Abstract
The polymorphism, KLF6-IVS1-27A, in the Krüppel-like factor 6 (KLF6) transcription factor gene enhances its splicing into antagonistic isoforms and is associated with delayed histological progression of non-alcoholic fatty liver disease (NAFLD). To explore a potential role for KLF6 in the development of insulin resistance, central to NAFLD pathogenesis, we genotyped KLF6-IVS1-27 in healthy subjects and assayed fasting plasma glucose (FPG) and insulin sensitivities. Furthermore, we quantified mRNA expression of KLF6 and glucokinase (GCK), as an important mediator of insulin sensitivity, in human livers and in liver tissues derived from a murine Klf6 knockdown model (DeltaKlf6). Klf6 overexpression studies in a mouse hepatocyte line were utilized to mechanistically link KLF6 with Gck promoter activity.
Results
KLF6-IVS1-27Gwt (ie., less KLF6 splicing) was associated with stepwise increases in FPG and insulin and reduced hepatic insulin sensitivity. KLF6 binds to the liver-specific Gck promoter and activates a GCK promoter-reporter, identifying GCK as a KLF6 direct transcriptional target. Accordingly, in DeltaKlf6 hepatocytes, Gck expression was reduced and stable transfection of Klf6 led to upregulation of Gck. GCK and KLF6 mRNAs correlate directly in human NAFLD tissues and immunohistochemistry studies confirm falling levels of both KLF6 and GCK in fat laden hepatocytes. In contrast to full length KLF6, splice variant KLF6-SV1 increases in NAFLD hepatocytes and inversely correlates with glucokinase regulatory protein, which negatively regulates GCK activity.
Conclusion
KLF6 regulation of GCK contributes to the development of hepatic insulin resistance. The KLF6-IVS1-27A polymorphism, which generates more KLF6-SV1, combats this, lowering hepatic insulin resistance and blood glucose.
Keywords: Krüppel-like Factor 6, non-alcoholic fatty liver disease, hepatic insulin sensitivity, insulin resistance, glucokinase
INTRODUCTION
The prevalence of non-alcoholic fatty liver disease (NAFLD), the hepatic manifestation of the metabolic syndrome, is rising dramatically (1). The factors conferring a risk of progression to cirrhosis include advanced age, increased body mass index (BMI), and an elevated fasting plasma glucose (FPG) associated with insulin resistance (IR) (2). In addition to metabolic and anthropometric factors, genetic factors, such as polymorphisms in adiponutrin (PNPLA3)(3, 4), are associated with the progression risk of NAFLD. Among these, we have identified the Krüppel-like Factor 6 (KLF6) genotype as a predictor of NAFLD histological stage (5). KLF6 is a ubiquitously expressed transcription factor (6) established as an immediate early gene in activated hepatic stellate cells (HSC) after liver injury (7, 8) and as a tumour suppressor gene in a range of tissues including the liver (9). A functional intron 1 single nucleotide polymorphism (SNP), KLF6–IVS1-27G>A (rs3750861) (10), creates a novel binding site for the splicing factor, SRp40, (11) and promotes alternative splicing of KLF6 into antagonistic, truncated isoforms (5). Among these isoforms, KLF6 splice variant 1 (KLF6-SV1) promotes cell growth while reducing differentiation in cancer cells and tumours in vivo. The roles of KLF6 and KLF-SV1 in non-malignant cellular homeostasis, however, are unknown (12).
To elucidate the mechanisms underlying KLF6’s contribution to NAFLD, we explored the association of KLF6-IVS1-27G>A with metabolic factors predictive of disease progression. Here we report a significant and independent association of the KLF6–IVS1-27A allele with lower FPG and increased hepatic insulin sensitivity. Glucokinase (GCK) is a major determinant of hepatic glucose metabolism and is closely associated with hepatic glucose disposal and production in response to glucose and insulin (13, 14). Further, we suggest that there is a mechanistic link between KLF6 isoforms and the regulation of GCK and the glucokinase regulatory protein (GCKR).
EXPERIMENTAL PROCEDURES
Characterization of human subjects
A total of 1276 healthy subjects were recruited from the RISC [Relationship between Insulin Sensitivity and Cardiovascular Disease] cohort, as approved by local ethics committees (15). Subjects with diabetes, hypertension, dyslipidemia or history of liver disease were excluded. Subjects underwent detailed anthropometric assessment and a 75g oral glucose tolerance test (OGTT) (16). Subjects underwent a hyperinsulinemic-euglycaemic clamp, as previously reported and standardized (15). Insulin sensitivity was assessed as the mean glucose infusion rate over the last 40min of the clamp, corrected for the mean plasma insulin levels achieved during the same period (M/I). Insulin secretion was calculated from the OGTT c-peptide concentrations by deconvolution (17). An estimate of the endogenous insulin clearance was obtained from the ratio of the areas under the insulin secretion and the insulin concentration curves. Baseline fasting serum measurements included glucose, insulin, triglycerides, and gamma glutamyl transferase. Genotype/phenotype relationships were studied by linear trend analysis corrected for age, sex and recruitment center. The KLF6-IVS1-27G>A SNP was genotyped by Kbiosciences, UK and a complete dataset was available in 1249 individuals. In a subset of 367 individuals from 9 centers, on the clamp day, basal endogenous glucose production (EGP) was determined by a primed-constant tracer infusion (6,6-2H2-glucose). A hepatic insulin resistance index was calculated as [EGP × fasting insulin].
Mouse model
Experiments were approved and conform to regulations of the Institutional Animal Care and Use Committee (IACUC). Mice with a floxed Klf6 targeting vector (C57BL/6;129Sv, Genentech, San Francisco, CA) (18) were crossed with mice expressing Cre recombinase (Cre) under control of the albumin promoter (B6.Cg-Tg(Alb-cre)21Mgn/J; Jackson Labs, Bar Harbor, ME). After backcrossing, male offspring expressing Cre with two floxed KLF6 alleles were used as the experimental group (‘DeltaKlf6’). Mice with two floxed alleles and no Cre expression were used as controls (wt). Temperature, humidity and light-dark cycle controlled conditions were controlled, mice were allowed food and water ad libitum and were euthanized at three months following an overnight fast. Caval blood was drawn and glucose quantified with a handheld device (LifeScan, Milpitas, CA). Liver tissue was conserved for histology, RNA and protein isolation. Serum insulin and colorimetric glycogen were quantified using commercial Elisa kits (Biovision, Mountain View, CA), while GCK was by IF-staining and Western Blot with an anti-GCK antibody (H-88) (Santa Cruz Biotechnology, Santa Cruz, CA).
Supplementary Methods
Supplementary methods describing stable Klf6 expression in mouse hepatocytes, liver GCK promoter characterization, real-time PCR analyses, studies on NAFLD liver tissues and statistical analyses are detailed as online supplementary information.
RESULTS
KLF6-IVS1-27G>A is associated with lower fasting plasma glucose and fasting plasma insulin
We first sought to expand our previous study on the KLF6-IVS1-27G>A SNP in NAFLD (5) in an independent cohort. In normal subjects there was a highly significant association between the KLF6 genotype and fasting blood glucose (FPG) based on linear trend analysis of the genotype groups (AA, AG and GG) (Table 1A). Additionally, fat mass was lower in the KLF6-IVS1-27G>A group by univariate analysis (AA vs. AG+GG). Importantly, the association between FPG and KLF6 genotype was significant even when corrected for fat mass or BMI, as well as age, sex and recruitment centre. Trends in fasting plasma insulin (FPI) levels were also significant and paralleled those of FPG.
