Abstract
MicroRNAs (miRNA), a class of ~22-nucleotide RNA molecules, are important gene regulators that bind to the target sites of mRNAs to inhibit the gene expressions either through translational inhibition or mRNA destabilization. There are growing evidences that miRNAs have played several regulatory roles in opioid pharmacology. Like other research fields such as cancer biology, the area where numerous miRNAs are found to be involved in gene regulation, we assume that in opioid studies including research fields of drug additions and opioid receptor regulation, there may be more miRNAs waiting to be discovered. This review will summarize our current knowledge of miRNA functions on opioids biology and briefly describe future research directions of miRNAs related to opioids.
Keywords: miRNA, GPCR, opioid receptor, fentanyl, morphine, addiction
Introduction
MicroRNAs (abbreviated miRNAs) are endogenously expressed and short non-coding RNAs (~22-nucleotides in length) that regulate gene expression at the post-transcriptional level through binding to a target complementary mRNA (Lee et al. 1993; Bartel 2004). The biological roles of miRNAs, although still largely unknown, are rapidly accumulating for their importance in development and diseases, such as cancers, drug addictions, metabolic diseases, neurological diseases, and viral infections. miRNAs are found in almost all bio-organisms and affect both the stability and translation of mRNAs (Valencia-Sanchez et al. 2006). Moreover, the biogenesis and activity of miRNAs are closely associated to those of small interfering RNAs (siRNAs) that mediate RNA interference, another ancient mechanism for post-transcriptional gene silencing (Kim et al. 2009). Bioinformatic predictions suggest that miRNAs target at least 30% of protein-coding genes (van Rooij 2011) and are involved in almost all cellular functions, and regulate gene expression mainly by binding to the 3′-UTRs of targeted mRNAs. Expression analysis of miRNAs using miRNA microarrays has been widely used to monitor tissue-specific miRNA expression and regulatory changes in tissues/cell types, developmental stages, diseases as well as tissues treated with opioid drugs (Lagos-Quintana et al. 2002; Liu and Kohane 2009; Schratt et al. 2006; Somel et al. 2010; Bartel 2004; Zheng et al. 2010c). It was suggested that miRNAs have an organ and/or cell type-specific function based on tissue and temporal specificity of miRNAs (Chen et al. 2004; Lee et al. 2006). Abnormal patterns of miRNA expression have been found in many disease states and drug addictions where expression of miRNA was either increased or decreased (Yang et al. 2007; Lee and Dutta 2006; Hollander et al. 2010). A number of experimental reports addressing the involvement of miRNAs in the opioid system clearly indicate that opioids modify the expression profile of a number of miRNAs in central nervous system (CNS) (Zheng et al. 2010c; Wu et al. 2009; He et al. 2010).
1) Regulation of miRNAs by opioids
Recent studies have demonstrated that miRNAs are highly expressed in the CNS including the brain and spinal cord where biological action of opioids, as well as nociception takes place (Dave and Khalili 2010; Sanchez-Simon et al. 2010; He et al. 2010; Zheng et al. 2010c). Although we are currently in the initial stages of understanding how this novel class of gene regulators is involved in opioid-related biological functions (Table 1), a growing body of exciting evidence suggests that miRNAs are important regulators of opioid-associated biological processes such as drug addiction (Zheng et al. 2010a; He et al. 2010), pain perception (Kusuda et al. 2011), neuron development (Gao 2010), viral infection (Wang et al. 2011b; Dave and Khalili 2010), and opioid receptor regulation (Wu et al. 2009).
Table 1.
List of miRNAs regulated by opioids.
| miRNA | Mechanisms affected | Factors (or genes) involved | References |
|---|---|---|---|
| miR-190 |
|
pERK, β-arrestin2, pYY1, and Talin2 | (Zheng et al. 2010c; Zheng et al. 2010a; Zheng et al. 2010b) |
| miR-133b |
|
ERK 1/2, TH, and DAT | (Sanchez-Simon et al. 2010) |
| miR-15b and -181b |
|
FGF-2, MCP-2, and IL-6 | (Dave and Khalili 2010) |
|
| |||
| miR-28, 125b, 150, and 382 |
|
IFN-α/β | (Wang et al. 2011b) |
|
| |||
| miR-23b |
|
MOR | (Wu et al. 2008; Wu et al. 2009) |
|
| |||
| let-7 |
|
MOR | (He et al. 2010) |
|
| |||
| miR-339 |
|
MOR | Unpublished |
CYP2D6, cytochrome P450 2D6; DAT, dopamine transporter; Drd3, dopamine D3R; FosB, FBJ murine osteosarcoma viral oncogene homolog B; IFN, interferon; IL-6, Interleukin-6; MCP-2, monocyte chemoattractant protein-2; MOR, mu opioid receptor; pERK, phosphorylated ERK; pYY1, phosphorylated YY1; TH, tyrosine hydroxylase.
A series of articles from our laboratory have shown good examples of miRNA involvement on opioid signaling (Zheng et al. 2010c; Zheng et al. 2010a; Zheng et al. 2010b). Also opioid agonists have been shown to affect the expression of several miRNA differentially (Table 1). Fentanyl and morphine are both opioid analgesics that act on the mu opioid receptor (MOR). Morphine is a natural opioid and mainly used as a pain medicine in clinics, while fentanyl is a fully synthetic opioid. In the clinic, patients chronically treated with morphine adversely develop drug unresponsiveness, defined as tolerance. Fentanyl is up to 100 times stronger than morphine and currently the most widely used synthetic opioid in clinical practice for relieving pain, especially in cancer patients and severe chronic pain management. Along with its stronger analgesic effect than morphine, fentanyl exhibits a much higher potential for developing addiction, dependence, and causes more severe respiratory depression (Jeal and Benfield 1997; Pergolizzi et al. 2008). However, compared to morphine, fentanyl produces less tolerance (Duttaroy and Yoburn 1995), and milder side-effects of nausea and itching (Mayes and Ferrone 2006). As a typical G protein-coupled receptor (GPCR), MOR signals are mediated by adenylyl cyclase, extracellular signal-regulated kinase (ERK) pathway, intracellular calcium stores, and ion channels on cell membrane (Law et al. 2000). In addition to both agonists’ difference in pharmacological actions, differential signaling of both agonists in cellular level suggested that the agonists of MOR induce ERK phosphorylation through different pathways: morphine uses the protein kinase C (PKC)-pathway, whereas fentanyl functions in a beta-arrestin2-dependent manner (Zheng et al. 2010c). These differences were also correlated with regulation of a miRNA expression differentially as described below in details and a detailed summary shown in Fig. 1 that was adopted and redrawn from a recent paper (Zheng et al. 2010a).
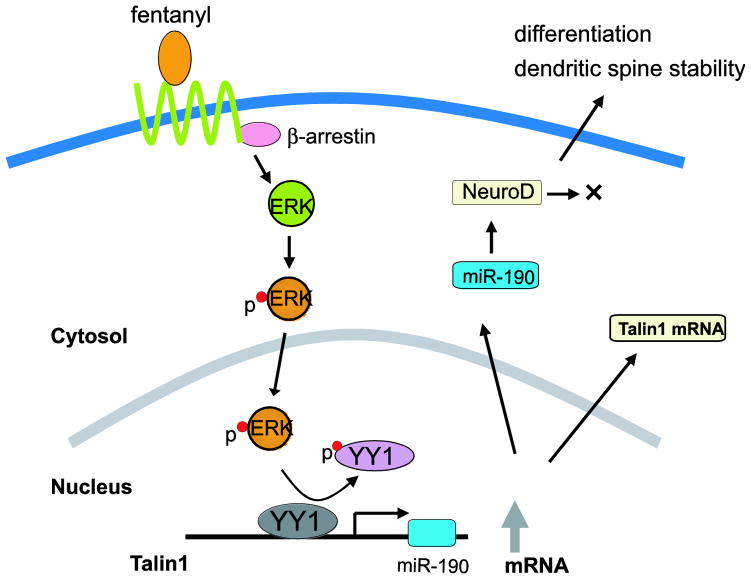
Fig. 1.
