Abstract
Controlling and preventing aggregation is critical to the development of safe and effective antibody drug products. The studies presented here test the hypothesis that Protein A and Protein G inhibit the agitation-induced aggregation of IgG. The hypothesis is motivated by the enhanced conformational stability of proteins upon ligand binding and the specific binding affinity of Protein A and Protein G to the Fc region of IgG. The aggregation of mixed human IgG from pooled human plasma was induced by agitation alone or in the presence of: (i) Protein A, (ii) Protein G or (iii) a library of 24 peptides derived from the IgG-binding domain of Protein A. Aggregation was assessed by UV spectroscopy, SDS-PAGE, high performance size-exclusion chromatography (HP-SEC), dynamic light scattering (DLS) and fluorescence spectroscopy. Additional information on IgG-ligand interactions was obtained using differential scanning fluorimetry (DSF) and competitive binding studies. The results demonstrate that Protein A provides near-complete inhibition of agitation-induced aggregation, while Protein G and two peptides from the peptide library show partial inhibition. The findings indicate that the IgG Protein A binding site is involved in the agitation-induced aggregation of IgG, and suggest a dominant role of colloidal interactions.
Keywords: protein formulation, biologics, IgG, aggregation, Protein A, Protein G, peptide library
INTRODUCTION
Antibodies are the fastest growing sector of the biopharmaceutical industry with more than thirty antibody drug products currently on the market and hundreds more in development 1–5. Antibody-based drugs include monoclonal antibodies (mAb), peptibodies, antibody-drug conjugates, single-chain variable fragment antibodies (scFv) and bispecific monoclonal antibodies (bimAb). Like other proteins, antibodies are subject to degradation processes that can damage their complex structure. Arguably the most serious of these is aggregation, since the presence of aggregates can alter potency and increase the potential for life-threatening immunogenic side effects. Aggregation involves the homotypic association of protein molecules to form oligomers and higher order aggregates, and may occur preferentially through partially or fully unfolded forms. While several models for aggregation have been proposed, two of the important limiting cases are: (i) aggregation controlled by protein unfolding or (ii) aggregation controlled by protein-protein interactions 6. In the former limit, aggregation is driven by changes in protein conformation and so is referred to as conformational instability. In the latter case, aggregation is driven by the intermolecular associations that lead to colloid formation, and so is termed colloidal instability. Attempts to prevent the aggregation of therapeutic proteins typically address one or both of these limits. For example, excipients have been added in an attempt to preserve the native conformation and thus improve conformational stability7, while antibody engineering has been used to modify aggregation-prone “hot spots” and enhance colloidal stability8.
The rational design of stable antibody formulations requires an understanding of the mechanisms of aggregation and the contributions of conformational and colloidal instability. In the studies reported here, we evaluated the ability of Protein A and Protein G to prevent aggregation of mixed human IgG. It is well known that protein native conformation is stabilized by ligand-binding interactions9, 10, and both Protein A and Protein G bind to IgG with high affinity11. Thus, we hypothesized that both Protein A and Protein G would improve the conformational stability of IgG and inhibit its aggregation. In addition, since Protein A and Protein G bind to a solvent-exposed hydrophobic site at the interface between the CH2 and CH3 domains12, we hypothesized that they could also improve the colloidal stability of IgG by blocking this site. To test these hypotheses, a solution of mixed human IgGs was subjected to agitation alone or in the presence of Protein A, Protein G or a library of peptides from the IgG-binding domain of Protein A. Protein A provided near-complete inhibition of aggregation, while Protein G and two of the peptides provided partial inhibition. The results indicate that the IgG Protein A binding site is involved in the agitation-induced aggregation of IgG, and further suggest a dominant role of colloidal interactions.
MATERIALS AND METHODS
Materials
A lyophilized mixed IgG from human serum (I4506, reagent grade, Sigma-Aldrich Corp., St. Louis, MO) was used as a model IgG. Recombinant Protein A (MW 44.6 kDa, 21184 and Protein G (MW 21.6 kDa, 21193) were purchased from Thermo Fisher Scientific (Rockford, IL). Protein A porous glass resin (ProSep Ultra Plus affinity chromatography media, Cat. No. 175118822) was purchased from Millipore (Billerica, MA). A library of 24 peptides derived from the IgG-binding domain of Protein A was synthesized by GenScript (Piscataway, NJ). Peptide sequences are provided in Table S1; peptides P1 (Ac-122NKFNKEQQNAFYEILHL138-NH2), P2 (Ac-143EEQRNGFIQSLKDD156-NH2) and P3 (Ac-123KFNKEQQNAFYEILHL138-NH2) will receive particular attention in the sections below. All peptides were modified by N-terminal acetylation and C-terminal amidation by the supplier, and were used in this form. All other materials were of the highest commercial grade and used as received.