Table 1. KLF6 Genotype and NAFLD related associations in normal individuals.
Table 1A includes variables classed by genotype for the cohort of 1250 individuals, while Table 1B presents subset data for those in which endogenous glucose production (EGP) was measured. Non-parametric comparisons for quantitative variables was by Mann-Whitney testing. Linear Trend analyses were corrected for differences in age, sex, center and BMI, as were combined group comparisons using the General Linear Model (GLM).
| KLF6 | Linear Trend | AA vs AG+GG | AA+AG vs GG | |||
|---|---|---|---|---|---|---|
| AA | GA | GG | ||||
| Number | 12 | 155 | 1082 | |||
| Age (years) | 42.08±2.95 | 45.74±0.68 | 43.57±0.25 | na | ns | 0.006 |
| Sex (M:F)* | 6:6 | 71:84 | 481:599 | na | ns | ns |
| BMI | 23.72±1.29 | 25.97±0.33 | 25.53±0.12 | ns | ns | ns |
| Fat mass (kg/m2) | 15.43±2.37 | 22.12±0.75 | 20.94±0.27 | ns | 0.023 | ns |
| Glucose (mmol/l) | 4.79±0.20 | 5.01±0.05 | 5.08±0.02 | 0.003 | ns | 0.009 |
| FPI (pmol/l) | 20.21±3.47 | 31.36±1.51 | 32.86±0.68 | 0.022 | ns(0.050) | ns |
| HOMA-IR | 0.95±0.14 | 1.37±0.07 | 1.36±0.03 | ns | ns | ns |
| OGIS | 486.1±19.7 | 442.8±5.11 | 441.5±2.04 | 0.042 | ns | ns |
| M/I μmol min−1 (kg fat-free mass)−1 nmol−1 | 159.8±25.6 | 143.5±5.48 | 137.7±2.21 | ns | ns | ns |
| icl_OGTT (l/min/m2) | 2.20±0.29 | 1.65±0.65 | 1.62±0.0.56 | ns | 0.015 | ns |
| TG (mmol/l) | 79.00±7.92 | 110.05±6.09 | 107.40±2.23 | ns | ns | ns |
| WC (cm) | 82.41±4.76 | 87.70±1.01 | 86.58±0.38 | ns | ns | ns |
| KLF6 | Linear Trend | AA vs AG+GG | AA+AG vs GG | |||
|---|---|---|---|---|---|---|
| AA | GA | GG | ||||
| Number | 5 | 38 | 327 | |||
| Age (years) | 36.40±2.65 | 45.76±1.49 | 42.75±8.08 | na | ns | ns |
| Sex (M:F)* | 5:0 | 17:21 | 163:164 | na | ns | ns |
| BMI | 25.04±1.67 | 27.00±0.62 | 26.01±0.23 | na | ns | ns |
| Fat_mass (kg/m2) | 14.14±3.38 | 23.26±1.47 | 21.51±0.51 | na | ns | ns |
| Glucose (mmol/l) | 4.72±0.14 | 4.93±0.08 | 4.99±0.03 | 0.027 | ns | ns |
| FPI (pmol/l) | 18.54±3.33 | 26.50±2.52 | 30.98±1.23 | 0.005 | ns | 0.015 |
| EGP | 17.06±2.34 | 16.33±0.79 | 16.36±0.26 | ns | ns | ns |
| Hepatic Insulin Resistance | 343.46±101.9 | 422.30±44.3 | 496.29±19.1 | 0.022 | ns | 0.029 |
Key: BMI – Body Mass Index; HOMA-IR – homeostatic model assessment of insulin resistance; OGIS – ‘oral glucose insulin sensitivity’ index; M/I – whole-body insulin sensitivity measured by hyperinsulinaemic clamp; FPI – Fasting Plasma Insulin; TG – triglycerides; WC – waist circumference; GCR_b – Basal glucose clearance (production/concentration); icl_OGTT – endogenous ‘pre-hepatic’ clearance during OGTT. ns: not significant; na: not appropriate; KW-Kruskal Wallis test.
KLF6-IVS1-27G>A is associated with enhanced insulin sensitivity and insulin clearance
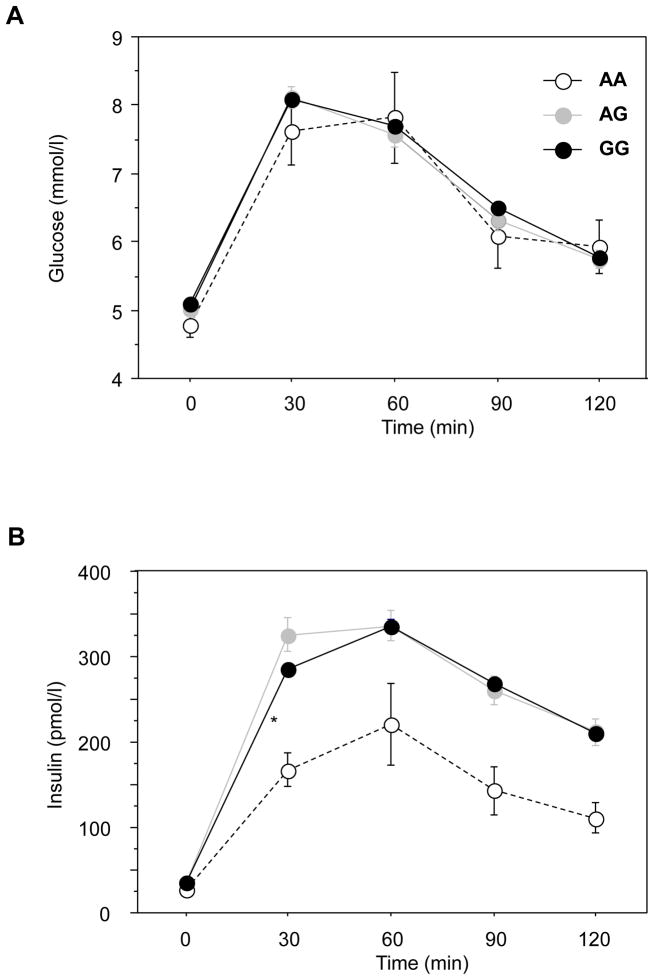
We next sought to elucidate the mechanisms underlying the protective effect of KLF6-IVS1-27G>A on glucose homeostasis. Insulin sensitivity assessed by the hyperinsulinemic clamp (M/I), a measure primarily of extrahepatic glucose disposal in euglycemic conditions, was not significantly different in the three genotypes. Because hepatic glucose disposal is markedly dependent on the route of glucose delivery and its concentration in the portal vein (19), the OGTT, and insulin sensitivity based on the OGTT data (OGIS) (16), provides greater insight into hepatic insulin sensitivity. The glucose profile during the OGTT was identical in all three KLF6 genotype groups (Figure 1A). Remarkably, however, the initial insulin plasma level was significantly lower in the KLF6-IVS1-27AA group (30min; p= 0.006 relative to GG; Figure 1B) and there was a trend towards lower insulin levels at later time points (n=12; area under curve p=0.07). Insulin secretion was the same across the genotype groups (data not shown), indicating that lower plasma insulin levels (Figure 1B) for the KLF6-IVS1-27AA genotype were not due to insulin secretion. This suggests a greater hepatic clearance of insulin, resulting in a lower peripheral level. Endogenous ‘hepatic’ clearance of insulin (icl-OGTT) has been estimated from insulin secretion and concentration during the OGTT; this parameter was significantly higher in KLF6-IVS1-27AA individuals (See Table 1A).
