Abstract
Toxoplasma gondii is an obligate intracellular protozoan parasite in which 36 predicted Hsp40 family members were identified by searching the T. gondii genome. The predicted protein sequence from the gene ID TGME49_065310 showed an amino acid sequence and domain structure similar to Saccharomyces cerevisiae Sis1. TgSis1 did not show differences in its expression profile during alkaline stress by microarray analysis. Furthermore, TgSis1 showed to be a cytosolic Hsp40 which co-immunoprecipitated with T. gondii Hsp70 and Hsp90. Structural modeling of the TgSis1 peptide binding fragment revealed structural and electrostatic properties different from the experimental model of human Sis1-like protein (Hdj1). Based on these differences; we propose that TgSis1 may be a potentially attractive drug target for developing a novel anti-T. gondii therapy.
Keywords: Toxoplasma, HSP40, HSP90
1. Introduction
Toxoplasma gondii is a ubiquitous obligate intracellular protozoan parasite that infects a large number of warm-blooded animals. T. gondii is a member of phylum Apicomplexa, which also includes Plasmodium spp, Eimeria spp, Babesia spp, Theileria spp, and Cryptosporidium spp. In humans and virtually all other warm-blooded vertebrates, T. gondii can cause severe infection, especially when the immune system is not fully developed or is seriously compromised [1–2]. In humans and other intermediate hosts, T. gondii follows an asexual replication cycle characterized by two stages: rapidly growing tachyzoites and latent bradyzoite tissue cysts. The pathology of toxoplasmosis is due to repeated cycles of host cell invasion and lysis by the actively dividing tachyzoites [3], which gives rise to acute disease and congenital birth defects. The slowly dividing bradyzoite form can remain latent for years, giving rise to recurrent, chronic infection. The interconversion process between tachyzoites and bradyzoites is characteristic of the asexual cycle, which is essential for disease propagation and causation [4]. Parasite invasion, replication, and differentiation involve complex biological pathways in which chaperones may play an important role [5].
Despite the importance of Hsp40s in cell biology, studies of this family in T. gondii have yet to be assessed. Hsp40s belong to a family of co-chaperones that participate in a variety of cellular processes that include protein folding, endocytosis, protein translocation across membranes, signal transduction, DNA replication and protein degradation [6–7]. Hsp40s functions by regulating the Hsp70 ATP hydrolytic cycle [8–9] and by acting as molecular chaperones that bind and target non-native proteins to the peptide binding site of Hsp70 [10–11]. To regulate Hsp70 ATPase activity, Hsp40 proteins utilize their DNAJ or J-domain [12–13], which is ~70 amino acids in length with a conserved HPD tripeptide that is the signature motif of this protein family [7]. The Hsp40 family is large, structurally and functionally diverse, with members classified into types I, II and III [14]. Recently, a novel J-domain protein was described in Plasmodium falciparum that was annotated as a type IV Hsp40 [15].
Sis1 (Hdj1 in humans) is a cytosolic type II J-domain protein associated with translating ribosomes, facilitating the assembly of translation initiation complexes [16–17]. Sis1 is essential in yeast and cannot be replaced by the cytosolic type I J-domain protein, Ydj1 [18]. Sis1 is also an essential chaperone in protozoan parasites [19–20]. Based on its importance in cell biology, the structure of Sis1 has been defined in yeast and human [21–24]. Sis1/Hdj1 J-domain proteins have a peptide-binding fragment located at the C-terminus [21, 23, 25–26]. The ability to bind non-native polypeptides is an essential function of cytosolic Hsp40 in vivo [27]. Sis1/Hdj1 chaperone also functions in concert with Hsp90/Hsp70 cycle [28]. In T. gondii, Hsp90/Hsp70 could be linked to parasite differentiation, an important process in the T. gondii pathology [29–30]. Taken all together, these studies indicate that Toxoplasma Sis1 could arise as a key molecule in the biology of the parasite.
The aim of this study is to characterize Toxoplasma Sis1-like (TgSis1) protein. Here we describe the predicted J-domain proteins in T. gondii as found in the T. gondii genome database (www.toxodb.org), and provide relative expression levels in stressed versus non-stressed parasites. TgSis1, first identified by in silico analysis based on the predicted subcellular localization, presence of characteristic motifs, and phylogenetic analysis, was characterized in a more detailed manner. Subcellular localization of TgSis1 was assessed by immunofluorescence analysis. Interactions between TgSis1 and other parasite proteins were determined by immunoprecipitation and Western blotting. Finally, a molecular and structural analysis of TgSis1 was performed in order to note the similarities and differences with its human counterpart Hdj1.
2. Experimental
2.1 In silico analysis
Sequences for putative Hsp40 proteins in T. gondii, which all have the corresponding J-domain, were found through screening of the T. gondii genome database (www.toxodb.org) and CD search (conserved domain search). Amino acids sequences and their respective DNA sequences were retrieved from ToxoDB. Sequences were further analyzed using Blast programs (www.ncbi.nlm.nih.gov.ar), Psort II (http://psort.ims.u-tokyo.ac.jp/cgi-bin/runpsort.pl), TargetP1.1 (http://www.cbs.dtu.dk/services/TargetP/), signalP 3.0 (http://www.cbs.dtu.dk/cgi-bin/webface?jobid=signalp,4C1A3214014BEFAF&opt=none), Plasmit (http://gecco.org.chemie.uni-frankfurt.de/plasmit/index.html), WolF Psort (http://wolfpsort.org/), MultiLoc/Target Loc (http://www-bs.informatik.uni-tuebingen.de/Services/MultiLoc/), and PATS (http://gecco.org.chemie.uni-frankfurt.de/pats/pats-index.php).
A neighbor-joining (NJ) tree was constructed with MEGA4 (Molecular Evolutionary Genetics Analysis Software version 4) as previously described [31]. Internal support was measured using 1000 replicates of the heuristic search bootstrap option.
2.2 Cloning of cDNA encoding putative Toxoplasma Sis1-like protein
Total RNA from RH strain (genotype I) tachyzoites was extracted using TRIzol®Reagent (Invitrogen Life Technologies) according to the manufacturer’s instructions. Between 1.0 and 5.0 μg of RNA were used for obtaining cDNA through reverse transcription reactions using 100 U of MMLV reverse transcriptase (Invitrogen Life Technologies) and oligo-dT as primers for mRNA. The following primers were used for the PCR reactions: TgSis1 sense (Fw): 5′-GGATCCATGGGGAAGGACTACTACAG-3′ BamHI site was added (underlined sequence) and antisense (Rv): 5′-AAGCTTCGCTCGAAGTGCAGA-3′ (recognizing a region on the 3 UTR sequence), where HindIII site was added (underlined sequence). The amplified product was cloned in the cloning vector pGEM-T easy (Promega) and sequenced (Macrogen Corp, USA). Likewise, the open reading frame (ORF) of the putative T. gondii Ydj1 (TGME49_111240) was cloned by using the following primers: sense (Fw), 5′-GGTACCATGTATTTTGGCAGCTTC-3′ and antisense (Rv), 5′-AAGCTTATTGGTGTATACGCGGTCTTCT-3′ (recognizing a region on the 3′ UTR sequence). KpnI and Hind III sites were added (underlined sequences).