Sample preparation and agitation
Stock solutions of IgG (1.92 mg/mL in phosphate buffered saline, PBS), Protein A (1 mg/mL in PBS) and Protein G (2.42 mg/mL in PBS) were dialyzed against PBS using Slide-A-Lyzer dialysis cassettes (Thermo Fisher Scientific) overnight at room temperature to remove unknown buffers, salts or other additives. Solutions were filtered before and after dialysis using 0.45 µm Nylaflow® membrane disc filters (Pall Life Sciences, East Hills, NY) to remove any aggregates or suspended solids. Samples prepared from the stock solutions (Table 1) contained IgG (1 mg/mL) and one or more additives (Protein A, Protein G or peptides) in a molar ratio of 1:4 in 1× PBS, pH 7.4. On preparation, the solutions were clear with no visible particulates. Freshly prepared samples were first incubated at room temperature for 2 h. Aliquots (1 mL) were placed in 2 mL microcentrifuge tubes (10011-744, VWR International, West Chester, PA) and shaken vertically on a VWR Signature™ Multi-Tube Vortexer (14005-826, VWR International) at 2500 rpm for 48 h at room temperature to induce protein aggregation. Solution samples held at room temperature for 48 h without agitation served as unstressed controls. Samples containing IgG alone and subjected to agitation in 2 mL tubes (without headspace) were used as controls to assess the effect of agitation. The temperature of the agitated samples was ~40 °C after 48 h agitation. Samples containing IgG alone and held at 60 °C for 48 h were used as an additional control. Following the agitation period, samples were analyzed by the methods described below to assess aggregate formation.
Table 1.
Aggregation index (A.I.), Z-average diameter, soluble protein recovery and precipitate for IgG samples
| Sample a | Agitation b | A.I. c, d | Z-average diameter (nm) d |
Soluble protein recovered (%) d, e |
Precipitate (%) d, f |
|---|---|---|---|---|---|
| IgG RT | No | 0.7 ± 0.1 | 48 ± 3 | N/A | N/A |
| IgG | Yes | 118 ± 5 | 2900 ± 200 | 41 ± 2 | 58 ± 2 |
| IgG + Protein A | Yes | 3 ± 1 | 31 ± 1 | 99.8 ± 0.8 | 0.2 ± 0.8 |
| IgG + Protein G | Yes | 87 ± 5 | 1500 ± 50 | 63 ± 2 | 37 ± 2 |
| IgG + P1 | Yes | 71 ± 3 | 970 ± 60 | 61 ± 4 | 39 ± 4 |
| IgG + P2 | Yes | 120 ± 8 | 2500 ± 100 | 42 ± 2 | 58 ± 1 |
| IgG + P3 | Yes | 74 ± 6 | 780 ± 50 | 54 ± 6 | 46 ± 6 |
| IgG + P1 + P2 | Yes | 65 ± 5 | 1420 ± 90 | 63 ± 3 | 37 ± 3 |
All samples contain 1 mg/mL IgG; molar ratio of IgG to each additive is 1:4.
Samples were agitated at 2500 rpm for 48h at room temperature. After agitation, the sample temperature is 40 ± 1 °C.
Calculated as OD350/(OD280–OD350)×100; see text for details.
n = 7, ± SEM
Recovered after removing insoluble protein; see text for details.
Percentage of precipitate was determined by subtracting the percentage of soluble protein recovered; see text for details.
UV spectroscopy
To monitor aggregation, samples were diluted four times in PBS and UV spectra were recorded between 230 and 400 nm using an Agilent 8453 UV-Vis spectrophotometer. Dilution was required because saturation was observed in undiluted samples containing aggregates; spectra were recorded immediately after dilution. Increased UV absorbance at 350 nm has been associated with the presence of IgG aggregates in solution, and an “aggregation index” based on UV absorbance has been reported 13, 14. Here, the aggregation index (A.I.) was determined in triplicate from the UV spectra using the equation A.I. = OD350/(OD280-OD350)×100, as reported by Katayama et al. 13.
To evaluate the relative IgG-binding affinities of Protein A, P1 and P2, 1 mL samples containing IgG (1 mg/mL) and one or more additives (Protein A or peptides) in a molar ratio of 1:4 (IgG:additive) were incubated with 90 µL Protein A resin for 2 h. Following a brief centrifugation (MiniFuge and MiniArray Microcentrifuges, VWR), UV absorbance spectra of the supernatants were recorded. Any decrease in absorbance was attributed to the removal of IgG from solution due to binding to the Protein A resin, indicating a higher binding affinity for IgG with resin-bound Protein A than with the ligand in solution.
