Abstract
Objective
Structural variations of DNA, such as copy number variations (CNVs), are recognized to contribute both to normal genomic variability and to risk for human diseases. For example, schizophrenia has an established connection with 22q11.2 deletions. Recent genome-wide studies have provided initial evidence that CNVs at other loci may also be associated with schizophrenia. In this article, the authors provide a brief overview of CNVs, review recent findings related to schizophrenia, outline implications for clinical practice and diagnostic subtyping, and make recommendations for future reports on CNVs to improve interpretation of results.
Method
The review included genome-wide surveys of CNVs in schizophrenia that included one or more comparison groups, were published before 2009, and used newer methods. Six studies were identified.
Results
Despite some limitations, these initial genome-wide studies of CNVs provide replicated associations of schizophrenia with rare 1q21.1 and 15q13.3 deletions. Collectively, the results point to a more general mutational mechanism involving rare CNVs that elevate risk for schizophrenia, especially more developmental forms of the disease. Including 22q11.2 deletions, rare risk-associated CNVs appear to account for up to 2% of schizophrenia.
Conclusions
The more penetrant CNVs have direct implications for clinical practice and diagnostic subtyping. CNVs with lower penetrance promise to contribute to our genetic understanding of pathogenesis. The findings provide insight into a broader neuropsychiatric spectrum for schizophrenia than previously conceived and indicate new directions for genetic studies.
In the ongoing search for genetic origins of schizophrenia, most of the focus has been on changes in DNA sequence that may elevate risk for the disorder. However, genomic copy number variations (CNVs) are increasingly recognized to contribute to risk for human diseases (1–3). One of the most exciting recent discoveries about the human genome is that, in addition to variations in sequence, such as single nucleotide polymorphisms (SNPs), individuals have variations in genomic structure (4–6). Structural variants, mainly CNVs involving loss (e.g., deletions) or gain (e.g., duplications) of up to several million base pairs of DNA sequence, are estimated to constitute upward of 5% of the human genome (4, 7, 8). CNVs can alter gene dosage and may involve multiple genes and/or regulatory regions (Figure 1). In general, CNV deletions show higher penetrance (more severe phenotype) than duplications and larger CNVs often have higher penetrance and/or more clinical features than smaller CNVs. It is now apparent that structural variants contribute to normal variability, disease risk, and developmental anomalies as well as acting as a major mutational mechanism in evolution (8, 9).
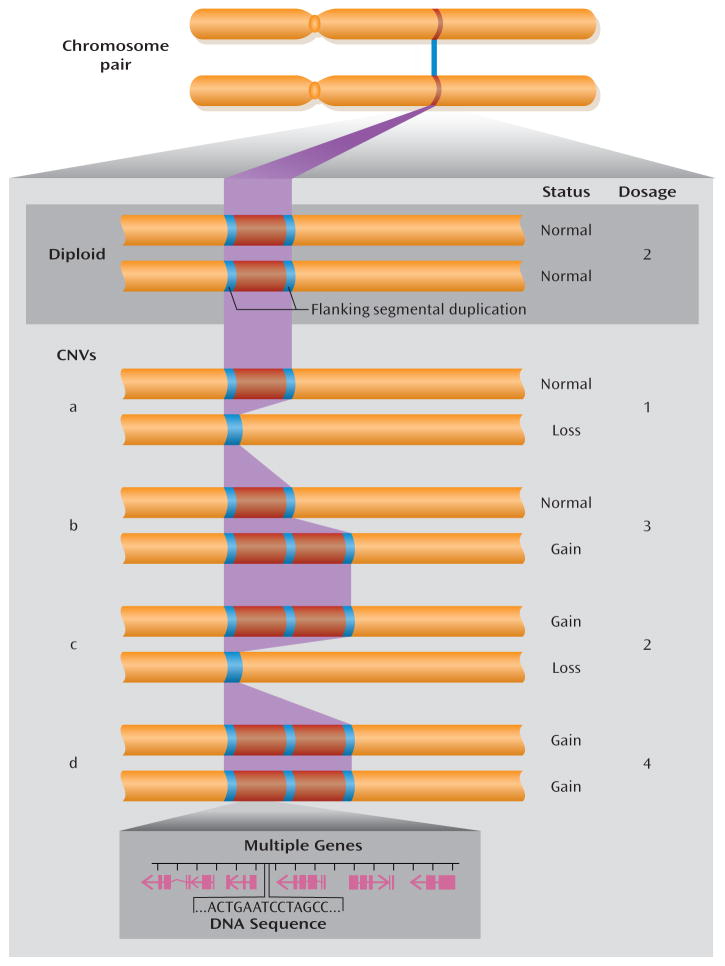
FIGURE 1. Examples of Chromosome Regions With Normal Diploid Status and With Copy Number Variations (CNVs)a.
a Flanking segmental duplications increase the risk for structural genomic changes, such as CNVs. Normal diploid status is shown in the example at the top. Examples of CNVs (a to d, below) show the change in copy number in various configurations. A simple loss (example a) is often called a microdeletion and a simple gain (example b) a microduplication. While CNVs associated with genomic disorders are more prone to higher mutation rates because of the highly identical sequences in segmental duplications, most CNVs in the genome do not arise from events mediated by segmental duplication. CNVs may involve no, one, or multiple genes (genomic extent including exons and introns shown in magenta).
Technological advances have driven much of the research on CNVs and allowed detection of smaller and smaller CNVs (1, 10, 11). As with most discoveries, however, those that are the most readily detectable and have the most severe effects are reported first. Thus, although the vast majority of CNVs are inherited, CNVs that have newly occurred as de novo (spontaneous) mutations have more readily been implicated in diseases. A significant part of the CNV-disease connection involves a new category of genetic diseases called genomic disorders (3, 12). The CNVs associated with genomic disorders involve microdeletions and microduplications typically too small to be detected by standard karyotype. These often arise as de novo mutations mediated by flanking segmental duplications (Figure 1), also known as low-copy-repeat sequences, comprising nearly identical DNA sequences (>90% nucleotide similarity) and involving unequal crossing over in meiosis (3, 12). The most common genomic disorder, 22q11.2 deletion syndrome, has an established association with schizophrenia (13). Individuals with 22q11.2 deletions have a 20-fold increase in risk for schizophrenia and account for about 0.9%–1.0% of schizophrenia in the general population (13). Recent reports (14–17) suggest that CNVs at other loci may also be associated with schizophrenia. Understanding how to interpret these findings and their implications is the goal of the current review. We begin with a brief overview of CNVs, then review the recent CNV findings related to schizophrenia, and conclude by outlining the implications of this research for clinical practice and diagnostic subtyping, making recommendations for future reports.
Overview of Copy Number Variations
Detection, Mechanisms, and Expression
CNVs may be detected by targeted or genome-wide methods. The latter include karyotype with a lower resolution of at best 5–10 megabases (Mb) of DNA (a typical band of a chromosome) and multiple genome-wide DNA microarrays with coverage and resolution that vary according to the individual technology platform, probe set, and methodology. Most microarrays use SNP-based or array comparative genomic hybridization (aCGH) methods. The former require algorithms to assist in determining copy number, and the latter rely on one or more “hybridization reference” genomes, the choice of which can influence results. These methods vary in their resolution and ability to determine the break points and extent of individual CNVs. There is also rapid progress to using whole-genome (“next-generation”) sequencing methods (18). Targeted methods include those often used in clinical laboratories that detect microdeletions or microduplications associated with genomic disorders, such as fluorescence in situ hybridization (FISH) using specific probes and molecular cytogenetic methods, and microarrays using various probe-screening sets that offer increasingly extensive coverage. Quantitative polymerase chain reaction (qPCR) is another targeted method often used to confirm a CNV identified by using array-based methods and to quickly screen large sample sets (1).
Much remains to be discovered about the mechanisms giving rise to CNVs and the pathways from CNVs to phenotypic expression recognizable as illness. Repeated elements in the genome have been implicated in many but not all CNV break points (8, 19). Mechanisms include meiotic unequal crossing over, or nonallelic homologous recombination, mediated by flanking repeated sequences or segmental duplications, and nonhomologous DNA repair mechanisms such as nonhomologous end joining or microhomology-mediated end joining (19). If DNA repair mechanisms during mitotic divisions of germ cells are involved, this could lead to new mutations arising in sperm through adulthood, possibly associated with late paternal age, or in maternal grandchildren, given the timing of these divisions in fetal development for females (19).
While CNVs make a significant contribution to observable variability between individuals, there is not always a straightforward correlation between gene dosage and expression (20, 21). Larger CNVs on average will encompass more genes, but they may also contain regulatory elements for genes and they can in addition exert effects on chromatin structure. Therefore, CNVs can have long-distance effects on expression of genes in neighboring chromosomal regions, even up to 1 Mb away from a particular structural variant (20–22).