2.3 Expression and purification of recombinant T. gondii proteins
Tgsis1 and the putative TgYdj1 ORFs were subcloned into the prokaryotic expression vector pRSET-A (TgSis1) and pRSET-B (TgYdj1) (Invitrogen Life Technology) in frame with a sequence that encodes for 6 N-terminal histidines (6Histag). Escherichia coli BL21 (DE3) pLys (Novagen) bacteria were freshly transformed with the expression plasmid. Cultures were grown to OD600 = 0.4–0.6 before protein expression induction by the addition of isopropyl thio-β-D-galactoside (2 mM). After overnight incubation at 37°C, the cells were harvested and purified using a commercial HisTrap 5ml column (GE Healthcare) according to the manufacturer’s instructions (under non-native conditions).
2.4 Antibody sources
In order to obtain anti-rTgSis1 polyclonal antibody, one rabbit was immunized with 250–300 μg of purified protein combined with 250–300 μl of complete Freund’s adjuvant (Sigma-Aldrich Argentina S. A.) and boosted every fifteen days with three successive injections with identical doses of the recombinant protein in incomplete Freund’s adjuvant (Sigma-Aldrich Argentina S. A.). Anti - T. gondii Hsp90 was previously described [29]. Anti - T. gondii H2AZNt (against N-terminal region of histone H2AZ) was previously described [32]. Obtention of the anti-T. gondii Hsp20 antibody was already described [33]. Anti-human Hsp70 antibody is commercially available (C92F3A-5, Stressgen) and showed to detect T. gondii HSP70 [34]
2.5 Parasite manipulation and co-localization analysis
Parasite growth and co-localization analyses have been described [29]. Briefly, tachyzoites of type I strain RH were grown in vitro in human foreskin fibroblasts (HFF). Following incubation with primary antibodies, cells were washed three times with PBS, and then incubated with the corresponding secondary antibodies (1:4000) Alexa fluor goat anti-rabbit 594 or Alexa fluor goat anti-mouse 488 (Invitrogen) as described [29]. Cover slips were washed three times and mounted in Fluoromont G (Southern Biotechnology Associates) and viewed using a Nikon Model Eclipse E600 (magnification 100X, numerical aperture 1,40 at 24°C). Green and red fluorescence were recorded separately and the images were analyzed by Image-Pro Plus version 5.1.0.20 and merged using Adobe Photoshop.
Soluble and insoluble material from tachyzoite lysate (Tg) were obtained by freeze/thaw in MOPS buffer (25 mM MOPS/KOH pH 7.5, 5 mM MgCl, 150 mM KCl) and further centrifuged for 10 min at 15,000 ×g (4°C) to obtain the pellet and the supernatant fractions. Samples were analyzed by Western blot using anti-T. gondii Hsp20 antibody (Inner membrane complex) as insoluble fraction marker (pellet) as described [33] and anti-TgSis1.
2.6 Immunoblot and co-immunoprecipitation (co-IP) analysis
In order to prepare tachyzoite lysates, between 5 × 108 to 1 × 109 parasites were collected from an infected HFF monolayer, centrifuged at 1,800 rpm for 10 min at room temperature, resuspended in sterile PBS and counted in a Neubauer hemocytometer chamber. Parasites were purified from the host cell material by passage through a 3 μm-pore size filter (Amersham Hybond, GE Healthcare Argentina S.A.).
For Western blot analysis, proteins from parasites and human foreskin fibroblasts (HFF) were extracted in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, electrophoresed and transferred to nitrocellulose. Non-specific binding sites were blocked with 5% non-fat-dried milk in PBS containing 0.1% Tween-20 (PBS-T) and the membranes were then incubated (1 h, room temperature) with polyclonal antibodies diluted 1:1,000 for rabbit antibodies and 1:500 for mouse antibodies. The membranes were washed with PBS-T prior to incubation with ECL-Plus detection reagent (Amersham Biosciences).
Co-immunoprecipitation (co-IP) assays were performed as follow: 5 × 108 to 1 × 109 tachyzoites were resuspended in 1–2 ml of ice cold receptor buffer (RB: 10mM Tris-HCl pH 7.5, 50mM NaCl, 1mM EDTA, 1mM DTT, 10% glycerol, 10mM Na-molybdate, protease inhibitor (optional): 1 μg/ml each of leupeptin, aprotinin, pepstatin A and 1mM PMSF) and immediately sonicated at 30% amplitude in cycles of 15 sec on and 3 sec off. Then, the samples were centrifuged in bench top for 10 min at maximal speed and the supernatant was recovered as the protein extract. The lysates were adjusted to 0.1% Triton X-100 and primary antibody was added (anti-TgHsp90 or anti-TgSis1 or anti-Hsp70) and incubated for 1–6 h at 4°C with rocking. Then 25 μl of 100 mg/ml Protein A-Sepharose (previously equilibrated in RB) (Santa Cruz Biotechnologies) were added and the incubation continued for 30–60 min. The beads were spun out, and the supernatant was carefully discarded. The beads were washed 3X with 1ml RB with Triton X-100 (in order to reduce the background, the beads were transfer to a fresh tube in the first wash). The last wash was performed with buffer RB without Triton X-100 and the supernatant was carefully removed. The beads were finally resuspended in SDS sample buffer, boiled for 3 min, spun for 5 min and loaded on SDS-PAGE.
2.7 Model generation and refinement
I-Tasser [35–37] was used to generate a threading model of Domain I of the peptide-binding fragment of TgSis1 (residues 155 to 239). The C-score of the Domain I model is 0.90 indicating high confidence of the proposed model.
The model obtained with I-Tasser was submitted to a simulated annealing protocol using Yasara Dynamics 10 [38]. The protocol and forcefield were especially designed to refine theoretical models. The lowest energy model was selected. The refined model is within the statistically expected values according to What_Check [39] stand-alone software, i.e. no abnormalities were detected.
2.8 Model analysis
The theoretical and experimental models were visualized using Pymol (The PyMOL Molecular Graphics System, Version 1.2r1, Schrödinger L, http://www.schrodinger.com/) and Spdb-Viewer [40]. Pymol also was used as a front-end of APBS [41] to resolve the Poisson-Boltzmann Equation. Default APBS parameters were used except for the concentration of monovalent ions, which was set at 0.150 M. The hydrogen bond desolvation was analyzed with dehydron calculator [42] using default parameters.