SDS-PAGE
The formation of large aggregates was evaluated using SDS-PAGE. The studies were designed to allow qualitative detection of the presence of large and/or insoluble aggregates as bands at the top of the stacking gel. A 150 µL aliquot of each sample was spun down at 14,000 rpm for 10 min and 140 µL of the supernatant was removed. The remaining protein solution (~10 µL) and precipitate were then re-suspended under reducing (β-mercaptoethanol) and nonreducing conditions and applied to a 4–15% Tris-HCl gradient gel. Because the amount of precipitate varied from sample to sample, the total protein loaded onto each lane was not identical and was not measured. A broad range molecular weight marker (Bio-Rad Laboratories, Hercules, CA) was used.
High performance size exclusion chromatography (HP-SEC)
HP-SEC was used to quantify soluble protein recovery using an Agilent 1200 system with UV detection at 280 nm (Agilent Technologies, Palo Alto, CA). IgG samples were centrifuged (Microfuge 22R refrigerated microcentrifuge, VWR) at 14,000 rpm for 10 min to remove insoluble aggregates, hereafter referred as “precipitate”. The supernatants were analyzed by HP-SEC as previously described 7, 14. Briefly, 50 µL aliquots were injected onto a Tosoh TSKgel G3000SWxl column (Tosoh Bioscience LLC, King of Prussia, PA; 3000 mm × 7.8 mm) and separated at a flow rate of 0.5 mL/min. The mobile phase contained 50 mM potassium phosphate and 100 mM K2SO4 at pH 7.1. The relative percentage of soluble protein recovered was calculated in triplicate by dividing the total chromatographic peak area at 280 nm for the stressed samples by that of the respective unstressed control. The percentage of precipitate in the sample was calculated by difference.
Dynamic light scattering (DLS)
DLS was carried out using a Zetasizer Nano ZS90 (Malvern Instruments, Ltd., Westborough MA). Undiluted samples (1 mL) were analyzed in polystryrene cuvettes (VWR) with a path length of 10 mm at 25 °C. Each sample was recorded 3 times with 7 subruns of 10 s. The Z-average diameter was calculated using the Dispersion Technology Software (Ver. 4.20, Malvern).
Fluorescence spectroscopy
Bis-ANS is a fluorescent dye that has been used to monitor protein folding and aggregation. Since Bis-ANS shows greater fluorescence in hydrophobic environments, an increase in fluorescence intensity is associated with an increase in protein aggregation. Fluorescence measurements were obtained using a Fluoromax-3 Fluorometer (Horiba Jobin Yvon, Inc., Edison, NJ) as described previously, with some modifications14. Briefly, IgG samples were diluted 10 times in PBS. Dilution was employed to avoid saturation in fluorescence readings and spectra were recorded immediately after dilution. A 20 µL aliquot of a 100 µM Bis-ANS solution (2 mM Bis-ANS stock solution in PBS) was added to 2 mL of the diluted samples. The samples were excited at 385 nm (slit width, 3 nm), and the emission was recorded from 410 to 600 nm (slit width 3 nm) using an average of 10 scans for each spectrum. All spectra were corrected by subtracting the emission spectrum of the buffer dye solution.
To confirm the binding of P1 and P2 to IgG, intrinsic fluorescence spectra of undiluted IgG samples were recorded as previously described 15, 16. Samples containing IgG and either Protein A, P1 or P2 were prepared at the concentrations used in the aggregation studies. The samples were subjected to fluorescence excitation at 280 nm (slit width, 2 nm) and emission spectra recorded between 310 and 420 nm (slit width, 2 nm) using the Fluoromax-3 Fluorometer. Intensities for the mixtures were corrected by subtracting the fluorescence emission intensity of Protein A, P1 or P2 alone.
Differential scanning fluorimetry (DSF)
DSF was performed to monitor thermally-induced protein unfolding as previously reported, with some modifications 17–19. Freshly prepared samples containing IgG (0.1 mg/mL) and different additives (Protein A, Protein G or derived peptides) in a molar ratio of 1:4 in PBS were incubated at room temperature for 2 h. A 40 uL aliquot of the protein solutions with 1× final concentration of SYPRO Orange (Invitrogen Inc., Calsbad, CA) was then added to each well of a 96-well microplate (MLL960, Bio-Rad Laboratories). The samples were heated from 25 to 95 °C at a rate of 1 °C/min using a CFX96 Real-Time PCR system (Bio-Rad Laboratories) and fluorescence signals were recorded. Fluorescence intensity was plotted as a function of temperature and normalized. The melting temperature, Tm, at which the concentrations of folded and unfolded protein are equal, was determined by fitting the normalized curves to the Boltzmann equation as described previously 19. Statistical analyses were conducted using Student’s paired t-test (KaleidaGraph 4.1, Synergy Software, Reading, PA).
RESULTS
Results are presented first for Protein A and Protein G, then for the peptides derived from the IgG-binding domain of Protein A. Figures and tables designated “S” are presented in on-line supplemental information and are available free of charge via the internet at http://pubs.acs.org.
