Abstract
The small GTPase H-Ras is a proto-oncogene that activates a variety of different pathways including the extracellular-signal-regulated kinase mitogen-activated protein kinase (ERK/MAPK) pathway. H-Ras is mutated in many human malignancies and these mutations cause the protein to be constitutively active. PEA-15 blocks ERK-dependent gene transcription and inhibits proliferation by sequestering ERK in the cytoplasm. We therefore investigated whether PEA-15 influences H-Ras mediated transformation. We found that PEA-15 does not block H-Ras activated proliferation when H-Ras is constitutively active. We show instead that in H-Ras transformed mouse kidney epithelial cells, co-expression of PEA-15 resulted in enhanced soft agar colony growth and increased tumor growth in vivo. Overexpression of both H-Ras and PEA-15 resulted in accelerated G1/S cell cycle transition and increased activation of the ERK signaling pathway. PEA-15 mediated these effects through activation of its binding partner phospholipase D1 (PLD1). Inhibition of PLD1 or interference with PEA-15/PLD1 binding blocked PEA-15’s ability to increase ERK activation. Our findings reveal a novel mechanism by which PEA-15 positively regulates Ras/ERK signaling and increases the proliferation of H-Ras transformed epithelial cells through enhanced PLD1 expression and activation. Thus, our work provides a surprising mechanism by which PEA-15 augments H-Ras driven transformation. These data reveal that PEA-15 not only suppresses ERK signaling and tumorigenesis but can alternatively enhance tumorigenesis in the context of active Ras.
Keywords: PEA-15, PLD, cell cycle, H-Ras, cell transformation
Introduction
The small GTPase Ras is frequently mutated in cancer and contributes to transformation and tumor progression (Dunn et al 2005, Tan et al 2005). Physiologically, Ras is involved in a variety of signaling pathways regulating different outcomes (Cordova-Alarcon et al 2005). GTP-loaded Ras activates many pathways including the mitogen-activated protein kinase extracellular-signal-regulated kinase (MAPK/ERK) pathway, the PI3 Kinase pathway and the RalGDS pathway all of which have known roles in cancer via diverse effects both on a cellular and physiological level. For example, Ras-MAPK/ERK signaling modulates transcription, cell cycle progression, proliferation and differentiation, as well as senescence and cell death (Castaneda et al 2010, Ferro and Trabalzini 2010, Ramos 2008).
Mutations in Ras genes such as K-Ras, N-Ras or H-Ras can be found in various tumors and are often associated with poor prognosis (Cordova-Alarcon et al 2005, Zeng et al 2010). Mutated H-Ras is associated with elevated expression of key cell cycle regulatory proteins such as cyclin D1 and cdk4 in in vivo models of oral squamous cell carcinoma of the head and neck region (HNSCC) (Sathyan et al 2007). Cordova-Alarcon et al. showed that mutational activation of H-Ras accelerates G1/S cell cycle transition in a human cervical cancer-derived cell line in vitro and plays an important role in the development of cervical tumors (Cordova-Alarcon et al 2005). Oncogenic H-Ras also transforms NIH3T3 fibroblasts at least in part through upregulation of cyclin D1 due to an overexpression of Krueppel-like factor 5 (Nandan et al 2004). Interestingly, mutations in H-Ras not only affect gene transcription but also contribute to tumor cell transformation by inducing anti-apoptotic signals. In immortal baby mouse kidney epithelial cells (iBMK) a constitutively activated form of H-Ras inhibited paclitaxel induced accumulation of the proapoptotic BH3-only protein BIM, thereby preventing BIM-dependent apoptosis. In this model, H-Ras to ERK signaling resulted in phosphorylation of BIM leading to its proteasomal degradation (Tan et al 2005).
The small death effector domain (DED)-containing protein PEA-15 (Phosphoprotein Enriched in Astrocytes, 15 kDa) is a molecular scaffold that regulates several pathways including the Ras-MAPK/ERK signaling cascade (Ramos 2008, Revet et al 2008). It has been shown to bind ERK and prevent ERK-mediated gene transcription (Formstecher et al 2001, Whitehurst et al 2004). This can result in a down-regulation of cell proliferation as shown in murine T cells (Pastorino et al 2010). Interaction of PEA-15 with specific partners is controlled by its phosphorylation and subcellular localization. These factors therefore determine the effects of PEA-15 on cellular signaling processes (Renganathan et al 2005). Phospholipase D (PLD) binds PEA-15 and is a widely expressed enzyme that catalyzes the hydrolysis of phosphatidylcholine to phosphatidic acid (PA) and choline (Wang et al 1999, Yang et al 2001). Although the physiological function of this interaction remains elusive, PEA-15 stabilizes PLD expression and may activate it by a yet undefined mechanism (Zhang et al 2000). PLD has been shown to modulate the Ras-ERK pathway and regulate the cell cycle and cell proliferation (Donaldson 2009, Yang et al 2001). PLD activity is crucial for Ras activation in NIH3T3 cells, where PLD-produced PA is necessary for the recruitment of Son of Sevenless (SOS), a direct activator of Ras, to the membrane (Zhao et al 2007). In addition, in Rat-1 fibroblasts PA can associate with Raf-1 and target it to the membrane. This Ras-independent recruitment is important for the subsequent activation of Raf-1 by Ras in these cells and is required for further downstream signaling (Rizzo et al 2000). Elevated PLD activity is found in several tumors including breast, gastric and renal cancers (Noh et al 2000, Uchida et al 1997, Uchida et al 1999, Zhao et al 2000). PLD has also been shown to have transforming properties in fibroblasts with aberrant tyrosine kinase activity and may contribute to tumor progression in this context (Joseph et al 2001, Lu et al 2000). Furthermore, in v-raf transformed NIH3T3 cells increased PLD activity was able to overcome Raf induced cell cycle arrest (Frankel et al 1999, Joseph et al 2002).
In this study, we analyzed the effects of PEA-15 expression on H-Ras transformed mouse kidney epithelial cells (iBMK). Surprisingly, we found that PEA-15 does not block H-Ras proliferation signals as previously described, but in this cellular context enhanced H-Ras driven transformation independent of apoptosis. Co-expression of H-Ras and PEA-15 resulted in enhanced tumor formation in vivo and increased colony formation in soft agar assays in vitro. Analysis of the cell cycle in iBMK cells revealed an accelerated G1/S cell cycle progression when both proteins were expressed. In this context activity of the Ras-MAPK/ERK signaling pathway, as well as the expression of major cell cycle regulatory proteins were elevated. At the same time, expression of both H-Ras and PEA-15 caused an increase in PLD1 expression and activity compared to the expression of H-Ras alone. Finally, treatment of these cells with inhibitors of PLD1 or expression of a PLD1 mutant preventing binding to PEA15 blocked activation of the ERK/MAPK pathway. We have thus identified a novel mechanism by which PEA-15 can positively regulate Ras- ERK/MAPK signaling and increase the proliferation of H-Ras transformed epithelial cells. The enhancement of Ras signaling was in part due to increased activity of PLD1 resulting from PEA-15 expression. This is the first evidence to link PEA-15 regulation of Ras signaling to effects on PLD.
Results
PEA-15 enhances H-Ras mediated epithelial cell transformation in vitro in an adhesion-independent manner
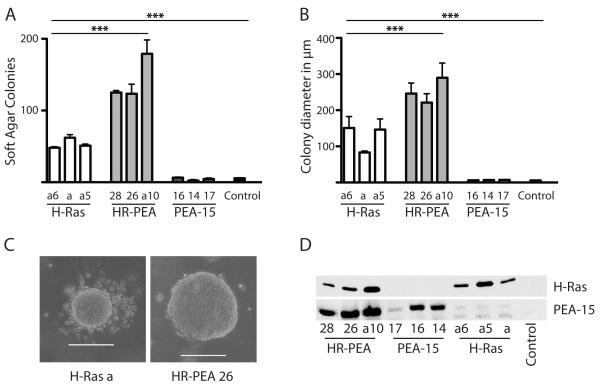
Expression of oncogenic H-Ras leads to a transformed phenotype in iBMK cells resulting in adhesion-independent colony formation in soft agar assays (Degenhardt and White 2006). PEA-15 negatively regulates H-Ras to ERK signaling and blocks H-Ras mediated proliferation signals by sequestering ERK in the cytoplasm (Formstecher et al 2001, Pastorino et al 2010). The ability of cells to grow in suspension is a hallmark of transformation. We therefore analyzed if co-expression of PEA-15 would abolish H-Ras mediated transformation in iBMK cells and inhibit Ras-induced growth in soft agar assays. Surprisingly, stable co-expression of PEA-15 in the presence of active H-Ras enhanced soft agar colony growth. Cells stably overexpressing both H-Ras and PEA-15 formed significantly (P < 0.0003) more colonies in soft agar compared to cells expressing H-Ras alone (Figure 1A). Neither the control cell line nor the cell lines stably expressing PEA-15 alone were able to form soft agar colonies. Co-expression of PEA-15 in cells expressing the constitutively active form of H-Ras not only augmented the number of soft agar colonies formed, but also increased the diameter of the colonies (Figure 1B, C). We verified the over-expression of PEA-15 and H-Ras after stable transfection by immunoblot (Figure 1D).