3. Results
3.1 Identification of T. gondii Sis1-like Hsp40
In higher eukaryotes, Sis1/Hdj1 is a cytosolic Type II Hsp40 that contains a J-domain, a G/F rich region (central domain), and a region termed the conserved carboxyl terminal domain (CTD) that includes a dimerization region, whereas others type II Hsp40s only contains the J-domain and G/F-rich region [43]. In addition, Type I Hsp40s present a similar sequence organization than Sis1-like proteins, but includes a zinc finger-like domain between G/F rich region and CTD domain. Type III Hsp40s only contains the J-domain [43] and type IV, is similar to type III but the tripeptide HPD motif is not conserved [15]. Based on this classification, 36 sequences containing J-domains were retrieved and grouped according their respective type (Table 1). The putative subcellular localization was also assessed based on different predictors as outlined in “2. Experimental” section (Table 1).
Table 1.
T. gondii J-family proteins and expression analysis in stressed vs non-stressed parasites
| Order | Annotation Release 5 | Annotation Release 3 | Type | HPD motif | cellular localization£ | Fold increase | P value | Chromosome |
|---|---|---|---|---|---|---|---|---|
| 1 | TGME49_007760 | 25.m01777* | I | Yes | nuclear | 1.23 | 0.029001 | Ib |
| 2 | TGME49_058390 | 55.m00016 | I | Yes | secretory | −1.01 | 0.716880 | VIIb |
| 3 | TGME49_111240 | 583.m05418** | I | Yes | cytoplasmic | 1.34 | 0.009482 | XI |
| 4 | TGME49_004480 | 20.m03869 | II | Yes | ER | 1.22 | 0.004479 | VIIa |
| 5 | TGME49_060630 | 55.m00017# | II | Yes | ERmem | 1.05 | 0.161529 | VIIb |
| 6 | TGME49_065310 | 57.m00015## | II | Yes | cytoplasmic | 1.13 | 0.000233 | IX |
| 7 | TGME49_039850 | 49.m03180§ | III | Yes | nuclear | −1.62 | 0.150720 | VI |
| 8 | TGME49_073630 | 59.m06079 | III | Yes | nucl/mito | 1.08 | 0.493139 | VIII |
| 9 | TGME49_090880 | 80.m02292 | III | Yes | nuclear | −1.21 | 0.278490 | IX |
| 10 | TGME49_113400 | 583.m05553 | III | Yes | nuclear | 1.19 | 0.025874 | XI |
| 11 | TGME49_014530 | 33.m02195 | III | Yes | nuclear | 1.14 | 0.387941 | X |
| 12 | TGME49_001670 | 20.m03665 | III | Yes | PM | 1.23 | 0.425140 | VIIa |
| 13 | TGME49_003380 | 20.m03793§§ | III | Yes | nuclear | −1.01 | 0.847117 | VIIa |
| 14 | TGME49_002810 | 20.m05961 | III | Yes | ERmem | 1.02 | 0.750840 | VIIa |
| 15 | TGME49_017710 | 37.m00768 | III | Yes | Golgi | −1.56 | 0.720086 | XII |
| 16 | TGME49_032050 | 44.m02699 | III | Yes | Elsewhere | 1.1 | 0.126574 | VIII |
| 17 | TGME49_046340 | 50.m00025 | III | Yes | Mito | −1.01 | 0.820159 | XII |
| 18 | TGME49_067430 | 57.m01857 | III | Yes | cytoplasmic | 1.3 | 0.000034 | IX |
| 19 | TGME49_083580 | 76.m01546 | III | Yes | Nucl/cyto | −1.37 | 0.000810 | V |
| 20 | TGME49_085270 | 76.m01596 | III | Yes | nuclear | −1.03 | 0.694224 | V |
| 21 | TGME49_089240 | 80.m02203 | III | Yes | cytoplasmic | 1.89 | 0.443950 | IX |
| 22 | TGME49_101340 | 162.m00513 | III | Yes | nuclear | −1.48 | 0.034398 | IV |
| 23 | TGME49_113310 | 583.m00680 | III | Yes | Mito | −1.34 | 0.002056 | XI |
| 24 | TGME49_044350 | 49.m00059 | III | Yes | cytoplasmic | 1.5 | 0.002328 | VI |
| 25 | TGME49_056810 | 55.m11052 | III | Yes | Mitochondria | 1.11 | 0.516288 | VIIb |
| 26 | TGME49_120060 | 641.m02560 | III | Yes | Mito/cyto | 1.21 | 0.004200 | IV |
| 27 | TGME49_115690 | 583.m05701 | III | Yes | Nuclear | 3.76 | 0.088219 | XI |
| 28 | TGME49_088690 | 80.m02164 | III | Yes | Nuclear | 1.15 | 0.212118 | IX |
| 29 | TGME49_010430 | 26.m00247 | III | Yes | Nucl/cyto | 2.01 | 0.011852 | IX |
| 30 | TGME49_061410 | 55.m00251§§§ | III | Yes | Nucl/cyto | 1.33 | 0.124509 | VIIb |
| 31 | TGME49_014330 | 33.m02674 | III | Yes | Nuclear | 1.07 | 0.214727 | X |
| 32 | TGME49_023420 | 42.m03277 | III | Yes | Cytoplasmic | −1.17 | 0.824685 | X |
| 33 | TGME49_071910 | 59.m03598 | III | Yes | Secretory | 1.5 | 0.030907 | VIII |
| 34 | TGME49_059980 | 55.m04854φ | III | Yes | Nuclear | 1.62 | 0.018412 | VIIb |
| 35 | TGME49_053650 | 52.m01602φφ | IV | No | cytoplasmic | 1.4 | 0.00155991 | III |
| 36 | TGME49_026070 | 42.m00068 | IV | No | PM | −1.25 | 0.006274 | X |
Analyzed by different software as mentioned in section 2. Experimental
Similar to GenBank Accesion numbers CAJ20508 (1 to 1032 out of 1064) and CAJ20509 (50 to the 539, the end of the protein). The first 50 residues of CAJ20509 does not present J-domain. Both CAJ20508 and CAJ20509 retrieved only TGME49_007760 locus.
Putative TgYdj1
Putative Sec63
Putative TgSis1 (AN: ABY21519)
Putative Ras-like GTPase superfamily
Putative Zuotin related factor 1 with C-terminal SANT domain
Putative auxilin-like protein
The analysis of the aminoacidic sequence showed a typical type I structure (J-domain,G/F rich región, zinc-finger motif and CTD).