Association With Disease
There are numerous complexities in phenotypic categorization, CNV detection and characterization, and statistical analyses that need to be considered in CNV-disease associations. Schizophrenia has strong evidence for neurodevelopmental origins (23, 24). Thus, schizophrenia may be expected to have CNV associations similar to those in disorders such as autism, where specific CNVs and conceptually similar but microscopically visible chromosomal anomalies detectable by karyotype are present in a substantial minority (approximately 10%) of cases (25–28). Hallmarks of developmental changes, e.g., mental retardation, birth defects, dysmorphic facies, and/or childhood-onset schizophrenia, could indicate individuals with schizophrenia for whom the a priori likelihood of having associated CNVs may be elevated. Sampling strategies can significantly influence the observed prevalence of CNVs in both individuals with the disorder and comparison subjects. In general, no or low prevalence in comparison subjects would be expected for highly penetrant CNVs and higher prevalence in comparison subjects may be expected with less-penetrant CNVs. Likewise, familial forms of schizophrenia may be less commonly associated with certain CNVs than “sporadic” forms of illness, depending on the likelihood of de novo mutations and penetrance of the disease for the specific CNV. Ideally, one would be able to assess all individuals with rare CNVs of interest to determine the full phenotypic spectrum and overt or cryptic relatedness to others who carry the same CNV.
In contrast to SNPs, for which one can expect a 99% sensitivity and specificity of detection of the targets on the microarray, the complex nature of CNVs (Figure 1) and related experimental issues do not allow the same level of confidence for CNV detection (1). Uncertainty about the presence, extent, and break points of CNVs affects analyses of the genes that may be involved. In general, with current commercial microarrays large CNVs (>100 kb) are more reliably detectable, have greater overlap between platforms, and are easier to assign pathological status, particularly if they arise as de novo mutations.
Recurrent de novo CNV events and/or multiple rare CNVs that show convergence on a single gene or set of genes (or networks or pathways of genes) strengthen the association with disease. Although SNP-based arrays have poor coverage of segmental duplications, large CNVs in intervals between such complex repeat regions are readily detectable (29). Sources of possible laboratory- and/or array-based variability include variable probe performance and measurement error, batch effects and “noise” from cell line artifacts, and/or poor-quality DNA samples. Platforms vary with respect to coverage across the genome; more recent arrays with 1 million or more SNPs increase detection down to CNVs spanning less than 20 kb. Smaller CNVs, which constitute the majority of those detected by using SNP-based platforms, are usually inherited (11). It is as yet unclear whether smaller CNVs will have properties similar to those of large CNVs with respect to association with diseases like schizophrenia.
Common CNVs (allele frequency >5%) have properties similar to those of SNPs, comprise the majority of CNV differences between individuals, and are almost always inherited (8, 11). Most are in strong linkage disequilibrium with SNPs and thus may be analyzed with standard genetic association methods. Genuine associations of CNVs with disease may be difficult to determine for rare events with low prior probability, however. Accurate estimates of such associations will depend on an individual with a rare CNV being reported only once or, if included in different studies, clearly demarcated as such. Parental data, i.e., phenotypes and genotypes, are valuable, although not always available given the late age at onset of schizophrenia. Data on family members carrying the CNV provide information relevant to estimates of penetrance and variable expression. Case-control studies rely on the comparison groups being free of disease and unrelated to the affected individuals. The relatively high prevalence of schizophrenia in the general population and reduced penetrance and/or variable expression of genetic variants present further challenges to interpreting CNV results. Given all of these factors, prevalence figures and odds ratios will be very approximate estimates at this early stage of CNV studies. Independent replication is essential.
Review Methods
For this review, we included genome-wide surveys of CNVs in schizophrenia that included one or more comparison groups and were published before Jan. 1, 2009. We excluded studies using earlier aCGH methods based on bacterial artificial chromosomes, as these platforms tend to lack precision because of the large size of the probes. We examined each report, all supplemental materials, and where possible, other reports of the samples and individuals with major CNVs. We made every attempt to determine subjects that overlapped between studies to supplement sampling descriptions and to be better able to discuss independent replication of CNV results. We abstracted methodological data, including ascertainment and study sites, phenotype, array platform, and selected recurring CNVs, focusing on the salient issues to consider in study design and how best to interpret emerging CNV data.
Results
We reviewed six genome-wide studies reporting on CNVs in schizophrenia, all of which were published in 2008. Table 1 presents the data on the major recurrent CNVs reported in these studies, with further details of sampling issues summarized in footnotes. In the four smaller studies (14, 15, 30, 34) we observed no overlap of study groups. The largest two studies, however, involved individuals originally collected for various purposes at multiple sites in several countries, predominantly in northern Europe (16, 17). Data on subjects from one of these sites (Aberdeen, Scotland) were included in both studies (16, 17), complicating evaluation of the results. Aberdeen had the highest number of subjects with 22q11.2 deletions and 1q21.1 deletions (Table 1). However, even if this site is excluded, the studies appear to support a previously established association with 22q11.2 deletions and two new associations: rare 1q21.1 and 15q13.2–15q13.3 deletions (Table 2). Results suggesting more CNVs overall in subjects with schizophrenia than in comparison subjects (so-called CNV load or burden) are more challenging to adjudicate given significant methodological variability, e.g., inclusion (or not) of groups that were filtered for karyotypic abnormalities (14), which may skew results. Results for smaller CNVs involving single genes, while requiring further independent reports, are beginning to provide evidence of convergence on genes and pathways of interest for neurodevelopment.
TABLE 1.
Studies of Genome-Wide Copy Number Variations (CNVs) in Schizophrenia
| Study | Ascertainment and Characteristics of Schizophrenia
|
Number of Subjects With Specific Recurrent Copy Number Losses
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of Schizophrenia Subjects |
Family History of Schizophrenia (%) |
Mental Retardation, Autism Spectrum Disorder, or Childhood-Onset Schizophrenia (<13 years) |
Dysmorphic Features (%) |
DNA Array Method |
22q11.2 Deletions
|
1q21.1 Deletions
|
15q13.2–q13.3 Deletions
|
15q11.2 Deletionsb
|
|||||
| Schizophrenia | Comparisona | Schizophrenia | Comparisona | Schizophrenia | Comparisona | Schizophrenia | Comparisona | ||||||
| Stefansson et al. (16): 11 major sitesc,d | 3,838–4,718 | —e | —e | —e | Multiple | 8f | 0 | 11g | 8g | 7h | 8h | 26i | 79i |
| International Schizophrenia Consortium (17): seven major sitescj | 3,391 | —e | —e | —e | 500K Affymetrix, 6.0 Affymetrix | 13f | 0 | 10g | 1g | 9h | 0 | —e | —e |
| Shi et al. (30): Han Chinesek | 155 | —e | —e | —e | 500K Affymetrix | 0 | 0 | 0 | 0 | 0 | 0 | —e | —e |
| Xu et al. (15, 31): Afrikanerl | 500K Affymetrix | ||||||||||||
| No family history of schizophrenia | 152 | 0 | Some mental retardation; autism spectrum not reported; some childhood-onset schizophrenia | Some | 3 | 0 | 0 | 0 | 0 | 0 | —e | —e | |
| Familial schizophrenia | 48 | 100 | —e | —e | 0 | 0 | 0 | 0 | 0 | 0 | —e | —e | |
| Walsh et al. (14, 32, 33): two U.S. sitesm | |||||||||||||
| Washington State | 150 | 13–18 | —e | —e | 85K representational oligonucleotide microarray analysis (ROMA) | Excludedm | 0 | 1 | 0 | 0 | 0 | —e | —e |
| NIMH (multiple U.S. sites) | 92 | 26 | 0% mental retardation; 30% autism spectrum disorders; 100% childhood-onset schizophrenia | 0 | 500K Affymetrix | 4 | 0 | 0 | 0 | 0 | 0 | —e | —e |
| Bassett et al. (34): Canadian subjects with 22q11.2 deletion syndromen | 44 | 4.5 | —e | 100 | 250K Affymetrix | 44 | 0 | 0 | 0 | 0 | 0 | 3 | 21 |
Comparison subjects were generally not phenotyped, and basic demographic features (e.g., age and sex) were usually unknown or unreported, especially in large studies, or were phenotyped for another disease (see information on individual studies for any available details).
15q11.2 deletions were considered not to be novel CNVs in most studies and therefore were usually not reported.
According to brief notes added in proof in the reports by Stefansson et al. (16) and the International Schizophrenia Consortium (17), there was overlap between those two studies in the subjects with schizophrenia and related psychotic disorders and in the comparison subjects from Aberdeen, Scotland. No details were provided.