A summary of proposed pathway for the regulation of miR-190 expression by fentanyl. This figure was adopted and redrawn from Zheng et al. (Zheng et al. 2010a).
In miRNA microarray analysis using the primary culture of rat hippocampal neurons and the mouse hippocampi treated either with morphine or fentanyl for 3 days, among the seven miRNAs regulated by one or two of the agonists, miR-190 was down-regulated by fentanyl but not by morphine (Zheng et al. 2010c). It also indicated that fentanyl increases neurogenic differentiation 1 level (NeuroD, one of the miR-190 targets), a transcription factor that is known to be involved in adult neurogenesis, by reducing the amount of miR-190. Hippocampal adult neurogenesis is important for learning and memory (Neves et al. 2008). However, morphine does not change NeuroD level, suggesting that the MOR could regulate the NeuroD pathways through the control of miR-190 expression. This agonist-dependent regulation of NeuroD was associated with dendritic spine stability in an agonist-dependent manner too (Zheng et al. 2010b). Reduction of the miR-190 level by fentanyl was due to its host gene, talin2, suggesting that fentanyl decreases the miR-190 level by inhibiting the transcription of talin2 (Zheng et al. 2010a). Fentanyl signals to induce ERK phosphorylation by mediating β-arrestin2 pathway, resulting in the phosphorylation of Yin Yang1 (YY1). The resulting YY1 phosphorylation leads to a decrease in the association of YY1 with the Talin2 promoter which is essential for YY1 to stimulate the transcription of talin2. Therefore, fentanyl decreased the transcription of talin2 and subsequently reduced the cellular level of miR-190 by inducing YY1 phosphorylation. The reduction of miR-190 by fentanyl could result in increase of NeuroD level as mentioned earlier (Zheng et al. 2010c). In contrast, because morphine induces ERK phosphorylation via the PKC pathway, morphine did not induce YY1 phosphorylation and had no effect on the transcription of talin2 and the cellular content of miR-190. Together, the agonist-selective regulation on miR-190 is related with the agonist-selective ERK phosphorylation (Zheng et al. 2010c). Therefore, the above researches in regard to miR-190 provide a new insight to understand the differential mechanisms of MOR signaling, such as regulating dendritic spine stability, by different MOR agonists.
A recent article has reported miRNA’s association in neuronal development induced by an opioid (Sanchez-Simon et al. 2010). Previously, it was shown that morphine regulates neuron differentiation in vivo (Kim et al. 2006). Sanchez-Simon’s group (Sanchez-Simon et al. 2010), using zebrafish embryos as the model, reported that morphine decreases miR-133b expression, resulting in increase of its target gene expression, Pitx3 (Pituitary homeobox 3). The Pitx3, the homeobox (a DNA sequence encoding a protein domain (the homeodomain) which can bind DNA, mostly a promoter region of its target gene) gene as a transcription factor, is directly involved in the differentiation of dopaminergic neurons by activating genes such as the tyrosine hydroxylase and the dopamine transporter (Kim et al. 2007). They demonstrated that reduction of miR-133b level by morphine in zebrafish embryos is mediated through its MOR receptor via ERK 1/2. Morphine-induced down-regulation of miR-133b was also observed in the immature but not in mature rat hippocampal neurons. Long-term exposure to morphine was known to disrupt the ERK 1/2 signaling, which is thought to enhance the development of tolerance (Macey et al. 2009). This miR-133b study indicates that zebrafish can serve as a good model to investigate the roles of miRNA in neuronal development affected by long-term morphine exposure (Sanchez-Simon et al. 2010).
Regarding miRNAs in neuroimmune systems, two research groups recently reported that several miRNAs were regulated by opioids in human immune cells (Dave and Khalili 2010; Wang et al. 2011b). It is known that opioid abusers infected with HIV-1 (human immunodeficiency virus type 1) often exhibit an aggressive form of HIV-1-associated dementia and enhanced neurological disorders (Bell et al. 2006; Fitting et al. 2010). Active HIV-1 infection and secretion of neurotoxic molecules by microglia and perivascular macrophages are believed to be contributing factors for the HIV-1 neuropathogenesis (Gonzalez-Scarano and Martin-Garcia 2005; Hauser et al. 2007). A study by Dave et al. (Dave and Khalili 2010) reported that in human monocyte-derived macrophages treated with morphine, two miRNAs (miR-15b and miR-181b) were either greatly increased or decreased in expression levels, respectively. Fibroblast growth factor-2 (FGF-2), identified as a miR-15b target gene, was decreased at the protein expression levels in response to morphine. Other target genes, such as those involved in inflammation and T-cell activation pathways, namely MCP-2 (monocyte chemoattractant protein-2) and IL-6 (Interleukin-6), were also induced by morphine. Both, MCP-2 and IL-6 have been reported to be secreted in response to HIV-1 infection of microglia in the CNS (Schwartz et al. 2000; Wang and Gabuzda 2006), indicating the similar phenomena of the two pro-inflammatory responses (MCP-2 and IL-6 secretion) that were observed during HIV-1 infection. Moreover, proteomic analysis revealed the induction of mitochondrial superoxide dismutase in response to morphine treatment (Dave and Khalili 2010). In HIV-1 infection mitochondrial superoxide dismutase was not induced, suggesting the unique aspects of morphine inducing the metabolic change in CNS. These results by Dave et al. (Dave and Khalili 2010) demonstrate that morphine induces inflammation and oxidative stress in immune cells through regulating the miRNAs (miR-15b and 181-b), thereby contributing to expansion of HIV-1 CNS reservoir and disease progression.
Another group’s works by Wang et al. (Wang et al. 2011b) found the different types of miRNAs regulated by opioids in human monocytes. Morphine treatment in monocytes led to decrease in several anti-HIV miRNAs (miR-28, 125b, 150, and 382) which were correlated with susceptibility of monocytes/macrophages to HIV-1 infection (Wang et al. 2009). Antagonists of the opioid receptors blocked the decrease of miRNAs mediated by morphine, indicating morphine’s function through its own receptors. However, type I interferon (IFN), IFN-α/β in monocytes could induce the expression of the anti-HIV miRNAs (miR-28, 125b, 150, and 382). Type I IFNs are known to play a crucial role in the host innate immune defense mechanism against viral infection, including HIV-1 infection (Perry et al. 2005; Mogensen et al. 2010). Other studies have also shown that type I IFNs modulate miRNA expression in several cell systems (Pedersen et al. 2007; O’Connell et al. 2007; Ohno et al. 2009), functioning as the potent inducer of miRNAs. However, morphine co-treatment with the IFN-α/β in monocytes inhibited the induction of IFN-mediated anti-HIV miRNAs (Wang et al. 2011b). These results support previous observations from several reports that morphine suppresses IFNs expression in various cell systems, including monocytes (Pedersen et al. 2007), lymphocytes (Nair et al. 1997), peripheral blood mononuclear cells (PBMCs) (Homan et al. 2002). It is known that opioid potentiates HIV-1 infection at least partly by attenuating the immune response and is, therefore, a cofactor in the pathogenesis of HIV-1 infection (Vallejo et al. 2004; Donahoe and Vlahov 1998). Interestingly, Wang et al. (Wang et al. 2011b) observed the similar in vivo results in human that heroin (a synthetic derivative of morphine)-dependent subjects had significantly lower levels of anti-HIV miRNAs (miR-28, 125b, 150, and 382) in PMBCs than those of the healthy subjects, suggesting the important role of the miRNAs in opioid-mediated immunosuppression of HIV-1.