Putative sphingomyelin phosphodiesterase
Cyto: cytoplasmic; ER: endoplasmic reticulum; ERmem: ER membrane; Nucl: nuclear; Mito: mitochondria; PM: plasma membrane
P values were calculated as described in [45]. P values <0.05 are considered significant
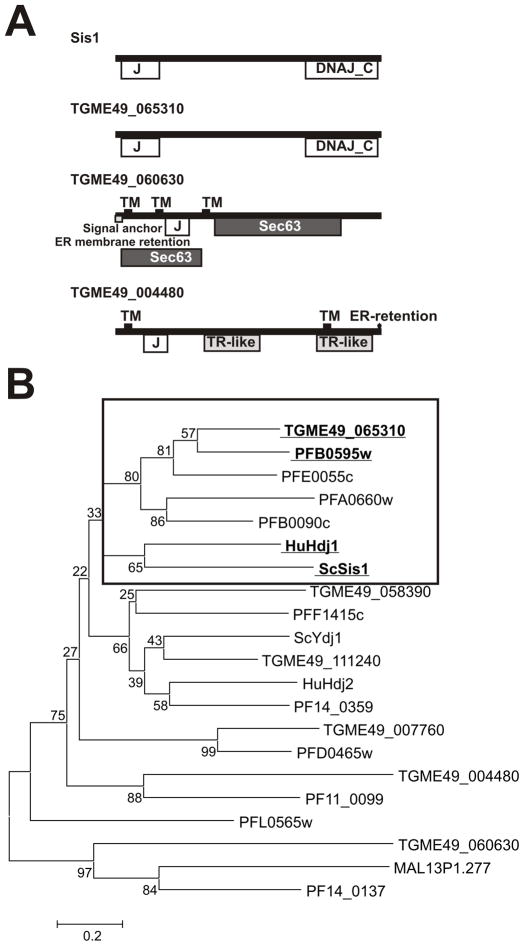
We noted that TGME49_065310 is the only type II protein predicted to have the requisites necessary to be classified as a Sis-like protein as described above. In fact, TGME49_065310 is the only cytosolic Type II Hsp40 harboring the typical Sis-like motif structure: the J-domain, the G/F-G/M rich region (central domain), and a CTD domain (Fig. 1A). To further confirm that TGME49_065310 is the Sis1-like chaperone in T. gondii, type II sequences were analyzed together with yeast Sis1 and human Hdj1 sequences to generate a Neighbor-joining (NJ) tree. TGME49_065310 clusters with Sis1 and Hdj1 sequences (Fig. 1B). TGME49_065310 is also closely related to four type II Plasmodium Hsp40 sequences, including PFA0660w (Fig. 1B), which is considered a Sis1-like Hsp40 [15]. The alignment of T. gondii with the yeast and human counterparts shows a high degree of conservation along the amino acid sequence (Fig 2). The identity values of TGME49_065310 vs ScSis1 or HuHdj1 are 35.1% and 42%, respectively. Comparison with P. falciparum sequences in the cluster is: PFA0660w, 40.5%; PFB0090c, 44.3%; PFE0055c, 48.8% and PFB0595w, 59.8%. Based on the collective evidence, we have named TGME49_065310 as TgSis1.
Fig. 1.
Domain organization and phylogenetic analysis of T. gondii Type I and II J-domain proteins. (A) Domain and motif organization of yeast Sis1 and T. gondii putative type II Hsp40s. Domains and motifs were identified by NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). (B) Neighbor joining (NJ) tree of Type I and II J-domain protein amino acid sequences of T. gondii, P. falciparum, S. cerevisiae Sis1, and human Hdj1. T. gondii sequences were included on the basis of gene ID release 4 (see Table 1). P. falciparum gene IDs begin with PF except Malp13P1.277. Frame indicates cluster of sequences related to Sis1/Hdj1. Type I and II sequences were analyzed because Sis1-like Hsp40 share the CTD domain present in type I Hsp40s.
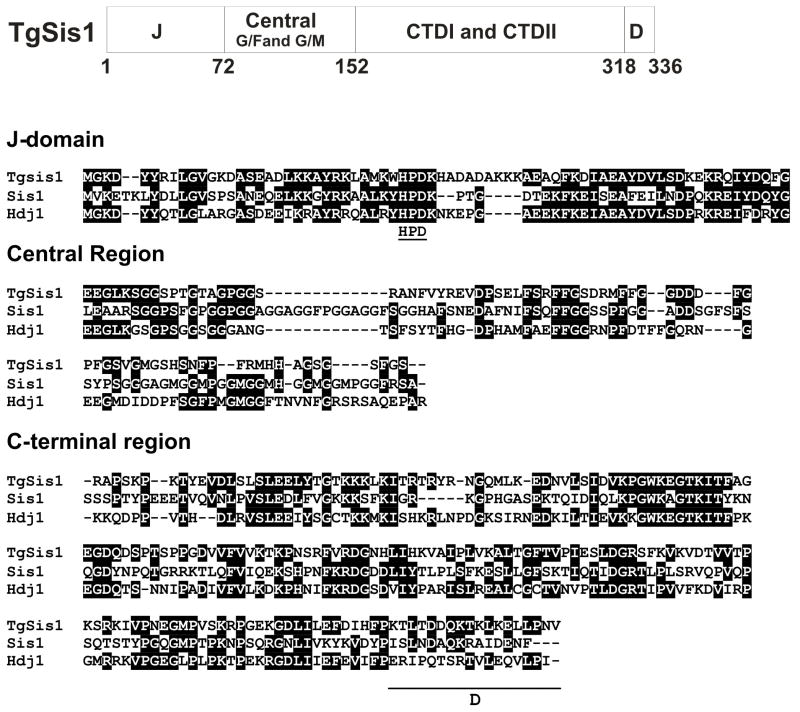
Fig. 2.
Sequence analysis of T. gondii Sis1 like protein (TgSis1). Upper panel: Domains of TGME49_065310 (putative TgSis1). J: J-domain; Central: central region that includes G/F- and G/M-rich regions; C-terminal I/II: C-terminal domain (CTD) containing Domain I (peptide binding fragment) and Domain II; D: dimerization motif. Middle and bottom panels: Sequence alignment generated by clustal W (Bioedit program). Letters highlighted in black indicate identical residues. Gaps are indicated by dashes and were introduced to improve the alignment. Sis1, S. cereviseae Sis1 (AN: CAA41366), HuHDJ1, Human HDJ1 (AN: P25685). The sequence was split in J-domain, Central region and C-terminal region (containing CTDI, CTDII and dimerization motif [underlined]) according Lee et al., [22] full length alignment.
3.2 Domain organization and sequence analysis of TgSis1
The predicted TgSis1 sequence displays the typical Sis1/Hdj1 domain organization (Fig. 2): J-domain, central region (G/F-G/M rich region), and C-terminal region (CTDI+CTDII+ dimerization motif). CTDI contains the peptide binding fragment [26]. The J-domain region shows identities ranging between 47– 56%, the central region shows identities ranging from 26.8–29.4%, and the CTD between 32.8–44.4% with Sis1 and Hdj1, respectively.