The number of comparison subjects varied from 39,299 to 41,199 (16). The number of subjects with schizophrenia was 3,838 for 22q11.2, 4,718 for 1q21.1 and 15q11.2 deletions and 4,213 for 15q13.3 deletions. The large comparison group (N=32,442) from Iceland included multiple groups with genetic diseases (N=31,522), including autism (N=199), dyslexia (N=700), ADHD (N=500), and anxiety disorders (N=1,190); there were apparently 900 population comparison subjects (16). One comparison subject with a 1q21.1 deletion originated from the autism group; the disease-related origins of the other Icelandic comparison subjects with CNVs were not reported. Icelandic subjects were related to each other in some cases; for instance, among the nine Icelandic subjects with 1q21.1 deletions (one schizophrenia and eight comparison subjects), there were six de novo events, but no other details were provided as to relationships. There were some differences between the data presented in Table 1 and Supplementary Table 3 of reference 16 for 15q11.2 deletions; we have assumed that Table 1 was correct. The Stefansson et al. study groups originated from 10 countries and two ethnic groups, predominantly Caucasian. The locations (with the responsible researchers) and numbers of schizophrenia and comparison subjects for the genome-wide studies used to detect the selected CNVs reported were as follows: Iceland (Stefansson), 648 schizophrenia and 32,442 comparison subjects; the Netherlands (Utrecht “GROUP”/Kahn), 806 schizophrenia and 4,039 comparison subjects; Aberdeen, Scotland (St. Clair), 622 schizophrenia and 670 comparison subjects; Munich (Rujescu) and Bonn (Cichon/Nothen), Germany, 1,106 schizophrenia and 1,489 comparison subjects; Norway (Andreassen), 237 schizophrenia and 272 comparison subjects; England (Collier), 105 schizophrenia and 96 comparison subjects; Italy (Ruggeri), 85 schizophrenia and 91 comparison subjects; Finland (Peltonen), 191 schizophrenia and 200 comparison subjects (including about half from a Finnish isolate). For two additional sites, targeted quantitative polymerase chain reaction (qPCR) gene dosage methods, not genome-wide methods, were used to detect selected CNVs at 22q11.2, 1q21.1, 15q13.3, and 15q11.2 in 375–442 schizophrenia and 501–1,437 comparison subjects from Denmark (Hanson) and at 1q21.2 and 15q11.2 in 438 schizophrenia and 463 comparison subjects from China.
Not reported or data not available.
In the Stefansson et al. study (16), the eight affected schizophrenia subjects were Scottish (Aberdeen, N=5), Icelandic (N=1), German (N=1), and Danish (N=1). In the International Schizophrenia Consortium study (17), the 13 affected schizophrenia subjects were Scottish (Aberdeen, N=6), English, Scottish, or Welsh (London, N=2), Swedish (N=2), of Portuguese descent (N=1), Irish (N=1), and Bulgarian (N=1).
In the Stefansson et al. study (16), the 11 affected schizophrenia subjects were Scottish (Aberdeen, N=4), German (N=3), Danish (N=3), and Icelandic (N=1); the eight affected comparison subjects were all Icelandic, with N=7 if the subject with autism is excluded. In the International Schizophrenia Consortium study (17), the 10 affected schizophrenia subjects were Scottish (Aberdeen, N=5), English, Scottish, or Welsh (London, N=2), of Portuguese descent (N=2), and Swedish (N=1); the one affected comparison subject was Scottish (Aberdeen).
In the Stefansson et al. study (16), the seven affected schizophrenia subjects were Icelandic (N=1), Scottish (Aberdeen, N=1), German (N=1), Dutch (N=3), and Norwegian (N=1); the eight affected comparison subjects were Icelandic (N=7) and Dutch (N=1). In the International Schizophrenia Consortium study (17), the nine affected schizophrenia subjects were English, Scottish, or Welsh (London, N=2), Swedish (N=2), Bulgarian (N=2), Scottish (Aberdeen, N=1), of Portuguese descent (N=1), and Irish (N=1).
In the Stefansson et al. study (16), the 26 affected schizophrenia subjects were Scottish (N=7), German (N=6), Icelandic (N=4), Dutch (N=4), Danish (N=4), and English (N=1); the 79 affected comparison subjects were Icelandic (N=58), Dutch (N=12), German (N=4), Danish (N=3), Scottish (N=1), and Finnish (N=1).
The subjects were Caucasian and included 3,181 comparison subjects. The locations (with the responsible researchers) and numbers of schizophrenia and comparison subjects were as follows: Aberdeen, Scotland (St. Clair), 727 schizophrenia and 694 comparison subjects; Sweden (Hultman), 622 schizophrenia and 437 comparison subjects (Swedish and other Nordic subjects); United Kingdom (Gurling), 547 schizophrenia and 0 comparison subjects (Scottish, English, and Welsh subjects); Bulgaria (Kirov/Owen), 479 schizophrenia and 646 comparison subjects; Ireland (Corvin/Gill), 280 schizophrenia and 914 comparison subjects; Edinburgh, Scotland (Blackwood), 403 schizophrenia and 290 comparison subjects; Portugal, the Azores, Madeira, and the United States (all of Portuguese descent), 333 schizophrenia (all with familial schizophrenia) and 200 comparison subjects. Some of the originating study groups explicitly excluded individuals with mental retardation, and others ascertained individuals with familial schizophrenia, i.e., with a positive family history of schizophrenia; however, details were not provided about these ascertainment issues for most study groups.
The Han Chinese subjects were recruited from Shanghai and included 187 comparison subjects.
The subjects were obtained from an Afrikaner founder group and included 152 of an original 251 probands with no family history of schizophrenia and 48 of 71 others with familial schizophrenia (15, 35). Of these subjects with schizophrenia, 85 were previously reported in a 2004 study of syndromic features (31), including six with dysmorphic features, of whom two had 22q11.2 deletions according to testing using standard fluorescence in situ hybridization (FISH) methods. Another of these subjects with mental retardation but no dysmorphic features was reported to have a 22q11.2 deletion in the Xu et al. study (15). Of the eight other schizophrenia subjects with de novo CNVs longer than 100 kb in the Xu et al. study, two (25%) had dysmorphic features identified in the 2004 study, one of whom also had childhood-onset schizophrenia according to Table S3 (15), although a different age at onset for this subject appears to be reported in the 2004 study (31). The Afrikaner comparison subjects were recruited from the same founder group.
Of the 150 multiethnic cases from Washington State, 120 were severely ill adult inpatients at a state psychiatric hospital, 64% of whom were forensic patients and none of whom had childhood-onset schizophrenia, although several had an age at onset of 13 years. Another 30 subjects were children and adolescents with early-onset schizophrenia, several of whom had childhood onsets (given a mean age at onset of 13.2 years) and/or mild mental retardation (14). One of these 150 subjects had XYY (i.e., entire Y chromosome duplicated). Subjects with 22q11.2 deletions were excluded. Comparison subjects (219 Caucasian and 49 African American) for these subjects with schizophrenia were obtained from multiple sources, including lymphoblastoid cell lines from unaffected Caucasian subjects older than 54 years in the National Institute of Neurological Diseases and Stroke’s Neurogenetics Repository, anonymous unaffected African American subjects older than 34 years from a Cold Spring Harbor Laboratory collection, and an academic consortium (Academy for Medical Development and Collaboration). In the well-characterized National Institute of Mental Health (NIMH) group of subjects with childhood-onset schizophrenia (32), from which the Walsh et al. group was derived; originally there were four subjects with 22q11.2 deletions and five previously reported patients with other chromosomal anomalies: X;16 unbalanced translocation involving a Xq24-qter deletion causing atypical Turner syndrome and a 17-Mb 16q22.2-qter duplication; XO/XX mosaic Turner syndrome; XXX (triple X syndrome); 1;7 balanced translocation; and 5q32-qter uniparental isodisomy (32). Thus, a total of 10% (nine of 92) of this group had chromosomal anomalies and/or typical 3-Mb 22q11.2 deletions (32). These nine subjects were excluded from the Walsh et al. genome-wide array study (14). The comparison group for this study involved 77 nontransmitted chromosomes from 154 parents of the subjects with childhood-onset schizophrenia.
The 44 Canadian subjects with 22q11.2 deletion syndrome and schizophrenia were observed to be 90.4% European, had a mean IQ of 68.6 (SD=9.7), and had a mean age at onset of psychosis of 20.9 years (SD=5.4). The comparison groups in this study comprised 118 unaffected parents of the subjects with 22q11.2 deletion syndrome (data presented here), studied with a 250K Affymetrix array, and two groups studied with a 500K Affymetrix array: 500 Europeans from a German population (27, 36) and 1,152 non-disease comparison subjects of European origin from Ontario, Canada (27, 37). CNV data were analyzed by using the same methods as for the schizophrenia group. Entries in the Database of Genomic Variants (5) were also used for comparison purposes. A CNV was considered to be novel in subjects if it was 10 kb or longer, it contained at least three probes, and at least 20% of its total length was unique in relation to population-based comparison subjects.
TABLE 2.
