid identification
Principal Component Analysis (PCA) based on the multilocus genotypes was carried out in GENETIX [48] in order to separate the species and identify any intermediate genotypes resulting from species admixing [49]. We also used the Bayesian assignment approach implemented in STRUCTURE 2.3.2 [50] assuming K = 2 (burn-in period of 25,000 steps and 100,000 MCMC iterations, 20 runs for each K), applying the admixture model with correlated allele frequencies. The results from the 20 replicates were averaged using the software CLUMPP [51] and the output was represented using DISTRUCT 1.1. Individuals were assigned on the basis of their membership coefficient Q. In order to assess the statistical power of the admixture analysis to detect hybrids, we used HYBRIDLAB [52] to simulate parental and hybrid genotypes. We used 100 Aplochiton zebra and 100 Aplochiton taeniatus (as classified in STRUCTURE by individual membership values of Q>0.9) to simulate the genotypes of 100 individuals from each of the parental and hybrid classes, repeated 10 times. Given the importance of threshold Q-values for identification of hybrids in STRUCTURE [53], we run the simulated purebred and hybrid individuals in STRUCTURE using an admixture model with no prior information and K = 2 to define the appropriate Q for individual assignment with our set of microsatellites. STRUCTURE was also used to compare the structuring within species. Finally, potential hybrids were checked for the presence of private alleles of A. zebra and A. taeniatus in their genotypes. Private alleles were defined as occurring only in one species or occurring in both species but with a frequency of less than 1% in one of them to incorporate the possibility of genotype and/or sampling error [54].
Results
mtDNA (COI and cyt b) sequence variation
We sequenced a total 367 Aplochiton sp., 335 of which had been identified as A. zebra and 32 as A. taeniatus using phenotypic criteria. All 367 individuals were sequenced for COI and 105 of these were also sequenced for cyt b.
Based on COI sequence variation, the 367 individuals were resolved into two distinct haplogroups consisting of 262 (haplogroup A) and 105 individuals (haplogroup B), respectively (Figure S1). All of the 262 individuals of haplogroup A had been identified as A. zebra based on phenotypic criteria, whereas 32 out of the 105 individuals of haplogroup B had been initially identified as A. taeniatus. On this basis, we classified fish in haplogroup A as A. zebra and fish in haplogroup B as A. taeniatus. We detected 6 unique haplotypes defined by 5 mutations amongst A. zebra (HA1 n = 237, HA2 n = 5, HA3 n = 6, HA4 n = 9, HA5 n = 4, HA6 n = 1 individual in each case) and 4 haplotypes differing in 3 mutations amongst A. taeniatus (HB1 n = 68; HB2 n = 1; HB3 n = 21; HB4 n = 15; GenBank accession numbers HQ540330-HQ540339). Nucleotide diversity was π = 0.00115 ± 0.00011 for A. taeniatus and π = 0.00044 ± 0.00008 for A. zebra.
For the cyt b region, amplification failed in 75 individuals identified as A. zebra based on phenotype and COI sequence, possibly as a result of mutation in one of the priming sites. Variation in the cyt b sequence resolved the remaining 105 individuals into two haplogroups, consisting of 22 and 83 fish respectively. Using the previous COI classification, fish in the first group corresponded to A. zebra (Haplogroup A), while fish in the second group corresponded to A. taeniatus (Haplogroup B). The most common haplotype of A. taeniatus showed 100% base agreement with the unique A. zebra cyt b haplotype present in GenBank [37], which was derived from a single specimen collected in the Falkland Islands (Waters pers. comm.). This suggests that the sample in Genbank may have been misidentified, given that A. taeniatus had not been described in the Falkland Islands previously [10], [17].
We detected 5 cyt b unique haplotypes defined by 4 mutations amongst A. zebra (HA1 n = 7; HA2 n = 8; HA3 n = 1; HA4 n = 5; HA5 n = 1) and 3 haplotypes differing in 3 mutations amongst A. taeniatus (HB1 n = 4; HB2 n = 78; HB3 n = 1; GenBank accession numbers HQ540340-HQ540347). As with COI, none of the haplotypes were shared between species. Nucleotide diversity was low for both species: π = 0.00292 ± 0.0004 for A. zebra and π = 0.00039 ± 0.00017 for A. taeniatus.
Extent of intraspecific and interspecific divergence
The two species differed in 38 fixed mutations at COI and 31 fixed mutations at the 354 bp fragment of cyt b, and did not share any haplotypes or mutations in either marker. Intraspecific divergence (K2P distance) at COI was 0.0012 ± 0.0002 for A. zebra and 0.0015 ± 0.0005 for A. taeniatus, while interspecific distance was 70 fold greater (0.0882 ± 0.0142). At cyt b, intraspecific K2P divergence was 0.0029±0.0018 for A. zebra and 0.0004±0.0003 for A. taeniatus. Again, divergence was approximately two orders of magnitude greater between species (0.1005 ± 0.0173) than within species. Furthermore, classification agreement for individuals amplified for both markers was 100%. Thus, both mtDNA markers provided complete and concordant species discrimination.
Microsatellite analysis
A total of 456 Aplochiton samples (367 of which were also sequenced for mtDNA) were genotyped using 13 nuclear microsatellite markers. Results from the admixture analysis conducted in STRUCTURE assuming K = 2 and based on 11 microsatellites (excluding two microsatellites with low amplification success in A. taeniatus) separated the samples into three distinct groups: a first group of 338 individuals classified as A. zebra by barcoding, a second group of 113 individuals classified as A. taeniatus by barcoding, and a third group of five individuals representing potential hybrids (Figure S2).