3.3 Expression analysis of Toxoplasma Sis1-like protein in stressed parasites
As heat shock proteins are often involved in the cellular stress response, it was of interest to determine the expression levels of the J-family proteins in stressed versus unstressed parasites. It is known that some stresses -like alkaline stress- induce bradyzoite gene induction [44]. A whole-genome microarray analysis of RH strain was performed previously using the Affymetrix ToxoGene Chip to compare parasites stressed for 3 days in pH 8.1 media relative to unstressed parasites cultured for 3 days in normal pH 7.0 media [32, 45]. The fold increase during stress was analyzed for the 36 putative Toxoplasma dnaJ genes (Table 1). TgSis1 (TGME49_065310) expression did not appear to be significantly altered under these stress conditions, suggesting that its role could not be related to alkaline stress.
Only two predicted J-proteins (TGME49_115690 and TGME49_010430) showed a fold increase value higher than 2.0 during stress (Table 1). TGME49_115690 is highly similar to Plasmodium vivax EDL45931.1 (31% identical) and Cryptosporidium parvum EAZ51503.1 (32% identical) sequences among others, suggesting that this gene is well-conserved across the phylum. No significant similarity was observed for the other gene ID in the GenBank database.
3.4 Analysis of the presence of the native proteins TgSis1 in T. gondii
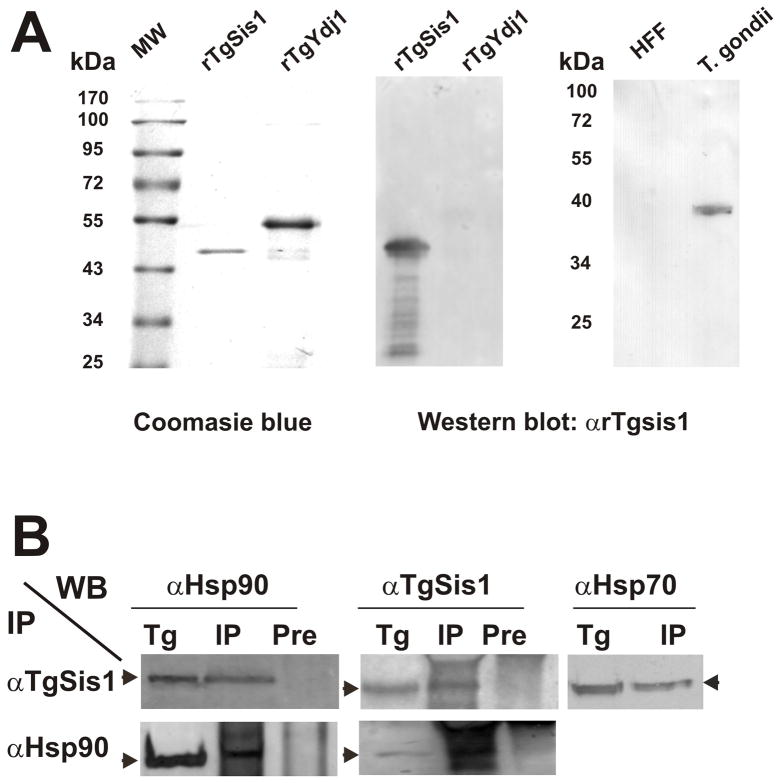
In order to characterize TgSis1, the gene predicted by TGME49_065310 was cloned and sequenced from cDNA obtained from T. gondii RH, a parasite genotype I strain, observing no differences with the correspondent sequence from GT1 strain (another genotype I strain). However, when compared with genotype II Me49 strain, few differences at nucleotide level can be observed, corresponding to 99% identity (data not shown). The TgSis1 cDNA is comprised of an open reading frame (ORF) of 1011 bp, which encodes a protein with a theoretical mass of 37.2 kDa. In order to produce polyclonal antibodies, TgSis1 was expressed as a recombinant protein (rTgSis1) in E. coli (Fig. 3A). The identity of the recombinant protein was confirmed by mass spectrometry (data not shown). The TgSis1 antiserum was able to recognize rTgSis1, but not the putative rTgYdj1 (Fig. 3A), Being TgYdj1 the closest related T. gondii protein to TgSis1 (identity between J-domains of 55%), the result obtained indicates that the anti-TgSis1 antibody produced is highly specific. The polyclonal anti-TgSis1 antibody (αTgSis1) recognized a band of expected molecular weight (~ 40 kDa) in T. gondii tachyzoite lysate and did not cross-react to host cell proteins (Fig. 3A).
Fig. 3.
Detection of native TgSis1-like protein and co-immunoprecipitation analysis. (A) Left panel, purified recombinant rTgSis1 and putative rTgYdj1 were electrophoresed in SDS-PAGE and stained with Coomassie Brilliant Blue. Middle and right panels, Western blot with rabbit α-TgSis1. T. gondii: parasite RH strain lysate. HFF: Protein extract from uninfected HFF cells. Pre-immune serum samples did not show reactivity (data not shown). Migration of molecular weight markers (PageRuler™Prestainded Protein Ladder, Fermentas International Inc.) is indicated in kilo Daltons (kDa). (B) Co-immunoprecipitation analysis. T. gondii RH strain lysates were used for immunoprecipitation (IP) using specific α-TgSis1 and T. gondii Hsp90 (α–Hsp90) antisera as indicated on the left side. IPs were analyzed by Western blotting with αTgSis1, α-Hsp90, and a commercial anti-human Hsp70 antibody (αHsp70). T. gondii RH strain lysate (Tg) was included in the Western blot analysis to identify the corresponding band. IP with pre-immune sera (Pre) was also analyzed with αTgSis1 and αHsp90 to test the specificity of the IP.
3.5 Co-Immunoprecipitation analysis of TgSis1 with Hsp70 and Hsp90
In order to determine whether TgSis1 forms complex with other chaperones, the anti-TgSis1 antibody was used to immunoprecipitate T. gondii lysates. As expected, anti-TgSis1 pulled down TgSis1 itself (Fig. 3B), suggesting that the antibody is adequate for co-immunoprecipitation assays combined with Western blot (coIP-WB). Immunoprecipitation of T. gondii lysate with αTgSis1 and Western blot with anti-Hsp70 and anti-TgHsp90, and viceversa, showed that in T. gondii, TgSis1 could be forming complexes with Hsp70 and Hsp90 (Fig 3B). Pre-immune sera did not pull down any of these proteins (Fig. 3B).