Examples of Recurring Interstitial Copy Number Variations (CNVs) in Schizophrenia
| Recurring CNV and Length (referencesa) |
Putative Candidate Gene(s) |
Estimated Rate of De Novo CNVs |
Estimated Prevalence in Schizophrenia |
Estimated Penetrance of Selected Phenotypes
|
||
|---|---|---|---|---|---|---|
| Schizophrenia | Autism | Any Phenotype | ||||
| 22q11.2 deletion, usually 1.5–3.0 Mb (13, 38, 39) | 25–45 genes, including PRODH, DGCR2, DGCR8, COMT, GNB1L, PIK4CA | >90% | ~0.9% | 20%–25% | Unknown | ~100% |
| 1q21.1 deletion, 860 kb to 2.8 Mb, usually 1.35 Mb (14, 16, 17, 26, 33, 40) | 8–24 genes | <40% | Unknown (rare; estimated to be 0.2%–0.3%) | Unknown | Reduced | Unknown |
| 15q13.2–q13.3 deletion, 500 kb to 3.8 Mb, usually 1.5 Mb (16, 17, 41, 42) | >7 genes, including CHRNA7 | <40% | Unknown (rare; estimated to be 0.2%–0.3%) | Unknown | Unknown | Unknown |
| 2p16.3 deletion, 25–375 kb (14, 17, 25, 26, 43–45) | NRXN1 gene | Unknown | Unknown (rare) | Unknown | Reduced | High |
| 7q35–q36.1 deletion, 220 kb to 1.5 Mb (17, 25, 26, 46) | CNTNAP2 gene | Unknown | Unknown (rare) | Unknown | ~70% | Unknown |
In some cases, multiple references indicate individual subjects reported in two or more articles (see text for details).
Newly Identified Major CNVs and Lower-Penetrance CNVs Associated With Schizophrenia
Up to 18 subjects with schizophrenia who had 1q21.1 deletions, originating from eight sites, were reported in three studies involving over 6,000 patients with schizophrenia (Table 1) (14, 16, 17). There were up to 15 schizophrenia subjects from 10 originating sites reported with 15q13.3 deletions in the studies reported by Stefansson et al. (16) and the International Schizophrenia Consortium (17) (Table 1). Both studies indicated that the proportion of subjects with schizophrenia and these CNVs was significantly different from the proportion of subjects with these CNVs in the comparison groups used. As expected for case-control studies with no capability of following up subjects, limited data are available on the phenotype or de novo versus inherited status of the individuals from either the schizophrenia or comparison groups with these deletions.
Results for the 15q11.2 locus are less clear. In one study (16), the 471-kb CNV microdeletion at 15q11.2 was reported, in fairly large numbers, in both the schizophrenia subjects and comparison subjects with unspecified disorders, including autism (Table 1). While the difference was reportedly significant, the effect size appeared smaller than that for the 1q21.1 and 15q13.3 deletion CNVs (16). A later publication that included data from the International Schizophrenia Consortium study supported the possibility that CNVs at the 15q11.2 locus are risk factors for schizophrenia with reduced penetrance (47). Other recurrent CNVs flanked by segmental duplications previously reported in autism and/or developmental delay (25–27, 48–50) that may also be risk factors for schizophrenia with reduced penetrance include 400–600-kb 16p11.2 deletions and duplications (14, 16) and duplications at 16p13.1 (1.16 Mb) (47, 51), 1q21.1 (1.4 Mb) (16, 51), and 22q11.2 (3–4 Mb) (47); the latter two are reciprocal to higher-penetrance deletions at the same loci.
Overall Prevalence of CNVs in Schizophrenia
To address the hypothesis that a general mechanism of increased spontaneous copy number mutations could be involved in causing schizophrenia, one would like to know the answer to the question, Is the overall prevalence of CNVs higher in schizophrenia than in the general population? Some of the studies reviewed suggest that this is the case, but to interpret the findings reported one needs to consider the methods used. As may be expected for such different sampling, molecular, and analytic methods (Table 1), there were no consistent CNV prevalence data reported in all six studies that would allow direct comparisons, for either the schizophrenia or the comparison groups; thus, they are considered individually.
The International Schizophrenia Consortium study considered CNVs that were more than 100 kb long and found at a frequency of less than 1% in the comparison groups used (17). The CNVs discovered were 1.15-fold more common in the total schizophrenia group than in the total comparison group, in which the base rate was 0.99 CNVs per subject. Multiple subanalyses were presented. A 1.67-fold increase in schizophrenia over the total comparison group was the largest reported, obtained by restricting the analysis to loss CNVs more than 500 kb in size that had the lowest base rate (0.03) per subject in the comparison group (17). Not surprisingly, especially given that the large, gene-rich 22q11.2 deletions, as well as 1q21.1 and 15q13.3 deletions, were included (Table 2), the schizophrenia group was reported to have on average 3.57-fold more genes involved in CNVs than the comparison group. Individual genome-wide CNV data were not provided.
The multisite study of Stefansson et al. (16) covered 66 CNVs that had been identified as de novo events in genome-wide surveys of Icelandic samples of parents and offspring from 7,718 parent-offspring pairs or trios. Samples from this genetic isolate had been assembled for various genetic studies of medical and psychiatric disorders, such as autism, but excluding schizophrenia. This strategy thus enhanced for recurrent CNVs with variable expression. Genome-wide CNVs, except for the 66 selected CNVs, were not reported, precluding comparisons with data from other populations.
Walsh et al. (14) considered CNVs greater than 100 kb that had not been described in the literature or in public databases as of November 2007 and that affected genes. They reported a threefold greater proportion with these CNVs in their forensic and adolescent groups with schizophrenia (22 of 150 subjects) than in the composite comparison subjects used (13 of 268 subjects). Inheritance or de novo status of the CNVs was unknown. The schizophrenia group excluded subjects with 22q11.2 deletions but included a subject with a sex chromosome anomaly, 47,XYY. This report also included a study using a different array platform and another group comprising 83 subjects with childhood-onset schizophrenia. This group excluded nine additional subjects with chromosomal anomalies or 22q11.2 deletions (32). For the same categories of CNVs, this group showed a 2.6-fold greater proportion (23 of 83) than did the parental comparison subjects used (10 of 77). It is not clear why the proportion of individuals studied who had CNVs longer than 100 kb was lower in the comparison group for the forensic and adolescent subjects than in both the subjects with childhood-onset schizophrenia and the parental comparison group, but array differences and small study group sizes could be factors. Two (2.4%) of the CNVs in patients with childhood-onset schizophrenia were de novo changes. The expected de novo CNV mutation rate is approximately 1% at this resolution of analysis in comparison populations. Some individual genome-wide CNV data were provided.
Xu et al. (15) considered rare de novo CNVs of all sizes and prevalences and reported an eightfold greater number of these CNVs per subject in the Afrikaner subjects with sporadic schizophrenia (15 of 152), including three with 22q11.2 deletions, than in the comparison group used (two of 159). In contrast, there was a barely significant difference between groups with respect to inherited CNVs (46 of 152 versus 32 of 159). With few exceptions, individual data on genome-wide CNVs were not provided, precluding comparisons with data from other populations.
Using a study group of similar size to the groups in the studies by Walsh et al. (14) and Xu et al. (15), Shi et al. (30) reported no significant difference in the proportion of subjects with rare (<1%) CNVs larger than 100 kb between individuals with schizophrenia (109 of 155) and a comparison group (132 of 187). Results were similar whether or not the CNVs involved genes (30). No individual genome-wide CNV data were provided.
Our study (34) focused on subjects with 22q11.2 deletions and included all other CNVs in the analyses. There were no significant differences between subjects with and without schizophrenia or between those with 22q11.2 deletions and unaffected parents on any CNV-related measure, regardless of size, rarity, or de novo status. The genome-wide CNV results were also similar to comparable CNV data available from large general population samples (34). Moreover, there was no evidence of an increase in de novo CNVs as an underlying mechanism for occurrence of the 22q11.2 deletion. This study provided individual CNV data for all subjects studied.
CNVs Implicating Individual Genes
In addition to the major CNVs just discussed, the studies reviewed provided some initial evidence for recurring smaller CNVs that may implicate individual candidate genes for schizophrenia (Table 2). Some methodological issues, however, obscure the published findings.
NRXN1
Three of the genome-wide studies included individuals with schizophrenia and 2p16.3 CNVs involving the NRXN1 gene, which encodes neurexin, a scaffolding protein. Walsh et al. (14) reported monozygotic twins concordant for childhood-onset schizophrenia and a 2p16.3 loss CNV. A supplementary figure in the findings of the International Schizophrenia Consortium included five subjects with schizophrenia and six comparison subjects with 2p16.3 CNVs (17). In a previous article, the consortium also reported a Bulgarian affected sibling pair with a similar CNV at one of the study sites (43). In the study by Stefansson et al. (16), the 2p16.3 loss CNV was one of the initial 66 de novo CNVs examined, which had been identified in a proband with autism (44). In total, there appear to have been 12 subjects with CNVs involving NRXN1 in that study, three of whom appear to have been previously reported in a Utrecht study group with schizophrenia (45). The multisite results were reported in a separate publication (44) that produced a significant result when it focused on a subset of affected subjects with CNVs (six with losses, one with a gain) that disrupted exons in NRXN1 and a comparison group that excluded subjects ascertained with major psychiatric illnesses.