Figure 1.
PEA-15 enhances H-Ras mediated epithelial cell transformation in vitro in an adhesion-independent manner. (A) The number of colonies formed by stably transfected iBMK cells in soft agar colony formation assays was analyzed after 14 days of incubation. (B) The colony diameter of colonies formed by stably transfected iBMK cells in soft agar after 14 days of incubation was measured. (C) Brightfield images shown depict average size and morphology of colonies formed by stably transfected cells (H-Ras and HR-PEA) in soft agar colony formation assays. White scale bars represent 200μm. (D) Immunoblot analysis of iBMK cell lines stably transfected with H-Ras and/or PEA-15. Membranes were probed for H-Ras and PEA-15 expression, respectively.
PEA-15 promotes G1- to S-phase transition and does not alter anoikis
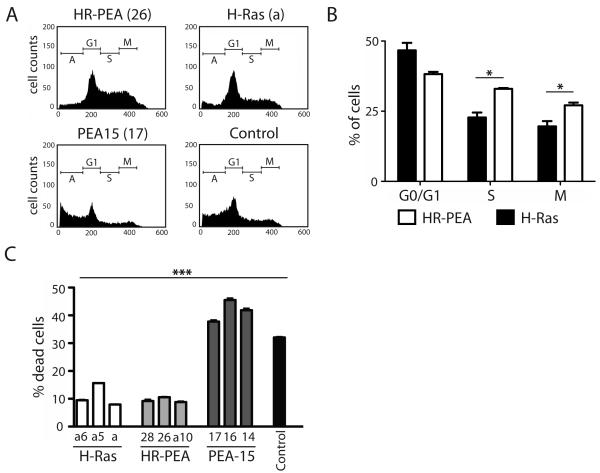
The Ras-MAPK/ERK signaling cascade has been shown to be involved in the regulation of cell cycle progression both in physiological and pathological conditions (Cordova-Alarcon et al 2005, Sathyan et al 2007, Viparelli et al 2008). We therefore analyzed if the increased colony formation in soft agar was due to influences of PEA-15 expression on cell cycle regulation. Interestingly, suspended iBMK cells stably expressing both H-Ras and PEA-15 had fewer cells at the G0/G1 phase and significantly increased cells in the S and mitotic phases compared to H-Ras expression alone. Cell lines stably expressing PEA-15 alone showed a decreased number of cells in G0/G1 compared to the control cell line, but no significant differences could be observed in S- and M-phase (Figure 2A,B).
Figure 2.
PEA-15 promotes G1- to S-phase transition and does not alter cell anoikis. (A) Histograms of flow cytometric analysis of DNA content in stably transfected iBMK cells related to their stage in the cell cycle (A, apoptotic; G1-, synthesis- or mitotic phase) after 48 h culture in suspension. (B) Quantitative distribution of H-Ras or H-Ras and PEA-15 stably co-expressing cells (HR-PEA) in G0/G1, S and M cell cycle phases after 48h incubation in suspension. Each bar represents the average percentage of three different clones; measurements were done in triplicates. (C) Percentage of dead cells in stably transfected iBMK cell lines after incubation in suspension for 48h shown as the percentage of cells in sub-G1 phase after staining with propidium iodide. Measurements were done in triplicates. Statistically significant differences are marked with asterisks (* p<0.05, ** p<0.01, *** p<0.001).
Previous reports indicate that PEA-15 can enhance tumorigenesis by inhibiting apoptosis (Formisano et al 2005). Therefore, we investigated whether stable PEA-15 expression in iBMK cells would lead to protection from anoikis. In suspension, PEA-15 expression did not alter anoikis significantly compared to control cell lines, as shown by the percentage of dead cells. The stable expression of H-Ras significantly decreased the number of cells undergoing anoikis. However, no significant differences were observed between cells stably expressing H-Ras alone and cells expressing both H-Ras and PEA-15 (Figure 2C).
PEA-15 alters expression of cell cycle regulatory proteins
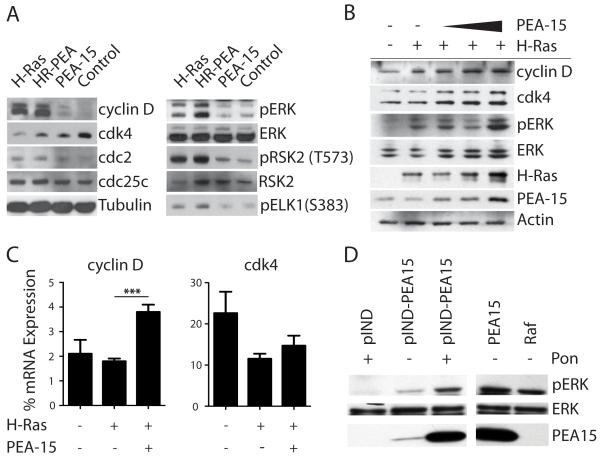
The observed differences in the cell cycle of stably transfected iBMK cell lines in suspension led us to analyze the expression levels of ERK regulated cell cycle proteins in these cells. As previously reported, H-Ras enhanced cyclin D expression levels compared to control cell lines (Lavoie et al 1996, Lovec et al 1994, Nandan et al 2004). Cells stably expressing both H-Ras and PEA-15 had a slight increase in cyclin D expression levels compared to cells expressing H-Ras alone. PEA-15 expression alone slightly induced cyclin D1 expression compared to control cells (Figure 3A).
Figure 3.
PEA-15 alters expression of cell cycle regulatory proteins and increases activation and expression levels of MAPK pathway members. (A) Western blot analysis of cell cycle regulatory proteins (left panel) or MAPK pathway members (right panel) in stably transfected iBMK cell lines kept in suspension for 48h. (B) Western blot analysis of cell cycle regulatory proteins and MAPK pathway members in transiently transfected iBMK cells kept in suspension for 18h. (C) mRNA expression analysis of cyclin D and cdk4 in transiently transfected iBMK cells kept in suspension for 18h. Shown are the relative mRNA expression levels compared to the housekeeping gene. Statistically significant differences are marked with asterisks (* p<0.05, ** p<0.01, *** p<0.001). (D) Immunoblot of CHOK1 cells transiently transfected with ponasterone inducible PEA-15 (pIND-PEA15), vector control (pIND), PEA-15 or Raf. Cells were serum-starved and then treated with ponasterone or vehicle control and immunoblotted for expression of phosphorylated ERK, total ERK and PEA-15.
The cyclin dependent kinase cdk4 is an important binding partner for cyclin D and is necessary for G1-S transition (Connell-Crowley et al 1997). As described previously, H-Ras transiently decreases cellular cdk4 expression (Lazarov et al 2002)(Figure 3A). Moreover, PEA-15 expression alone slightly lowered cdk4 levels when cells were in suspension. Interestingly, PEA-15 was able to maintain cdk4 expression when co-expressed with H-Ras in suspended cells (Figure 3A). No significant differences were observed with cdc25c and cdc2.
To exclude effects selected for during the generation and culture of the stable transfected cell lines we analyzed the expression of two key cell cycle regulatory proteins downstream of ERK namely cyclin D and cdk4 in a transient model. iBMK cells only carrying the control vector were transiently transfected with H-Ras alone or H-Ras and increasing amounts of PEA-15. Expression of the transfected constructs was verified by immunoblotting. The findings in the transient model confirmed the results from the experiments with stable transfected iBMK cells. Compared to control cells, transient transfection with H-Ras increased the expression of cyclin D but not the level of cdk4 (Figure 3B). Transient co-expression of increasing amounts of PEA-15 elevated cyclin D expression levels and in addition induced increased cdk4 expression (Figure 3B).
To further elucidate the mechanism by which PEA-15 alters the expression of cell cycle regulatory proteins in the H-Ras transformed iBMK background we analyzed the mRNA expression levels of the affected proteins cyclin D and cdk4. Transient co-expression of PEA-15 and H-Ras significantly increased cyclin D transcription (Figure 3C). Transient transfection of H-Ras alone did not alter cyclin D mRNA expression compared to control cells. In contrast, cdk4 mRNA expression was not affected by the transient transfection of PEA-15. Transient H-Ras expression decreased the amount of cdk4 mRNA transcription both when transfected alone and in combination with PEA-15 (Figure 3C). Moreover, co-transfection of PEA-15 with H-Ras did not augment the effect of H-Ras on cdk4.