In future studies, it will be interesting to know whether opioid-mediated miRNA regulation, in addition to the above mentioned routes, is involved in HIV-1 infection through different factors. One factor would be a chemokine receptor 5 (CCR5), which is the main route to gain HIV-1 entry into host cells (Tuttle et al. 1998; Keele et al. 2008; Gottlieb et al. 2008). Several researches suggest that opioids may facilitate HIV-1 infection in human immune cells by modulating CCR5 (Tuttle et al. 1998; Guo et al. 2002; Vallejo et al. 2004) as well as cytokines. Opioids, morphine and methadone (a synthetic opioid and generally used in the treatment of opioid dependence), were shown to increases CCR5 expression in human immune cells (Guo et al. 2002; Li et al. 2002) and enhanced viral activation and replication. The effect of methadone on viral replication and entry was blocked by opioid antagonists, naltrexone and methylnaltrexone (Guo et al. 2002; Ho et al. 2003; Li et al. 2002). Other viral infections, including addicted patients with hepatitis C, were modified by opioids as well (Jeffrey et al. 2007). Therefore, in those model systems, it is reasonable to suggest that opioids may regulate miRNAs and contribute to regulatory effects of HIV-1 infectivity through CCR5 or cytokines.
In addition, various miRNAs were differentially regulated by xenobiotic drugs, including dexamethasone, methadone, and cocaine, in several human cell lines (Rodrigues et al. 2011). Methadone as well as cocaine down-regulated miR-124a, the most abundant microRNA expressed in neuronal cells and is involved in neuronal differentiation (Makeyev et al. 2007; Yu et al. 2008; Visvanathan et al. 2007), in human neuroblastoma (BE(2)-M17 and SH-SY5Y) cells (Rodrigues et al. 2011). Dexamethasone is a potent synthetic member of steroid drugs (a glucocorticoid receptor agonist), generally is known to act as an anti-inflammatory and immunosuppressant. Dexamethasone is also an inducer of the drug-metabolizing enzyme cytochrome P450 2D6 (CYP2D6) which converts codeine to a bioactive morphine in the liver (Gasche et al. 2004; Madadi et al. 2011). Codeine is one of the most commonly used pain medicines and is used to treat mild to moderate pain and to relieve cough (Drugs.com 2011). Since codeine is considered as a prodrug because of its conversion to active analgesic (morphine as well as codeine-6-glucuronide), drug-metabolism differences by enzymes, like the CYP2D6, significantly affect the pharmacokinetics of the codeine drug (Gasche et al. 2004). Several studies demonstrated differential analgesic response in individuals with genetic polymorphisms in the CYP2D6 gene (Flores and Mogil 2001; Ali et al. 2010), life-threatening opioid intoxication in patients for the treatment of a cough associated with bilateral pneumonia (Gasche et al. 2004), and differential prevalence rates of functional CYP2D6 alleles in ethnic groups with distinct codeine metabolism (Ingelman-Sundberg 2005). Rodrigues et al. (Rodrigues et al. 2011) reported that the inducer of CYP2D6, dexamethasone, in human cells (Caco-2), suppressed the expression of miR-27b, miR-148a, and miR-451 that regulate genes involving in adipocyte differentiation (Lin et al. 2009; McGregor and Choi 2011), epigenetic modification (Duursma et al. 2008; Sato et al. 2011; Lehmann et al. 2008), and multidrug resistance (drug-transporter P-glycoprotein) (van Jaarsveld et al. 2010). Dexamethasone, combined with other drugs, inhibited miR-21 expression in myeloma cells that adhered to bone marrow stromal cells (Wang et al. 2011a) and dexamethasone treatment in human corneal fibroblasts showed changes of global miRNA profiles analyzed by miRNA microarrays (Liu et al. 2011). The detailed mechanism underlying the induction of miRNAs by dexamethasone in different biosystems is still unclear and whether miRNAs directly involve in the up-regulation of CYP2D6 enzyme. However, it is possible that the regulation of CYP2D6 expression may be due to post-translational regulation by miRNAs.
All those forerunning studies (see Table 1) of miRNA regulation by opioid and its-related drugs strongly support potential and significant roles for miRNAs in opioid pharmacobiology (Kalow 2001) and human diseases. Future studies may or will find more direct networks of opioids in related to miRNAs and target genes, and may provide better insights in clinical pharmacology of opioids for pain, and their side effects including opioid dependence, withdrawal, and tolerance, as well as in opioid-related human diseases.
2) Opioid receptor gene regulation by miRNAs
Involvement of miRNA, currently considered as a new epigenetic factor, has begun to emerge on opioid receptor gene regulation, especially for mu opioid receptor (MOR) gene. In general, the MOR mRNA is uniquely distributed in brain and correlates with its protein expression patterns, as previously defined by autoradiography and immunohistochemical studies (Mansour et al. 1995; Brodsky et al. 1995). However, there are some discrepancies in certain brain regions between its mRNA and protein levels, as high levels of mRNA with little mu opioid binding sites or vice versa (Mansour et al. 1995). The discrepancy was also shown in antagonist-treated brain regions too (Unterwald et al. 1995). Although there are many possible reasons to explain the discrepancies, such as different sensitivities of the detection methods, RNA transport, existence of MOR isoforms (Pan et al. 2001), it is also possible that MOR at the brain region could be regulated at the post-transcriptional level—including different translation rates by miRNAs, RNA binding factors or through mRNA stability.