Eleven of the thirteen microsatellites (85%) originally designed for A. zebra reliably amplified in A. taeniatus and nine (69%) were polymorphic. Successful cross-amplification was observed for all of the 13 A. zebra microsatellites analysed, although two of them (Aze11 and Aze13) amplified only in 33% and 6% of the A. taeniatus samples, respectively. Two of the microsatellites, Aze4 (allelic size 99 bp) and Aze14 (allelic size 92 bp) were monomorphic for A. taeniatus (Table 2 and Table S1). These alleles were private (Aze14-92) to A. taeniatus or present only at a very low frequency in A. zebra (1.2%; Aze4-99), and therefore in combination these could be used to discriminate between A. taeniatus and A. zebra. In total 70% of the alleles were private to one of the two species.
Table 2. Sample size (N), allele size ranges, number of alleles (Na), expected heterozygosity (He) and observed heterozygosity (Ho) for the microsatellite markers Aze1-Aze14 for Aplochiton taeniatus and Aplochiton zebra.
| A. taeniatus | A. zebra | |||||||||
| Locus | N | Size range | Na | He | Ho | N | Size range | Na | He | Ho |
| Aze1 | 99 | 125–139 | 5 | 0.12 | 0.12 | 336 | 121–139 | 8 | 0.47 | 0.44 |
| Aze2 | 106 | 127–169 | 13 | 0.43 | 0.26 | 333 | 123–169 | 13 | 0.76 | 0.68 |
| Aze3 | 110 | 87–89 | 2 | 0.15 | 0.19 | 337 | 75–91 | 5 | 0.46 | 0.39 |
| Aze4 | 108 | 99 | 1 | 0.00 | 0.00 | 260 | 89–115 | 10 | 0.54 | 0.26 |
| Aze5 | 106 | 122–217 | 29 | 0.91 | 0.84 | 333 | 122–321 | 88 | 0.97 | 0.88 |
| Aze6 | 109 | 151–177 | 10 | 0.67 | 0.69 | 324 | 157–191 | 16 | 0.84 | 0.70 |
| Aze8 | 109 | 201–225 | 7 | 0.47 | 0.29 | 329 | 173–307 | 43 | 0.94 | 0.70 |
| Aze9 | 106 | 79–175 | 20 | 0.90 | 0.84 | 324 | 79–267 | 45 | 0.97 | 0.88 |
| Aze10 | 110 | 166–172 | 4 | 0.53 | 0.50 | 333 | 154–190 | 16 | 0.86 | 0.78 |
| Aze11 | 38 | 124–164 | 13 | 0.87 | 0.49 | 337 | 108–172 | 20 | 0.69 | 0.56 |
| Aze12 | 107 | 127–253 | 32 | 0.80 | 0.61 | 323 | 151–241 | 40 | 0.93 | 0.84 |
| Aze13 | 7 | 134–176 | 6 | 0.80 | 0.10 | 336 | 124–174 | 18 | 0.69 | 0.63 |
| Aze14 | 108 | 92 | 1 | 0.00 | 0.00 | 337 | 100–112 | 7 | 0.32 | 0.21 |
These observed rates of cross-amplification (85%) and polymorphism (69%) fall within the expected range of cross-species microsatellite amplification/polymorphism success, given the evolutionary distance of the two Aplochiton species estimated by pairwise cyt b genetic distances. Thus, using our estimated rate of cyt b divergence between A. zebra and A. taeniatus (0.1005), the expected rates of amplification and polymorphism would be 84% and 42%, or 56% and 33%, using the relationship found for cetaceans and frogs, respectively [55].
Pairwise population differentiation comparison showed high levels of divergence between both species with FST values ranging from 0.24 to 0.32 (P<0.001). Analysis of molecular variance (AMOVA) of the two species of Aplochiton (excluding hybrids) showed that 25.2% of genetic variance was distributed between species (FST = 0.300; P<0.001), while 4.8% of the variance was due to differences between populations within species (FSC = 0.065; P<0.001), and 70% of the variance was due to variation among individuals within populations (FCT = 0.252; P<0.001). Average observed heterozygosity (excluding the Az13 locus) and allelic richness were lower for A. taeniatus (H o = 0.37; Ar = 3.5) than for A. zebra (H o = 0.61; Ar = 2.5).
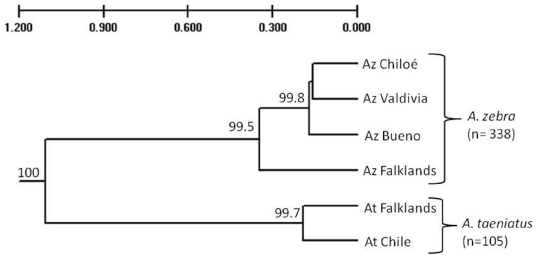
The UPGMA tree clustered the individuals in two main groups supported by high bootstrap values (higher than 99%). This is in agreement with results from mtDNA analyses and revealed some further regional structuring (Figure 2). In order to compare the relative structuring of both species, FST analyses of genetic distance were carried out among populations within species when the sample size allowed it. Among populations of A. zebra in Chile, the estimated genetic distance was FST = 0.045 but a similar analysis could not be carried out in the Falklands due to the limited number of individuals per population (Table 1). Genetic distance for A. taeniatus in the Falklands was estimated by grouping the individuals regionally in East and West Falklands (FST = 0.039 P<0.001), while for Chile only the two populations with enough A. taeniatus (R. Huincha and U30) were compared (FST = 0.010 P = 0.025). Genetic differentiation between Chilean Patagonia and the Falkland Islands was highly significant for both species (Figures 2, 3), being greater for A. taeniatus (FST = 0.135, P<0.0010) than for A. zebra (FST = 0.084, P<0.001).