Figure 1. KLF6_IVS1-27AA individuals have a normal glucose profile during the OGTT, despite lower plasma insulin levels.
The profile of plasma glucose levels during the oral glucose tolerance test (OGTT) was unchanged between the three KLF6_IVS1-27G>A genotype groups, depicted as homozygous variant (-○- AA), heterozygous variant (-●- GA) and wild type (-●- GG) (A). In contrast, the plasma insulin levels were lower, significantly so at 30 minutes, in the homozygous variant AA individuals (* = p<0.05) (B).
Despite the reduced insulin concentration attributed to increased insulin clearance in KLF6-IVS1-27AA individuals, the glucose concentrations during the OGTT were not increased, suggesting increased insulin sensitivity. While the hyperinsulinemic clamp data reflecting extrahepatic glucose disposal showed no significant differences in peripheral insulin sensitivity, insulin sensitivity estimated by OGIS during the OGTT was significantly associated with KLF6 genotype (Table 1A). The association of KLF6 genotype with OGIS but not M/I is again suggestive of a difference in the hepatic insulin clearance and sensitivity, rather than peripheral reaction to insulin.
KLF6-IVS1-27G>A is associated with increased hepatic insulin sensitivity
To explore an association with hepatic insulin sensitivity, endogenous glucose production (EGP) was measured after an overnight fast by infusion of 6,6-2H2-glucose in a subgroup of 367 patients from 9 centers. EGP was identical in the KLF6 genotype groups (Table 1B). However, when EGP was corrected for the insulin concentration (EGP × Fasting Insulin), which is a measure of hepatic insulin resistance, there was a stepwise change indicating increased insulin sensitivity associated with the KLF6 IVS1-27<A allele (KLF6_AA 343 ± 102; KLF6_GA 422 ± 44; KLF6_GG 494 ± 19; p=0.023). Thus, KLF6-IVS1-27AA individuals have both increased hepatic insulin sensitivity and insulin clearance. The hepatic insulin resistance index was correlated with the insulin clearance estimate (Spearman rho −0.44, p<0.0001).
Glucokinase expression is reduced in DeltaKlf6 mouse livers
Glucokinase (GCK) is the rate-limiting enzyme in glycolysis and its expression in the liver is closely associated with hepatic glucose metabolism in response to glucose and insulin (20, 21). An hepatic GCK promoter polymorphism is associated with altered hepatic insulin resistance and underscores the potential role of GCK as a KLF6-regulated target (22). Moreover, GCK activity is closely linked to the expression of GCKR, which determines GCK activity, subcellular location and GCK protein stability (23).
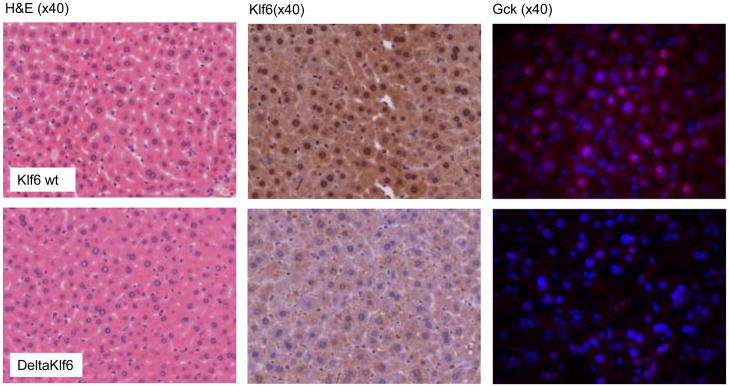
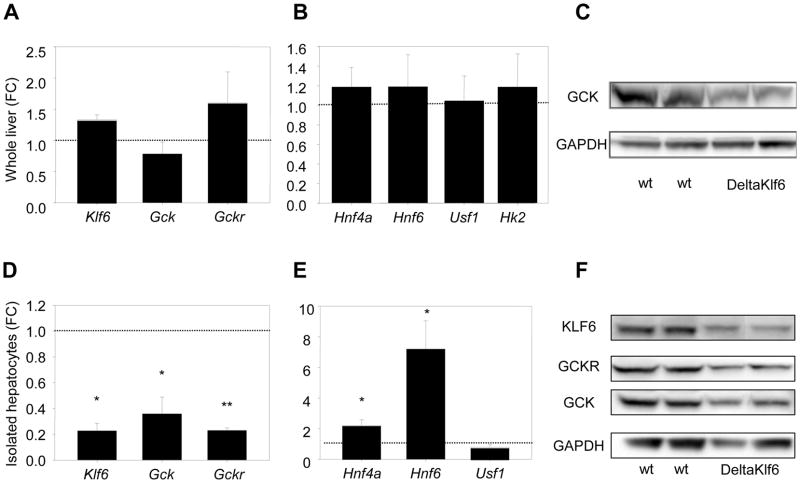
To explore the role of KLF6 in the regulation of GCK, we generated mice with reduced expression of Klf6 (DeltaKlf6) in hepatocytes by crossing Klf6 floxed mice (18) with animals expressing albumin-Cre. Cre expression was gradually increased during the first six weeks, and tissues were harvested at 11–12 weeks of age. Immunohistochemistry confirmed a clear reduction in hepatocyte expression of Klf6 (Figure 2). There were no clear phenotypic differences (haemotoxylin and eosin, Figure 2), although there was a trend towards higher FBG, HOMA-IR and triglyceride levels (Supplementary Table 3). In whole liver lysates, Klf6 mRNA was not in fact reduced in the DeltaKlf6 mice (Figure 3A) and this was attributed to the reduction in hepatocytes being masked by a relative increase in KLF6 protein in hepatic perisinusoidal cells, as evident from immunohistochemistry of liver sections (Figure 2) and as previously reported for other genes targeted in the albumin-Cre model (24, 25). In contrast, in hepatocytes isolated from DeltaKlf6 mice, Klf6 was markedly reduced at mRNA and protein levels (3D & 3F). Both in vivo and in hepatocytes in vitro, Gck mRNA and protein were markedly downregulated in the DeltaKlf6 mice relative to wild type littermates (Figures 2, 3D & F). Notably, in whole tissues there were no significant changes in the expression of other transcription factors that regulate Gck, including Hnf4a, Hnf6 or Usf1, or in hexokinase 2, which is expressed at low levels in hepatocytes (26) (Figure 3B & E). In isolated hepatocytes, the negative regulator Gckr was reduced (Figure 3D, 3F), while significant increases in Hnf4α and Hnf6 mRNAs were observed. It is unlikely, therefore, that these factors contributed to the Gck decrease in the DeltaKlf6 mice.