Effects of Protein A and Protein G on IgG aggregation
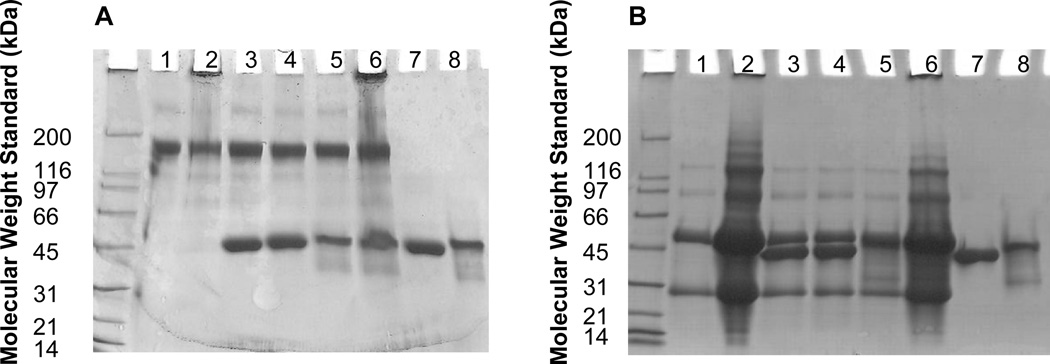
Initial studies evaluated the ability of Protein A and Protein G to inhibit the agitation-induced aggregation of IgG. SDS-PAGE was used to qualitatively detect large aggregates. IgG samples agitated alone (IgG, Table 1) and in the presence of Protein G (IgG + Protein G, Table 1) were opalescent and showed precipitates on visual examination. A band of high molecular weight was noted at the top of the stacking gel under non-reducing conditions for these samples (Figure 1A, Lanes 2 and 6). The bands were also present under reducing conditions, though with lower intensity (Figure 1B, Lanes 2 and 6). These results suggest that large and/or insoluble aggregates had been formed. In contrast, high molecular weight bands were not observed on SDS-PAGE for the agitated sample containing IgG and Protein A (IgG + Protein A, Table 1), nor were they observed in unstressed controls (Figure 1, Lanes 1, 3, 4 and 5). These results suggest that Protein A inhibits the formation of large insoluble IgG aggregates on agitation. Samples containing Protein A alone or Protein G alone did not show aggregate bands after storage at RT for 48 h (Figure 1, Lanes 7 and 8) or following agitation at 2500 rpm for 48 h (data not shown).
Figure 1.
SDS-PAGE of IgG samples containing Protein A or Protein G under non-reducing conditions (A) and reducing conditions (B). Lane 1: IgG at RT for 48h; Lane 2: IgG subjected to agitation at 2500 rpm for 48 h; Lane 3: IgG + Protein A at RT for 48h h; Lane 4: IgG subjected to agitation with Protein A at 2500 rpm for 48 h; Lane 5: IgG + Protein G at RT for 48h h; Lane 6: IgG subjected to agitation with Protein G at 2500 rpm for 48 h; Lane 7: Protein A at RT for 48h; Lane 8: Protein G at RT for 48h.
UV spectra were also recorded to assess the formation of insoluble IgG aggregates (Figure S1). Stressed samples containing IgG alone and IgG + Protein G showed altered UV spectra with significantly increased absorbance at 350 nm (Figure S1, A and C), consistent with greater light scattering intensity as a result of greater particle size. These samples also had increased values of the aggregation index (A.I.) (Table 1), suggesting greater insoluble aggregate content. After centrifugation at 14,000 rpm for 10 min, the supernatants from these samples had UV spectra and A.I. values similar to the unstressed control (data not shown), indicating that most of the aggregates were removed by centrifugation. In contrast, the agitated IgG + Protein A samples had UV spectra (Figure S1B) and A.I. values (Table 1) similar to those of unstressed controls. Neither opalescence nor visible precipitates were observed in samples that contained IgG alone and were agitated without head space or held at 60 °C for 48 h. These samples also had low A.I. values (4.1 ± 0.1; see Figure S1G) similar to the unstressed controls (Table 1), suggesting that aggregation is caused by agitation.
DLS was used to estimate the size of the large aggregates. Agitation significantly increased the Z-average diameter for the IgG and IgG + Protein G samples, corresponding to the formation of large aggregates (Table 1). However, the Z-average diameter of the agitated IgG + Protein A samples was comparable to that of unstressed controls (Table 1), consistent with protection from aggregation.