CNTNAP2
In a supplementary figure, the International Schizophrenia Consortium (17) reported three subjects with schizophrenia who had 7q35 loss CNVs and one comparison subject with a nonoverlapping, smaller 7q35 loss CNV involving the CNTNAP2 gene, which codes for contactin-associated protein 2. One of these three subjects with schizophrenia and a 220-kb 7q35 deletion CNV may have been included previously in the Friedman et al. study of CNTNAP2 gene dosage, which used subjects from Utrecht (46). A small, 80-kb 7q35 loss CNV involving this gene was reported in a nonpsychotic subject with a 22q11.2 deletion (34). The other multisite study reported a 7q35 loss CNV affecting the CNTNAP2 gene as one of the 66 de novo CNVs in an Icelandic proband with an unspecified condition (16). This group has not subsequently reported results for this CNV.
Discussion
Despite some methodological issues, the recent genome-wide CNV studies reviewed provide evidence that certain recurring CNVs are associated with schizophrenia. This confirms a mechanism of genetic mutation for schizophrenia that has implications in both clinical and research domains. Individually, these CNVs are rarely associated with susceptibility for schizophrenia (Table 2). The total number of individuals with schizophrenia and these CNVs reported to date is small, and any effect sizes calculated must be considered very exploratory (52). Nevertheless, the data suggest that, in addition to 22q11.2 deletions, rarer 1q21.1 and 15q13.3 deletions are associated with schizophrenia (52). Although further well-designed studies (Table 3) will be needed to determine true prevalence, the available data suggest that these three deletions may account for about 1%–2% of all cases of schizophrenia. Studies published in 2009 provide support for these deletions and several large lower-penetrance CNVs but not for a general increased prevalence of CNVs in schizophrenia (47, 51).
TABLE 3.
Recommendations for Reports on Copy Number Variations (CNVs) in Neuropsychiatric Diseasesa
| Methodological or Reporting Issue | Reporting Recommendations |
|---|---|
| Individuals affected by disease | Group size, demographic variables (including age, sex, ethnicity [if unknown, provide this information]), DNA source (blood, cell lines, tissue; whole genome amplified), previous genetic testing (including karyotype), previous reports on the study group or individuals from the group, ability to go back to obtain more detailed phenotypic and/or genetic data |
| Ascertainment | Ascertainment method, sources and inclusion/exclusion criteria, proportion of subjects with key features relevant to CNV detection, e.g., mental retardation, young age at onset, familial disease, birth defects, seizures/epilepsy (if unknown, provide this information) |
| Phenotyping | Psychiatric assessment method, medical/developmental/physical assessments, intellect/cognitive assessment, family history assessment (if not done or unknown, provide this information) |
| Comparison (control) group(s) | Group size, demographic variables (including age, sex, ethnicity [if unknown, provide this information]), DNA source (blood, cell lines, tissue), previous genetic testing, previous reports on the group or individuals from the group |
| Ascertainment | As for affected individuals (if unknown, provide this information) |
| Phenotyping | As for affected individuals (if not done or unknown, provide this information) |
| CNV technological and analytic issues | CNV loss/gain status, size, coordinates of start and stop (specifying the annotation source/reference genome sequence used) |
| Detection and analysis | CNV detection methods and limitations (e.g., whether different platforms were used for various groups), array details and resolution, quality control and filtering methods, number of algorithms used, definitions of “rare” and “novel” used for CNV; upon publication, release of underlying data to databases such as Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) so that others can compare/contrast results |
| Validation | Validation methods used, validation results specifying CNVs validated or not (if not done, provide this information); comparison to CNV database, e.g., Database of Genomic Variants (http://projects.tcag.ca/variation/) |
| Genetics | Inherited or de novo status of CNV (if unknown, provide this information) |
| Individuals with CNV of interest Origin and phenotype details | Ascertainment origin of subjects, age, sex, ethnicity, age at disease onset, intellectual level/mental retardation, dysmorphic features, birth defects, seizures/epilepsy, family history of disease (if unknown, provide this information), previous publications of the CNV and/or individual |
| CNV details | As for technological and analytic issues above, including loss/gain and inheritance or de novo status, size, chromosomal locus (e.g., 22q11.2), coordinates of start and stop (including annotation source), validation, other evidence implicating the CNV (loss or gain) in neuropsychiatric diseases, previous publications on the CNV and/or individual |
| Possible genes involved | CNV coordinates and chromosomal locus, CNV overlapping gene(s) in whole or in part, exons/introns, specific genome reference source/build, pertinent information on gene families and function (including specific information sources), other evidence implicating the gene(s) in neuropsychiatric diseases |
More details may be found in a previous publication (1).
Major Recurrent CNVs Associated With Schizophrenia
22q11.2 deletions
A consistent finding of these CNV studies was the absence of any 22q11.2 deletions in any of the comparison groups investigated, supporting the generally pathogenic nature of these deletions and the high penetrance of observable phenotypes (Table 2). The phenotypes associated with 22q11.2 deletions are known to be highly variable in number and severity, comprising congenital and later-onset physical and neuropsychiatric conditions, including epilepsy in a minority and learning difficulties in the majority. What new data do these genome-wide CNV studies offer on the well-established association of 22q11.2 deletions and schizophrenia? The prevalence of 22q11.2 deletions in schizophrenia in the multisite studies (16, 17) was about one-tenth as high (about 0.2%–0.4%) as that in the Afrikaner study group (2%) (31) used in the Xu et al. study (15) or in the original group of subjects with childhood-onset schizophrenia (4%) (32) from which one of the Walsh et al. study groups was derived (14). Estimates suggest that the overall prevalence of 22q11.2 deletions in schizophrenia is 0.9%–1% (13). Reasons for these varying prevalence estimates may involve the number and selection of subjects (13). Truly inclusive population-based prevalence samples of schizophrenia are difficult to obtain, and it is unclear whether any of the originating groups in the studies reviewed could be considered representative of a general schizophrenia population. With notable exceptions, e.g., the Aberdeen study group (16, 17), many of the originating subject groups for the consortium studies may have implicitly or explicitly excluded subjects with dysmorphic features, birth defects, learning difficulties, or known genetic syndromes. Age may have been another factor; the average age at death in 22q11.2 deletion syndrome is in the 40s (53). These sampling issues may similarly have affected the observed prevalence of other CNVs.
1q21.1 deletions
The data available suggest that the approximate prevalence of 1q21.1 deletions in schizophrenia is about 0.2%–0.3%, comparable to prevalence estimates in mental retardation (33, 40). Genome-wide studies indicate that 1q21.1 deletions may be de novo or inherited and that the phenotype is variable, including mental retardation, autism, attention deficit hyperactivity disorder (ADHD), and seizures (33, 40). Among 5,218 patients with mental retardation and congenital anomalies at 12 centers, there were 25 with 1q21.1 deletions (33). Although they may be inherited from apparently unaffected parents (33), no 1q21.1 deletions were found in 4,737 comparison subjects (33), a finding consistent with results for the schizophrenia CNV studies (Table 2) and indicating these rare deletions are often pathogenic in nature. This is supported by another study of 16,557 patients from multiple centers whose samples were sent to a single clinical laboratory, of whom 21 showed 1q21.1 deletions (40). No schizophrenia was reported in either study, but few adults were included. Minor dysmorphic features were variable, with a suggestion that microcephaly may be associated (33, 40). Few such details were available for the estimated 18 subjects in the schizophrenia studies (14, 16, 17).
As with 22q11.2 deletions (34), the phenotype of 1q21.1 deletions showed no correspondence to the extent of the deletion. The 1q21.1 deletion region is highly complex, with at least four large segmental duplications (4). The 1q21.1 deletion associated with a genomic disorder appears to most commonly involve the 1.35-Mb region between two of these sequences and less commonly a larger deletion that extends proximally (4, 33, 40), similar to those in the schizophrenia CNV studies (16, 17) and confirmed for one subject (14, 33).
15q13.3 deletions
The 15q13.3 deletions associated with genomic disorders are, most commonly, the 1.5-Mb region between two large segmental duplications (breakpoints 4 and 5) and, less frequently, a larger, 3.8-Mb deletion extending more proximally to another segmental duplication (breakpoint 3) (41, 54). These deletions have now been reported in as many as 15 subjects with schizophrenia (Table 2), including some with mental retardation and/or seizures or epilepsy (16, 17), nine subjects with mental retardation, including one with autism (41), 14 children with cognitive and autistic or attention disorders and six transmitting parents (42), and 14 subjects with idiopathic generalized epilepsy, including three with mental retardation (54). Subject overlaps between some of these studies are possible. Although the data are insufficient, there is no evidence that phenotypic severity corresponds to the extent of the deletion. Where assessed, many individuals with 15q13.3 deletions were noted to have mild and variable dysmorphic features (41). The preliminary prevalence data available suggest that these CNVs are rarer in schizophrenia or mental retardation (about 0.2%–0.3%) than they are in epilepsy (about 1%) (54). Notably, the approximate prevalence of 15q13.3 deletions in idiopathic generalized epilepsy is about the same as that of 22q11.2 deletions in schizophrenia, each CNV representing the most prevalent major risk factor identified to date for the respective complex disorder (13, 54).