Figure 2. Reduced Klf6 and glucokinase expression in vivo DeltaKlf6 mice.
Haematoxylin and Eosin (H&E) stains for wt and DeltaKlf6 mice were similar with no apparent phenotypic differences at 3 months of age. KLF6 immunohistochemistry revealed a decreased expression of KLF6 within hepatocytes (4.1 fold fewer positive nuclei by manual counting; p=0.002) rather than non-parenchymal cells. GCK immunofluorescent staining (1300ms exposure time, DAPI blue, GCK red) confirmed a relative abundance (1.92 fold by integrated density quantification, p=0.038) of hepatocyte Gck expression in wt versus DeltaKlf6 mice.
Figure 3. Reduced Klf6 and glucokinase expression in DeltaKlf6 mice.
While expression of Klf6, Gck and Gckr mRNA was not significantly suppressed in whole liver tissues in DeltaKlf6 mice compared to wild type littermates (n=7) (n=7) (A), levels were markedly suppressed in primary hepatocytes isolated from DeltaKlf6 mice (n=3) (D). Data are presented as fold change (FC) relative to wt littermates. Known transcriptional activators of Gck were not reduced in the presence of reduced Gck in deltaKlf6 mice (B & E). Western blot of both whole tissues and isolated hepatocytes confirmed decreased Gck protein in association with reduced Klf6. (* denotes p-value<0.05; **p-value<0.005)
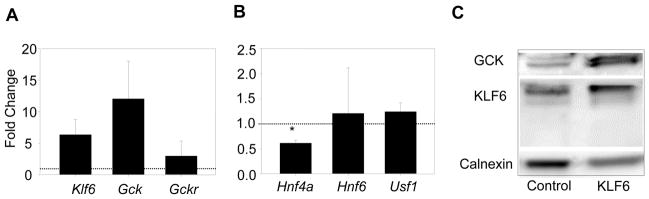
Klf6 expression in mouse hepatocytes increases Gck
To further link Klf6 directly to Gck expression in hepatocytes, we examined the impact of stable Klf6 cDNA expression on Gck expression in the AML12 mouse hepatocyte cell line, which retains major hepatocyte characteristics (27). In cells stably expressing full length Klf6 cDNA, an increase in Gck mRNA and protein was evident (Figure 4A, 4C). Consistent with the observations in DeltaKlf6 mice, Hnf4α expression was significantly decreased in these cells, most likely as part of a counter regulatory mechanism (Figure 4B). Immunofluorescence confirmed hepatocyte accumulation of GCK protein (Supplementary Figure 1).
Figure 4. Increased Klf6 and glucokinase expression in vitro in Klf6 transfected AML12 cells.
Increased Klf6 and Gck expression were present in the murine hepatoma cell line, AML12 after transfection with a plasmid vector expressing Klf6 (A, C). Known transcriptional activators of Gck remained unchanged. Hnf4a mRNA was downregulated indicating counter-regulatory mechanisms (B). (* = p<0.05).
The Gck promoter is a direct transcriptional target of Klf6
To test for direct transcriptional activation of the GCK promoter by Klf6, AML12 cells were co-transfected with the Klf6 cDNA and a Gck-luc promoter reporter containing candidate KLF6 binding sites (28). Co-transfection with Klf6 increased Gck promoter activity (Supplementary Figure 2A). Interaction of KLF6 with the endogenous Gck promoter at two defined sites containing putative KLF6 binding sites was confirmed (Supplementary Figure 2B) by chromosomal immunoprecipitation (ChIP).
KLF6 and GCK expression are directly correlated in NAFLD livers
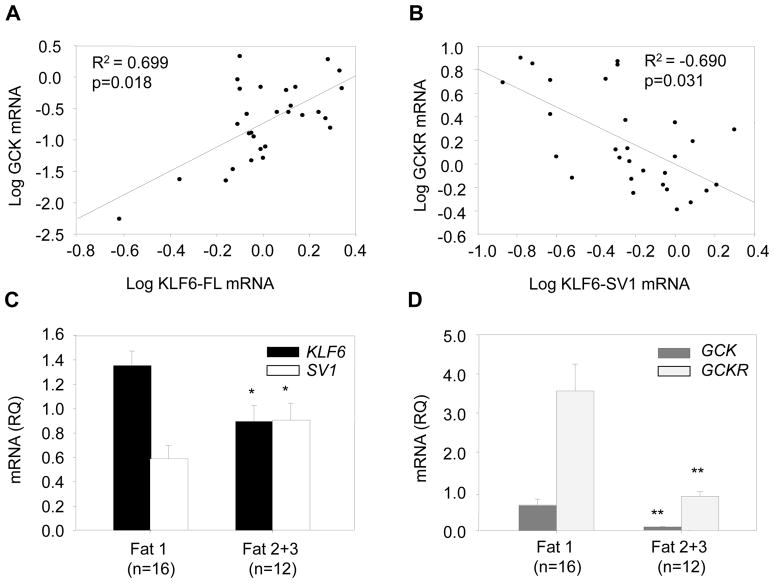
We have analyzed KLF6 and GCK expression in 28 pre-cirrhotic NAFLD biopsy samples classed as having mild (grade 1) or severe (grades 2 and 3) hepatocyte steatosis. While we have previously shown increased KLF6 expression in NAFLD livers in association with inflammation and fibrosis (5), the data presented here demonstrate a significant reduction of the full-length isoform in association with increased steatosis (Pearson correlation KLF6-FL and steatosis: −0.445; p=0.018; 1.34±0.126 versus 0.91±0.122; t-test =0.018, Figure 5C). This apparent discrepancy is clarified by immunohistochemistry (IHC) data which reveal differential KLF6 expression in distinct populations of liver cells. IHC studies in normal liver using KLF6 monoclonal 1A9 antibody (detecting all KLF6 isoforms) demonstrate predominantly nuclear KLF6 expression in hepatocytes, portal tract cells and occasional sinusoidal cells (Figure 6Ai). In more advanced disease (Figure 6Ci), there is a dramatic increase in KLF6 expression, but this is notably in a mixed inflammatory cell population within portal tracts and in sinusoidal cells, rather than hepatocytes. Parallel IHC with a KLF6-SV1 isoform specific monoclonal antibody (2A2) shows the opposite, with no parallel increase in inflammatory cells, but with much more prominent expression in hepatocytes (Figure 6Ci+ii). These data parallel isoform specific real-time PCR data in which, unlike KLF6-FL, the KLF6-SV1 isoform is significantly increased with more advanced steatosis (Figure 5C).
Figure 5. KLF6-FL and GCK are reduced with advanced steatosis, while elevated KLF6-SV1 is associated with suppressed GCKR.
Expression of the KLF6 full-length isoform (KLF6-FL) correlated positively with GCK expression (A). In contrast, expression of KLF6-SV1 was associated with a dramatic reduction in expression of GCKR (B). KLF6-FL expression was significantly suppressed with more advanced steatosis (grade 2+3) while KLF6-SV1 was significantly upregulated (C). Both GCK and GCKR were significantly downregulated in patients with advanced steatosis (grade 2+3) (D). (* = p<0.05; ** = p<0.01).