Figure 2. UPGMA clustering of A. zebra and A. taeniatus based on microsatellite markers using Nei's original distance [46].
Numbers at each branch node represent % bootstrap support derived from 1,000 replicates.
Figure 3. Species discrimination and identification of hybrids based on Principal Component Analysis of Aplochiton taeniatus and Aplochiton zebra microsatellite genotypes.
PC1 and PC2 represent the first two factorial components. Three putative F1 hybrids are indicated by their ID sample codes.
Identification of hybrids
Principal Component Analysis (PCA) based on microsatellite genotypes of individual Aplochiton revealed a clear segregation between Aplochiton species and between geographical regions within species (Figure 3). Three individuals were identified as hybrids, and of them had mtDNA haplotypes typical of A. taeniatus (both for cyt b and COI), but microsatellite alleles unique to A. zebra. Results from admixture analysis using STRUCTURE and assuming K = 2 assigned 451 individuals as A. taeniatus or A. zebra while 5 individuals showed genotypes admixed between both clusters (Figure S2). A. zebra membership coefficients (Q1) ranged between 0.84 and 0.99 (mean: 0.99±0.02) and A. taeniatus membership coefficients (Q2) ranged from 0.89 to 0.99 (mean: 0.99±0.01). Pure individuals from each species with Q>0.9 were used to simulate parental and hybrid classes in HYBRIDLAB. Using STRUCTURE, the simulated parental classes (A. zebra and A. taeniatus) were correctly assigned to their species (Figure S2) with a minimum Q threshold of 0.88 (average Q = 0.96). F1 hybrids were also correctly identified with Qmax = 0.67 and so were the F2 hybrids and backcrosses showing a Qmax = 0.79.
None of the hybrids showed a multilocus genotype which could be classified as parental. However, we did not observe strong differences in Q values for the different hybrid classes (averages Qmin/max = 0.46–0.54). On this basis, we identified three potential F1 hybrids from the admixture analysis that corresponded to those individuals previously classified as hybrids by PCA analysis, with the following relative admixture values: Q1/Q2 = 0.37/0.63; Q1/Q2 = 0.60/0.40; and Q1/Q2 = 0.67/0.33 (Figure S2). Two further individuals were identified by STRUCTURE (but not by PCA) as potential hybrids, one with A. taeniatus mtDNA (AtChile-36) with admixture value Q1/Q2 = 0.26/0.74 and a second with A. zebra mtDNA (AzU29-36) and admixture coefficients Q1/Q2 = 0.84/0.16 (Figure S2). Based on their Q values, these fish could represent F2 or backcrosses, but results from HYBRIDLAB suggest that our combination of microsatellites did not allow for accurate discrimination.
The three F1 hybrids all possessed A. taeniatus mtDNA and showed between four and six clearly introgressed microsatellite alleles (Table 3). Two of the three hybrids had been phenotypically identified as A. zebra, and one as A. taeniatus. Thirty per cent of the alleles were shared between species. The three identified hybrids possessed two A. taeniatus alleles fixed for Aze 4 (99) and Aze 14 (92), and were heterozygous for Aze 14 (92) and one of the A. zebra private alleles (108). Two of the hybrids (ATChile-44 and ATFalklands-41) were homozygous for the A. taeniatus characteristic 99 allele (Aze 14) that only appears at low frequency in A. zebra (1.34%), and were homozygous for alleles private to A. zebra for one microsatellite locus (Aze 10; ATFalklands-41) and two microsatellite loci (Aze 2 and Aze 10; ATChile-44), respectively. The remaining hybrid was heterozygous for private alleles of each species in five of the eleven markers, and had shared alleles in the remaining six markers.
Table 3. Population frequencies of A. zebra alleles present in the three hybrids with A. taeniatus mitochondrial DNA haplotype.
| Locus | Allele | A. taeniatus (%) | A. zebra (%) |
| Aze2 | 127 | 0.47 | 40.57 |
| 129 | 0 | 14.67 | |
| Aze4 | 103 | 0 | 56.51 |
| Aze6 | 173 | 0 | 11.84 |
| Aze10 | 174 | 0 | 5.53 |
| 178 | 0 | 15.86 | |
| 180 | 0 | 18.26 | |
| Aze12 | 183 | 0.91 | 11.88 |
| Aze14 | 108 | 0 | 78.4 |
Phenotypic misidentification
The incidence of phenotypic misidentification was relatively high (85/454 = 19%) and, perhaps more importantly, highly asymmetric (Table 4; McNemar symmetry test χ2 = 85, df = 1, P<0.001). Thus, whereas all the 339 fish genetically classified as A. zebra were correctly identified as such based on their phenotype, most (85/115 or 74%) of the A. taeniatus were wrongly identified as A. zebra. Misidentification was particularly evident in the case of the Falkland Islands, where only one species (Aplochiton zebra) was thought to exist.