Smaller Individual CNVs Implicating Specific Genes
Although the data are preliminary and involve few individuals, CNV studies provide some evidence to support the neurexin superfamily, specifically the NRXN1 and CNTNAP2 genes, as potentially important in schizophrenia. These genes cover large genomic extents, over 1 Mb for NRXN1 and over 2 Mb for CNTNAP2. Recurring CNVs may more likely affect such genes than genes of smaller size. Altogether, there appear to be 18 or 19 individual subjects with schizophrenia reported in a total of six articles to have 2p16.3 loss CNVs involving NRXN1 (44), providing some evidence that the NRXN1 gene may be involved in schizophrenia. As is the case with the larger CNVs associated with schizophrenia, the phenotype of CNVs involving these genes embraces other conditions, including autism and epilepsy (25–27, 46). In the case of CNTNAP2, Tourette’s syndrome, obsessive-compulsive disorder, and ADHD may also be part of the expression profile (55, 56). The relevance of these findings awaits further data.
Limitations and Recommendations for Future CNV Studies
Relatively few investigators have yet reported their complete CNV data sets or released their raw data, minimizing the opportunity for comparing and contrasting data or for meta-analyses. As a result, some commonalities (or differences) in the data may be missed. We anticipate that the results presented are just the beginning of a rich literature that will broaden our understanding of structural variants as risk factors for schizophrenia and their possible role in genetic interactions.
We propose some recommendations (Table 3) that will help in the evaluation of CNV results and appreciation of their implications. Methodological and reporting issues significantly limit the interpretations possible from the genome-wide CNV studies reported to date. This review highlights ascertainment and phenotypic details and subject overlap that hampered adjudication of results. More rigorous phenotyping is key to genetic discoveries and to understanding implications of potential discoveries, including those related to CNVs (1, 2, 57). As for most mutations, expression associated with CNVs is variable and severe phenotypes are usually the first to be discovered (Figure 2). Early penetrance estimates for specific CNVs may therefore be higher than those available after further study, and more low-penetrance CNVs will be reported over time (47, 51). Apart from 22q11.2 deletions (34), little has been reported about the chromosomal parent-of-origin status of de novo deletions. This may be particularly important for imprinted regions, such as 15q11–q13, where a specific phenotype may be observed only in a maternally (or paternally) transmitted chromosome. There is a paucity of data on parents and other family members of individuals reported to have these CNVs, leaving the inheritance or de novo status and full range of expression largely unknown. Differing methods were used to determine rates of novel CNVs, de novo CNVs, rare CNVs, and genes, including CNS genes, that were disrupted, involved, and/or affected (Table 1). Differences in array platforms and algorithms used between study groups and between studies further limit interpretation of results. With few exceptions (14, 34), there were no or only limited validation studies of CNVs (Table 3) reported as recurring and/or of interest. The studies to date mostly involve Caucasians, and the effects of ethnicity on the prevalence of CNVs are largely unknown (4; also see the Database of Genomic Variants: http://projects.tcag.ca/variation/). Despite the limitations, however, the replicated findings provide new perspectives for the genetics of schizophrenia and genetic concepts of neuropsychiatric disease, and they have important clinical implications.
FIGURE 2. Neuropsychiatric Phenotypes Associated With Copy Number Variations (CNVs)a.
a The top diagram illustrates the discovery of rare CNVs, which may be discovered in studies of both adult-onset and developmental diseases; studying individual CNVs can delineate the spectra of their respective phenotypic expressions. The charts below represent three CNVs having elevated prevalences in schizophrenia. These indicate the most common core phenotype of ascertainment based on current knowledge; the expression spectrum may change as more data accumulate. Proportions would differ if combined phenotypes of ascertainment (e.g., schizophrenia and mental retardation) were considered.
Clinical Relevance of Major CNVs Associated With Genomic Disorders
What is the general relevance of these CNV findings for clinicians? The 22q11.2, 1q21.1, and 15q13.3 deletions are the only major genetic risk factors identified to date for schizophrenia—and for which clinical genetic assessment and testing are widely available. The relatively modest prevalence of these anomalies suggests to us that, at this time, expensive genomic investigations are not warranted for all individuals with schizophrenia. Clinicians should, however, have a raised index of suspicion for genomic disorders or de novo CNVs in individuals with schizophrenia who have significant learning difficulties, dysmorphic facial features, birth defects, and/or unprovoked seizures (58). Family history, e.g., in offspring, of birth defects, dysmorphic features, developmental delay, and/or autism should also prompt consideration of a genomic disorder. If a syndromic form of schizophrenia is suspected, a referral to a genetics specialist would usually be recommended for diagnostic assessment (58).
What is the clinical relevance of these CNV findings for patients determined to have one of these major CNVs? For 22q11.2 deletions, the relevance is clear: detection significantly changes genetic counseling and anticipatory care from that for other patients with schizophrenia (58). For the 1q21.1 and 15q13.3 deletions, much less is known and there are challenges to providing genetic counseling for individuals with emerging genomic disorders (1, 57). However, detection of these anomalies would be clinically relevant to individuals with these CNVs, their families, and clinicians.
For the patient, genetic counseling would include the 50% risk of transmitting the deletion at each pregnancy, with the caveat that the severity of the phenotype cannot be predicted (58). As for 22q11.2 deletions, prenatal detection would be available (58). Parents of affected individuals should be tested because the deletion may be inherited from a parent. Specific anticipatory care recommendations for 1q21.1 and 15q13.3 deletions await further clinical data and, preferably, larger studies of population comparison subjects. However, history and physical examination, including neurological examination, echocardiogram, and abdominal ultrasound to investigate organ involvement, and routine blood work would also appear warranted (58). For lower-penetrance CNVs, many of which may be inherited from apparently unaffected parents, genetic counseling and clinical recommendations would be even less certain at this time (1, 57). In all cases, a genetics specialist would have the most up-to-date knowledge about these issues.
Neuropsychiatric Perspectives on Phenotype and Implications for Diagnostic Classification
In addition to being directly relevant to clinical practice, these CNV findings challenge some widely held beliefs and point to new research strategies. The data explode a common myth related to the diagnostic specificity of genetic findings. While there has been growing acceptance of the genetic relatedness of schizophrenia and bipolar disorder, there may be some resistance to accepting that expression of an individual CNV may also include autism and other developmental disorders (Figure 2, Table 2). The CNV results suggest a broader neuropsychiatric spectrum of phenotypes. In fact, there is little evidence in medicine for diagnostic purity associated with individual genetic changes. Genetic heterogeneity (many different genetic variants leading to the same phenotype), reduced penetrance (presence of the genetic variant not always expressed as the full disease), and variable expressivity (the same genetic variant leading to various phenotypic expressions) are the norm in human diseases and are apparent with the CNVs associated with schizophrenia. Studies of familial connections between schizophrenia and mental retardation (59, 60) and studies of childhood-onset schizophrenia, a rare clinical subtype in which comorbid autism spectrum disorders are common, also support a neuropsychiatric spectrum involving these developmental conditions (24). On the other hand, the data suggest that although there is some overlap, the pattern of associated CNVs may differ between schizophrenia and autism (Figure 2). For example, while autistic features are commonly reported in children with 22q11.2 deletions who receive psychiatric assessments, 22q11.2 deletions are rarely reported in subjects with autism who are studied (25–27). Also, current data suggest that 1q21.1 duplications are more often present than 1q21.1 deletions in autism (26, 61), whereas the reverse may be the case for schizophrenia (16, 51).
With respect to psychiatric diagnosis, these CNV findings suggest that, as for the dementias, the schizophrenias are beginning to yield to classification based on major causal factors. Comparable to an Alzheimer-type dementia associated with a beta-amyloid precursor protein (APP) gene mutation, a schizophrenia that is related to a 22q11.2 deletion (and perhaps a 1q21.1 or 15q13.3 deletion) may be considered a subtype with distinct management implications.
The findings may also prompt consideration of historical concepts of schizophrenia as an “epiphenomenon” of mental retardation. In fact, schizophrenia is not associated with most forms of mental retardation. The majority of individuals with 22q11.2 deletions, including those with schizophrenia, do not have mental retardation (34, 62), and this will probably also be the case for 1q21.1 and 15q13.3 deletions and lower-penetrance CNVs. Nonetheless, dual-diagnosis populations with schizophrenia and mental retardation are likely to be enriched for these recurrent CNVs, providing a much greater window on the etiology of pfropfschizophrenie than have chromosomal abnormalities detectable by karyotype.