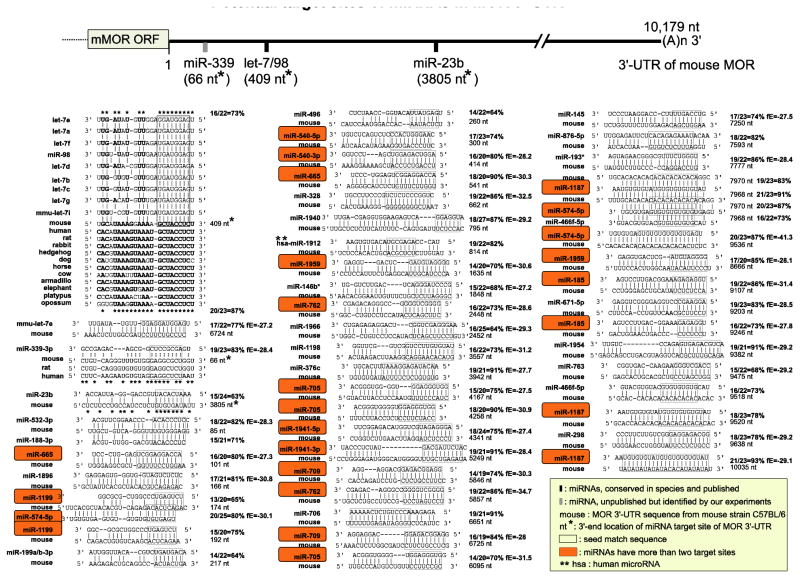
From our studies to determine whether miRNAs, that have their target sites especially in 3′-UTR of MOR gene, and are stimulated or inhibited by opioids or other stimulants to regulate MOR at the post-transcriptional level, we have found that two miRNAs were involved to regulate MOR gene: miR-23b and miR-339. The first one, miR-23b interacts with the MOR 3′-UTR via a K box motif (5′-UGUGAU-3′), a conserved 3′-UTR sequence motif (Fig. 2). In Drosophila, miRNAs directly bind to the K box of Notch target genes and regulate the expression of the Notch genes at the post-transcriptional level (Lai 2002; Lai et al. 1998; Lai et al. 2005). In mouse cells (P19 mouse embryonal carcinoma cells), miR-23b, via K box binding, regulates a notch gene Hairy/enhancer of split (Hes1) (Kimura et al. 2004), the transcription factor whose expression is initiated by the Notch signaling pathway. The Notch pathway is important for cell-cell communication involved in gene regulation mechanisms that control multiple cell differentiation processes during embryonic and adult life (Artavanis-Tsakonas et al. 1999). Several studies have demonstrated alteration of miR-23 expression (including its genome clustered miRNAs) in human diseases (Chhabra et al. 2009; Chhabra et al. 2010; Hassan et al. 2010), thereby indicating that miR-23 controls several processes during health and diseases.
Fig. 2.
Potential target sites of miRNAs in MOR 3′-UTR from mouse strain C57BL/6 by bioinformatic analysis (MicroInspector: bioinfo.uni-plovdiv.bg/microinspector/) (Rusinov et al. 2005). Homology percentage between miRNA and target site is shown on right side of each sequence alignment. Cutoff value of free energy (fE) for binding is −27 (kcal/mol) and each fE is indicated on right side. Sequence alignments without fE value were either obtained from previous studies (He et al. 2010; Wu et al. 2008) or analyzed by other web-based softwares, such as TargetScan (http://www.targetscan.org/mmu_50/) (Lewis et al. 2005), miRBase (http://www.mirbase.org/) (Kozomara and Griffiths-Jones 2011), or miRDB (http://mirdb.org/miRDB/) (Wang and El Naqa 2008; Wang 2008).
By knocking down endogenous miR-23b in mouse neuroblastoma NS20Y cells using miR-23b inhibitor (Wu et al. 2008), we confirmed that miR-23b inhibits MOR protein expression in vivo. This was the first study reporting a translationally repressive role for the MOR 3′-UTR by miRNA. The regulatory mechanism of miR-23b was that miR-23b blocks the association of MOR mRNA with polysomes, thereby arresting its translation and suppressing the production of MOR protein. Later in another article (Wu et al. 2009), we demonstrated that long-term morphine treatment increases miR-23b expression in a dose- and time-dependent manner and represses the polysomal association of MOR mRNA through the MOR 3′-UTR. The suppressive effect of morphine was further proven by the translational luciferase reporter assay that requires the MOR 3′-UTR. This suggests a potential link between MOR expression and morphine treatment at the post-transcriptional level in which a specific miRNA, miR-23b, is involved.
The second miRNA, miR-339, was identified by a microarray data analysis using mice treated chronically with either morphine or fentanyl, and was found to be consistently increased in hippocampal brain region, about 2- or 4-fold, respectively, compared to non-treated and agonist-treated cerebellum controls (unpublished data). This miRNA was bound to its target site in MOR 3′-UTR, resulting in suppression of the MOR gene. The suppression was attenuated by addition of miR-339 inhibitor, as well as when the target site was mutated. In MOR-expressing N2A-MOR cells, fentanyl treatment induced a 4-fold increase in the expression of mature miR-339 through increasing its primary miRNA production, indicating the involvement of transcriptional regulation of the miRNA. The entire length of its primary RNA and the promoter were identified. It was revealed that mature miR-339 was embedded in non-coding 3′-UTR region of an unknown host gene and was regulated under control of the host promoter. The resulting promoter activity was increased by fentanyl treatment and the activity was blocked by antagonist co-treatment. These results suggest that miR-339 play a key role in agonist-mediated MOR down-regulation through the binding of MOR 3′-UTR.
There is additional information that is valuable to mention here about what we obtained from this miR-339 work (unpublished data). To determine a role of this miRNA in vitro, we co-transfected miR-339 expression plasmid (pSuper-339) with the luciferase reporter construct, pMUTR, containing MOR promoter and the full-length of 3′-UTR of MOR gene, into HEK293T cells. The luciferase reporter activity began to decrease at the limited amount (50 ng) of pSuper-339 which is relevant to the amount of endogenous miRNA, suggesting an interaction between miR-339-3p and MOR 3′-UTR. However, when the MOR promoter in pMUTR was replaced by SV40 promoter (pSVUTR), the inhibition of pSuper-339 on reporter activity was blunted. Similar effect was observed in a reporter with SV40 promoter fused with MOR 5′-UTR (pSV5MUTR), suggesting a need to use different promoters in miR-339’s effect. When we used a construct containing miR-339 target site in pmirGLO vector (Promega) which is under the control of human phosphoglycerate kinase promoter (pPGK) and provides low translational expression, we could observe the subtle changes led by the miRNA. If, in many cases, miRNA target sites are controlled under strong viral promoters such as SV40 promoter, effects of miRNA may be harder to determine because miRNA-mediated effects often come with subtle changes in their target genes. The viral promoter produces too strong transcription that might hinder observing the regulatory effect by miRNA, especially on SV40 promoter-controlled constructs.
Recently, another research group reported that miRNA let-7 targets MOR gene (see also our in silico analysis in Fig. 2) and regulates opioid tolerance (He et al. 2010). let-7 binds to its target site in the MOR 3′-UTR and represses MOR expression. Morphine upregulates let-7 expression both in SH-SY5Y cells and a mouse model of opioid tolerance. The let-7 inhibitor decreases let-7 levels in brain and partially attenuated opioid antinociceptive tolerance in mice. Association of polysomes with MOR mRNA was decreased in a let-7-dependent manner. The miRNA let-7 functions as a mediator sequestering MOR mRNA to P-bodies leading to translational repression. These results suggest that let-7 may play a significant role in opioid tolerance.
For other two members of opioid receptors, delta and kappa, DOR and KOR, respectively, there is no report about miRNA-mediated regulation. However, based on our unpublished in silico analysis, there are several miRNA target sites in their DNA sequences. Therefore, it will be worthy to study on miRNA-mediated regulation about the above two genes.
Although the above three miRNAs were identified as regulators of MOR expression, in silico analysis to search miRNA target sites in MOR 3′-UTR (Fig. 2) showed potential target sites of several more miRNAs with highly complementary sequence (over 80% compared to full sequences of miRNAs), low free energy values, and some miRNAs had more than two target sites in MOR 3′-UTR. These suggest that further research works may find more miRNAs targeting to MOR gene and provide details for better understanding in miRNA-mediated MOR regulation.