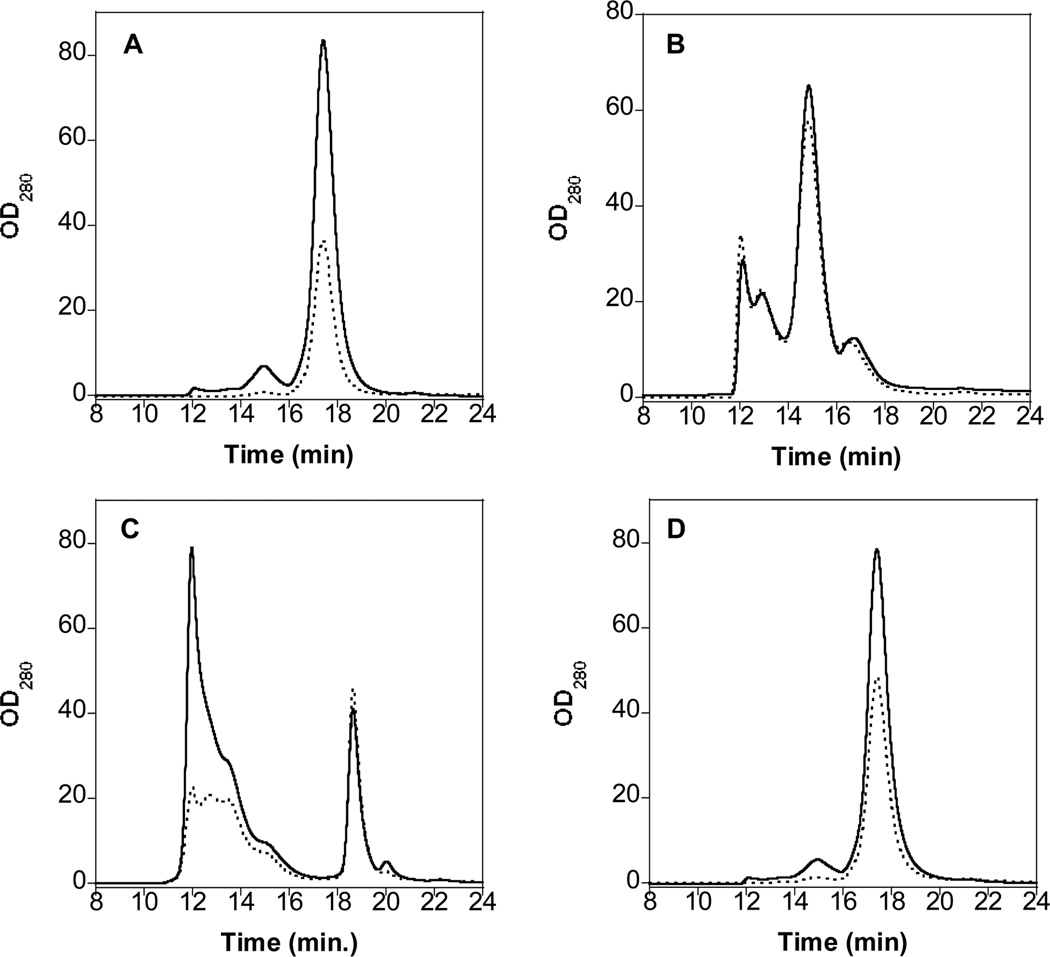
HP-SEC was used to determine the percentage of soluble protein recovered after centrifugation to remove insoluble aggregates. On HP-SEC, Protein A alone and Protein G alone eluted at ~17 and ~19 min, respectively (Figure S2). HP-SEC chromatograms of IgG alone showed a monomer peak at ~18 min, as well as a smaller peak at ~15 min attributed to the dimmer (Figure 2A). Both peaks decreased following agitation, consistent with the loss of soluble protein. The absence of peaks eluting prior to ~ 15 min suggests that soluble IgG multimers are not formed by agitation or are not detected by HP-SEC. On the basis of HP-SEC peak area, the recovery of soluble protein in samples containing IgG alone was 42% following agitation (Table 1). Samples containing IgG + Protein A showed four HP-SEC peaks (Figure 2B), all eluting earlier than the IgG monomer (Figure 2A). These peaks are attributed to multimeric complexes containing IgG and/or Protein A. The peak profile was essentially unaltered following agitation (Figure 2B) and recovery of soluble protein was near complete (Table 1), indicating that insoluble aggregates have not been formed and the complexes are unaltered by agitation. In contrast, the IgG + Protein G sample showed changes in the HP-SEC chromatogram following agitation (Figure 2C). Prior to agitation, the chromatogram showed a peak at ~19 min attributed to Protein G and a broad peak at ~12 min that can be attributed to Protein G-IgG complexes. Following agitation, the area of the ~12 min peak decreased markedly (Figure 2C), suggesting loss of soluble protein through the aggregation and precipitation of the IgG-Protein G complexes. On the basis of HP-SEC peak area, approximately 63% of soluble protein was recovered for the IgG + Protein G sample (Table 1).
Figure 2.
HP-SEC chromatograms with UV detection at 280 nm of IgG samples held at room temperature for 48 h (solid line) and subjected to agitation at 2500 rpm for 48 h (dotted line). A: IgG; B: IgG + Protein A; C: IgG + Protein G; D: IgG + P1.