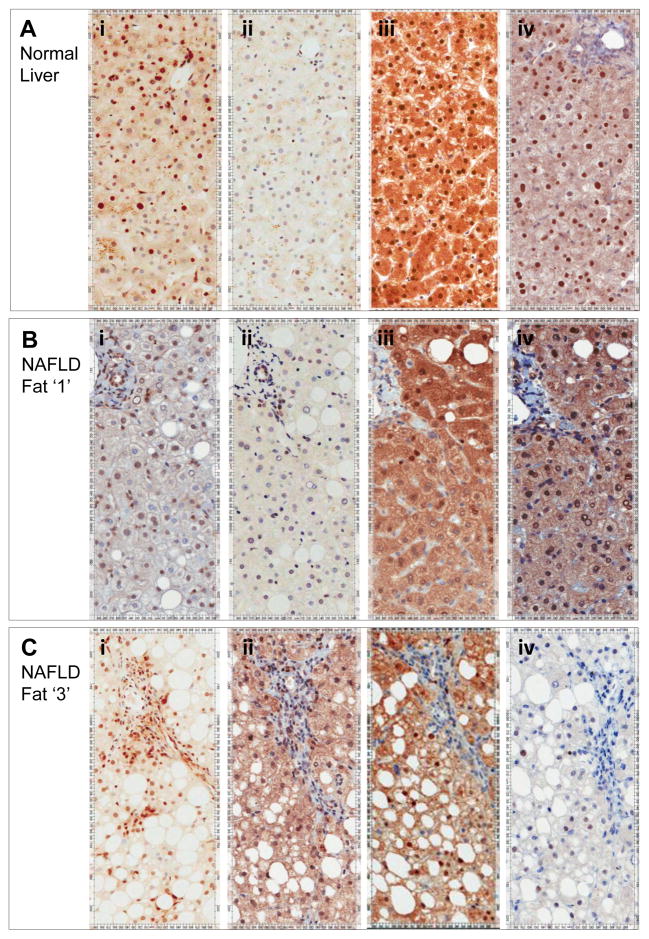
Figure 6. KLF6-FL falls in hepatocytes with advanced steatosis, but increases in inflammatory and sinusoidal cells.
KLF6 expression detected by the 1A9 monoclonal (all isoforms) is evident in hepatocytes and non-parenchymal cells in normal liver (Ai). A fall in hepatocytes in the presence of steatosis is contrary to an increase in inflammatory and sinusoidal cells (Bi+Ci). In contrast, KLF6-SV1 increases preferentially in hepatocytes in the presence of more advanced steatosis (Aii–Cii). These changes in representative sections parallel mRNA data in Figure 5, as do changes in GCK and GCKR. Both are present in normal liver (Aiii+iv), with notable nuclear localisation in this fasted state. The pattern is similar but less dramatic in the presence of fat grade 1 simple steatosis (Biii+iv). In the presence of more severe steatosis, expression of both is notably reduced (Ciii+iv). Quantification of nuclear expressions have been performed using imaging software and these additional data are included as Supplementary Figures.
We explored the relationship between KLF6 and its target gene, GCK in these NAFLD biopsies. Bivariate analysis of real time PCR analyses of mRNA extracted from whole human liver confirmed a highly significant association between KLF6 and GCK (Pearson correlation 0.687; p<0.0001). By linear regression, corrected for other significantly associated variables (Supplementary Table 4: center, fat and GCKR) the association between GCK and KLF6 remained independently significant (Figure 5A). Immunohistochemistry studies in normal liver confirm GCK expression in hepatocytes, which is notably sequestered in the nucleus in association with nuclear GCKR. Individuals with severe steatosis had decreased expression of KLF6 and GCK in hepatocytes (Figure 6Aiii–Ciii). While the role of KLF6-SV1 in NAFLD is presently unknown, our SNP association studies suggest a protective role in both chronic liver disease progression (5) and enhanced hepatic sensitivity to insulin. It is interesting therefore, that KLF6-SV1 is increasingly expressed in hepatocytes in the presence of steatosis - a marker not just of the severity of the insult, but possibly also of the livers ability to tolerate it. Further, there is a highly significant negative correlation between KLF6-SV1 and GCKR at mRNA levels (Figure 5B). This is apparent also at the protein level, as when KLF6-SV1 expression is evident in the presence of increased steatosis, GCKR is not detectable (Figures 6Cii+iv).
DISCUSSION
Although NAFLD prevalence is increasing in Western societies, established predictors of disease progression to advanced stages are scarce (2). We previously demonstrated that the functional KLF6-IVS1-27G>A polymorphism correlates inversely with disease progression in NAFLD. Individuals with KLF6-IVS1-27G>A were less likely to have significant fibrosis when compared to those with the wildtype allele (5). In the work presented here, we have functionally linked KLF6 to components of the hepatic insulin response, which is central the progression of NAFLD.
The association between the KLF6 genotype groups and FPG is novel and striking. While the KLF6-IVS1-27A allele was also associated with a lower FPI, the euglycemic clamp data discount peripheral insulin resistance as the cause of the variation with genotype. Together, the OGIS and OGTT derived data and absence of a difference in pancreatic beta cell insulin secretion, support increased hepatic clearance of insulin and increased hepatic insulin sensitivity in KLF6-IVS1-27A individuals. This was subsequently confirmed by assessing endogenous glucose production relative to fasting insulin levels.
IR is an established key feature of the metabolic syndrome and increased resistance associated with fasting hyperglycaemia is believed to contribute to hepatic steatosis in obese individuals (29). There is evidence, however, that insulin resistance is not essential for the development of steatosis. In hypobetalipoproteinaemia, for example, fatty liver develops because of defective VLDL export in the absence of IR (30). Furthermore, while a fatty liver GIWAS study identified the rs738409 C>G single nucleotide polymorphism in PNPLA3 - which leads to a missense mutation (I148M) - as associated with increased liver fat, it is not associated with IR.(3, 31, 32) Additional GWAS data in patients with fatty liver disease has further highlighted our incomplete understanding of the relationship between fatty liver disease and features of the metabolic syndrome. While some of the steatosis-associated variations of candidate genes are associated with high triglycerides and plasma LDL-cholesterol, for example, the opposite is true of other variants.(33, 34) Furthermore, the PP1R3B and GCKR polymorphisms are associated with a lower rather than a higher fasting glucose, in keeping with a relatively enhanced, rather than reduced, sensitivity to insulin. Therefore, while IR indisputably contributes to the progression of NAFLD to fibrosis and cirrhosis, its role in the development of steatosis is not as clear. Rather than being indicative of disease severity, in some individuals steatosis may instead be a biomarker of an enhanced ability to convert glucose to hepatic fat, conferring protection from an elevated blood glucose and reflecting enhanced insulin sensitivity.
Our own study has focused on GCK, whose expression in the liver is closely associated with hepatic insulin sensitivity (20, 21). In conjunction with GCKR it is a major determinant of insulin responsive hepatic glucose metabolism, catalyzing the production of glucose-6-phosphate (23). While the hepatic expression of GCK protein is reduced in cirrhosis attributed to both alcohol excess and primary biliary cirrhosis (35, 36), its contribution to NAFLD has not previously been characterised. GCK’s vital role in determining blood glucose is underscored by the discoveries of over 600 hereditary mutations of GCK associated with glycemic disease (37, 38). Subjects with a single inactivating mutant allele have a mild form of diabetes, associated with elevated FPG (39, 40). Furthermore, recent data suggests a role for KLF6 in response to glucose-stimulation (41). GCK activity is closely linked to the abundance of its regulator, GCKR, and small changes in the molar ratio of GCK/GCKR protein markedly impact hepatic glucose metabolism (42). In the fasting state GCK is sequestered in an inactive state in the nucleus, bound to GCKR. However, after a meal, glucose and insulin act synergistically in causing rapid dissociation of GCK from GCKR and translocation to the cytoplasm (43).