Table 4. Classification of matched samples of Aplochiton zebra and Aplochiton taeniatus based on morphometric traits (phenotype) and molecular markers (COI, cytochrome b and microsatellites).
| Classification by molecular markers | Identification by phenotypic criteria | |||
| A. zebra | A. taeniatus | Total | % agreement | |
| A. zebra | 338 | 0 | 338 | 100 |
| A. taeniatus | 85 | 30 | 115 | 26.1 |
| Hybrids | 2 | 1 | 3 | |
| Total | 426 | 31 | 456 | |
| % agreement | 79.6 | 96.8 | ||
Distributional range and abundance of A. zebra and A. taeniatus
Our results indicate that the two Aplochiton species occur sympatrically in Chilean Patagonia and also in the Falkland Islands, although their relative abundances differed significantly across sample sites (Table 1; G-test = 183.59, df = 3, P<0.001). In Chile, A. taeniatus appears to be more abundant on the Island of Chiloé (98% of individuals), whereas A. zebra was more abundant in mainland samples (58% of individuals; Fisher exact test, P<0.001). Likewise, in the Falkland Islands, the relative abundance of the two species appears to differ significantly between the two islands (Fisher exact test, P = 0.014): A. taeniatus appears to be the dominant species in East Falkland (81%), whereas the two species appear to be equally common in West Falkland (52% vs. 48%).
A. taeniatus was significantly larger than A. zebra in both Chile and the Falklands (Figure 4; Species effect F 1,441 = 233.4, P<0.001), though the size difference was more pronounced in the Falkland Islands than in Chile (Species x Location interaction F 1,441 = 38.39, P<0.001). Aplochiton in the Falklands were significantly larger than in Chile, regardless of species identity (Location effect F 1,441 = 4.62, P = 0.032). Analysis of condition factor indicates that A. taeniatus has a thinner, more streamlined body than A. zebra (F 1,435 = 23.5, P<0.001), regardless of location (F 1,435 = 0.93, P = 0.334).
Figure 4. Size variation (fork length, mm) of A. taeniatus and A. zebra in Chilean Patagonia and the Falkland Islands, as inferred from molecular identification.
Discussion
By using two different mtDNA markers commonly employed for species barcoding, our phylogenetic reconstruction of Aplochiton reveals two distinct, non-overlapping haplogroups corresponding to A. zebra and A. taeniatus. The average intraspecific distance among individuals was 0.12% for A. zebra and 0.15% for A. taeniatus, compared to 8.8% between the two Aplochiton species. The observed divergence is thus 60–100 times higher between groups than among individuals within groups, supporting the contention that A. zebra and A. taeniatus are indeed two different species, and that DNA barcoding correctly identified them as such [30].
Furthermore, results using 11 microsatellite markers were fully consistent with the groups previously identified by DNA barcoding.
The observed ratio of intra to interspecific divergence in Aplochiton is very similar to that reported for many other fish species across several families [30], [56], where the average intraspecific distance was 0.39% and the average interspecific distance was 8.11–9.93%. On the other hand, we found no evidence to support the existence of a third species (A. marinus), whose presence in Chilean Patagonia has been suggested by some workers [8]–[9], [16], [57]. Although our Chilean samples were collected from 20 different locations, the sites were largely concentrated in the northern part of the species' range. With this caveat in mind, we suggest that molecular evidence is needed to clarify the taxonomic status of A. marinus. As indicated by McDowall [17], [24]–[25], it is possible that A. marinus is simply the migratory form of A. taeniatus.
Our data suggest that misidentification of Aplochiton could be common. Indeed, 19% of A. taeniatus in our study were misidentified as A. zebra based on phenotypic traits, suggesting that A. taeniatus could occur as two ecotypes, one which is readily identified by workers in the field, and another, more cryptic form, which is often mistaken for A. zebra. In contrast, all individuals classified as A. zebra using genetic data were correctly identified as such using phenotypic traits. Despite the threat that invasive salmonids pose to the conservation of Aplochiton [17]–[19], it is not clear to what extent the two species are equally vulnerable to salmonid invasions, or whether one species has been more impacted than the other. Both species appear to occupy similar fast-flowing habitats [16]–[17], but may have different diets, as A. taeniatus is thought to have a specialised piscivorous diet, while A. zebra appears to feed mostly on invertebrates. Our results indicate that A. taeniatus tends to have a more streamlined body and attain a larger size than A. zebra, which is consistent with previous findings [17], [24] and a greater dependence on piscivory, as reported initially by McDowall & Nakaya [12]. As invasion success in fish often depends on invader body size [58], a larger body size may make A. taeniatus more resilient than A. zebra to salmonid invasions, but its more specialised diet may also make it a more vulnerable to competition from ecologically similar salmonids.
Whatever the precise nature of salmonid impacts, misidentification could have important consequences for the conservation of endangered Aplochiton, if it leads to inappropriate protection measures or fails to recognise the species' distinct needs [59]. Misidentification can also have important implications for ex-situ conservation [60], as captive breeding could inadvertently produce hybrids and impact on the very same species targeted for conservation.