3.6 Subcellular localization of TgSis1
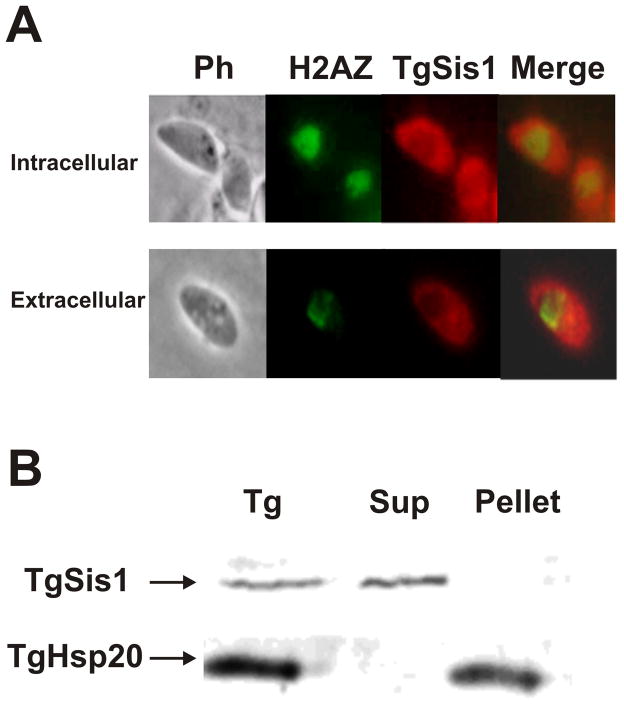
In order to determine the subcellular localization of TgSis1, the rabbit antibody αTgSis1 was used in immunofluorescence studies using the murine anti-H2AZNt antibody. The localization of TgSis1 was clearly cytosolic (excluding the nucleus) in intracellular as well as in extracellular tachzyoites (Fig. 4A). In concordance with cytosolic localization, when parasites were disrupted by sonication, TgSis1 was found in the soluble fraction (Fig. 4B), unlike the membrane associated chaperone Hsp20 [46].
Fig. 4.
Subcellular localization of TgSis1. (A) Indirect immunofluorescence assays (IFA) and subcellular localization of TgSis1-like protein in tachyzoites (Tz). IFA was performed on fixed intracellular parasites with rabbit αTgSis1 and murine anti-H2AZNt (H2AZ serves as a marker of the parasite’s nucleus). Merged, merge of TgSis1 and H2AZ images. (B) Tachyzoite lysate (Tg) was obtained by freeze/thaw in MOPS buffer, centrifuged to obtain the pellet and the supernatant (Sup). Samples were analyzed by Western blot with anti-T. gondii Hsp20 antibody and αTgSis1.
3.7 Structural analysis of TgSis1
Since Sis1-like proteins are essential in other species, TgSis1 could be considered as a good candidate for future drug design. To explore this possibility, it is necessary to analyze the structural and functional differences between TgSis1 and its human homologue. The peptide binding site at Domain I was previously identified in the human Hdj1 (from 162 to 241) and S. cerevisiae Sis1 (from 180 to 255) and shown to be essential in vivo [21, 23, 26]. Here we generated a threading model of Domain I of the peptide-binding fragment of TgSis1 (residues 155 to 239) that correspond to Domain I of Hdj1. The results suggest that important differences exist between TgSis1 and Hdj1. In our model, the peptide binding site of Domain I is deeper, a little narrower, and longer than the peptide binding site of Hdj1 [PDB ID 2QLD; http://www.pdb.org/pdb/explore/sequenceCluster.do?structureId=2QLD&entity=1&cluster=3715&seqid=30 [23] (Fig. 5A). These geometrical differences are mainly the result of a different global position of strands 2 and 3 with respect to the rest of the protein. These strands also display important differences at the secondary structural level; the strands 2 and 3 are longer and connected with a shorter loop in our model than in the Hdj1 X-ray model. This feature is the result of conformational preferences of the residues involved and not the result of insertion/deletion at the sequence level. Another important difference is the conformation of the K179 residue, which, in our model, is pointing towards the solvent instead of inside the peptide binding site as in the Hdj1 X-ray model, likely leaving more room for a peptide to bind.
Fig. 5.
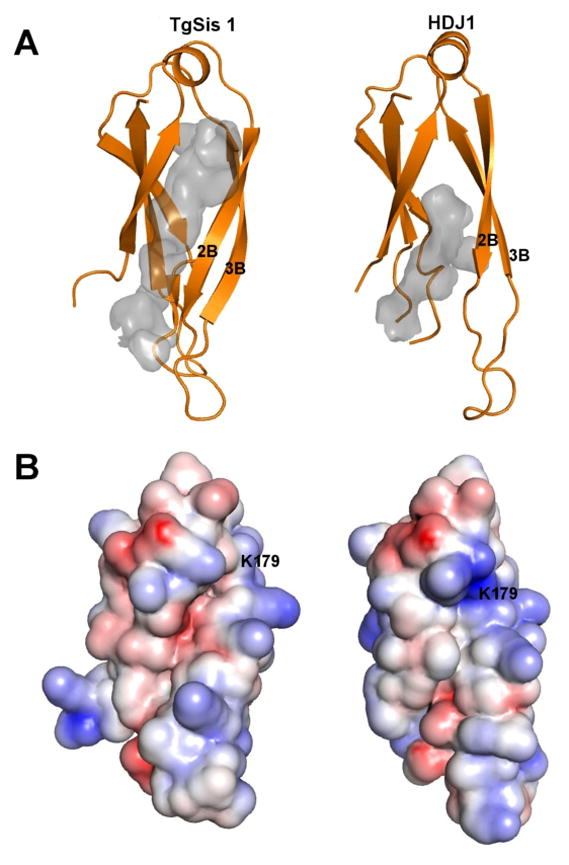
Structural comparison of peptide binding fragment (Domain I) of TgSis1 and of HjD1. (A) Both structures were drawn with PyMol using cartoon representation. It is clearly seen that the largest differences reside at the secondary structure and position of strand 2 and 3. The peptide binding sites are shown as gray cavities. The actual shape of the cavities depends on several user-adjustable parameters, but the peptide binding site of TgHSP40 is consistently deeper, a bit narrower and longer than the peptide binding site of Hdj1. (B) Electrostatic potential of Domain I of the peptide binding fragment of Hsp40 in PyMol plotted on the solvent-accessible surface from −5 kT/e (basic residue, Red) to +5 kT/e (acid residue, Blue).
Electrostatic features of the peptide binding site also show some subtle differences between our model and the X-ray model of human Hdj1 (Fig. 5B). While is true that both sites are mainly hydrophobic there are some basic and acid residues participating in the peptide-binding-site. As show in figure 5 (panel B) the charge distribution of the binding site differs between the two proteins.
We found three dehydrons at the tip of the peptide binding site that only exist on our model of TgSis1. These dehydrons include one hydrogen bond formed by T185-D220 and two formed by G119-G129. The dehydrons are present at the I-Tasser model and in the Yasara refined model.