Recurrent De Novo CNVs and a General Mutational Mechanism
Recurrent rare CNVs such as the 22q11.2, 1q21.1, and 15q13.3 deletions may also provide some concrete proof of a long-suspected mutational mechanism in schizophrenia. Over 50 years ago, geneticists Lionel Penrose (63) and Jan Böök (64) proposed that elevated rates of new mutations were likely in schizophrenia. Decreased reproductive fitness in schizophrenia (negative selection) in the face of a steady population prevalence of schizophrenia supports this possibility (65). The mechanism of the recurrence of the associated major CNVs, involving segmental duplications and nonallelic homologous recombination (3, 19), can account for the persistence of these mutations despite the biological disadvantage (reduced fitness) associated with their full expression. It is tantalizing to speculate about accumulating rare CNVs in schizophrenia and CNVs disrupting multiple genes in relevant pathways (14), but these possibilities remain to be proven. Certainly, the relationship of CNVs to reproductive fitness, natural selection, disease prevalence, and human evolution is of intense interest in genetics (9).
Implications for Gene Identification
Multiple independent reports of CNVs overlapping specific genes in patients with schizophrenia may show convergence on individual genes and/or pathways of interest that could assist in understanding pathogenesis. The mechanism of action of CNVs on the expressed phenotype is hypothesized to include effects of the copy number change, e.g., dosage effects, on genes within the CNV and perhaps extending as far as several megabases adjacent (“position” effect). There may be disruption of genes at the CNV break points. Sequence changes, e.g., on the intact chromosome in the case of deletions, may also be a factor. It is important to recognize that copy number gains such as duplications can affect gene expression, and in many cases these effects are similar to the effects of deletions involving the same chromosomal region.
In contrast to the situations for autism and other neuropsychiatric disorders, identifying rare mutations in single genes implicated by multigene CNVs remains a hypothetical possibility for schizophrenia. Could family studies be redesigned to help localize such rare mutations in CNV-related single genes? The phenotypic spectrum associated with the CNVs may provide a clue. One could predict that inherited (familial) forms of “pure” schizophrenia would differ in associated regions from those identified by de novo CNV mutations. The findings of a meta-analysis of linkage studies of familial schizophrenia are consistent with this prediction (66). Rare families with both schizophrenia and autism or mental retardation segregating (59, 60) may represent a better strategy for subtypes of schizophrenia that are associated with CNVs and/or with mutations in key genes affected by such CNVs.
Conclusions
We are in an exciting new era of identifying specific etiologies for individual subtypes of schizophrenia that have important implications for clinical practice in the genomic era. Even though compelling mutations in individual genes have not yet been identified, the campaign to understand the genetic heterogeneity of this complex disorder has begun. Just as for autism, diagnostic classification systems may now begin to delineate genetic subtypes associated with CNVs that represent up to 2% of schizophrenia. Despite significant limitations, the recent genome-wide studies of copy number add to established results for 22q11.2 deletions and point to a more general mutational mechanism involving rare CNVs that elevate risk for schizophrenia, especially more developmental forms of the disease. The findings provide insight into a broader neuropsychiatric spectrum for schizophrenia than previously conceived and indicate new directions for genetic studies. Future studies of CNVs that focus as much on phenotype as on technological advances promise further clinically relevant results and discoveries of genetic pathways to schizophrenia and other psychiatric diseases.
Acknowledgments
Supported by Canadian Institutes of Health Research grants (MOP-97800 and MOP-79518) to Dr. Bassett, NIMH grants (R01 MH-62440 and R01 MH-80429) to Dr. Brzustowicz, and the Schizophrenia Research Award from the National Alliance for Research on Schizophrenia and Depression (NARSAD) and Staglin Family Music Festival to Dr. Brzustowicz. Dr. Bassett holds the Canada Research Chair in Schizophrenia Genetics and Genomic Disorders. Dr. Scherer holds the GlaxoSmithKline–Canadian Institutes of Health Research Chair in Genetics and Genomics at the University of Toronto, and the Hospital for Sick Children.
The authors thank Sean Bekeschus for assistance with the figures and Gladys Wong for assistance with preparing the manuscript and tables.
Footnotes
All authors report no financial relationships with commercial interests.
References
- 1.Scherer SW, Lee C, Birney E, Altshuler DM, Eichler EE, Carter NP, Hurles M, Feuk L. Challenges and standards in integrating surveys of structural variation. Nat Genet. 2007;39(7 suppl):S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali-Khan SE, Daar AS, Shuman C, Ray PN, Scherer SW. Whole genome scanning: resolving clinical diagnosis and management amidst complex data. Pediatr Res. 2009;66:357–363. doi: 10.1203/PDR.0b013e3181b0cbd8. [DOI] [PubMed] [Google Scholar]
- 3.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 4.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:941–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 6.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 7.Sharp AJ. Emerging themes and new challenges in defining the role of structural variation in human disease. Hum Mutat. 2009;30:135–144. doi: 10.1002/humu.20843. [DOI] [PubMed] [Google Scholar]
- 8.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME. Origins and functional impact of copy number variation in the human genome. Nature. doi: 10.1038/nature08516. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen DQ, Webber C, Hehir-Kwa J, Pfundt R, Veltman J, Ponting CP. Reduced purifying selection prevails over positive selection in human copy number variant evolution. Genome Res. 2008;18:1711–1723. doi: 10.1101/gr.077289.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet. 2007;39(7 suppl):S48–S54. doi: 10.1038/ng2092. [DOI] [PubMed] [Google Scholar]
- 11.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, Elliott AL, Parkin M, Hubbell E, Webster T, Mei R, Veitch J, Collins PJ, Handsaker R, Lincoln S, Nizzari M, Blume J, Jones KW, Rava R, Daly MJ, Gabriel SB, Altshuler D. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 12.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- 13.Bassett AS, Chow EWC. Schizophrenia and 22q11.2. deletion syndrome. Curr Psychiatry Rep. 2008;10:148–157. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 15.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 16.Stefansson H, Rujescu D, Cichon S, Pietilainen OPH, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusodottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Forti MD, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K GROUP. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Schizophrenia Consortium: Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mardis ER. New strategies and emerging technologies for massively parallel sequencing: applications in medical research (editorial) Genome Med. 2009;1:40. doi: 10.1186/gm40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet. 2009;84:339–350. doi: 10.1016/j.ajhg.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrichsen CN, Vinckenbosch N, Zollner S, Chaignat E, Pradervand S, Schutz F, Ruedi M, Kaessmann H, Reymond A. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 21.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, Tyler-Smith C, Carter N, Scherer SW, Tavare S, Deloukas P, Hurles ME, Dermitzakis ET. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 23.Bassett AS, Chow EWC, O’Neill S, Brzustowicz LM. Genetic insights into the neurodevelopmental hypothesis of schizophrenia. Schizophr Bull. 2001;27:417–430. doi: 10.1093/oxfordjournals.schbul.a006884. [DOI] [PubMed] [Google Scholar]
- 24.Rapoport JL, Addington AM, Frangou S, Psych MRC. The neurodevelopmental model of schizophrenia: update 2005. Mol Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 25.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 27.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper GM, Zerr T, Kidd JM, Eichler EE, Nickerson DA. Systematic assessment of copy number variant detection via genome-wide SNP genotyping. Nat Genet. 2008;40:1199–1203. doi: 10.1038/ng.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi YY, He G, Zhang Z, Tang W, Zhang J, Jr, Zhao Q, Zhang J, Sr, Li XW, Xi ZR, Fang C, Zhao XZ, Feng GY, He L. A study of rare structural variants in schizophrenia patients and normal controls from Chinese Han population. Mol Psychiatry. 2008;13:911–913. doi: 10.1038/mp.2008.69. [DOI] [PubMed] [Google Scholar]
- 31.Wiehahn GJ, Bosch GP, du Preez RR, Pretorius HW, Karayiorgou M, Roos JL. Assessment of the frequency of the 22q11 deletion in Afrikaner schizophrenic patients. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:20–22. doi: 10.1002/ajmg.b.20168. [DOI] [PubMed] [Google Scholar]