PEA-15 increases activation and expression levels of ERK/MAPK pathway members
The Ras-MAPK cascade is closely integrated into the regulation of the cell cycle and can influence the expression and activity of cell cycle regulatory proteins (Viparelli et al 2008). In addition, PEA-15 has been shown to regulate the ERK/MAPK signaling pathway (Formstecher et al 2001, Pastorino et al 2010). Intriguingly, PEA-15 also potentiates Ras activation of ERK by an adhesion independent mechanism (Ramos et al 2000). We therefore analyzed if the changes in cyclin D and cdk4 expression coincide with increased ERK signaling in the stably transfected iBMK cell lines. We determined the activation and expression levels of ERK, its cytoplasmic target RSK2 and the nuclear ERK target Elk-1. PEA-15 and H-Ras individually as well as in combination were able to activate ERK equally in adherent cells as reported previously (not shown) (Haling et al 2010, Ramos et al 2000). Interestingly, cells in suspension stably expressing both H-Ras and PEA-15 showed higher ERK activation compared to cell lines expressing H-Ras or PEA-15 alone (Figure 3A). Expression levels of ERK were unaltered in all iBMK cell lines (Figure 3A). Transiently transfected cells in suspension showed similar results (Figure 3B). Transfection of H-Ras alone induced phosphorylation of ERK and co-transfection of PEA-15 further enhanced this effect. Indeed the more PEA-15 transfected the greater the increase in phosphorylated ERK (Figure 3B).
In parallel to our previous reports, we observed differences in the phosphorylation of the cytoplasmic ERK target RSK2 in the different iBMK cell lines (Vaidyanathan et al 2007). Suspended cells stably co-expressing H-Ras and PEA-15 showed enhanced RSK2 phosphorylation compared to cells expressing H-Ras alone (Figure 3A). However, PEA-15 overexpression alone also augmented RSK2 phosphorylation compared to control cell lines in suspension. PEA-15 expression not only increased phosphorylation of RSK2, but also caused a modest increase in RSK2 expression when cells were in suspension.
Elk-1 is a nuclear target of ERK in Ras-MAPK signaling. We previously reported that PEA-15 can restrict ERK to the cytoplasm and prevent phosphorylation of nuclear targets in astrocytes and T lymphocytes (Formstecher et al 2001, Pastorino et al 2010). As expected expression of oncogenic H-Ras alone increased phosphorylation of Elk-1 in suspended stably transfected iBMK cells (Figure 3A). Surprisingly, co-expression of PEA-15 and H-Ras further enhanced the phosphorylation of Elk-1 in these cells. No phosphorylation was detected when only PEA-15 was expressed.
PEA-15 induces phosphorylation of ERK in the absence of growth factors
PEA-15 can potentiate H-Ras activation of ERK (Ramos et al 2000). PEA-15 can also increase phosphorylation of ERK in part through inhibiting a feedback loop that inactivates the Ras/MAPK pathway (Haling et al 2010). In contrast, our results showed that transfection of PEA-15 in iBMK cells expressing active H-Ras increases phosphorylation of ERK (Figure 3A,B). We therefore sought to determine if PEA-15 is capable of activating ERK independently of prior activation of the Ras/Raf signaling cascade. We analyzed the effects of inducible PEA-15 expression in transiently transfected CHO cells in which Ras/MAPK signaling was shut down through starvation. Starvation of the cells resulted in the expected shut down of Ras-MAPK signaling as seen in the absence of phosphorylated ERK in the vector control. Starvation of the cells did not alter the phosphorylation of ERK when a constitutively active form of Raf was transfected or PEA-15 was constitutively overexpressed. Importantly, induction of PEA-15 by ponasterone in the serum-starved cells resulted in ERK activation to a similar level as the constitutively active controls (Figure 3D). Minimal activation of ERK was observed in the absence of ponasterone induction-likely due to low un-induced expression of PEA-15 in these cells.
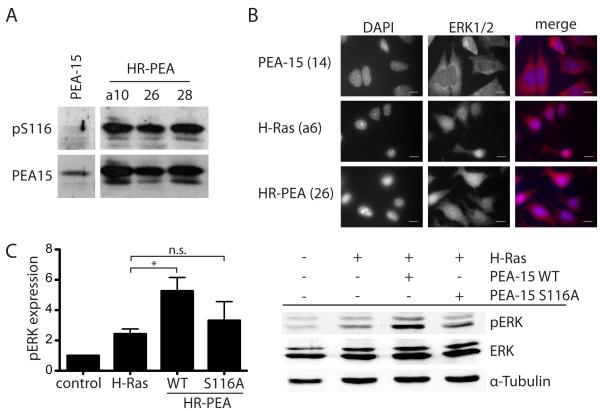
PEA-15 is phosphorylated at Ser116 and does not sequester ERK in the cytoplasm when co-expressed with constitutively active H-Ras
The elevated level of phosphorylated Elk-1 in cells stably co-expressing H-Ras and PEA-15 led us to the conclusion that in these cells PEA-15 is not able to prevent phosphorylation of nuclear ERK targets as described previously in other cell types (Formstecher et al 2001, Pastorino et al 2010, Whitehurst et al 2004). PEA-15 loses its ability to bind ERK upon phosphorylation at Ser116 (Renganathan et al 2005). We therefore analyzed the phosphorylation status of PEA-15 at Ser116 in PEA-15 expressing iBMK cell lines. All three independent cell lines stably co-expressing H-Ras and PEA-15 showed phosphorylation of PEA-15 at Ser116 (Figure 4A). Minimal phosphorylation at this site was observed in cells expressing PEA-15 alone. The presence of PEA-15 in all cell lines was confirmed by immunoblotting for total PEA-15.
Figure 4.
PEA-15 is phosphorylated at Ser116 and does not sequester ERK in the cytoplasm when co-expressed with constitutively active H-Ras. (A) Western blot analysis of PEA-15 phosphorylation at Ser116 and total PEA-15 expression in stably transfected iBMK cells. (B) Immunofluorescence staining of ERK1/2 in stably transfected iBMK cell lines plated on fibronectin showing the cellular localization of ERK1/2 in these cells. Nuclei are visualized by DAPI staining. Both signals are also shown in a merged image. White scale bars represent 20μm.
Ser116 phosphorylation of PEA-15 is necessary for its enhancing effect on H-Ras induced activation of ERK. (C) Western blot analysis of pERK and total ERK in iBMK cells transiently expressing combinations of H-Ras, PEA-15 wildtype (WT) or a S116A mutant of PEA-15 that cannot be phosphorylated at Serine116. Cells were kept in suspension for 18h. Phosphorylation of ERK was normalized to total ERK expression. Shown is the mean and SEM of three independent experiments.
To confirm that the phosphorylation at Ser116 resulted in a loss of the ability to sequester ERK in the cytoplasm we analyzed the cellular localization of ERK by immunofluorescence staining of the stably transfected iBMK cells. In cells expressing PEA-15 alone, ERK was localized predominantly in the cytoplasm as previously reported (Figure 4B). The expression of H-Ras alone led to an increased translocation of ERK into the nucleus. PEA-15 co-expression with H-Ras did not alter the translocation. In these cells, ERK was found both in the cytoplasm and the nucleus with a slightly higher concentration of nuclear ERK (Figure 4B).
Ser116 phosphorylation of PEA-15 is necessary for its enhancing effect on H-Ras induced activation of ERK
The finding that PEA-15 is phosphorylated at Ser116 when stably co-transfected with H-Ras into iBMK cells, made us further analyze the importance of this phosphorylation for the enhancement of H-Ras mediated cell transformation. We analyzed the ability of a constitutively unphosphorylated PEA-15 mutant (S116A) to increase H-Ras induced activation of ERK. This mutant has an amino acid substitution at the serine-116 phosphorylation site that prevents phosphorylation. In a transient system we transfected iBMK cells with H-Ras in combination with wild type (WT) PEA-15 or the Ser116 phosphorylation deficient (S116A) mutant. As expected transfection of H-Ras increased activation of ERK. PEA-15 co-transfection further enhanced this effect. Interestingly, co-transfection of S116A failed to show significant enhancement of the H-Ras induced pERK elevation (Figure 4 C). Levels of activated ERK were still higher then in control cells, but not significantly increased compared to H-Ras only expressing cells.
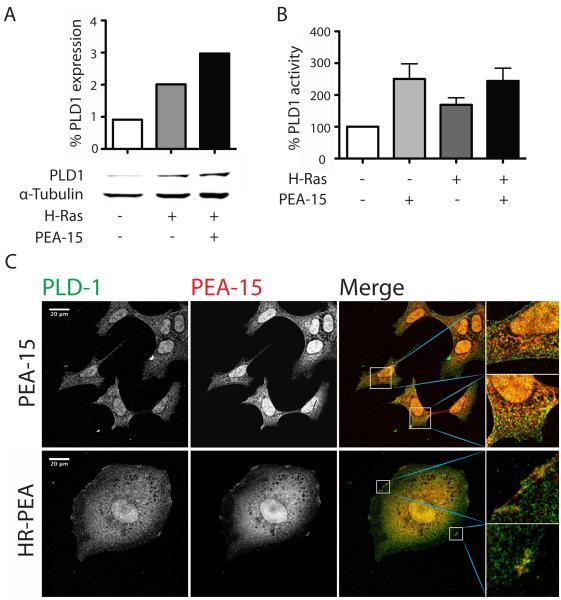
PEA-15 increases PLD1 expression and activity
PEA-15 does not appear to block inactivation of ERK in iBMK cells by impairing a feedback loop and so an alternative mechanism must explain the increased ERK activity. PEA-15 also binds and stabilizes PLD1 and can lead to increased PLD activity (Zhang et al 2000). PLD1 is involved in regulating both Ras-MAPK signaling and cell cycle progression (Donaldson 2009, Yang et al 2001). PLD1 therefore was a promising candidate to mediate PEA-15’s effects on ERK activity and cyclin D/cdk4 expression. In transiently transfected iBMK cells expression of H-Ras doubled the amount of PLD1 compared to control cells (Figure 5A). Co-expression of both PEA-15 and H-Ras further enhanced this effect and resulted in a more than 3-fold higher expression compared to the control cell line and 1.5-times higher than in cells expressing only H-Ras. Transient overexpression of PEA-15 and H-Ras also affected PLD1 activity. H-Ras and PEA-15 both individually as well as in combination increased the PLD1 activity of iBMK cells compared to control cells (Figure 5B). Co-expression of PEA-15 and H-Ras resulted in higher PLD1 activity increases than H-Ras expression alone.