4. Discussion
Here we have shown that T. gondii parasites contain a large number of predicted J-family members (36 ORFs), analogous to what has been found for P. falciparum (43–44 sequences) [15, 46]. Botha et al. [15], identified 32 T. gondii J-domain containing proteins. Since the Toxoplasma genome database is periodically updated, we consider that the differences between ours and Botha’s analysis are due to searching the database at different times. We also found that the transcription of some of these TgHsp40 genes is increased during alkaline stress, indicating a possible role in the parasite stress response, a treatment that triggers bradyzoite gene expression and parasite encystment in vitro [44]. Although Toxoplasma RH strain does not form cyst walls in vitro, it does express detectable levels of bradyzoite-specific marker genes in response to stresses that induce cyst formation [32, 45]. As such, the relationship of these three Hsp40s and the bradyzoite differentiation should be further studied. Regarding other protozoa, a genomic analysis revealed the existence of at least 70 distinct HSP40s in Trypanosomatids [47]. In S. cerevisiae, 22 open reading frames exist with conserved J-domains and 3 bear marginal similarity with J-domains [43], whereas human genome present 50 hsp40 genes [48]. It is clear that protozoan parasites have maintained a relative high number of highly differentiated HSP40s despite the reduction in their total genome size. As mentioned Shonhai et al. [49], the data suggests an important role for molecular chaperones in different biological processes of intracellular protozoan parasites like survival, differentiation and virulence. In the case of Hsp40s, the large number of J-domain proteins in these intracellular parasites may be related to different aspects of the complex life cycle of parasites, which involves rapid adaptation to environmental changes including fluctuation between different hosts, as well as having both intracellular with extracellular stages. The wide variety of Hsp40 proteins could interact with a diverse variety of substrates that could be assisted by the key chaperone Hsp70 either from the parasite or the host. Some Plasmodium spp. Hsp40s are good examples of chaperones that are exported to erythrocyte cytoplasm, possibly interacting with human Hsp70 [20, 46, 50–52].
TgSis1 shares Sis1/Hdj2 organization, possessing an N-terminal J-domain, a central region with G/F-G/M motifs, and a C-terminal region that contains a carboxyterminal Domain I, a carboxyterminal Domain II and a dimerization motif. While the J-domain of TgSis1 shows high identity level compared with those from S. cerevisisae and human, the central region is the most divergent region on the three Hsp40s (TgSis1, Sis1 and Hdj1). Yan and Craig [53] demonstrated that the J-domain is essential and the G/F region could be involved in determining the specificity of the chaperone. Yeast mutants transformed with a truncated version of Sis1 (from residue 1 to 206) which includes essential J-domain, the G-M region and part of the Domain I, restored wild-type growth rates. Residues 108–257 of Sis1 were shown to play an important function in maintaining the quaternary structure in an arrangement where the J domains point away from the compact C-terminal core [24]. Based on these observations, it would be interesting to investigate how this divergent sequence alters the Sis1 quaternary structure and Hsp40 specificity. In this regard, future analysis can include yeast complementation studies, by using a SIS1 mutant [53], which was used to analyze Trypanosoma TcJ2 chaperone [54].
The C-terminal region of Sis1 is comprised of three parts: domain I, domain II, and a dimerization region. While domain II and the dimerization region are not required for cell viability, domain I is essential [27, 53]. TgSis1 and Hdj1 Domain I (peptide binding fragment) share general structure, suggesting a similar function in vivo. However, from a pharmacological perspective, it is important to define differences between TgSis1 and Hdj1 that could be selectively targeted. Our results revealed several differences between the Hdj1 X-ray model and our theoretical model of TgSis1. It is interesting to note that differences at sequence and structural levels of strands 2 and 3 are also observed between H. sapiens Hsp40, Hdj1, and S. cerevisiae Hsp40, Sis1 [23]. These differences may reflect adaptation of these chaperons to specific environments or functions across different species. We also searched for dehydrons in both models. Dehydrons are partially dehydrated intramolecular hydrogen bonds that result from an incomplete clustering of non-polar side-chain groups. Solvated hydrogen bonds are energetically less favorable than desolvated ones, hence dehydrons promote the removal of surrounding water through protein associations or ligand binding. These packing defects are generally not conserved among related proteins. Collectively, these features render the identification of dehydrons as a useful tool during rational drug design [55]. Both I-Tasser and the refinement protocol implemented in Yasara have proven to be successful at the last Critical Assessment of Techniques for Protein Structure Prediction (CASP) competition [37–38]. While it is important to keep in mind that in the absence of experimental data, theoretical models can be considered valid and refined solely on statistical terms, the differences we have observed in this preliminary structural analysis prompt us to perform a deeper study.
Our results demonstrated that TgSis1 co-immunoprecipitates with parasite Hsp70 and Hsp90. While the interaction with Hsp70 was previously described [12], the complex with Hsp90 was only recently documented [56]. Hsp90 functions in concert with a well-defined set of cofactors, including an Hsp40, which are essential to drive the cycle of Hsp90–substrate interaction [28], all of which have been identified in T. gondii [30]. Cintron and Toft [56] observed that both Ydj1 and Sis1 are able to promote the binding of progesterone receptor to the Hsp90-substrate interaction cycle. Despite Ydj1 binding tightly to progesterone receptor, the authors suggested that both Ydj1 and Sis1 protein can facilitate the maturation of hormone receptor. Interestingly, the Hsp90-heterocomplex is also linked to the Hepatitis B reverse transcriptase function, whereby Hdj1 (Sis1-like) protein was the most active Hsp40 [23]. As suggested by Cintron and Toft [56], it is possible that the different Hsp90 client proteins can differ in their Hsp40 requirements. If that is the case, it is possible that TgSis1 also participates in a version of the parasite Hsp90-heterocomplex cycle.
In conclusion, we identified a large number of T. gondii J-domain proteins belonging to the three Hsp40 types (I, II and III) as well as the recently described Type IV class described for P. falciparum. T. gondii possesses a Sis1/Hdj1-like chaperone that we named TgSis1 that is present in the tachyzoite cytosol and associates with both Hsp70 and Hsp90. The Domain I (peptide binding fragment) of TgSis1 displays some geometrical, electrostatic and hydrogen bond wrapping differences with its human counterpart. These differences may be exploited in future rational drug design aimed to selectively disrupt the normal function of the TgSis1 protein.
Research highlights.