- 32.Eckstrand K, Addington AM, Stromberg T, Merriman B, Miller R, Gochman P, Long R, Dutra A, Chen Z, Meltzer P, Nelson SF, Rapoport JL. Sex chromosome anomalies in childhood onset schizophrenia: an update. Mol Psychiatry. 2008;13:910–911. doi: 10.1038/mp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, Bongers EM, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Raber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, Knight SJ, Sebat J, Romano C, Schwartz CE, Veltman JA, de Vries BB, Vermeesch JR, Barber JC, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21. 1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bassett A, Marshall C, Lionel A, Chow E, Scherer S. Copy number variations and risk for schizophrenia in 22q11. 2 deletion syndrome. Hum Mol Genet. 2008;17:4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karayiorgou M, Gogos JA. The molecular genetics of the 22q11-associated schizophrenia. Brain Res Mol Brain Res. 2004;132:95–104. doi: 10.1016/j.molbrainres.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 36.Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;16:168–173. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- 37.Zogopoulos G, Ha KC, Naqib F, Moore S, Kim H, Montpetit A, Robidoux F, Laflamme P, Cotterchio M, Greenwood C, Scherer SW, Zanke B, Hudson TJ, Bader GD, Gallinger S. Germline DNA copy number variation frequencies in a large North American population. Hum Genet. 2007;122:345–353. doi: 10.1007/s00439-007-0404-5. [DOI] [PubMed] [Google Scholar]
- 38.Ogilvie CM, Moore J, Daker M, Palferman S, Docherty Z. Chromosome 22q11 deletions are not found in autistic patients identified using strict diagnostic criteria, IMGSAC. International Molecular Genetics Study of Autism Consortium. Am J Med Genet. 2000;96:15–17. [PubMed] [Google Scholar]
- 39.Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heinemande Boer JA, Beemer FA, Swaab H, Kahn RS, van Engeland H. The 22q11. 2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child Adolesc Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 40.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SH, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21. 1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities (letter) Nat Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, Broomer A, Casuga I, Wang Y, Xiao C, Barbacioru C, Gimelli G, Bernardina BD, Torniero C, Giorda R, Regan R, Murday V, Mansour S, Fichera M, Castiglia L, Failla P, Ventura M, Jiang Z, Cooper GM, Knight SJ, Romano C, Zuffardi O, Chen C, Schwartz CE, Eichler EE. A recurrent 15q13. 3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ben-Shachar S, Lanpher B, German JR, Qasaymeh M, Potocki L, Nagamani SC, Franco LM, Malphrus A, Bottenfield GW, Spence JE, Amato S, Rousseau JA, Moghaddam B, Skinner C, Skinner SA, Bernes S, Armstrong N, Shinawi M, Stankiewicz P, Patel A, Cheung SW, Lupski JR, Beaudet AL, Sahoo T. Microdeletion 15q13. 3: a locus with incomplete penetrance for autism, mental retardation, and psychiatric disorders. J Med Genet. 2009;46:382–388. doi: 10.1136/jmg.2008.064378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O’Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 44.Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, Murray R, Ruggeri M, Tosato S, Bonetto C, Steinberg S, Sigurdsson E, Sigmundsson T, Petursson H, Gylfason A, Olason PI, Hardarsson G, Jonsdottir GA, Gustafsson O, Fossdal R, Giegling I, Moller HJ, Hartmann A, Hoffmann P, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Andreassen OA, Djurovic S, Hansen T, Werge T, Melle I, Kiemeney LA, Franke B, Buizer-Voskamp JE, Ophoff RA, Rietschel M, Nothen MM, Stefansson K, Peltonen L, St Clair D, Stefansson H, Collier DA. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, Geurts van Kessel A, Brunner HG, Ophoff RA, Veltman JA. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman JI, Vrijenhoek T, Markx S, Janssen IM, van der Vliet WA, Faas BH, Knoers NV, Cahn W, Kahn RS, Edelmann L, Davis KL, Silverman JM, Brunner HG, Geurts van Kessel A, Wijmenga C, Ophoff RA, Veltman JA. CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- 47.Need AC, Ge D, Weale ME, Maia J, Feng S, Heinzen EL, Shianna KV, Yoon W, Kasperaviciute D, Gennarelli M, Strittmatter WJ, Bonvicini C, Rossi G, Jayathilake K, Cola PA, McEvoy JP, Keefe RS, Fisher EM, St Jean PL, Giegling I, Hartmann AM, Moller HJ, Ruppert A, Fraser G, Crombie C, Middleton LT, St Clair D, Roses AD, Muglia P, Francks C, Rujescu D, Meltzer HY, Goldstein DB. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5(2):e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez BA, Roberts W, Chung B, Weksberg R, Meyn S, Szatmari P, Joseph-George AM, Mackay S, Whitten K, Noble B, Vardy C, Crosbie V, Luscombe S, Tucker E, Turner L, Marshall CR, Scherer SW. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2009 Sep 24; doi: 10.1136/jmg.2009.069369. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 49.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Association between microdeletion and microduplication at 16p11. 2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 50.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, Christian SL. Recurrent 16p11. 2 microdeletions in autism. Hum Mol Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 51.Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Itsara A, Cooper GM, Baker C, Girirajan S, Li J, Absher D, Krauss RM, Myers RM, Ridker PM, Chasman DI, Mefford H, Ying P, Nickerson DA, Eichler EE. Population analysis of large copy number variants and hotspots of human genetic disease. Am J Hum Genet. 2009;84:148–161. doi: 10.1016/j.ajhg.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bassett AS, Chow EWC, Husted J, Hodgkinson KA, Oechslin E, Harris L, Silversides C. Premature death in adults with 22q11.2. deletion syndrome. J Med Genet. 2009;46:324–330. doi: 10.1136/jmg.2008.063800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, Kron KL, Steinich I, Kleefuss-Lie AA, Leu C, Gaus V, Schmitz B, Klein KM, Reif PS, Rosenow F, Weber Y, Lerche H, Zimprich F, Urak L, Fuchs K, Feucht M, Genton P, Thomas P, Visscher F, de Haan GJ, Moller RS, Hjalgrim H, Luciano D, Wittig M, Nothnagel M, Elger CE, Nurnberg P, Romano C, Malafosse A, Koeleman BP, Lindhout D, Stephani U, Schreiber S, Eichler EE, Sander T. 15q13. 3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verkerk AJ, Mathews CA, Joosse M, Eussen BH, Heutink P, Oostra BA. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 56.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’Arcy M, Deberardinis R, Frackelton E, Kim C, Lantieri F, Muganga BM, Wang L, Takeda T, Rappaport EF, Grant SF, Berrettini W, Devoto M, Shaikh TH, Hakonarson H, White PS. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2009 Jun 23; doi: 10.1038/mp.2009.57. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Ravenswaaij-Arts CM, Kleefstra T. Emerging microdeletion and microduplication syndromes: the counseling paradigm. Eur J Med Genet. 2009;52:75–76. doi: 10.1016/j.ejmg.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 58.Bassett AS, Chow EWC, Hodgkinson KA. Schizophrenia and psychotic disorders. In: Smoller JW, Sheidley BR, Tsuang MT, editors. Psychiatric Genetics: Applications in Clinical Practice. Arlington, Va: American Psychiatric Publishing; 2008. pp. 99–130. [Google Scholar]
- 59.Greenwood CMT, Husted J, Bomba MD, Hodgkinson KA, Bassett AS. Elevated rates of schizophrenia in a familial sample with mental illness and intellectual disability. J Intellect Disabil Res. 2004;48:531–539. doi: 10.1111/j.1365-2788.2004.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doody GA, Johnstone EC, Sanderson TL, Cunningham Owens DG, Muir WJ. ‘Pfropfschizophrenie’ revisited: schizophrenia in people with mild learning disability. Br J Psychiatry. 1998;173:145–153. doi: 10.1192/bjp.173.2.145. [DOI] [PubMed] [Google Scholar]
- 61.Autism Genome Project Consortium. Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Rogé B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bölte S, Feineis-Matthews S, Herbrecht E, Schmötzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, Hijmans C, Staal WG, Baird G, Bolton PF, Rutter ML, Weisblatt E, Green J, Aldred C, Wilkinson JA, Pickles A, Le Couteur A, Berney T, Mc-Conachie H, Bailey AJ, Francis K, Honeyman G, Hutchinson A, Parr JR, Wallace S, Monaco AP, Barnby G, Kobayashi K, Lamb JA, Sousa I, Sykes N, Cook EH, Guter SJ, Leventhal BL, Salt J, Lord C, Corsello C, Hus V, Weeks DE, Volkmar F, Tauber M, Fombonne E, Shih A, Meyer KJ. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. correction, 39:1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassett AS, Chow EW, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R. The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry. 2003;160:1580–1586. doi: 10.1176/appi.ajp.160.9.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Penrose LS. Mutation in man. Acta Genetica. 1956;6:169–182. doi: 10.1159/000150820. [DOI] [PubMed] [Google Scholar]
- 64.Böök JA. Schizophrenia as a gene mutation. Acta Genetica. 1953;4:133–139. [PubMed] [Google Scholar]
- 65.Bassett AS, Bury A, Hodgkinson KA, Honer WG. Reproductive fitness in familial schizophrenia. Schizophr Res. 1996;21:151–160. doi: 10.1016/0920-9964(96)00018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng M, Levinson D, Wise L, DeLisi L, Straub R, Hovatta I, Williams N, Schwab S, Pulver A, Faraone S, Brzustowicz L, Kaufmann C, Garver D, Gurling H, Lindholm E, Coon H, Moises H, Byerley W, Shaw S, Mesen A, Sherrington R, O’Neill F, Walsh D, Kendler K, Ekelund J, Paunio T, Lönnqvist J, Peltonen L, O’Donovan M, Owen M, Wildenauer D, Nestadt G, Blouin J, Antonarakis S, Mowry B, Silverman J, Crowe R, Cloninger C, Tsuang M, Malaspina D, Harkavy Friedman J, Svrakic D, Bassett A, Holcomb J, Kalsi G, McQuillin A, Curtis D, Brynjolfson J, Sigmundsson T, Petursson H, Jazin E, Zoëga T, Helgason T, Lewis C. Meta-analysis of 32 genomewide linkage studies of schizophrenia. Mol Psychiatry. 2009;14:774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]