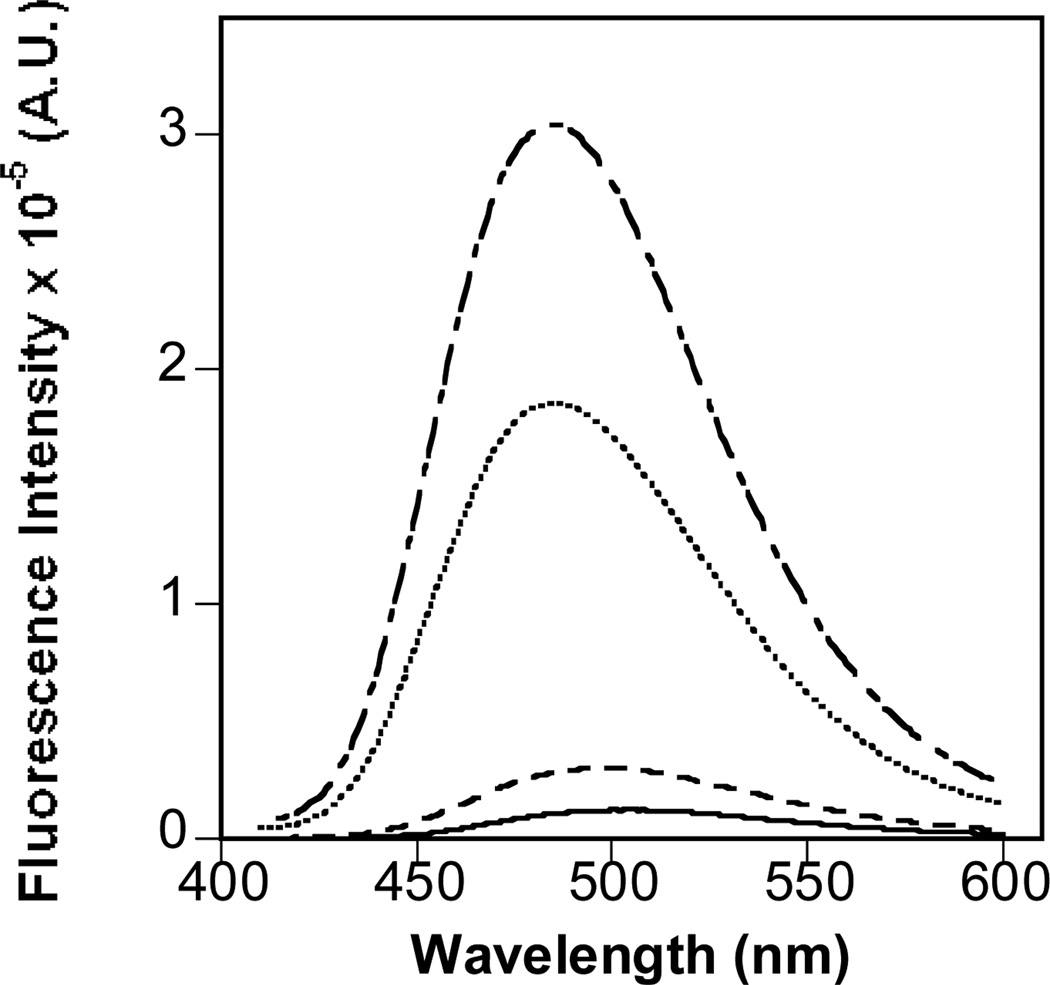
Fluorescence spectroscopy of Bis-ANS was used to monitor protein folding and aggregation. Fluorescence spectra were similar for unstressed samples (IgG, IgG + Protein A, IgG + Protein G) (data not shown). In the agitated IgG and IgG + Protein G samples, fluorescence emission spectra of Bis-ANS showed a greater intensity and a blue shift of ~20 nm relative to unstressed IgG sample, suggesting increased exposure of hydrophobic surfaces as a result of aggregation and/or unfolding (Figure 3, Figure S3). In contrast, the agitated IgG + Protein A sample had a fluorescence intensity comparable to the unstressed control (Figure 3), consistent with low exposure of hydrophobic surfaces and limited aggregate formation.
Figure 3.
Fluorescence emission spectra of IgG samples in the presence of Bis-ANS at room temperature for 48 h (solid line) or subjected to agitation at 2500 rpm for 48 h: IgG (dash-dotted line), IgG + Protein A (dashed line), IgG + Protein G (dotted line).