3) miRNAs in opioid drug addiction and future directions
Opioid drug addiction is a major public health issue, despite its usefulness of pain-killing effects. Addictive drugs, such as cocaine, opioids (including heroin) and amphetamines, trigger strong and persistent neuroadaptive changes in the brain through a series of gene regulatory mechanisms leading to addiction. Recent studies have reported involvement of miRNAs in drug addiction (Hollander et al. 2010; Dreyer 2010; Schaefer et al. 2010), mainly associated with cocaine-related drug addiction, although there is no direct report of miRNAs involved in opioid-related addiction so far. It is worth to review various articles related to other drugs regarding miRNAs which will guide miRNA study further for opioid addiction. A recent report (Hollander et al. 2010) showed that miR-212, which is closely related to miR-132, is upregulated in the dorsal striatum, the brain region known to regulate the development of compulsive drug use in drug-abused rats. Surprisingly, even more overexpression of the miRNA using lentiviral vector system in the same brain region caused the abused rats to eliminate the compulsive drug-seeking behavior. The report demonstrated that miR-212 regulate the activity of CREB (cAMP-response element binding protein), a transcription factor known to play a role in decreasing the rewarding properties of cocaine. CREB, in turn, increases the expression of miR-212. Interestingly, the expression of CREB is stimulated by cocaine intake, and may play a key role of a self-limiting mechanism of drug use. This may provide a new direction for addiction therapeutics by miRNA expression.
Previously it was reported that MeCP2 (methyl CpG binding protein 2) expression is regulated by a miRNA miR-132 (a same family member of miR-212), which is also induced by CREB (Klein et al. 2007). The MeCP2 protein (Lewis et al. 1992) binds to forms of DNA that have been methylated at the cytosine of CpG islands, which frequently occur at the gene promoters. The MeCP2 protein then interacts with other repressor proteins, such as histone deacetylases and histone methyltransferases, and the co-repressor Sin3a, to form a complex that repress the target gene. Mutations of MeCP2 gene cause most cases of the neurodevelopmental disorder Rett syndrome and one of the most common causes of mental retardation in females (Amir et al. 2005; Amir et al. 1999). Loss of function as well as increased expression of the MeCP2 gene also cause several neuropsychiatric disorders (Chahrour et al. 2008), indicating MeCP2 is a key contributor to neurological diseases.
MeCP2 translation was decreased by miR-132 binding to its own target site of MeCP2 long variant’s 3′-UTR (Klein et al. 2007). Blocking miR-132-mediated repression led to increased MeCP2 and brain-derived neurotrophic factor (BDNF) levels. The increased MeCP2 enhanced BDNF and miR-132 levels in vivo. Increase in miR-132 will then decrease the levels of MeCP2 and restore the balance. This feedback circuit may provide a mechanism for homeostatic control of MeCP2 expression in the brain. Failure to regulate MeCP2 levels is connected to neurological disorders including Rett syndrome. The transcription of this cluster was enhanced by CREB. BDNF activates CREB through kinases (ERK1/2 and MSK) and the activated CREB then enhances production of miR-132. Enhancers of CREB phosphorylation (i.e., forskolin and KSHV) can also enhance miR-132 production in vitro. In vivo data also showed that the miR-132 levels were increased in the in vivo activation of neurons of brain regions that were induced by long-term potentiation, bicuculline, pilocarpine-induced seizures, contextual fear conditioning, odor-exposure, and cocaine injection (Wayman et al. 2008; Nudelman et al. 2010; Wibrand et al. 2010). These in vivo studies suggest a strong causal relationship between neuronal activation and miR-132 transcription. miR-132 is also associated with synaptic function as it increases post-synaptic protein levels (BDNF-related increase of miR-132) (Kawashima et al. 2010) and is associate with Fragile X Mental Retardation Protein (FMRP) (Edbauer et al. 2010), which is important for learning and memory.
In addition, it was recently reported the involvement of the MeCP2 in cocaine drug addiction with the same type miRNAs (Albulescu et al. 2011). Cocaine increases MeCP2 expression in dorsal striatum, which suppresses cocaine-stimulated increases of miR-212 and miR-132, and thereby increasing expression to promote further cocaine intake (Albulescu et al. 2011). However, there were contradicting/complicated network functions between those factors reported from several previous works (Feng and Nestler 2010); as miR-212/miR-132 repress MeCP2 translation; MeCP2 represses the BDNF gene directly; CREB, which is also activated by cocaine, stimulates BDNF as well as miR-212/miR-132 expression; miR-212/miR-132 can enhance CREB activation; and CREB can compete with MeCP2 for promoter binding sites. These complicated/coordinated epigenetic networks remain to be resolved to better understand the role of MeCP2 in drug addiction. MeCP2 was also reported that it modulates amphetamine (AMPH)-induced behaviors (Deng et al. 2010) and the psychostimulant induces phosphorylation of MeCP2 at Ser421 which is associated with the repressor function of MeCP2. The phosphorylation (pMeCP2) is selectively induced in GABAergic interneurons of nucleus accumbens and pMeCP2 is correlated with behavioral sensitization to AMPH.
In vivo administration of cocaine in rat can have profound effects on the expression of opioid receptors. Chronic repeated administration of cocaine resulted in increased binding to mu opioid receptor (Unterwald et al. 1992; Unterwald et al. 1994). Level of MOR mRNA was also transiently elevated following chronic continuous cocaine administration. Cocaine-induced MOR mRNA upregulation was blocked by co-administration of either D1, D2, or D3 dopamine receptor antagonists (Azaryan et al. 1996b; Azaryan et al. 1996a). Furthermore, elevations in MOR mRNA were also found after continuous administration of other direct and indirect dopamine agonists (Azaryan et al. 1996a). These data confirm the involvement of dopaminergic mechanisms in the effects of cocaine on MOR regulation. Importantly, specific miRNAs have emerged as key regulators leading to addiction, and could serve as valuable targets for more efficient therapies. As mentioned the above, a miRNA miR-212 inhibit the development of addiction in rats exposed to cocaine. It will be interesting to know whether the same miRNA miR-212 is involved in opioid drug addiction or are there other miRNAs too?
There is another example of miRNA’s involvement to cocaine addiction that may lead to a new direction of opioid addiction research regarding miRNA. Argonaute 2 (Ago2), which is known to control miRNA expression, in dopamine 2 receptor (Drd2)-expressing neurons, is involved in regulation of cocaine addiction (Schaefer et al. 2010). Deficiency of Ago2 in Drd2-expressing neurons greatly reduces the motivation to self-administer cocaine in mice. The researchers identified several miRNAs that are specifically regulated by Ago2 in the striatum and suggested that those miRNAs may likely play a role in cocaine addiction.
In the near future, we expect more miRNA researches to let us know better about the opioid addiction as we have seen in cocaine drug addiction for the last few years. Altogether, if future studies find more miRNAs’ involvements in opioid pharmacobiology, it is possible that miRNA may serve as a therapeutic intervention for patients who abuse opioids. Although the therapeutic use of miRNA/siRNA is in their early stages, the miRNA field has evolved remarkably fast since the original discovery of miRNA molecules and appears to be one of the most promising research areas for therapeutic strategy and drug discovery. Further research should be conducted to examine the role of this valuable gene regulator, miRNA in opioid pharmacology.
Acknowledgments
This work was supported by the National Institutes of Health National Institutes of Drug Abuse [Grants DA000564, DA001583, DA011806, K05-DA070554, DA011190, DA013926]; and by the A & F Stark Fund of the Minnesota Medical Foundation.