Our in vivo and in vitro murine data demonstrate that down-regulation of Klf6 in hepatocytes is associated with reduced Gck, whereas Klf6 over-expression increases Gck. ChIP and reporter studies confirm direct transactivation of the Gck promoter by Klf6, leading us to propose Klf6 as an additional mediator of glucose homeostasis. The normal phenotype of DeltaKlf6 mice is consistent with published data from liver specific Gck knockout mice as well as Gckr knockout mice, in which no differences in fasting glucose levels were detected (44–46). Biopsy studies from patients with histologically scored NAFLD confirm a significant association between KLF6-FL and GCK mRNA expression in human tissues. While, the significant reductions of KLF6-FL and GCK in individuals with more advanced versus mild steatosis may simply be representative of more advanced disease, they may also represent effectors of the development of resistance of hepatocytes to insulin.
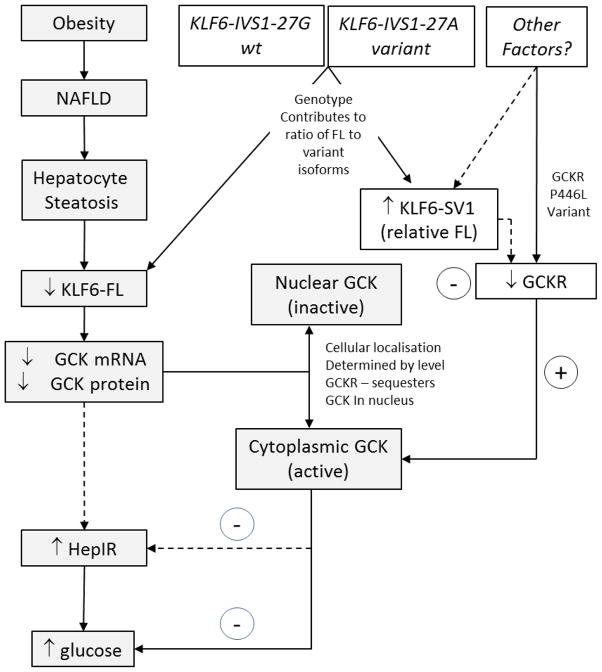
In human tissues, several alternative splice forms of KLF6 have been identified, and the presence of the KLF6-IVS1-27G>A allele promotes their generation (5). To date Klf6 splicing in the mouse has not been described, precluding our ability to study the effects of murine dominant negative splice variants on Gck or Gckr in an in vivo model. The mechanism underlying the enhanced hepatic insulin sensitivity in human individuals with the KLF6-SV1-promoting KLF6-IVS1-27G>A SNP is presently unknown. However, our expression data identifying a highly significant negative correlation between KLF6-SV1 and GCKR suggests that antagonism of GCKR, the negative regulator of GCK, is one potential mechanism. Here we are able to draw a link to the recently published data on SNPs in GCKR. The GCKR rs780094 GWAS identified SNP (34, 47) was previously associated with higher triacylglycerol, reduced insulin levels and a reduced risk of type 2 diabetes.(48, 49) GCKR rs780094 is commonly inherited with a GCKR coding SNP, rs1260326 (Pro446Leu), which codes for a mutant and inactive form of GCKR.(50) The consequences include enhanced GCK activity and reduced FPG, as well as increased de novo lipogenesis attributed to enhanced production of malonyl coA, the substrate for fatty acid synthesis.(51) Similar to this phenotypically altered GCKR variant, we hypothesize that KLF6-SV1 contributes to the lowering of FPG as a result of increased GCK activity brought about by the antagonism of GCKR, as summarised in Figure 7.
Figure 7. Proposed Mechanisms for KLF6 Regulation of Hepatic Glucose Control.
KLF6 is a ubiquitously expressed immediate early gene expressed in response to stressful stimuli. It is regulated by alternative splicing to dominant negative splice forms. The KLF6-IVS1-27A polymorphism favours alternative splicing. In the obesity related metabolic syndrome, fat accumulates in hepatocytes. While KLF6 expression increases in inflammatory and non-parenchymal cells in NASH, it falls in fat laden hepatocytes. Consequently expression of its transcriptional target GCK falls, leading to reduced GCK protein in hepatocytes, an increase in hepatic insulin resistance and an elevated plasma glucose. Importantly, however, the activity of the GCK that is expressed is determined by its localisation – being active in the cytoplasm, but inactive in the nucleus. GCKR is its negative regulator, responsible for its nuclear sequestration. A relative increase in KLF6-SV1 expression in steatotic hepatocytes is associated with a fall in GCKR, which we propose facilitates GCK activity, antagonising the KLF6-FL (or wt genotype) association with elevated serum glucose and HepIR.
In conclusion, we propose KLF6 as an additional regulator of fasting plasma glucose and hepatic insulin sensitivity. Understanding the interactions between KLF6 and KLF6-SV1, as well as their roles in regulation of expression of GCK and GCKR, may help us to clarify the role(s) of NAFLD in the metabolic syndrome, as well as its progression to more advanced disease.
Supplementary Material
Detection of DNA immunofluorescence was using 4′6-diamidino-2-phenylindole (DAPI, blue; 1800ms, red channel). An anti GCK antibody (H-88; Santa Cruz Biotechnology, Santa Cruz, CA) was detected with Texas Red. Independent and merged images are shown. Increased GCK protein expression was present in Klf6 expressing AML12 clones.
Co-transfection of a Klf6 expressing plasmid with a Gck promoter reporter shows a significant increase in reporter activity detected by Firefly Luciferase (A). Binding of KLF6 to the Gck promoter was confirmed by chromatin immunoprecipitation assay for two putative GC boxes within the mouse Gck promoter (B) (** = p<0.01).
Morphometric analyses, a software algorithm (Aperio Technologies) recognizing hepatocyte nuclei was optimized and applied to scanned images created from paraffin embedded tissues analysed by immunohistochemistry. Examples of the nuclear algorithm application for normal liver, as well as steatotic livers is shown in Figure 3. Negative nuclei are blue, while positive, moderately positive and strongly positive are yellow, orange and red respectively. The entire biopsy from representative cases has been captured in 8–14 fields and analysed in this way. As the background staining is not uniform across biopsies and antibodies used, the percentage of total nuclei being positive (rather than grades positive) have been compared. These data are presented in Supplementary Figure 4. Clear differences in expression are confirmed, as summarized in Figure 6 (Kruskal Wallis p<0.001 for each of KLF6, KLF6-SV1, GCK and GCKR across the three groups studied).