DNA barcoding in our study provides the first evidence of A. taeniatus in the Falkland Islands, and shows that the species is more widely distributed than previously thought, being present in both East and West Falkland. Molecular data also suggest that A. zebra might be less common - and its distribution more restricted - than reported in recent studies [16]. The two Aplochiton species appear to occur sympatrically across their entire range (Figure 1) and our study provides clear evidence, for the first time, that they also hybridise in the wild. Three hybrids were detected by both PCA and admixture analyses, and based on their membership coefficients (Q), these are most likely F1 hybrids. Two further hybrids were also identified by admixture analyses, but simulations carried out with HYBRIDLAB could not unambiguously resolve their origin as either F2 or backcrosses. If we consider only the unambiguous F1 hybrids identified by both PCA and STRUCTURE, the presence of A. taeniatus mtDNA indicates that the direction of hybridisation was in all cases via female A. taeniatus and male A. zebra. Such asymmetric hybridisation could have resulted from prezygotic barriers [61]–[62], postzygotic effects [63]–[64] or a combination of both [65]. Given that A. zebra will normally be smaller than A. taeniatus, this may have facilitated sneaking behaviour by male A. zebra during reproduction, leading to asymmetrical hybridisation, as observed in other fish species ([66]). Asymmetrical hybridisation can also arise from differences in the relative abundance of parental species, with the less common species typically becoming the female parent [67], and from Dobzhansky-Muller incompatibilities, whereby reciprocal interspecific crosses produce different rates of fertilization and/or sterility [68].
Although the low number of hybrids in our study precludes further testing of potential causes of Aplochiton hybridisation, the hypothesis that the less abundant species provides the female parent does not appear to be consistent with the data. Thus, A. zebra was dominant in one river, A. taeniatus in another river, and the two species were found in roughly the same proportions in the third river where hybrids were detected. When gene flow varies amongst hybridising species, interspecific introgression is more likely to occur in the more fragmented species [69]. In that sense, the degree of differentiation between Chilean and Falklands populations was greater for A. taeniatus than for A. zebra, but the structuring of Chilean A. zebra was similar in magnitude to the structuring of A. taeniatus within the Falklands, and hybrids occurred in both regions. Further studies, particularly in the Falkland Islands, are needed in order to clarify the roles of population fragmentation and mating behaviour on asymmetrical hybridisation in Aplochiton. Sexing would also reveal whether the two Aplochiton species conform to Haldane's rule, whereby the heterogametic sex is usually absent, rare, or sterile amongst F1 hybrids [70], and whether populations exhibit fluctuating sex ratios, as found in other Salmoniformes [71].
Recent work suggests that hybridisation is relatively frequent in animals, even if it tends to occur at low rates [72] and is typically less widespread than among plants. Hybridisation can play an important role in species' evolution [73], either enhancing or reducing the adaptive persistence of hybridising species. Thus, interspecific gene introgression could increase the genetic diversity and evolutionary potential of hybrids, while outbreeding depression could render them unviable or infertile [74]. Anthropogenic stressors, such as environmental degradation or introduction of exotic species, have been found to increase hybridisation rates, though the mechanisms can be subtle and not readily apparent. For example, increased water turbidity in Lake Victoria seems to have disrupted visually-mediated mate choice and reproductive isolation among cichlids [75], while stocking with hatchery-reared fish may have facilitated salmonid hybridisation in the Iberian peninsula [76]. Hybridisation can also occur when one species expands into the other species' range and there are no reproductive barriers [49]. In this sense, it is unclear what role, if any, exotic salmonids may have played in Aplochiton hybridisation. Invasive salmonids are widespread in Chilean Patagonia [77] and the Falkland Islands [10], where they displace and outcompete native galaxiids [17]–[20], but whether they may have facilitated Aplochiton hybridisation by increasing secondary contact [78] is not clear.
Like most galaxiids, the biology and conservation needs of Aplochiton are poorly known [17], and this probably constitutes one of the biggest obstacles to their conservation [18]. Almost 20% of galaxiids have only been identified over the last 25 years, and in most cases their conservation status has either not been evaluated (NE −55%) or suffers from data deficiency (DD −14%). Given that most of the remaining galaxiids are listed by the IUCN Red List as being critically endangered (CR, 8%), or vulnerable (VU, 18%), the implications of taxonomic misclassification could be serious because under such data deficient scenarios management may be acting upon the wrong species. Such uncertainty, termed epistemic uncertainty [79], results from lack of knowledge and represents a property of the observer, and therefore extrinsic to the scientific problem being addressed. Our study illustrates how molecular markers can help to decrease epistemic uncertainty in the identification of Aplochiton, paving the way for more efficient conservation programmes.
In summary, we show for the first time that the two Aplochiton species occur in sympatry and hybridise in Chilean Patagonia and also in the Falkland Islands, where only A. zebra was thought to be present and where our study indicates that A. taeniatus might in fact be the most common species. We also show that some microsatellite markers are diagnostic for Aplochiton, and provide a first estimate of genetic diversity and regional differentiation for these species. Finally, we reveal through DNA barcoding that phenotypic misidentification is common and caution against sole reliance on morphological traits for species delimitation of Aplochiton and other poorly known galaxiid fishes.
Supporting Information
Nucleotide sequence alignment of mitochondrial DNA of Aplochiton zebra (A) and Aplochiton taeniatus (B) according to (a) COI haplogroups and (b) Cyt b haplogroups.
(DOC)
Hybrid assignments based on (a) simulated membership proportions of 100 multilocus genotypes per class using HYBRIDLAB and (b) results of admixture analysis using STRUCTURE: A. zebra parentals, A. taeniatus parentals, F1 hybrids, F2 hybrids and backcrosses. Results from the admixture analysis in STRUCTURE are for K = 2, averaged from 20 runs. Each bar constitutes an individual genotype. Y-axis represents the proportion of each individual attributable to each cluster, which can be deduced from the colour of the bars. The horizontal line represents the upper limit for pure bred individuals estimated with HYBRYDLAB. Pure A. zebra genotypes are represented in pale grey; pure A. taeniatus genotypes are represented in dark grey. Potential hybrids are identified by an asterisk (*).