Figure 5.
PEA-15 increases expression and activity of PLD1 and co-localizes with it. (A) Western blot analysis of PLD1 expression in transiently transfected iBMK cells kept in suspension for 18h. Shown are Western blot (lower panel) and quantified expression levels normalized to α-tubulin. (B) PLD1 activity in transiently transfected iBMK cells after incubation in suspension for 18h. Shown is the percentage of PLD1 activity compared to control cells. (C) Immunofluorescence staining of stably transfected iBMK cells plated on fibronectin coated glass coverslips. Binding of specific primary antibodies against PLD-1 or PEA-15 was visualized using fluorescence labeled secondary antibodies (A488, green and Alexa647, red). Slides were analyzed using a Leica TCS-SP5 confocal microscope. White scale bars represent 20μm. Co-localization is shown in yellow.
PEA-15 co-localizes with PLD1
PEA-15’s ability to regulate PLD1 expression levels or activity has been shown to be dependent on the interaction of these two proteins and targeting this interaction can impair PLD1 functions (Viparelli et al 2008, Zhang et al 2000). We analyzed the expression pattern of PLD1 and PEA-15 in iBMK cells stably expressing PEA-15 or H-Ras and PEA-15 to analyze H-Ras mediated effects on the localization of these proteins and to determine their co-localization. Immunofluorescence images showed a similar pattern of expression of both proteins (Figure 5C). High levels of PLD1 and PEA-15 could be found in the nuclear and cytoplasmic regions surrounding the nucleus. The proteins co-localize in those regions (yellow color) and that is independent from H-Ras expression. Upon overexpression of H-Ras both PEA-15 and PLD1 can be found in patches at the cell membrane in addition to the nuclear and perinuclear localization. Within these spots both proteins co-localize as seen in the magnified images (Figure 5C, right panel).
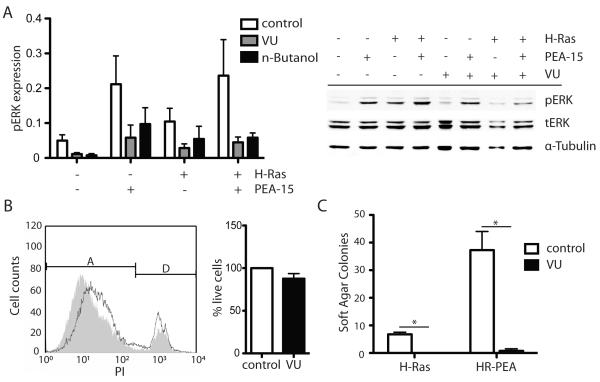
Inhibition of PLD1 reduces PEA-15 induced increase in ERK activation
To verify that PEA-15’s influence on ERK activation is mediated through PLD1 we inhibited PLD1 and analyzed the effects on PEA-15 mediated activation of ERK. PLD was either inhibited with n-Butanol or a small molecule inhibitor (VU 0155069) that has been described previously to selectively inhibit enzyme function of the PLD1 isoform (Scott et al 2009). To evaluate the effects of this treatment on ERK activation we analyzed the phosphorylation status of ERK by immunoblotting. Transient expression of either PEA-15 or H-Ras resulted in elevated levels of phosphorylated ERK compared to control cells (Figure 6A). Co-expression of both proteins increased ERK activation even further than either protein individually. Treatment with either PLD1 inhibitor VU 0155069 or n-Butanol impaired pERK in cells transiently transfected with PEA-15, H-Ras or both proteins. The greatest effects were observed in cells expressing both PEA-15 and H-Ras, where the inhibitor treatment resulted in about 60% less ERK activity. Both inhibitors had similar effects on the treated cell lines (Figure 6A). To exclude cytotoxic effects as main reason for the decreased ERK activation we analyzed the impacts of the PLD1 inhibitor VU 0155069 on the cell viability. Staining of VU treated iBMK cells with propidium iodide (PI) showed only a slight decrease in cell viability. Compared to control cells the inhibitor treatment resulted only in about 8% loss of live cells (Figure 6B). The histogram shows similar curves for control cells (grey) or VU treated cells (black) with similar populations of dead cells (D) and cells alive (A) after 30min inhibitor treatment.
Figure 6.
Inhibition of PLD1 suppresses PEA-15’s effect on ERK phosphorylation, but only slightly impairs cell viability. (A) Normalized expression of pERK in transiently transfected cell lines treated with 1-Butanol, VU 0155069 or the carrier control EtOH. Expression levels were determined by Western blotting, levels of pERK are normalized to the expression of total ERK and α-tubulin. Shown are the mean expression levels from three independent experiments. Western blot analysis of the phosphorylation status of ERK in transiently transfected iBMK cells treated with the PLD1 specific inhibitor VU 0155069 (50μM, 30min) or with the carrier control EtOH. (B) Histogram of flow cytometric analysis of viability of iBMK cells treated with VU 0155069 (black line) compared to cells treated with the carrier control EtOH (gray, solid) based on their propidium iodide uptake. Relative fluorescence emission of PI corresponds to live [A] or dead [D] cells. Right panel: Percentage of live cells in iBMK cell population after incubation with VU 0155069 or carrier control. Measurements were done in triplicates. (C) Inhibition of PLD1 inhibits soft agar colony formation. iBMK cells stably expressing H-Ras or H-Ras and PEA-15 were 30min pre-incubated with 50μM VU 0155069 or carrier control and then plated in soft agar. Formed colonies were counted after 14 days. Experiment was done in duplicates and repeated twice. Statistically significant differences are marked with an asterisk (* p<0.05).
Inhibition of PLD1 inhibits soft agar colony formation
As shown before, oncogenic H-Ras is able to transform iBMK cells (Degenhardt and White 2006). We analyzed if PLD1 activity is necessary for this transformation. We determined the effect of PLD1 inhibitor on soft agar colony formation using stably transfected iBMK cells. As previously shown, cells expressing H-Ras were able to form colonies independent from adhesion to a substrate and PEA-15 co-expression greatly magnified this effect (Figure 6C). Pre-treatment of cells with the PLD1 specific inhibitor VU 0155069 significantly inhibited the formation of colonies in cells expressing PEA-15 and H-Ras.
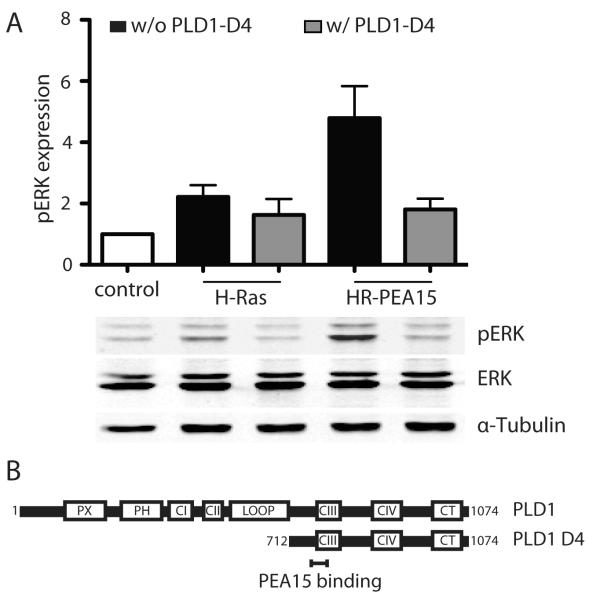
Binding of PEA-15 to PLD1 is required for PEA-15’s effect on H-Ras mediated enhancement of ERK activity
PEA-15 can bind to PLD1 and it has been shown that disruption of the PEA-15-PLD1 interaction can impair downstream PLD1 effects like PKC-α activation (Viparelli et al 2008). These findings in addition to our finding that PLD1 co-localizes with PEA-15 in iBMK cells raised the question if binding of the two proteins is necessary for the observed enhancement of ERK activity in H-Ras expressing iBMK cells. The PEA-15 binding region of PLD1 has already been mapped to a C-terminal domain D4 (AA 712-1074) and the residues 762-801 within this domain have been identified as the shortest PEA-15 binding segment (Figure 7A) (Doti et al 2010, Zhang et al 2000). Here we used a PLD1 construct only consisting of the D4 domain (Figure 7B) and lacking catalytic activity (PLD1-D4). When overexpressed this construct competes with the endogenous PLD1 and keeps it from binding to PEA-15. We analyzed if transient overexpression of this mutant in addition to transient expression of H-Ras and PEA-15 would affect the previously observed changes in pERK. Without overexpression of mutant PLD1-D4 H-Ras expression led to increased ERK activation and PEA-15 enhanced this effect as expected (Figure 7A). Interestingly, co-expression of PLD1-D4 prevented the increase in pERK upon H-Ras and PEA-15 co-transfection. pERK levels in these cells are not significantly different from cells only expressing H-Ras. There was no statistically significant effect of the PLD-D4 mutant on H-Ras only transfected cells (Figure 7A).