Together, the data show that Protein A and Protein G have different effects on the agitation-induced aggregation of IgG. As assessed by SDS-PAGE, UV spectroscopy, DLS, HPSEC and fluorescence spectroscopy, Protein A effectively inhibits the agitation-induced formation of insoluble IgG aggregates, while Protein G shows only partial protection. Protein A and Protein G bind to overlapping sites near the interface between the CH2 and CH3 domains of IgG-Fc, with similar high affinity11, 12. This suggests that the Protein A-binding site of IgG may be involved in the agitation-induced aggregation of IgG. The interaction of human IgG-Fc with Protein A has been mapped in detail (PDBid: 1FC2)20 and two α-helices from Protein A have been shown to be essential for IgG binding. In the binding interaction, eleven residues from fragment B of Protein A interact with nine residues from the CH2 and CH3 domains of IgG-Fc (Figure S4). Accordingly, to test the hypothesis that the Protein A-binding site is critical in preventing IgG aggregation, two peptides spanning the two α-helices of Protein A that are essential for IgG binding were synthesized and tested for their ability to inhibit aggregation using the methods described above. The peptides are termed P1 (Ac-122NKFNKEQQNAFYEILHL138-NH2) and P2 (Ac-143EEQRNGFIQSLKDD156-NH2).
Effect of Protein-A-derived peptides on IgG aggregation
Following agitation at 2500 rpm for 48h, IgG samples containing P1 (IgG + P1), P2 (IgG + P2) or both P1 and P2 (IgG + P1 + P2) all showed opalescence or visible precipitates. On SDS-PAGE, a band of high molecular weight aggregates was noted at the top of the stacking gel under non-reducing conditions for each of these samples (Figure S5, Lanes 4, 5, and 6), which remained under reducing conditions. Following agitation, the UV spectra of both the IgG + P1 and IgG + P2 samples showed increased absorbance at 350 nm (Figure S1, D and E). The UV spectrum of the IgG + P1 + P2 sample was similar to that of the IgG + P1 sample (data not shown). The agitated IgG + P2 sample had A.I. values similar to that of agitated IgG alone, while agitated IgG + P1 and IgG + P1 + P2 samples had much lower A.I. values (Table 1). The Z-average diameter of the agitated IgG + P1 and IgG + P1 + P2 samples was greater than that of unstressed IgG, corresponding to the formation of large aggregates (Table 1). However, Z-average diameters following agitation were less for IgG + P1 than for IgG and IgG + P2 samples (Table 1), indicating partial protection from aggregation by P1.
On HP-SEC, the chromatogram for the agitated IgG + P2 sample (data not shown) was similar to that of agitated IgG (Figure 2A), with ~ 40% soluble protein recovery. The agitated IgG + P1 sample showed greater soluble protein recovery (~ 60%) and less precipitate than the agitated IgG and IgG + P2 samples (Table 1, Figure 2D). The agitated IgG + P1 + P2 had soluble protein recovery (Table 1) and an HP-SEC chromatogram (data not shown) similar to that of the agitated IgG + P1 sample. In the presence of Bis-ANS, all of the agitated IgG + P1, IgG + P2 and IgG + P1 + P2 samples displayed increased fluorescence emission intensity, suggesting increased exposure of hydrophobic surfaces as a result of aggregate formation (Figure S3).
Together, these results indicate that P1 provides partial protection from IgG aggregation. The degree of protection is less than that afforded by Protein A and comparable to that provided by Protein G. P2 alone provided little or no protection by any metric, and P1+P2 showed protection comparable to P1 alone. The results support the hypothesis that Protein A inhibits aggregation by blocking an aggregation-prone site on IgG, and further suggest that the P1 sequence is directly involved in this interaction. The reduced effectiveness of P1 relative to Protein A may be due to different binding affinities, differences in conformational stability upon ligand binding, or both. To test these possibilities, the binding affinity of P1 and P2 to IgG was assessed using fluorescence spectroscopy, DSF, and competitive binding assays with UV analysis.
Fluorescence quenching was observed following ligand binding, confirming that both P1 and P2 bind to IgG. The addition of Protein A to a solution of IgG caused the intrinsic IgG fluorescence to be quenched (Figure S6), indicating that Protein A binds to IgG. Similarly, P1 and P2 also quenched IgG fluorescence, albeit to a lesser extent than Protein A (Figure S6). This result confirms the interaction between the peptides and IgG.