List of Toxoplasma Hsp40 members including expression profile under alkaline stress
Identification and characterization of Toxoplasma Sis1
TgSis1 pulled down with Hsp70 and hsp90
TgSis1 is located at cytosol in tachyzoite
Molecular modeling of TgSis1 showed differences with human hdj1
Acknowledgments
SO Angel (Researcher), MM Corvi (Researcher), N De Miguel (Researcher), MJ Figueras (Fellow) and O. Martin (Fellow) are members of National Research Council of Argentina (CONICET). This work was supported by: UNSAM SA08/006; ANPCyT grant BID OC-AR PICT 05-34415 (to S.O.A); NIH-NIAID 1R01AI083162-01 (to S.O. A.) and NIH R01 AI077502 (to W.J.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Israelski DM, Chmiel JS, Poggensee L, Phair JP, Remington JS. J Acquir Immune Defic Syndr. 1993;6:414–418. [PubMed] [Google Scholar]
- 2.Luft BJ, Remington JS. Clin Infect Dis. 1992;15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 3.Black MW, Boothroyd JC. Microbiol Mol Biol Rev. 2000;64:607–623. doi: 10.1128/mmbr.64.3.607-623.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tenter AM, Heckeroth AR, Weiss LM. Int J Parasitol. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vonlaufen N, Kanzok SM, Wek RC, Sullivan WJ., Jr Cell Microbiol. 2008;10:2387–2399. doi: 10.1111/j.1462-5822.2008.01210.x. [DOI] [PubMed] [Google Scholar]
- 6.Cyr DM, Langer T, Douglas MG. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 7.Qiu XB, Shao YM, Miao S, Wang L. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cyr DM, Lu X, Douglas MG. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- 9.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 11.Wickner S, Hoskins J, McKenney K. Proc Natl Acad Sci U S A. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgopoulos CP, Lundquist-Heil A, Yochem J, Feiss M. Mol Gen Genet. 1980;178:583–588. doi: 10.1007/BF00337864. [DOI] [PubMed] [Google Scholar]
- 13.Zylicz M, Georgopoulos C. J Biol Chem. 1984;259:8820–8825. [PubMed] [Google Scholar]
- 14.Cheetham ME, Caplan AJ. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Botha M, Pesce ER, Blatch GL. Int J Biochem Cell Biol. 2007;39:1781–1803. doi: 10.1016/j.biocel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Horton LE, James P, Craig EA, Hensold JO. J Biol Chem. 2001;276:14426–14433. doi: 10.1074/jbc.M100266200. [DOI] [PubMed] [Google Scholar]
- 17.Zhong T, Arndt KT. Cell. 1993;73:1175–1186. doi: 10.1016/0092-8674(93)90646-8. [DOI] [PubMed] [Google Scholar]
- 18.Luke MM, Sutton A, Arndt KT. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmon D, Montero-Lomeli M, Goldenberg S. J Biol Chem. 2001;276:43970–43979. doi: 10.1074/jbc.M102427200. [DOI] [PubMed] [Google Scholar]
- 20.Maier AG, Rug M, O'Neill MT, Brown M, Chakravorty S, Szestak T, Chesson J, Wu Y, Hughes K, Coppel RL, Newbold C, Beeson JG, Craig A, Crabb BS, Cowman AF. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sha B, Lee S, Cyr DM. Structure. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 22.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Protein Sci. 2005;14:1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, Wu Y, Li J, Qian X, Fu Z, Sha B. BMC Struct Biol. 2008;8:3. doi: 10.1186/1472-6807-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos CH, Oliveira CL, Fan CY, Torriani IL, Cyr DM. J Mol Biol. 2008;383:155–166. doi: 10.1016/j.jmb.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sha B, Cyr D. Acta Crystallogr D Biol Crystallogr. 1999;55:1234–1236. doi: 10.1107/s090744499900476x. [DOI] [PubMed] [Google Scholar]
- 26.Lee S, Fan CY, Younger JM, Ren H, Cyr DM. J Biol Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JL, Craig EA. J Cell Biol. 2001;152:851–856. doi: 10.1083/jcb.152.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheibel T, Buchner J. Biochem Pharmacol. 1998;56:675–682. doi: 10.1016/s0006-2952(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 29.Echeverria PC, Matrajt M, Harb OS, Zappia MP, Costas MA, Roos DS, Dubremetz JF, Angel SO. J Mol Biol. 2005;350:723–734. doi: 10.1016/j.jmb.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 30.Echeverria PC, Figueras MJ, Vogler M, Kriehuber T, de Miguel N, Deng B, Dalmasso MC, Matthews DE, Matrajt M, Haslbeck M, Buchner J, Angel SO. Mol Biochem Parasitol. 2010;172:129–140. doi: 10.1016/j.molbiopara.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Dudley J, Nei M, Kumar S. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 32.Dalmasso MC, Onyango DO, Naguleswaran A, Sullivan WJ, Jr, Angel SO. J Mol Biol. 2009;392:33–47. doi: 10.1016/j.jmb.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Miguel N, Echeverria PC, Angel SO. Eukaryot Cell. 2005;4:1990–1997. doi: 10.1128/EC.4.12.1990-1997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss LM, Ma YF, Takvorian PM, Tanowitz HB, Wittner M. Infect Immun. 1998;66:3295–3302. doi: 10.1128/iai.66.7.3295-3302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A, Kucukural A, Zhang Y. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y. Proteins. 2009;77(Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. Proteins. 2009;77(Suppl 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hooft RW, Vriend G, Sander C, Abola EE. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 40.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez A, Scott R. Biophys J. 2003;85:1914–1928. doi: 10.1016/S0006-3495(03)74619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soete M, Camus D, Dubremetz JF. Exp Parasitol. 1994;78:361–370. doi: 10.1006/expr.1994.1039. [DOI] [PubMed] [Google Scholar]
- 45.Naguleswaran A, Elias EV, McClintick J, Edenberg HJ, Sullivan WJ., Jr PLoS Pathog. 2010;6:e1001232. doi: 10.1371/journal.ppat.1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Miguel N, Lebrun M, Heaslip A, Hu K, Beckers CJ, Matrajt M, Dubremetz JF, Angel SO. Biol Cell. 2008;100:479–489. doi: 10.1042/BC20080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pesce ER, Cockburn IL, Goble JL, Stephens LL, Blatch GL. Infect Disord Drug Targets. 2010;10:147–157. doi: 10.2174/187152610791163417. [DOI] [PubMed] [Google Scholar]
- 48.Folgueira C, Requena JM. FEMS Microbiol Rev. 2007;31:359–377. doi: 10.1111/j.1574-6976.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- 49.Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shonhai A, Maier AG, Przyborski JM, Blatch GL. Protein Pept Lett. 2011;18:143–157. doi: 10.2174/092986611794475002. [DOI] [PubMed] [Google Scholar]
- 51.Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 52.Vincensini L, Richert S, Blisnick T, Van Dorsselaer A, Leize-Wagner E, Rabilloud T, Braun Breton C. Mol Cell Proteomics. 2005;4:582–593. doi: 10.1074/mcp.M400176-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharjee S, van Ooij C, Balu B, Adams JH, Haldar K. Blood. 2008;111:2418–2426. doi: 10.1182/blood-2007-09-115279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan W, Craig EA. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edkins AL, Ludewig MH, Blatch GL. Int J Biochem Cell Biol. 2004;36:1585–1598. doi: 10.1016/j.biocel.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 56.Fernandez A, Crespo A. Chem Soc Rev. 2008;37:2373–2382. doi: 10.1039/b804150b. [DOI] [PubMed] [Google Scholar]
- 57.Cintron NS, Toft D. J Biol Chem. 2006;281:26235–26244. doi: 10.1074/jbc.M605417200. [DOI] [PubMed] [Google Scholar]