(DOC)
Microsatellite allele frequencies per locus per species (Aze1-Aze4).
(DOC)
Acknowledgments
We are grateful to Ben Perry, Jessica Stephenson, Jane MacDonald, Ana Maria Cerda, Alexandre Terreau, Anne-Flore Thailly and a number of volunteers for help in collecting the samples. We also thank Brendan Gara for logistic support in the Falklands.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding for this study was provided by a DEFRA Darwin Initiative (Grant No. 162/15/020) and a Darwin post-project award ‘Protecting galaxiids from salmonid invasions in Chile and the Falklands’ (Grant No. EIDPO-041; www.biodiversity.cl); with additional support from the University of Los Lagos (Chile) and Falkland Islands Development Corporation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hendry AP, Lohmann LG, Conti E, Cracraft J, Crandall KA, et al. Evolutionary biology in biodiversity science, conservation, and policy: a call to action. Evolution. 2010;64:1517–1528. doi: 10.1111/j.1558-5646.2010.00947.x. [DOI] [PubMed] [Google Scholar]
- 2.Milne DJ, Jackling FC, Sidhu M, Appleton BR. Shedding new light on old species identifications: morphological and genetic evidence suggest a need for conservation status review of the critically endangered bat, Saccolaimus saccolaimus. Wildlife Research. 2009;36:496–508. [Google Scholar]
- 3.Oliveira R, Castro D, Godinho R, Luikart G, Alves P. Species identification using a small nuclear gene fragment: application to sympatric wild carnivores from South-western Europe. Conservation Genetics. 2010;11:1023–1032. [Google Scholar]
- 4.Speziale K, Lambertucci S. A call for action to curb invasive species in South America. Nature. 2010;467:153–153. doi: 10.1038/467153c. [DOI] [PubMed] [Google Scholar]
- 5.Clavero M, García-Berthou E. Invasive species are a leading cause of animal extinctions. Trends in Ecology & Evolution. 2005;20:110–110. doi: 10.1016/j.tree.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Cambray JA. Impact on indigenous species biodiversity caused by the globalisation of alien recreational freshwater fisheries. Hydrobiologia. 2003;500:217–230. [Google Scholar]
- 7.Miller RR, Williams JD, Williams JE. Extinctions of North American fishes during the past century. Fisheries. 1989;14:22–38. [Google Scholar]
- 8.Dyer B. Systematic review and biogeography of the freshwater fishes of Chile. Estudios Oceanológicos. 2000;19:77–98. [Google Scholar]
- 9.Cussac V, Ortubay S, Iglesias G, Milano D, Lattuca ME, et al. The distribution of South American galaxiid fishes: the role of biological traits and post-glacial history. Journal of Biogeography. 2004;31:103–121. [Google Scholar]
- 10.McDowall R, Allibone R, Chadderton W. Issues for the conservation and management of Falkland Islands freshwater fishes. Aquatic Conservation: Marine and Freshwater Ecosystems. 2001;11:473–486. [Google Scholar]
- 11.McDowall RM, Nakaya K. Identity of the galaxioid fishes of the genus Aplochiton Jenyns from Southern Chile. Japanese Journal of Ichthyology. 1987;34:377–383. [Google Scholar]
- 12.McDowall RM, Nakaya K. Morphological divergence in the two species of Aplochiton Jenyns (Salmoniformes, Aplochitonidae) - a generalist and a specialist. Copeia. 1988;1:233–236. [Google Scholar]
- 13.Lattuca ME, Brown D, Castineira L, Renzi M, Luizon C, et al. Reproduction of landlocked Aplochiton zebra Jenyns (Pisces, Galaxiidae). Ecology of Freshwater Fish. 2008;17:394–405. [Google Scholar]
- 14.McDowall RM. The evolution of diadromy in fishes (revisited) and its place in phylogenetic analysis. Reviews in Fish Biology and Fisheries. 1997;7:443–462. [Google Scholar]
- 15.Pascual MA, Cussac V, Dyer B, Soto D, Vigliano P, et al. Freshwater fishes of Patagonia in the 21st Century after a hundred years of human settlement, species introductions, and environmental change. Aquatic Ecosystem Health & Management. 2007;10:212–227. [Google Scholar]
- 16.Habit E, Piedra P, Ruzzante DE, Walde SJ, Belk MC, et al. Changes in the distribution of native fishes in response to introduced species and other anthropogenic effects. Global Ecology and Biogeography. 2010;19:697–710. [Google Scholar]
- 17.McDowall RM. Crying wolf, crying foul, or crying shame: alien salmonids and a biodiversity crisis in the southern cool-temperate galaxioid fishes? Reviews in Fish Biology and Fisheries. 2006;16:233–422. [Google Scholar]
- 18.Garcia de Leaniz C, Gajardo G, Consuegra S. From best to pest: changing perspectives on the impact of exotic salmonids in the southern hemisphere. Systematics and Biodiversity. 2010;8:447–459. [Google Scholar]
- 19.Young KA, Dunham JB, Stephenson JF, Terreau A, Thailly AF, et al. A trial of two trouts: comparing the impacts of rainbow and brown trout on a native galaxiid. Animal Conservation. 2010;13:399–410. [Google Scholar]
- 20.Young K, Stephenson J, Terreau A, Thailly A-F, Gajardo G, et al. The diversity of juvenile salmonids does not affect their competitive impact on a native galaxiid. Biological Invasions. 2009;11:1955–1961. [Google Scholar]
- 21.McDowall R. Fighting the flow: downstream–upstream linkages in the ecology of diadromous fish faunas in West Coast New Zealand rivers. Freshwater Biology. 1998;40:111–122. [Google Scholar]
- 22.McDowall R. Diadromy and genetic diversity in Nearctic and Palearctic fishes. Molecular Ecology. 1999;8:527–528. [Google Scholar]