Abbreviations
- Ago2
Argonaute 2
- BDNF
brain-derived neurotrophic factor
- CNS
central nervous system
- CREB
cAMP-response element binding protein
- Drd2
dopamine 2 receptor
- ERK
extracellular signal-regulated kinase
- FGF-2
Fibroblast growth factor-2
- GPCR
G protein-coupled receptor
- HEK
human embryonic kidney
- HIV-1
human immunodeficiency virus type 1
- MeCP2
methyl CpG binding protein 2
- MOR
mu opioid receptor
- NeuroD
neurogenic differentiation 1 level
- PKA
protein kinase A
- PKC
protein kinase C
Footnotes
The authors declare no conflict of interest.
References
- Albulescu R, Neagu M, Albulescu L, Tanase C. Tissular and soluble miRNAs for diagnostic and therapy improvement in digestive tract cancers. Expert Rev Mol Diagn. 2011;11 (1):101–120. doi: 10.1586/erm.10.106. [DOI] [PubMed] [Google Scholar]
- Ali S, Drendel AL, Kircher J, Beno S. Pain management of musculoskeletal injuries in children: current state and future directions. Pediatr Emerg Care. 2010;26 (7):518–524. doi: 10.1097/PEC. quiz 525–518. [DOI] [PubMed] [Google Scholar]
- Amir RE, Sutton VR, Van den Veyver IB. Newborn screening and prenatal diagnosis for Rett syndrome: implications for therapy. J Child Neurol. 2005;20 (9):779–783. doi: 10.1177/08830738050200091401. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23 (2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284 (5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Azaryan AV, Clock BJ, Cox BM. Mu opioid receptor mRNA in nucleus accumbens is elevated following dopamine receptor activation. Neurochem Res. 1996a;21 (11):1411–1415. doi: 10.1007/BF02532382. [DOI] [PubMed] [Google Scholar]
- Azaryan AV, Coughlin LJ, Buzas B, Clock BJ, Cox BM. Effect of chronic cocaine treatment on mu- and delta-opioid receptor mRNA levels in dopaminergically innervated brain regions. J Neurochem. 1996b;66 (2):443–448. doi: 10.1046/j.1471-4159.1996.66020443.x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116 (2):281–297. doi: 10.1016/s0092-8674(04)00045-5. S0092867404000455. [DOI] [PubMed] [Google Scholar]
- Bell JE, Arango JC, Anthony IC. Neurobiology of multiple insults: HIV-1-associated brain disorders in those who use illicit drugs. J Neuroimmune Pharmacol. 2006;1 (2):182–191. doi: 10.1007/s11481-006-9018-2. [DOI] [PubMed] [Google Scholar]
- Brodsky M, Elliott K, Hynansky A, Jenab S, Inturrisi CE. Quantitation of mu-opioid receptor (MOR-1) mRNA in selected regions of the rat CNS. Neuroreport. 1995;6 (5):725–729. doi: 10.1097/00001756-199503270-00005. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320 (5880):1224–1229. doi: 10.1126/science.1153252. 320/5880/1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303 (5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chhabra R, Adlakha YK, Hariharan M, Scaria V, Saini N. Upregulation of miR-23a-27a-24-2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PLoS One. 2009;4 (6):e5848. doi: 10.1371/journal.pone.0005848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. 2010;9:232. doi: 10.1186/1476-4598-9-232. 1476-4598-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave RS, Khalili K. Morphine treatment of human monocyte-derived macrophages induces differential miRNA and protein expression: impact on inflammation and oxidative stress in the central nervous system. J Cell Biochem. 2010;110 (4):834–845. doi: 10.1002/jcb.22592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13 (9):1128–1136. doi: 10.1038/nn.2614. nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahoe RM, Vlahov D. Opiates as potential cofactors in progression of HIV-1 infections to AIDS. J Neuroimmunol. 1998;83 (1–2):77–87. doi: 10.1016/s0165-5728(97)00224-5. [DOI] [PubMed] [Google Scholar]
- Dreyer JL. New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome Med. 2010;2 (12):92. doi: 10.1186/gm213. gm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugs.com. [Accessed October 18 2011];Codeine Information from Drugs.com. 2011 Web site from Drugs.com. http://www.drugs.com/monograph/codeine.html.
- Duttaroy A, Yoburn BC. The effect of intrinsic efficacy on opioid tolerance. Anesthesiology. 1995;82 (5):1226–1236. doi: 10.1097/00000542-199505000-00018. [DOI] [PubMed] [Google Scholar]
- Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14 (5):872–877. doi: 10.1261/rna.972008. rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65 (3):373–384. doi: 10.1016/j.neuron.2010.01.005. S0896-6273(10)00010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Nestler EJ. MeCP2 and drug addiction. Nat Neurosci. 2010;13 (9):1039–1041. doi: 10.1038/nn0910-1039. nn0910-1039. [DOI] [PubMed] [Google Scholar]
- Fitting S, Xu R, Bull C, Buch SK, El-Hage N, Nath A, Knapp PE, Hauser KF. Interactive comorbidity between opioid drug abuse and HIV-1 Tat: chronic exposure augments spine loss and sublethal dendritic pathology in striatal neurons. Am J Pathol. 2010;177 (3):1397–1410. doi: 10.2353/ajpath.2010.090945. S0002-9440(10)60193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores CM, Mogil JS. The pharmacogenetics of analgesia: toward a genetically-based approach to pain management. Pharmacogenomics. 2001;2 (3):177–194. doi: 10.1517/14622416.2.3.177. [DOI] [PubMed] [Google Scholar]
- Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. 1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasche Y, Daali Y, Fathi M, Chiappe A, Cottini S, Dayer P, Desmeules J. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351 (27):2827–2831. doi: 10.1056/NEJMoa041888. 351/27/2827. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5 (1):69–81. doi: 10.1038/nri1527. nri1527. [DOI] [PubMed] [Google Scholar]
- Gottlieb GS, Heath L, Nickle DC, Wong KG, Leach SE, Jacobs B, Gezahegne S, van ‘t Wout AB, Jacobson LP, Margolick JB, Mullins JI. HIV-1 variation before seroconversion in men who have sex with men: analysis of acute/early HIV infection in the multicenter AIDS cohort study. J Infect Dis. 2008;197 (7):1011–1015. doi: 10.1086/529206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo CJ, Li Y, Tian S, Wang X, Douglas SD, Ho WZ. Morphine enhances HIV infection of human blood mononuclear phagocytes through modulation of beta-chemokines and CCR5 receptor. J Investig Med. 2002;50 (6):435–442. doi: 10.1136/jim-50-06-03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Gordon JA, Beloti MM, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc Natl Acad Sci U S A. 2010;107 (46):19879–19884. doi: 10.1073/pnas.1007698107. 1007698107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser KF, El-Hage N, Stiene-Martin A, Maragos WF, Nath A, Persidsky Y, Volsky DJ, Knapp PE. HIV-1 neuropathogenesis: glial mechanisms revealed through substance abuse. J Neurochem. 2007;100 (3):567–586. doi: 10.1111/j.1471-4159.2006.04227.x. JNC4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Yang C, Kirkmire CM, Wang ZJ. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J Neurosci. 2010;30 (30):10251–10258. doi: 10.1523/JNEUROSCI.2419-10.2010. 30/30/10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WZ, Guo CJ, Yuan CS, Douglas SD, Moss J. Methylnaltrexone antagonizes opioid-mediated enhancement of HIV infection of human blood mononuclear phagocytes. J Pharmacol Exp Ther. 2003;307 (3):1158–1162. doi: 10.1124/jpet.103.056697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466 (7303):197–202. doi: 10.1038/nature09202. nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan JW, Steele AD, Martinand-Mari C, Rogers TJ, Henderson EE, Charubala R, Pfleiderer W, Reichenbach NL, Suhadolnik RJ. Inhibition of morphine-potentiated HIV-1 replication in peripheral blood mononuclear cells with the nuclease-resistant 2–5A agonist analog, 2-5A(N6B) J Acquir Immune Defic Syndr. 2002;30 (1):9–20. doi: 10.1097/00042560-200205010-00002. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5 (1):6–13. doi: 10.1038/sj.tpj.6500285. 6500285. [DOI] [PubMed] [Google Scholar]