Figure 7.
Binding of PEA-15 to PLD1 is required for PEA-15’s effect on H-Ras mediated enhancement of ERK activity. (A) Western blot analysis of pERK and total ERK in iBMK cells transiently expressing combinations of H-Ras, PEA-15 and the PEA-15 binding domain mutant of PLD1 (PLD1-D4). Cells were kept in suspension for 18h. pERK levels are normalized to the total ERK expression. Shown is the mean and SEM of three independent experiments. n.s. not statistically significant (B) Structure of PLD1 and its PEA-15 binding site containing D4 domain used in the experiment. Boxes show functional domains, the PEA-15 binding site is highlighted. PX: phox homology domain, PH: pleckstrin homology domain, CI-CIV: conserved regions, CT: C-terminus.
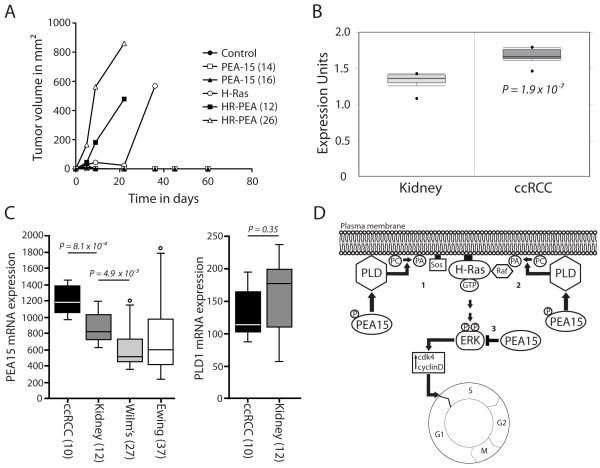
PEA-15 promotes earlier formation of tumors in vivo
To elucidate if the observed effects of PEA-15 on H-Ras mediated cell transformation are in vitro artifacts or also apply in vivo we analyzed how PEA-15 expression alters active H-Ras transformation in the presence of a tumor micro environment in nude mice. No tumors developed when the control cell line or the cell line stably expressing PEA-15 alone was injected. The injection of cells stably expressing H-Ras led to the formation of tumors in mice. Importantly, we observed on average larger tumors when injecting H-Ras and PEA-15 co-expressing cells compared to an injection of cells expressing H-Ras alone. Furthermore, tumors resulting from injection of H-Ras and PEA-15 co-expressing cells formed earlier than those arising from H-Ras expression alone (Figure 8A).
Figure 8.
PEA-15 co expression promotes earlier H-Ras tumor formation in nude mice in vivo; PEA-15 expression is elevated in human ccRCC samples. (A) Tumor formation in nude mice was followed after injection of iBMK cells stably expressing H-Ras, PEA-15, H-Ras/PEA-15 (HR-PEA) or control vector. Graphs represent changes in tumor volume from up to 60 days. Mice were sacrificed once the tumor burden was too great. (B) Differential expression analysis of PEA-15 levels in human kidney samples and human clear cell renal cell carcinoma samples (ccRCC) showing the relative protein expression units. Data was extracted from the Oncomine cancer microarray database. As a corrected measure of significance t-statistics with false discovery rates were used. (C) PEA15 and PLD1 expression in different tumor types and normal kidney. The data shown are from the Francesconi-37 Ewing sarcoma series (for Ewing), or from the Kort-79 mixed kidney series (for all other samples). Statistical analysis was performed using the non-parametric Kruskal-Wallis test, P < 0.05 was considered significant. The error bars represent the SEM. (D) Model of a proposed mechanism by which PEA-15 can increase H-Ras driven activation of ERK. Overexpression of PEA-15 in iBMK cells overexpressing constitutively active H-Ras causes higher PLD1 activity either through higher PLD1 expression levels or a yet undefined activation mechanism. Subsequently, PLD1 produces increased amounts of the signaling molecule phosphatidic acid (PA). PA then increases recruitment of the guanine nucleotide exchange factor SOS (1) and/or the kinase Raf (2) to the plasma membrane. H-Ras is constitutively bound to the plasma membrane and is immediately able to induce higher signaling rates upon GTP loading. This leads to the observed downstream effects like increased ERK activity and cell cycle progression. In this scenario PEA-15 is phosphorylated at Ser116. Unphosphorylated PEA-15 (3) is able to bind ERK and sequester it in the cytoplasm as previously reported.
PEA-15 is significantly overexpressed in human ccRCC tissue samples
In order to further investigate the in vivo relevance of our findings we analyzed the expression levels of PEA-15 and PLD1 in human tumor tissue samples and normal kidney tissue. PEA-15 expression was significantly higher in clear cell renal cell carcinoma (ccRCC) compared to normal kidney tissue (Figure 8B and C). This elevated PEA-15 expression is peculiar to clear cell renal cell carcinoma, since the unrelated renal Wilm’s tumor, or Ewing sarcoma, show only low PEA-15 expression, which is not significantly correlated to important clinical parameters (results not shown). PLD1 expression is not significantly different in ccRCC and normal kidney tissue (Figure 8C). However, PLD1 expression is lower in Wilm’s tumor and Ewing sarcoma (results not shown). Our data was further confirmed by results obtained with two other tumor series in the public domain (results not shown): Li-39 (for Ewing), and EXPO-261 (for kidney).
Discussion
Our findings reveal a novel mechanism by which PEA-15 can enhance H-Ras/MAPK signaling and contribute to H-Ras mediated transformation of immortalized baby mouse kidney (iBMK) cells. In this epithelial cell line, co-expression of H-Ras and PEA-15 increased expression of cell cycle regulatory proteins cyclin D and cdk4 as well as activity of the ERK/MAPK pathway in an adhesion-independent manner. This resulted in accelerated G1/S cell cycle transition and a higher tumor formation rate both in vitro and in vivo. PEA-15 acted in part through PLD1 whose expression level and activity were elevated in cells overexpressing PEA-15. Inhibition of PLD1 catalytic activity or interference with PEA-15 binding to PLD1 abolished the enhancing effect of PEA-15 on ERK activation. This suggests that PEA-15 influences H-Ras mediated cell transformation through increased PLD1 signaling.
Previous reports have shown the ability of H-Ras to contribute to cell transformation. Ras has been reported in various cell types to affect transcription, drive cell cycling and proliferation or to block apoptosis and anoikis (Cordova-Alarcon et al 2005, Macaluso et al 2002, Viparelli et al 2008). Degenhardt et al. described that overexpression of constitutively active H-Ras (G12V) is enough to transform iBMK cells (Degenhardt and White 2006). H-Ras promotes the proteasomal degradation of the pro-apoptotic Bcl-2 family protein BIM through the Raf/MAPK pathway in these cells (Tan et al 2005).
Conversely, PEA-15 has generally been described to block Ras/MAPK mediated gene transcription and to inhibit proliferation by sequestering ERK in the cytoplasm (Formstecher et al 2001, Pastorino et al 2010). Moreover, in ovarian cancer cells PEA-15 has been shown to exert tumor-suppressive effects at least in part through induction of ERK-dependent autophagy. However, the mechanism of PEA-15’s involvement is not fully understood and it is unclear if H-Ras signaling is involved in this regulation (Bartholomeusz et al 2008).
In contrast to reports suggesting a tumor-suppressor function of PEA-15, our findings confirm that PEA-15 expression can also potentiate ERK activation as we have previously described (Ramos et al 2000). However, oncogenic H-Ras expression remains necessary for full transformation, since overexpression of PEA-15 alone did not lead to a transformed phenotype. PEA-15 overexpression in iBMK cells prevents ERK translocation to the nucleus as previously reported (Formstecher et al 2001). However, when co-transfected with H-Ras we could not observe this effect. ERK is not sequestered in the cytoplasm and can translocate into the nucleus. This is likely due to phosphorylation of PEA-15 at Ser116, a phosphorylation that decreases PEA-15’s binding to ERK (Renganathan et al 2005). A phosphorylation deficient mutant of PEA-15 was not able to further enhance H-Ras driven enhancement of ERK activity. Thus, phosphorylation at Ser116 is required for PEA-15 effects on H-Ras/ERK MAP kinase signaling.