- 23.Milano D, Ruzzante DE, Cussac VE, Macchi PJ, Ferriz RA, et al. Latitudinal and ecological correlates of morphological variation in Galaxias platei (Pisces, Galaxiidae) in Patagonia. Biological Journal of the Linnean Society. 2006;87:69–82. [Google Scholar]
- 24.McDowall RM. Fishes of the family Aplochitonidae. Journal of the Royal Society of New Zealand. 1971a;1:31–52. [Google Scholar]
- 25.McDowall R. The galaxiid fishes of South America. Zoological Journal of the Linnean Society. 1971b;50:33–73. [Google Scholar]
- 26.Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360:1805–1811. doi: 10.1098/rstb.2005.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witt JDS, Threloff DL, Hebert PDN. DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: implications for desert spring conservation. Molecular Ecology. 2006;15:3073–3082. doi: 10.1111/j.1365-294X.2006.02999.x. [DOI] [PubMed] [Google Scholar]
- 29.Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia's fish species. Philosophical Transactions of the Royal Society of London B Biological Sciences. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kochzius M, Seidel C, Antoniou A, Botla SK, Campo D, et al. Identifying fishes through DNA barcodes and microarrays. PLoS One. 2010;5:Article No: e12620. doi: 10.1371/journal.pone.0012620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggarwal RK, Allainguillaume J, Bajay MM, Barthwal S, Bertolino P, et al. Permanent Genetic Resources added to Molecular Ecology Resources Database 1 August 2010–30 September 2010. Molecular Ecology Resources. 2011;11:219–222. doi: 10.1111/j.1755-0998.2010.02944.x. [DOI] [PubMed] [Google Scholar]
- 33.Blackwell BG, Brown ML, Willis DW. Relative weight (Wr) status and current use in fisheries assessment and management. Reviews in Fisheries Science. 2000;8:1–44. [Google Scholar]
- 34.Paabo S. Amplifying Ancient DNA. Academic Press, San Diego; 1990. [Google Scholar]
- 35.Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, et al. Dynamics of mitochondrial-dna evolution in animals - amplification and sequencing with conserved primers. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 37.Waters JM, White RWG. Molecular phylogeny and biogeography of the Tasmanian and New Zealand mudfishes (Salmoniformes : Galaxiidae). Australian Journal of Zoology. 1997;45:39–48. [Google Scholar]
- 38.Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- 39.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 40.Kimura M. A simple method of estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal Of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 42.Raymond M, Rousset F. GENEPOP (version-1.2) - Population-genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- 43.Goudet J. FSTAT (Version 1.2): A computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- 44.Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 45.Excoffier L, Laval G, Schneider S. Arlequin ver. 3.1: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 46.Nei M. Genetic distance between populations. American Naturalist. 1972;106:283. [Google Scholar]
- 47.Miller MP. Tools for population genetic analyses (TFPGA) 1.3: A Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author 1997 [Google Scholar]
- 48.Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.02, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR. Université de Montpellier II, Montpellier (France) 2001 [Google Scholar]
- 49.Pastorini J, Zaramody A, Curtis D, Nievergelt C, Mundy N. Genetic analysis of hybridisation and introgression between wild mongoose and brown lemurs. BMC Evolutionary Biology. 2009;9:32.49. doi: 10.1186/1471-2148-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. . Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen EEG, Bach LA, Kotlicki P. HYBRIDLAB (version 1.0): a program for generating simulated hybrids from population samples. Molecular Ecology Notes. 2006;6:971–973. [Google Scholar]
- 53.Vaha JP, Primmer CR. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridisation scenarios and with different numbers of loci. Molecular Ecology. 2006;15:63–72. doi: 10.1111/j.1365-294X.2005.02773.x. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira R, Godinho R, Randi E, Ferrand N, Alves PC. Molecular analysis of hybridisation between wild and domestic cats (Felis silvestris) in Portugal: implications for conservation. Conservation Genetics. 2008;9:1–11. [Google Scholar]
- 55.Primmer CR, Painter JN, Koskinen MTU, Palo J, Merilä J. Factors affecting avian cross-species microsatellite amplification. Journal of Avian Biology. 2005;36:348–360. [Google Scholar]
- 56.Ward RD, Hanner R, Hebert PDN. The campaign to DNA barcode all fishes, FISH-BOL. Journal of Fish Biology. 2009;74:329–356. doi: 10.1111/j.1095-8649.2008.02080.x. [DOI] [PubMed] [Google Scholar]
- 57.Campos H. Distribution of the fishes in the Andean rivers in the south of Chile. Archiv Fur Hydrobiologie. 1985;104:169–191. [Google Scholar]
- 58.Schröder A, Nilsson K, Persson L, Van Kooten T, Reichstein B. Invasion success depends on invader body size in a size-structured mixed predation-competition community. Journal of Animal Ecology. 2009;78:1152–1162. doi: 10.1111/j.1365-2656.2009.01590.x. [DOI] [PubMed] [Google Scholar]
- 59.Daugherty CH, Cree A, Hay JM, Thompson MB. Neglected taxonomy and continuing extinctions of tuatara (Sphenodon). Nature. 1990;347:177–179. [Google Scholar]
- 60.Frankham R, Ballou JD, Briscoe DA. 2002. Introduction to conservation genetics Cambridge: Cambridge University Press.