Acknowledgments
Financial support: Supported by grants from GSK (HLR); Newcastle-upon-Tyne Hospitals Special Trustees (HLR); the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement Health-F2-2009-241762, for the project FLIP; (HLR, CPD, ADB, GLP; AG; EB); European Commission FP5 and AstraZeneca (AG, LP, MW and the RISC consortium); European Foundation for the Study of Diabetes (AG), European Association for Study of the Liver Sheila Sherlock fellowship (LM, LPB); and the National Institutes of Health (SLF): DK37340, DK56621 and AA017067; IFORES program of the University of Essen (LPB); Swiss National Fund (DV).
List of abbreviations
- NAFLD
Nonalcoholic Fatty Liver Disease
- KLF6
Krüppel-like Factor 6
- KLF6-FL
KLF6 Full Length
- KLF6-SV1
KLF6 Splice Variant 1
- NIDDM
Non-Insulin Dependent Diabetes Mellitus
- SNP
Single Nucleotide Polymorphism
- IR
Insulin resistance
- Hep(IR)
Hepatic Insulin Resistance
- GCK
Glucokinase
- GCKR
Glucokinase Regulatory Protein
Footnotes
There is no conflict of interest to disclose
References
- 1.Farrell GC. The liver and the waistline: Fifty years of growth. J Gastroenterol Hepatol. 2009;24 (Suppl 3):S105–118. doi: 10.1111/j.1440-1746.2009.06080.x. [DOI] [PubMed] [Google Scholar]
- 2.Angulo P, Hui JM, Marchesini G, Bugianesi E, George J, Farrell GC, Enders F, et al. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846–854. doi: 10.1002/hep.21496. [DOI] [PubMed] [Google Scholar]
- 3.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miele L, Beale G, Patman G, Nobili V, Leathart J, Grieco A, Abate M, et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology. 2008;135:282–291. e281. doi: 10.1053/j.gastro.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koritschoner NP, Bocco JL, Panzetta-Dutari GM, Dumur CI, Flury A, Patrito LC. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J Biol Chem. 1997;272:9573–9580. doi: 10.1074/jbc.272.14.9573. [DOI] [PubMed] [Google Scholar]
- 7.Lalazar A, Wong L, Yamasaki G, Friedman SL. Early genes induced in hepatic stellate cells during wound healing. Gene. 1997;195:235–243. doi: 10.1016/s0378-1119(97)00159-5. [DOI] [PubMed] [Google Scholar]
- 8.Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, et al. Zf9, a Kruppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci U S A. 1998;95:9500–9505. doi: 10.1073/pnas.95.16.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narla G, Kremer-Tal S, Matsumoto N, Zhao X, Yao S, Kelley K, Tarocchi M, et al. In vivo regulation of p21 by the Kruppel-like factor 6 tumor-suppressor gene in mouse liver and human hepatocellular carcinoma. Oncogene. 2007;26:4428–4434. doi: 10.1038/sj.onc.1210223. [DOI] [PubMed] [Google Scholar]
- 10.Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213–1222. doi: 10.1158/0008-5472.CAN-04-4249. [DOI] [PubMed] [Google Scholar]
- 11.Narla G, DiFeo A, Yao S, Banno A, Hod E, Reeves HL, Qiao RF, et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005;65:5761–5768. doi: 10.1158/0008-5472.CAN-05-0217. [DOI] [PubMed] [Google Scholar]
- 12.Narla G, DiFeo A, Fernandez Y, Dhanasekaran S, Huang F, Sangodkar J, Hod E, et al. KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J Clin Invest. 2008;118:2711–2721. doi: 10.1172/JCI34780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clement K, Pueyo ME, Vaxillaire M, Rakotoambinina B, Thuillier F, Passa P, Froguel P, et al. Assessment of insulin sensitivity in glucokinase-deficient subjects. Diabetologia. 1996;39:82–90. doi: 10.1007/BF00400417. [DOI] [PubMed] [Google Scholar]
- 14.Tappy L, Dussoix P, Iynedjian P, Henry S, Schneiter P, Zahnd G, Jequier E, et al. Abnormal regulation of hepatic glucose output in maturity-onset diabetes of the young caused by a specific mutation of the glucokinase gene. Diabetes. 1997;46:204–208. doi: 10.2337/diab.46.2.204. [DOI] [PubMed] [Google Scholar]
- 15.Hills SA, Balkau B, Coppack SW, Dekker JM, Mari A, Natali A, Walker M, et al. The EGIR-RISC STUDY (The European group for the study of insulin resistance: relationship between insulin sensitivity and cardiovascular disease risk): I. Methodology and objectives Diabetologia. 2004;47:566–570. doi: 10.1007/s00125-004-1335-5. [DOI] [PubMed] [Google Scholar]
- 16.Mari A, Pacini G, Murphy E, Ludvik B, Nolan JJ. A model-based method for assessing insulin sensitivity from the oral glucose tolerance test. Diabetes Care. 2001;24:539–548. doi: 10.2337/diacare.24.3.539. [DOI] [PubMed] [Google Scholar]
- 17.Van Cauter E, Mestrez F, Sturis J, Polonsky KS. Estimation of insulin secretion rates from C-peptide levels. Comparison of individual and standard kinetic parameters for C-peptide clearance. Diabetes. 1992;41:368–377. doi: 10.2337/diab.41.3.368. [DOI] [PubMed] [Google Scholar]
- 18.Leow CC, Wang BE, Ross J, Chan SM, Zha J, Carano RA, Frantz G, et al. Prostate-specific Klf6 inactivation impairs anterior prostate branching morphogenesis through increased activation of the Shh pathway. J Biol Chem. 2009;284:21057–21065. doi: 10.1074/jbc.M109.001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dardevet D, Moore MC, Remond D, Everett-Grueter CA, Cherrington AD. Regulation of hepatic metabolism by enteral delivery of nutrients. Nutr Res Rev. 2006;19:161–173. doi: 10.1017/S0954422407315175. [DOI] [PubMed] [Google Scholar]
- 20.Bebernitz GR, Beaulieu V, Dale BA, Deacon R, Duttaroy A, Gao J, Grondine MS, et al. Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes. J Med Chem. 2009;52:6142–6152. doi: 10.1021/jm900839k. [DOI] [PubMed] [Google Scholar]
- 21.Agius L. Targeting hepatic glucokinase in type 2 diabetes: weighing the benefits and risks. Diabetes. 2009;58:18–20. doi: 10.2337/db08-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu KC, Chuang LM, Yoon C, Saad MF. Hepatic glucokinase promoter polymorphism is associated with hepatic insulin resistance in Asian Indians. BMC Genet. 2000;1:2. doi: 10.1186/1471-2156-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agius L. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem J. 2008;414:1–18. doi: 10.1042/BJ20080595. [DOI] [PubMed] [Google Scholar]
- 24.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 25.Gkretsi V, Apte U, Mars WM, Bowen WC, Luo JH, Yang Y, Yu YP, et al. Liver-specific ablation of integrin-linked kinase in mice results in abnormal histology, enhanced cell proliferation, and hepatomegaly. Hepatology. 2008;48:1932–1941. doi: 10.1002/hep.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyes A, Cardenas ML. All hexokinase isoenzymes coexist in rat hepatocytes. Biochem J. 1984;221:303–309. doi: 10.1042/bj2210303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci U S A. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim TH, Kim H, Park JM, Im SS, Bae JS, Kim MY, Yoon HG, et al. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J Biol Chem. 2009;284:15071–15083. doi: 10.1074/jbc.M109.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCullough AJ. Update on nonalcoholic fatty liver disease. J Clin Gastroenterol. 2002;34:255–262. doi: 10.1097/00004836-200203000-00013. [DOI] [PubMed] [Google Scholar]