Ras-ERK signaling is regulated through a system of positive and negative feedback loops (Ramos 2008). Haling and colleagues reported that PEA-15 can increase ERK activation by interrupting a negative feedback loop in which ERK phosphorylates fibroblast receptor substrate 2α (FRS2α) to shut down Ras signals in fibroblasts (Haling et al 2010). We report an alternative mechanism by which PEA-15 can activate ERK through direct activation of PLD1, which leads to activation of Ras/MAPK signaling upstream of ERK. We found that even under serum-starved conditions lacking any initial Ras signaling, PEA-15 is still capable of activating ERK to a similar level as Ras.
The PEA-15 binding partner PLD1 played a part in this context. PLD1 has been described previously to be involved in the regulation of many signaling pathways including the Ras-ERK pathway, thereby driving cell cycling and proliferation (Donaldson 2009, Yang et al 2001). PLD1 is activated by H-Ras and has been proven to be crucial for the transformation of rat fibroblasts by H-Ras (Buchanan et al 2005, Shi et al 2007). PLD1 has been reported to regulate Ras-ERK signaling both upstream and downstream of Ras through the generation of phosphatidic acid (PA). PA is crucial for epidermal growth factor (EGF)-induced activation of Ras because of its recruitment of the guanine nucleotide-exchange factor SOS to the plasma membrane (Hancock 2007). However, PA seems to also exert effects downstream of Ras where it is necessary to allow Ras binding to Raf-1 (Rizzo et al 2000). Both studies show that PLD1 is an important co-factor in Ras-MAPK signaling and acts as a positive regulator.
We observed higher levels of PLD1 upon over-expression of PEA-15 in iBMK cells. This may have been due to a stabilization of PLD1 levels by PEA-15 as previously described (Zhang et al 2000). In iBMK cells PEA-15 and PLD1 show co-localization. PEA-15 co-localizes with PLD1 in the nucleus and in perinuclear regions, and upon H-Ras co-expression also in regions at the cell membrane. Additionally, over-expression of PEA-15 or H-Ras increased PLD1 activity. Possibly due to the increased levels of protein, PLD1 activity was higher when both H-Ras and PEA-15 were expressed as compared to overexpression of H-Ras alone.
To assess the involvement of PLD1 in H-Ras mediated transformation in iBMK cells we used two different inhibitor approaches. We blocked PLD1 catalytic activity with the commonly used but less specific inhibitor n-Butanol, and also looked at the effects of a PLD1 specific inhibitor VU0155069. This potent small molecule inhibitor has been described to selectively inhibit the PLD1 isoform (Scott et al 2009). We analyzed phosphorylation of ERK as a major transforming signaling pathway in these cells (Tan et al 2005) and used it as a read-out after interfering with PLD1 catalytic activity. Treatment of iBMK cells with inhibitors of PLD1 hydrolytic activity blocked ERK phosphorylation upon both H-Ras and H-Ras/PEA-15 co-overexpression resulting in up to a 60% decrease in ERK activity. PLD1 inhibition in this experiment only slightly reduced the viability of the cells. A loss of PLD1 catalytic activity also resulted in inhibition of soft agar growth. Neither H-Ras nor H-Ras/PEA-15 co-expressing cells formed colonies after pre-treatment with the specific PLD1 inhibitor. We further analyzed the involvement of PLD1 by putting special focus on its binding to PEA-15. We used the PLD1-D4 domain as a mutant construct that specifically keeps functional endogenous PLD1 from binding to PEA-15. This allowed us to analyze the importance of this protein interaction for the activation of ERK in the iBMK system. Co-expression of mutant PLD1-D4 blocked the increase of pERK upon H-Ras and PEA-15 co-expression, further confirming the necessity of functional PLD1 and an intact PLD1-PEA15 interaction for PEA-15’s ability to increase H-Ras mediated ERK activation.
Based on our findings and previous reports, PEA-15 could act at least in part through two different ways to enhance H-Ras mediated cell transformation through PLD1 (Figure 8D). Overexpression of PEA-15 results in higher PLD1 activity which leads to increased amounts of the signaling molecule phosphatidic acid (PA). Elevated signaling through PA can enhances levels of the guanine nucleotide exchange factor SOS recruited to the plasma membrane as described by Hancock et al. (Hancock 2007). H-Ras is bound to the plasma membrane and activated upon GTP loading by SOS (Figure 8D, #1). Alternatively, increased PA can result in a higher recruitment of the kinase Raf to the membrane and enhance Ras-Raf binding (Figure 8D, #2) as previously shown by Rizzo et al. (Rizzo et al 2000). Both mechanisms can lead to the observed downstream effects and result in elevated ERK activity. Although both pathways are possible an enhancement of Ras-MAPK signaling at the Raf level seems more likely in our study. Transfected oncogenic H-Ras G12V already is constitutively active making further enhancement of the signaling cascade by PEA-15 upstream of H-Ras less likely. In the presence of oncogenic H-Ras, PEA-15 is phosphorylated at serine 116 and does not restrict ERK to the cytoplasm. Note that unphosphorylated PEA-15 has been previously shown to negatively regulate Ras-MAPK signaling by preventing ERK translocation into the nucleus (Figure 8D, #3) (Formstecher et al 2001, Pastorino et al 2010).
Based on these results we propose a novel function of PEA-15 in enhancing H-Ras driven transformation. This effect was also observed in an in vivo mouse model, where PEA-15 co-expression promoted earlier and larger tumor formation. H-Ras activating mutations are common among various kinds of human malignancies and about 30% of human cancers show mutations in the Ras gene that cause the gene product to be constitutively active (Dunn et al 2005). Additionally, other common mutations in tumors, such as activating EGF receptor (EGFR) mutations, can induce increased H-Ras activity as shown in human renal cell carcinomas (RCC) (Barrett et al 2009). Interestingly, these tumors also show elevated PEA-15 expression levels compared to normal kidney tissues, mainly in clear cell RCC (Figure 8B and C). Renal Wilm’s tumor or Ewing sarcoma, tumors in which no Ras mutations are involved in the pathogenesis, show only low PEA-15 expression (Figure 8C) (Radig et al 1998, Waber et al 1993).
These findings together with our results in mouse kidney epithelial cells indicate that PEA-15 can potentiate tumorigenesis in the context of oncogenic Ras signaling. Thus, PEA-15 can either enhance or impair tumorigenesis depending on the signaling pathways active in the specific tumor cell.
Materials and Methods
Cell lines and reagents
Parent immortalized baby mouse kidney epithelial cell lines (iBMK cells) were developed in the laboratory of Eileen White and described previously (Tan et al 2005). Stable iBMK cells expressing PEA-15, H-Ras, H-Ras and PEA-15 (HR-PEA) or the vector control were derived by electroporation with pcDNA1.H-rasV12 (Lin et al 1995), pIRESpuro3-PEA-15, or pcDNA3.1 (Tan et al 2005) followed by selection with G418 (vector control, H-Ras) or puromycin (PEA-15, HR-PEA). For every type three independent cell lines were generated and analyzed. Cells were grown in D-MEM (Thermo Fisher Scientific Inc.) supplemented with 4 mM L-glutamine, 10 % fetal bovine serum, nonessential amino acids, penicillin/streptomycin and 800 μg/ml G418 or 10 μg/ml puromycin. The successful transfection was further confirmed by Western blotting. In addition, iBMK cells carrying the vector control pcDNA3.1 were transiently transfected with the constructs mentioned above using Lipofectamine (Invitrogen, Carlsbad, CA) as described by the manufacturer’s protocol. For suspension cultures cells were harvested and plated on cell culture dishes coated with 3% agarose for the indicated amount of time.
Antibodies used were specific for H-Ras (C-20), RSK2 (C-19), cdk4 (C-22), cdc25c (C-20), α-tubulin (DM1A), actin (C-11), PEA-15 (H-80) (Santa Cruz Biotech., Santa Cruz, CA); cdc2, (eBioscience, San Diego, CA); cyclin D (DCS6), p44/42 MAPK (Erk1/2), phospho-p44/42 MAPK (Erk1/2) (Thr202/ Tyr204), phospho p90RSK (Thr573), PLD1 (Cell Signaling, Danvers, MA); phospho-Elk1 (S383; New England Biolabs, Ipswich, MA); phospho PED/PEA 15 (pS116; Invitrogen, Carlsbad, CA); PLD1 (N-term) (Epitomics, Burlingame, CA), and PEA-15 (from Dr. Hervé Chneiweiss, Institut National de la Santé et de la Recherche Medicalé, Paris, France). Ponasterone was purchased from Sigma-Aldrich Corp. (St. Louis, MO), VU 0155069 from Tocris Bioscience (Ellisville, MO).
Western blotting
Cells were cultured as described. After 48 h of incubation, cells were lysed with ice-cold M2 buffer as previously described (Ramos et al 2000). Cell lysate protein was resolved by SDS-PAGE. Expression and phosphorylation of proteins was determined by Western blotting using specific antibodies. Binding of primary antibodies was detected using either horseradish peroxidase-conjugated secondary antibodies (Cell signaling) or IRDye 680 goat anti-mouse and IRDye 800 goat anti-rabbit antibodies. Bands were visualized by autoradiography after incubation with enhanced chemiluminescence reagents (PerkinElmer, Waltham, MA) or detected using an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). Scanned blots were cropped to improve clarity and conciseness of presentation using Adobe Photoshop CS (Adobe Systems Inc., San Jose, CA).