- 61.Lamb T, Avise JC. Directional introgression of mitochondrial DNA in a hybrid population of tree frogs: The influence of mating behavior. Proceedings of the National Academy of Sciences. 1986;83:2526–2530. doi: 10.1073/pnas.83.8.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peter W. Mother species-father species: unidirectional hybridisation in animals with female choice. Animal Behaviour. 1999;58:1–12. doi: 10.1006/anbe.1999.1144. [DOI] [PubMed] [Google Scholar]
- 63.Welch JJ. Accumulating Dobzhansky-Müller incompatibilities: reconciling theory and data. Evolution. 2004;58:1145–1156. doi: 10.1111/j.0014-3820.2004.tb01695.x. [DOI] [PubMed] [Google Scholar]
- 64.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scribner KT, Avise JC. Population cage experiments with a vertebrate: the temporal dmography and ctonuclear gnetics of hbridisation in Gambusia fishes. Evolution. 1994;48:155–171. doi: 10.1111/j.1558-5646.1994.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 66.Taylor EB, Foote CJ, Wood CC. Molecular genetic evidence for parallel life-history evolution within a Pacific salmon (sockeye salmon and kokanee, Oncorhynchus nerka). Evolution. 1996;50:401–416. doi: 10.1111/j.1558-5646.1996.tb04502.x. [DOI] [PubMed] [Google Scholar]
- 67.Avise JC, Saunders NC. Hybridisation and introgression among species of sunfish (Lepomis): analysis by mitochondrial DNA and allozyme markers. Genetics. 1984;108:237–255. doi: 10.1093/genetics/108.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arntzen JW, Jehle R, Bardakci F, Burke T, Wallis GP. Asymmetric viability of reciprocal-cross hybrids between crested and marbled newts (Triturus cristatus and T. marmoratus). Evolution. 2009;63:1191–1202. doi: 10.1111/j.1558-5646.2009.00611.x. [DOI] [PubMed] [Google Scholar]
- 69.Petit RMJ, Excoffier L. Gene flow and species delimitation. Trends in Ecology & Evolution. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 70.Haldane J. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics. 1922;12:101–109. [Google Scholar]
- 71.Consuegra S, Garcia de Leániz C. Fluctuating sex ratios, but no sex-biased dispersal, in a promiscuous fish. Evolutionary Ecology. 2007;21:229–245. [Google Scholar]
- 72.Arnold ML, Meyer A. Natural hybridisation in primates: One evolutionary mechanism. Zoology. 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 73.Salzburger W, Baric S, Sturmbauer C. Speciation via introgressive hybridisation in East African cichlids? Molecular Ecology. 2002;11:619–625. doi: 10.1046/j.0962-1083.2001.01438.x. [DOI] [PubMed] [Google Scholar]
- 74.Roberts DG, Gray CA, West RJ, Ayre DJ. Evolutionary impacts of hybridisation and interspecific gene flow on an obligately estuarine fish. Journal of Evolutionary Biology. 2009;22:27–35. doi: 10.1111/j.1420-9101.2008.01620.x. [DOI] [PubMed] [Google Scholar]
- 75.Seehausen O, Alphen JJMv, Witte F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science. 1997;277:1808–1811. [Google Scholar]
- 76.Garcia de Leaniz C, Verspoor E. Natural hybridisation between Atlantic salmon (Salmo salar) and brown trout (Salmo trutta) in northern Spain. Journal of Fish Biology. 1989;34:41–46. [Google Scholar]
- 77.Consuegra S, Phillips NC, Gajardo G, Garcia de Leaniz C. Winning the invasion roulette: escapes from fish farms increase admixture and facilitate establishment of non-native rainbow trout. Evolutionary Applications. 2011;4:660–671. doi: 10.1111/j.1752-4571.2011.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brown R, Jordan W, Faulkes C, Jones C, Bugoni L, et al. Phylogenetic relationships in Pterodroma petrels are obscured by recent secondary contact and hybridisation. PloS One. 2011;6:e20350. doi: 10.1371/journal.pone.0020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown V, Gutknecht J, Harden L, Harrison C, Hively D, et al. Understanding and engaging values in policy relevant science. Bulletin of the British Ecological Society. 2010;41:48–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequence alignment of mitochondrial DNA of Aplochiton zebra (A) and Aplochiton taeniatus (B) according to (a) COI haplogroups and (b) Cyt b haplogroups.
(DOC)
Hybrid assignments based on (a) simulated membership proportions of 100 multilocus genotypes per class using HYBRIDLAB and (b) results of admixture analysis using STRUCTURE: A. zebra parentals, A. taeniatus parentals, F1 hybrids, F2 hybrids and backcrosses. Results from the admixture analysis in STRUCTURE are for K = 2, averaged from 20 runs. Each bar constitutes an individual genotype. Y-axis represents the proportion of each individual attributable to each cluster, which can be deduced from the colour of the bars. The horizontal line represents the upper limit for pure bred individuals estimated with HYBRYDLAB. Pure A. zebra genotypes are represented in pale grey; pure A. taeniatus genotypes are represented in dark grey. Potential hybrids are identified by an asterisk (*).
(DOC)
Microsatellite allele frequencies per locus per species (Aze1-Aze4).
(DOC)