- Jeal W, Benfield P. Transdermal fentanyl. A review of its pharmacological properties and therapeutic efficacy in pain control. Drugs. 1997;53 (1):109–138. doi: 10.2165/00003495-199753010-00011. [DOI] [PubMed] [Google Scholar]
- Jeffrey GP, MacQuillan G, Chua F, Galhenage S, Bull J, Young E, Hulse G, O’Neil G. Hepatitis C virus eradication in intravenous drug users maintained with subcutaneous naltrexone implants. Hepatology. 2007;45 (1):111–117. doi: 10.1002/hep.21470. [DOI] [PubMed] [Google Scholar]
- Kalow W. Pharmacogenetics, pharmacogenomics, and pharmacobiology. Clin Pharmacol Ther. 2001;70 (1):1–4. doi: 10.1067/mcp.2001.116714. S0009-9236(01)51246-1. [DOI] [PubMed] [Google Scholar]
- Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, Kunugi H, Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165 (4):1301–1311. doi: 10.1016/j.neuroscience.2009.11.057. S0306-4522(09)01968-X. [DOI] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105 (21):7552–7557. doi: 10.1073/pnas.0802203105. 0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Clark AL, Kiss A, Hahn JW, Wesselschmidt R, Coscia CJ, Belcheva MM. Mu- and kappa-opioids induce the differentiation of embryonic stem cells to neural progenitors. J Biol Chem. 2006;281 (44):33749–33760. doi: 10.1074/jbc.M603862200. M603862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317 (5842):1220–1224. doi: 10.1126/science.1140481. 317/5842/1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10 (2):126–139. doi: 10.1038/nrm2632. nrm2632. [DOI] [PubMed] [Google Scholar]
- Kimura H, Kawasaki H, Taira K. Mouse microRNA-23b regulates expression of Hes1 gene in P19 cells. Nucleic Acids Symp Ser (Oxf) 2004;(48):213–214. doi: 10.1093/nass/48.1.213. 48/1/213. [DOI] [PubMed] [Google Scholar]
- Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10 (12):1513–1514. doi: 10.1038/nn2010. nn2010. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39 (Database issue):D152–157. doi: 10.1093/nar/gkq1027. gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuda R, Cadetti F, Ravanelli MI, Sousa TA, Zanon S, De Lucca FL, Lucas G. Differential expression of microRNAs in mouse pain models. Mol Pain. 2011;7:17. doi: 10.1186/1744-8069-7-17. 1744-8069-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12 (9):735–739. doi: 10.1016/s0960-9822(02)00809-6. S0960982202008096. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30 (4):363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 1998;125 (20):4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19 (9):1067–1080. doi: 10.1101/gad.1291905. gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law PY, Wong YH, Loh HH. Molecular mechanisms and regulation of opioid receptor signaling. Annu Rev Pharmacol Toxicol. 2000;40:389–430. doi: 10.1146/annurev.pharmtox.40.1.389. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Palkovits M, Young WS., 3rd miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc Natl Acad Sci U S A. 2006;103 (42):15669–15674. doi: 10.1073/pnas.0605781103. 0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75 (5):843–854. doi: 10.1016/0092-8674(93)90529-y. 0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7 (6):560–564. [PubMed] [Google Scholar]
- Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214 (1):17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120 (1):15–20. doi: 10.1016/j.cell.2004.12.035. S0092867404012607. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69 (6):905–914. doi: 10.1016/0092-8674(92)90610-o. 0092-8674(92)90610-O. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Tian S, Guo CJ, Douglas SD, Ho WZ. Methadone enhances human immunodeficiency virus infection of human immune cells. J Infect Dis. 2002;185 (1):118–122. doi: 10.1086/338011. JID010390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276 (8):2348–2358. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kohane IS. Tissue and process specific microRNA-mRNA co-expression in mammalian development and malignancy. PLoS One. 2009;4 (5):e5436. doi: 10.1371/journal.pone.0005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Walker EA, Kissane S, Khan I, Murray PI, Rauz S, Wallace GR. Gene Expression and miR Profiles of Human Corneal Fibroblasts in Response to Dexamethasone. Invest Ophthalmol Vis Sci. 2011;52 (10):7282–7288. doi: 10.1167/iovs.11-7463. iovs.11-7463. [DOI] [PubMed] [Google Scholar]
- Macey TA, Bobeck EN, Hegarty DM, Aicher SA, Ingram SL, Morgan MM. Extracellular signal-regulated kinase 1/2 activation counteracts morphine tolerance in the periaqueductal gray of the rat. J Pharmacol Exp Ther. 2009;331 (2):412–418. doi: 10.1124/jpet.109.152157. jpet.109.152157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madadi P, Ciszkowski C, Gaedigk A, Leeder JS, Teitelbaum R, Chitayat D, Koren G. Genetic transmission of cytochrome P450 2D6 (CYP2D6) ultrarapid metabolism: implications for breastfeeding women taking codeine. Curr Drug Saf. 2011;6 (1):36–39. doi: 10.2174/157488611794479991. CDS ABS-63. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27 (3):435–448. doi: 10.1016/j.molcel.2007.07.015. S1097-2765(07)00488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Akil H, Watson SJ. Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci. 1995;18 (1):22–29. doi: 10.1016/0166-2236(95)93946-u. [DOI] [PubMed] [Google Scholar]
- Mayes S, Ferrone M. Fentanyl HCl patient-controlled iontophoretic transdermal system for the management of acute postoperative pain. Ann Pharmacother. 2006;40 (12):2178–2186. doi: 10.1345/aph.1H135. 40/12/2178. [DOI] [PubMed] [Google Scholar]
- McGregor RA, Choi MS. microRNAs in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11(4):304–316. doi: 10.2174/156652411795677990. CMM # 105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH, Melchjorsen J, Larsen CS, Paludan SR. Innate immune recognition and activation during HIV infection. Retrovirology. 2010;7:54. doi: 10.1186/1742-4690-7-54. 1742-4690-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair MP, Schwartz SA, Polasani R, Hou J, Sweet A, Chadha KC. Immunoregulatory effects of morphine on human lymphocytes. Clin Diagn Lab Immunol. 1997;4 (2):127–132. doi: 10.1128/cdli.4.2.127-132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9 (1):65–75. doi: 10.1038/nrn2303. nrn2303. [DOI] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20 (4):492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104 (5):1604–1609. doi: 10.1073/pnas.0610731104. 0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Natsume A, Kondo Y, Iwamizu H, Motomura K, Toda H, Ito M, Kato T, Wakabayashi T. The modulation of microRNAs by type I IFN through the activation of signal transducers and activators of transcription 3 in human glioma. Mol Cancer Res. 2009;7 (12):2022–2030. doi: 10.1158/1541-7786.MCR-09-0319. 1541-7786.MCR-09-0319. [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci U S A. 2001;98 (24):14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449 (7164):919–922. doi: 10.1038/nature06205. nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergolizzi J, Boger RH, Budd K, Dahan A, Erdine S, Hans G, Kress HG, Langford R, Likar R, Raffa RB, Sacerdote P. Opioids and the management of chronic severe pain in the elderly: consensus statement of an International Expert Panel with focus on the six clinically most often used World Health Organization Step III opioids (buprenorphine, fentanyl, hydromorphone, methadone, morphine, oxycodone) Pain Pract. 2008;8 (4):287–313. doi: 10.1111/j.1533-2500.2008.00204.x. PPR204. [DOI] [PubMed] [Google Scholar]
- Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15 (6):407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- Rodrigues AC, Li X, Radecki L, Pan YZ, Winter JC, Huang M, Yu AM. MicroRNA expression is differentially altered by xenobiotic drugs in different human cell lines. Biopharm Drug Dispos. 2011;32 (6):355–367. doi: 10.1002/bdd.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33 (Web Server issue):W696–700. doi: 10.1093/nar/gki364. 33/suppl_2/W696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Simon FM, Zhang XX, Loh HH, Law PY, Rodriguez RE. Morphine regulates dopaminergic neuron differentiation via miR-133b. Mol Pharmacol. 2010;78 (5):935–942. doi: 10.1124/mol.110.066837. mol.110.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278 (10):1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Im HI, Veno MT, Fowler CD, Min A, Intrator A, Kjems J, Kenny PJ, O’Carroll D, Greengard P. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J Exp Med. 2010;207 (9):1843–1851. doi: 10.1084/jem.20100451. jem.20100451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439 (7074):283–289. doi: 10.1038/nature04367. nature04367. [DOI] [PubMed] [Google Scholar]
- Schwartz C, Catez P, Rohr O, Lecestre D, Aunis D, Schaeffer E. Functional interactions between C/EBP, Sp1, and COUP-TF regulate human immunodeficiency virus type 1 gene transcription in human brain cells. J Virol. 2000;74 (1):65–73. doi: 10.1128/jvi.74.1.65-73.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, Yuan Y, Ning Z, Hu Y, Menzel C, Hu H, Lachmann M, Zeng R, Chen W, Khaitovich P. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20 (9):1207–1218. doi: 10.1101/gr.106849.110. gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle DL, Harrison JK, Anders C, Sleasman JW, Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol. 1998;72 (6):4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterwald EM, Horne-King J, Kreek MJ. Chronic cocaine alters brain mu opioid receptors. Brain Res. 1992;584 (1–2):314–318. doi: 10.1016/0006-8993(92)90912-s. [DOI] [PubMed] [Google Scholar]
- Unterwald EM, Rubenfeld JM, Imai Y, Wang JB, Uhl GR, Kreek MJ. Chronic opioid antagonist administration upregulates mu opioid receptor binding without altering mu opioid receptor mRNA levels. Brain Res Mol Brain Res. 1995;33 (2):351–355. doi: 10.1016/0169-328x(95)00143-g. 0169328X9500143G. [DOI] [PubMed] [Google Scholar]
- Unterwald EM, Rubenfeld JM, Kreek MJ. Repeated cocaine administration upregulates kappa and mu, but not delta, opioid receptors. Neuroreport. 1994;5 (13):1613–1616. doi: 10.1097/00001756-199408150-00018. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20 (5):515–524. doi: 10.1101/gad.1399806. 20/5/515. [DOI] [PubMed] [Google Scholar]
- Vallejo R, de Leon-Casasola O, Benyamin R. Opioid therapy and immunosuppression: a review. Am J Ther. 2004;11 (5):354–365. doi: 10.1097/01.mjt.0000132250.95650.85. 00045391-200409000-00005. [DOI] [PubMed] [Google Scholar]
- van Jaarsveld MT, Helleman J, Berns EM, Wiemer EA. MicroRNAs in ovarian cancer biology and therapy resistance. Int J Biochem Cell Biol. 2010;42 (8):1282–1290. doi: 10.1016/j.biocel.2010.01.014. S1357-2725(10)00032-4. [DOI] [PubMed] [Google Scholar]
- van Rooij E. The Art of MicroRNA Research. Circ Res. 2011;108 (2):219–234. doi: 10.1161/circresaha.110.227496. [DOI] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21 (7):744–749. doi: 10.1101/gad.1519107. 21/7/744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gabuzda D. Reconstitution of human immunodeficiency virus-induced neurodegeneration using isolated populations of human neurons, astrocytes, and microglia and neuroprotection mediated by insulin-like growth factors. J Neurovirol. 2006;12 (6):472–491. doi: 10.1080/13550280601039659. GW1126U62300520V. [DOI] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14 (6):1012–1017. doi: 10.1261/rna.965408. rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24 (3):325–332. doi: 10.1093/bioinformatics/btm595. btm595. [DOI] [PubMed] [Google Scholar]
- Wang X, Li C, Ju S, Wang Y, Wang H, Zhong R. Myeloma cell adhesion to bone marrow stromal cells confers drug resistance by microRNA-21 up-regulation. Leuk Lymphoma. 2011a;52 (10):1991–1998. doi: 10.3109/10428194.2011.591004. [DOI] [PubMed] [Google Scholar]
- Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113 (3):671–674. doi: 10.1182/blood-2008-09-175000. blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ye L, Zhou Y, Liu MQ, Zhou DJ, Ho WZ. Inhibition of anti-HIV microRNA expression: a mechanism for opioid-mediated enhancement of HIV infection of monocytes. Am J Pathol. 2011b;178 (1):41–47. doi: 10.1016/j.ajpath.2010.11.042. S0002-9440(10)00089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105 (26):9093–9098. doi: 10.1073/pnas.0803072105. 0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KO, Lee CS, Pena JT, Tuschl T, Bramham CR. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci. 2010;31 (4):636–645. doi: 10.1111/j.1460-9568.2010.07112.x. EJN7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Law PY, Wei LN, Loh HH. Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3′ untranslated region: a role for microRNA23b. FASEB J. 2008;22 (12):4085–4095. doi: 10.1096/fj.08-108175. fj.08-108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang L, Law PY, Wei LN, Loh HH. Long-term morphine treatment decreases the association of mu-opioid receptor (MOR1) mRNA with polysomes through miRNA23b. Mol Pharmacol. 2009;75 (4):744–750. doi: 10.1124/mol.108.053462. mol.108.053462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13 (4):486–491. doi: 10.1038/nm1569. nm1569. [DOI] [PubMed] [Google Scholar]
- Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314 (14):2618–2633. doi: 10.1016/j.yexcr.2008.06.002. S0014-4827(08)00232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chu J, Zeng Y, Loh HH, Law PY. Yin Yang 1 phosphorylation contributes to the differential effects of mu-opioid receptor agonists on microRNA-190 expression. J Biol Chem. 2010a;285 (29):21994–22002. doi: 10.1074/jbc.M110.112607. M110.112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zeng Y, Chu J, Kam AY, Loh HH, Law PY. Modulations of NeuroD activity contribute to the differential effects of morphine and fentanyl on dendritic spine stability. J Neurosci. 2010b;30 (24):8102–8110. doi: 10.1523/JNEUROSCI.6069-09.2010. 30/24/8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zeng Y, Zhang X, Chu J, Loh HH, Law PY. mu-Opioid receptor agonists differentially regulate the expression of miR-190 and NeuroD. Mol Pharmacol. 2010c;77 (1):102–109. doi: 10.1124/mol.109.060848. mol.109.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]