- 30.Visser ME, Lammers NM, Nederveen AJ, van der Graaf M, Heerschap A, Ackermans MT, Sauerwein HP, et al. Hepatic steatosis does not cause insulin resistance in people with familial hypobetalipoproteinaemia. Diabetologia. 2011;54:2113–2121. doi: 10.1007/s00125-011-2157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Zhao Q, Wu K, Fan D. I148M variant of PNPLA3 confer increased risk for nonalcoholic fatty liver disease not only in European population, but also in Chinese population. Hepatology. 2011 doi: 10.1002/hep.24567. [DOI] [PubMed] [Google Scholar]
- 32.Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chalasani N, Guo X, Loomba R, Goodarzi MO, Haritunians T, Kwon S, Cui J, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology. 2010;139:1567–1576. 1576 e1561–1566. doi: 10.1053/j.gastro.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, et al. Genome-wide association analysis identifies variants associated with nonalcoholic Fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sotaniemi EA, Keinanen K, Lahtela JT, Arranto AJ, Kairaluoma M. Carbohydrate intolerance associated with reduced hepatic glucose phosphorylating and releasing enzyme activities and peripheral insulin resistance in alcoholics with liver cirrhosis. J Hepatol. 1985;1:277–290. doi: 10.1016/s0168-8278(85)80055-6. [DOI] [PubMed] [Google Scholar]
- 36.Lowes W, Walker M, Alberti KG, Agius L. Hexokinase isoenzymes in normal and cirrhotic human liver: suppression of glucokinase in cirrhosis. Biochim Biophys Acta. 1998;1379:134–142. doi: 10.1016/s0304-4165(97)00092-5. [DOI] [PubMed] [Google Scholar]
- 37.Davis EA, Cuesta-Munoz A, Raoul M, Buettger C, Sweet I, Moates M, Magnuson MA, et al. Mutants of glucokinase cause hypoglycaemia- and hyperglycaemia syndromes and their analysis illuminates fundamental quantitative concepts of glucose homeostasis. Diabetologia. 1999;42:1175–1186. doi: 10.1007/s001250051289. [DOI] [PubMed] [Google Scholar]
- 38.Massa O, Meschi F, Cuesta-Munoz A, Caumo A, Cerutti F, Toni S, Cherubini V, et al. High prevalence of glucokinase mutations in Italian children with MODY. Influence on glucose tolerance, first-phase insulin response, insulin sensitivity and BMI. Diabetes Study Group of the Italian Society of Paediatric Endocrinology and Diabetes (SIEDP) Diabetologia. 2001;44:898–905. doi: 10.1007/s001250100530. [DOI] [PubMed] [Google Scholar]
- 39.Miller SP, Anand GR, Karschnia EJ, Bell GI, LaPorte DC, Lange AJ. Characterization of glucokinase mutations associated with maturity-onset diabetes of the young type 2 (MODY-2): different glucokinase defects lead to a common phenotype. Diabetes. 1999;48:1645–1651. doi: 10.2337/diabetes.48.8.1645. [DOI] [PubMed] [Google Scholar]
- 40.Cuesta-Munoz AL, Tuomi T, Cobo-Vuilleumier N, Koskela H, Odili S, Stride A, Buettger C, et al. Clinical heterogeneity in monogenic diabetes caused by mutations in the glucokinase gene (GCK-MODY) Diabetes Care. 2010;33:290–292. doi: 10.2337/dc09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holian J, Qi W, Kelly DJ, Zhang Y, Mreich E, Pollock CA, Chen XM. Role of Kruppel-like factor 6 in transforming growth factor-beta1-induced epithelial-mesenchymal transition of proximal tubule cells. Am J Physiol Renal Physiol. 2008;295:F1388–1396. doi: 10.1152/ajprenal.00055.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farrelly D, Brown KS, Tieman A, Ren J, Lira SA, Hagan D, Gregg R, et al. Mice mutant for glucokinase regulatory protein exhibit decreased liver glucokinase: a sequestration mechanism in metabolic regulation. Proc Natl Acad Sci U S A. 1999;96:14511–14516. doi: 10.1073/pnas.96.25.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu CA, Fujimoto Y, Igawa K, Grimsby J, Grippo JF, Magnuson MA, Cherrington AD, et al. Rapid translocation of hepatic glucokinase in response to intraduodenal glucose infusion and changes in plasma glucose and insulin in conscious rats. Am J Physiol Gastrointest Liver Physiol. 2004;286:G627–634. doi: 10.1152/ajpgi.00218.2003. [DOI] [PubMed] [Google Scholar]
- 44.Grimsby J, Coffey JW, Dvorozniak MT, Magram J, Li G, Matschinsky FM, Shiota C, et al. Characterization of glucokinase regulatory protein-deficient mice. J Biol Chem. 2000;275:7826–7831. doi: 10.1074/jbc.275.11.7826. [DOI] [PubMed] [Google Scholar]
- 45.Postic C, Shiota M, Magnuson MA. Cell-specific roles of glucokinase in glucose homeostasis. Recent Prog Horm Res. 2001;56:195–217. doi: 10.1210/rp.56.1.195. [DOI] [PubMed] [Google Scholar]
- 46.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 47.Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparso T, Andersen G, Nielsen T, Burgdorf KS, Gjesing AP, Nielsen AL, Albrechtsen A, et al. The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 2008;51:70–75. doi: 10.1007/s00125-007-0865-z. [DOI] [PubMed] [Google Scholar]
- 49.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 50.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, Roos C, Tewhey R, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, Gloyn AL. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of DNA immunofluorescence was using 4′6-diamidino-2-phenylindole (DAPI, blue; 1800ms, red channel). An anti GCK antibody (H-88; Santa Cruz Biotechnology, Santa Cruz, CA) was detected with Texas Red. Independent and merged images are shown. Increased GCK protein expression was present in Klf6 expressing AML12 clones.
Co-transfection of a Klf6 expressing plasmid with a Gck promoter reporter shows a significant increase in reporter activity detected by Firefly Luciferase (A). Binding of KLF6 to the Gck promoter was confirmed by chromatin immunoprecipitation assay for two putative GC boxes within the mouse Gck promoter (B) (** = p<0.01).
Morphometric analyses, a software algorithm (Aperio Technologies) recognizing hepatocyte nuclei was optimized and applied to scanned images created from paraffin embedded tissues analysed by immunohistochemistry. Examples of the nuclear algorithm application for normal liver, as well as steatotic livers is shown in Figure 3. Negative nuclei are blue, while positive, moderately positive and strongly positive are yellow, orange and red respectively. The entire biopsy from representative cases has been captured in 8–14 fields and analysed in this way. As the background staining is not uniform across biopsies and antibodies used, the percentage of total nuclei being positive (rather than grades positive) have been compared. These data are presented in Supplementary Figure 4. Clear differences in expression are confirmed, as summarized in Figure 6 (Kruskal Wallis p<0.001 for each of KLF6, KLF6-SV1, GCK and GCKR across the three groups studied).