Soft agar colony formation assay
Equal cell numbers (1×104) of each stably transfected iBMK cell line were cultured for 14 days suspended in D-MEM in a 0.3 % agar solution on agar (0.6% in D-MEM) coated culture plates. Formed colonies in the soft agar assay were counted and analyzed using a Biorad imager and Quantity One software v4.6 (Bio-Rad, Hercules, CA). For the inhibitor assay cells were pre-incubated with 50μM VU0155069 for 30min and an equal inhibitor concentration was added to the soft agar solution.
Cell cycle analysis and Viability Assay
Equal numbers of iBMK cells (2×105) from each cell line were plated on agarose coated dishes to keep the cells in suspension. Cells were cultured for 48 h as described above. Subsequently, cells were fixed in 70 % EtOH and stained in permeabilization buffer containing 100 μg/ml propidium iodide (PI), 50 μg/ml RNAase and 0.2 % Triton X. For the viability assay cells were cultured as indicated and stained with PI. Cells were analyzed using a FASCcan flow cytometer and CellQuest software (Becton Dickinson, San Jose, CA). Each cell line was examined in triplicate and for each measurement approximately 20,000 cells were analyzed.
mRNA expression analysis
iBMK cells carrying the vector control pcDNA3.1 were transiently transfected with 2μg of H-Ras, PEA-15 or both constructs. 48h after transfection cells were harvested, plated on agarose coated dishes (to prevent adherence) and incubated for 18h. Samples were lysed with Qiagen RLT lysis buffer (Qiagen Inc., Valencia, CA). The lysates were homogenized using QIAShredder columns (Qiagen). RNA was extracted using Qiagen’s RNeasy Mini kit according to the manufacturer’s protocol. RNA integrity was assessed on RNA Nano 6000 chips and 2100 Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA was converted to cDNA using the High Capacity cDNA RT Kit with RNase Inhibitor (Life Technologies Corporation, Carlsbad, CA). cDNA levels were quantified using qPCR on an HT 7900 FAST real-Time PCR System. qPCR was performed on a 96-well plate in triplicates for the 2 genes of interest (GOI; CCND1 and cdk4) and ß-actin as housekeeping gene (HKP). Primer assays for GOI and HKP (CCND1 and cdk4 QuantiTect SYBRGreen assays) were purchased from Quiagen.
Immunofluorescence
Each iBMK cell line was grown on glass coverslips coated with 10 μg/ml fibronectin under regular cell culture conditions as described above. When about 50 % confluent, cells were fixed in 2 % paraformaldehyde, permeabilized in 0.1 % Triton X-100 in PBS, and blocked with 1 % normal goat serum. Cells were stained with primary antibody against ERK1/2 (p44/42; Cell signalling), PEA-15 (Santa Cruz, #sc-166678) or PLD1 (Epitomics, #2315-1). Alexa Fluor 488, 594 and 647 secondary antibodies (Invitrogen) were used to fluorescently label bound primary. Coverslips were mounted on slides using 4′,6-diamidino-2-phenylindole containing mounting medium (Vector Labs, Burlingame, CA) or Fluoromount G (Electron Microscopy Sciences #17984-25). Images were acquired on a Leica TCS-SP5 confocal microscope using a 62x oil-immersion objective or a Zeiss Axiovert 100M fluorescent microscope using a 100X oil-immersion objective and Zeiss AvioVision acquisition software (Carl Zeiss MicroImaging Inc., Thornwood, NY).
PLD1 activity assay
Transiently transfected iBMK cells carrying the vector control pcDNA3.1 were lysed by three freeze and thaw cycles in ice-cold Tris (50 mM, pH 8.0). PLD activity was measured using the Amplex Red Phospholipase D Assay Kit (Invitrogen) following the manufacturer’s protocol.
Tumor formation in nude mice
Tumor formation in nude mice was performed as previously described (Streit et al 1999). In brief, equal cell numbers of each iBMK cell line were injected subcutaneously in the abdominal region of 6-week-old nude mice (Taconic, Germantown, NY). We used 5 mice for each iBMK cell line and followed tumor growth for 60 days. Tumor growth was monitored weekly by measurements with a 6-inch dial caliper (General Tools Mfg. Co., New York, NY). Mice were sacrificed once the tumor had reached 500mm2 size. All experiments were approved by the Institutional Review Board of Rutgers, The State University of New Jersey.
Analysis of micro-array expression arrays in the public domain
CEL data from the public Affymetrix data-sets (Barrett et al 2009) for the Francesconi-37 (GSE12102), Kort-79 (GSE11024), Li-39 (GSE6120), and Tumor Kidney EXPO-261 (GSE2109) series were downloaded from the NCBI GEO site and analyzed as described in (Revet et al 2008). Annotations and clinical data for these series are available from http://www.ncbi.nlm.nih.gov/geo/query/. Affymetrix probe-sets were selected using the R2 bio-informatic platform (see below). All gene transcript levels were determined from data image files using GeneChip operating software (MAS5.0 and GCOS1.0, from Affymetrix). Samples were scaled by setting the average intensity of the middle 96% of all probe-set signals to a fixed value of 100 for every sample in the dataset, allowing comparisons between micro-arrays. The TranscriptView genomic analysis and visualization tool was used to check if the probe-set selected had an anti-sense position in an exon of the gene (http://bioinfo.amc.uva.nl/human-genetics/transcriptview/). The probe-sets selected for a gene showed the highest expression in samples containing a present call for that gene: 200788_s_at (PEA15) and 226636_at (PLD1). All essential conclusions remain the same for the additional valid probe-sets 200787_s_at (for PEA15) and 177_at or 205203_at (for PLD1). All analyses were performed using R2; an Affymetrix analysis and visualization platform developed in the Department of Human Genetics at the Academic Medical Center – University of Amsterdam. R2 can be accessed at: http://r2.amc.nl.
Statistical Analysis
All results are shown as mean ± SEM. Statistical significance was determined using 1way ANOVA followed by Bonferroni’s Multiple Comparison test for all data sets, except for the human tumor expression analysis, where the non-parametric Kruskal-Wallis test was used. P values < 0.05 were considered significant.
Acknowledgements
We thank Dr. Maarit Tiirikainen and the University of Hawai’i Cancer Center Genomics Shared Resource (GSR) laboratory for their assistance with the mRNA expression analysis. We also thank Anna Knapinska, Shirley Young-Robbins and Marci Takemoto for excellent technical assistance. This work was supported by the National Institutes of Health National Cancer Institute (R01CA93849 to JWR) and National Institute of General Medicine (R01GM088266 to JWR) and the Victoria S. and Bradley Geist Foundation (to J.W.R.). M.L.M was supported by NIH National Center for Research Resources Grant RR016453. D.A.N and E.P.W were supported by a grant from the National Institutes of Health (R37CA53370 to EPW).
Footnotes
Conflict of interest The authors declare no conflict of interest.
References
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomeusz C, Rosen D, Wei C, Kazansky A, Yamasaki F, Takahashi T, et al. PEA-15 induces autophagy in human ovarian cancer cells and is associated with prolonged overall survival. Cancer Res. 2008;68:9302–9310. doi: 10.1158/0008-5472.CAN-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan FG, McReynolds M, Couvillon A, Kam Y, Holla VR, Dubois RN, et al. Requirement of phospholipase D1 activity in H-RasV12-induced transformation. Proc Natl Acad Sci U S A. 2005;102:1638–1642. doi: 10.1073/pnas.0406698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda CA, Cortes-Funes H, Gomez HL, Ciruelos EM. The phosphatidyl inositol 3-kinase/AKT signaling pathway in breast cancer. Cancer Metastasis Rev. 2010;29:751–759. doi: 10.1007/s10555-010-9261-0. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L, Harper JW, Goodrich DW. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova-Alarcon E, Centeno F, Reyes-Esparza J, Garcia-Carranca A, Garrido E. Effects of HRAS oncogene on cell cycle progression in a cervical cancer-derived cell line. Arch Med Res. 2005;36:311–316. doi: 10.1016/j.arcmed.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, White E. A mouse model system to genetically dissect the molecular mechanisms regulating tumorigenesis. Clin Cancer Res. 2006;12:5298–5304. doi: 10.1158/1078-0432.CCR-06-0439. [DOI] [PubMed] [Google Scholar]
- Donaldson JG. Phospholipase D in endocytosis and endosomal recycling pathways. Biochim Biophys Acta. 2009;1791:845–849. doi: 10.1016/j.bbalip.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doti N, Cassese A, Marasco D, Paturzo F, Sabatella M, Viparelli F, et al. Residues 762-801 of PLD1 mediate the interaction with PED/PEA15. Mol Biosyst. 2010;6:2039–2048. doi: 10.1039/c005272h. [DOI] [PubMed] [Google Scholar]
- Dunn KL, Espino PS, Drobic B, He S, Davie JR. The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol. 2005;83:1–14. doi: 10.1139/o04-121. [DOI] [PubMed] [Google Scholar]
- Ferro E, Trabalzini L. RalGDS family members couple Ras to Ral signalling and that’s not all. Cell Signal. 2010;22:1804–1810. doi: 10.1016/j.cellsig.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Formisano P, Perruolo G, Libertini S, Santopietro S, Troncone G, Raciti GA, et al. Raised expression of the antiapoptotic protein ped/pea-15 increases susceptibility to chemically induced skin tumor development. Oncogene. 2005;24:7012–7021. doi: 10.1038/sj.onc.1208871. [DOI] [PubMed] [Google Scholar]
- Formstecher E, Ramos JW, Fauquet M, Calderwood DA, Hsieh JC, Canton B, et al. PEA-15 mediates cytoplasmic sequestration of ERK MAP kinase. Dev Cell. 2001;1:239–250. doi: 10.1016/s1534-5807(01)00035-1. [DOI] [PubMed] [Google Scholar]
- Frankel P, Ramos M, Flom J, Bychenok S, Joseph T, Kerkhoff E, et al. Ral and Rho-dependent activation of phospholipase D in v-Raf-transformed cells. Biochem Biophys Res Commun. 1999;255:502–507. doi: 10.1006/bbrc.1999.0234. [DOI] [PubMed] [Google Scholar]
- Haling JR, Wang F, Ginsberg MH. Phosphoprotein enriched in astrocytes 15 kDa (PEA-15) reprograms growth factor signaling by inhibiting threonine phosphorylation of fibroblast receptor substrate 2alpha. Mol Biol Cell. 2010;21:664–673. doi: 10.1091/mbc.E09-08-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF. PA promoted to manager. Nat Cell Biol. 2007;9:615–617. doi: 10.1038/ncb0607-615. [DOI] [PubMed] [Google Scholar]
- Joseph T, Wooden R, Bryant A, Zhong M, Lu Z, Foster DA. Transformation of cells overexpressing a tyrosine kinase by phospholipase D1 and D2. Biochem Biophys Res Commun. 2001;289:1019–1024. doi: 10.1006/bbrc.2001.6118. [DOI] [PubMed] [Google Scholar]
- Joseph T, Bryant A, Frankel P, Wooden R, Kerkhoff E, Rapp UR, et al. Phospholipase D overcomes cell cycle arrest induced by high-intensity Raf signaling. Oncogene. 2002;21:3651–3658. doi: 10.1038/sj.onc.1205380. [DOI] [PubMed] [Google Scholar]
- Lavoie JN, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Lazarov M, Kubo Y, Cai T, Dajee M, Tarutani M, Lin Q, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med. 2002;8:1105–1114. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- Lin HJ, Eviner V, Prendergast GC, White E. Activated H-ras rescues E1A-induced apoptosis and cooperates with E1A to overcome p53-dependent growth arrest. Mol Cell Biol. 1995;15:4536–4544. doi: 10.1128/mcb.15.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovec H, Sewing A, Lucibello FC, Muller R, Moroy T. Oncogenic activity of cyclin D1 revealed through cooperation with Ha-ras: link between cell cycle control and malignant transformation. Oncogene. 1994;9:323–326. [PubMed] [Google Scholar]
- Lu Z, Hornia A, Joseph T, Sukezane T, Frankel P, Zhong M, et al. Phospholipase D and RalA cooperate with the epidermal growth factor receptor to transform 3Y1 rat fibroblasts. Mol Cell Biol. 2000;20:462–467. doi: 10.1128/mcb.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso M, Russo G, Cinti C, Bazan V, Gebbia N, Russo A. Ras family genes: an interesting link between cell cycle and cancer. J Cell Physiol. 2002;192:125–130. doi: 10.1002/jcp.10109. [DOI] [PubMed] [Google Scholar]
- Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, et al. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer Lett. 2000;161:207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- Pastorino S, Renganathan H, Caliva MJ, Filbert EL, Opoku-Ansah J, Sulzmaier FJ, et al. The death effector domain protein PEA-15 negatively regulates T-cell receptor signaling. FASEB J. 2010 doi: 10.1096/fj.09-144295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radig K, Schneider-Stock R, Rose I, Mittler U, Oda Y, Roessner A. p53 and ras mutations in Ewing’s sarcoma. Pathol Res Pract. 1998;194:157–162. doi: 10.1016/S0344-0338(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Ramos JW, Hughes PE, Renshaw MW, Schwartz MA, Formstecher E, Chneiweiss H, et al. Death effector domain protein PEA-15 potentiates Ras activation of extracellular signal receptor-activated kinase by an adhesion-independent mechanism. Mol Biol Cell. 2000;11:2863–2872. doi: 10.1091/mbc.11.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JW. The regulation of extracellular signal-regulated kinase (ERK) in mammalian cells. Int J Biochem Cell Biol. 2008;40:2707–2719. doi: 10.1016/j.biocel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Renganathan H, Vaidyanathan H, Knapinska A, Ramos JW. Phosphorylation of PEA-15 switches its binding specificity from ERK/MAPK to FADD. Biochem J. 2005;390:729–735. doi: 10.1042/BJ20050378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revet I, Huizenga G, Chan A, Koster J, Volckmann R, van Sluis P, et al. The MSX1 homeobox transcription factor is a downstream target of PHOX2B and activates the Delta-Notch pathway in neuroblastoma. Exp Cell Res. 2008;314:707–719. doi: 10.1016/j.yexcr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Shome K, Watkins SC, Romero G. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J Biol Chem. 2000;275:23911–23918. doi: 10.1074/jbc.M001553200. [DOI] [PubMed] [Google Scholar]
- Sathyan KM, Nalinakumari KR, Kannan S. H-Ras mutation modulates the expression of major cell cycle regulatory proteins and disease prognosis in oral carcinoma. Mod Pathol. 2007;20:1141–1148. doi: 10.1038/modpathol.3800948. [DOI] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Zheng Y, Garcia A, Xu L, Foster DA. Phospholipase D provides a survival signal in human cancer cells with activated H-Ras or K-Ras. Cancer Lett. 2007;258:268–275. doi: 10.1016/j.canlet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit M, Velasco P, Brown LF, Skobe M, Richard L, Riccardi L, et al. Overexpression of thrombospondin-1 decreases angiogenesis and inhibits the growth of human cutaneous squamous cell carcinomas. Am J Pathol. 1999;155:441–452. doi: 10.1016/S0002-9440(10)65140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Uchida N, Okamura S, Nagamachi Y, Yamashita S. Increased phospholipase D activity in human breast cancer. J Cancer Res Clin Oncol. 1997;123:280–285. doi: 10.1007/BF01208639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer Res. 1999;19:671–675. [PubMed] [Google Scholar]
- Vaidyanathan H, Opoku-Ansah J, Pastorino S, Renganathan H, Matter ML, Ramos JW. ERK MAP kinase is targeted to RSK2 by the phosphoprotein PEA-15. Proc Natl Acad Sci U S A. 2007;104:19837–19842. doi: 10.1073/pnas.0704514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viparelli F, Cassese A, Doti N, Paturzo F, Marasco D, Dathan NA, et al. Targeting of PED/PEA-15 molecular interaction with phospholipase D1 enhances insulin sensitivity in skeletal muscle cells. J Biol Chem. 2008;283:21769–21778. doi: 10.1074/jbc.M803771200. [DOI] [PubMed] [Google Scholar]
- Waber PG, Chen J, Nisen PD. Infrequency of ras, p53, WT1, or RB gene alterations in Wilms tumors. Cancer. 1993;72:3732–3738. doi: 10.1002/1097-0142(19931215)72:12<3732::aid-cncr2820721228>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, et al. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- Whitehurst AW, Robinson FL, Moore MS, Cobb MH. The death effector domain protein PEA-15 prevents nuclear entry of ERK2 by inhibiting required interactions. J Biol Chem. 2004;279:12840–12847. doi: 10.1074/jbc.M310031200. [DOI] [PubMed] [Google Scholar]
- Yang H, Masters SC, Wang H, Fu H. The proapoptotic protein Bad binds the amphipathic groove of 14-3-3zeta. Biochim Biophys Acta. 2001;1547:313–319. doi: 10.1016/s0167-4838(01)00202-3. [DOI] [PubMed] [Google Scholar]
- Zeng X, Shaikh FY, Harrison MK, Adon AM, Trimboli AJ, Carroll KA, et al. The Ras oncogene signals centrosome amplification in mammary epithelial cells through cyclin D1/Cdk4 and Nek2. Oncogene. 2010;29:5103–5112. doi: 10.1038/onc.2010.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Redina O, Altshuller YM, Yamazaki M, Ramos J, Chneiweiss H, et al. Regulation of expression of phospholipase D1 and D2 by PEA-15, a novel protein that interacts with them. J Biol Chem. 2000;275:35224–35232. doi: 10.1074/jbc.M003329200. [DOI] [PubMed] [Google Scholar]
- Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat Cell Biol. 2007;9:706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y, et al. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochem Biophys Res Commun. 2000;278:140–143. doi: 10.1006/bbrc.2000.3719. [DOI] [PubMed] [Google Scholar]