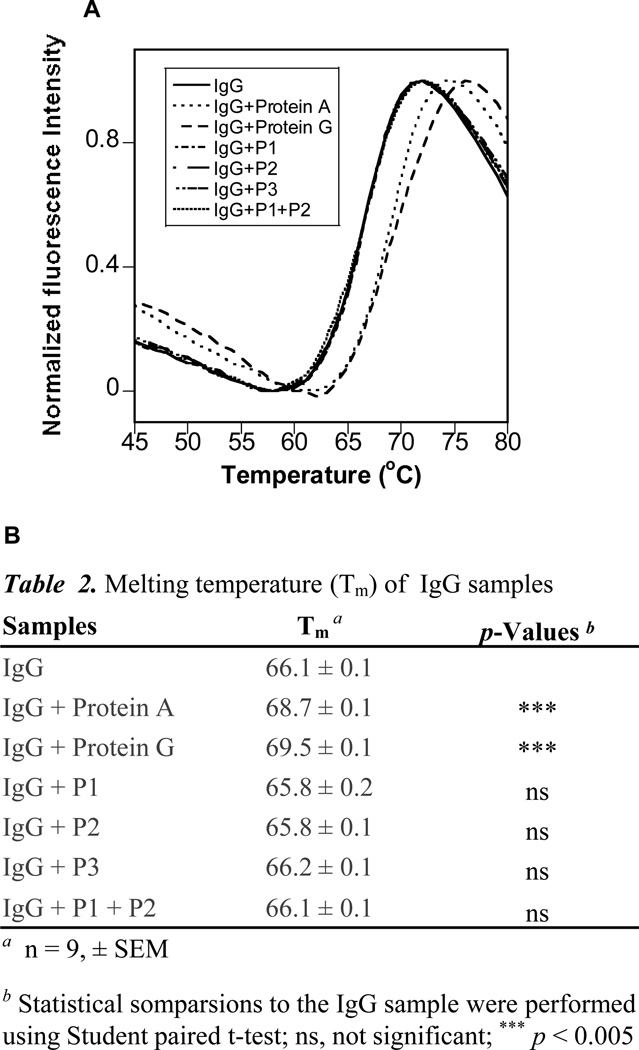
Tm was measured by DSF to assess the thermal stability of IgG in the presence of the ligands. Ligand binding generally improves protein thermal stability and results in an increased Tm 21, 22. DSF unfolding curves for both IgG + Protein A and IgG + Protein G samples shifted to higher temperature relative to IgG alone (Figure 4A), indicating higher Tm and enhanced thermal stability. The difference in Tm (~3 °C) was small but significant (p < 0.005) (Figure 4B). Interestingly, Protein G showed a slightly higher Tm (~0.8 °C) than Protein A, though it inhibited IgG aggregation less than Protein A. Neither P1 nor P2 caused a shift in Tm, indicating that these peptides do not affect IgG thermal stability, although P1 partially protected IgG from aggregation.
Figure 4.
DSF profiles (A) and Tm values (B) for IgG samples with and without Protein A, Protein A or peptides P1, P2 and P3.
Competitive binding was assessed using a Protein A porous glass resin and UV analysis of the supernatant. Adding the Protein A beads to an IgG solution removed IgG from solution due to the IgG-Protein A interaction, as indicated by a loss of UV absorbance in the 250–300 nm range (Figure S7A). Protein A beads added to an IgG + Protein A solution only partially removed IgG from solution, suggesting that Protein A in solution competed effectively with Protein A immobilized on the beads (Figure S7A). However, in the IgG + P1 + P2 sample, removal of IgG from solution by Protein A beads was nearly complete, indicating that P1 and P2 did not compete effectively with immobilized Protein A for binding to IgG (Figure S7B). These results indicate that P1 and P2 have weaker binding affinities for IgG than Protein A.
Finally, in an attempt to further localize the IgG aggregation-prone domain indicated by P1, a library of shorter peptides was synthesized (Table S1) and screened to for their ability to prevent aggregation. In this screen, aggregation was assessed by UV spectroscopy only. The results demonstrated that only P3 prevented aggregation as effectively as P1 (Table 1, Figure S8). The sequence of P3 is identical to that of P1 except that one amino acid (N122) has been truncated (Table S1). Thus, none of the peptides in this limited library allows the aggregation-prone region to be localized more narrowly.
DISCUSSION
In these studies, we tested the hypothesis that protein-ligand interactions prevent IgG aggregation by enhancing conformational and/or colloidal stability. Our initial studies focused on the effects of intact Protein A and Protein G on IgG aggregation. Protein A and Protein G bind to with high affinity to overlapping but not identical regions near the interface between the CH2 and CH3 domains11, 12, resulting in an increase in Tm (Figure 4). The binding site has been mapped12, 20 and is presented in several Protein Data Bank entries (e.g., 1FC220, 1FCC23). Interestingly, Protein A provided near-complete inhibition of IgG aggregation while Protein G provided only partial protection. These results suggested that the inhibition of IgG aggregation is not only due to improved conformational stability as a result of ligand binding, but also due to enhanced colloidal stability conferred by blocking an aggregation-prone site on IgG. The results further suggested that the aggregation-prone site is blocked more effectively by Protein A than by Protein G.
Based on these results, we hypothesized that the Protein A-binding site of IgG is critical to aggregation. To test this hypothesis, we examined the ability of peptides derived from the IgG-binding domain of Protein A to inhibit aggregation. P1 is a 17-amino acid peptide that spans one of the two α-helices essential for IgG-binding (Figure S4). P1 showed inhibition of IgG aggregation comparable to that of Protein G (Table 1), though it did not increase IgG thermal stability (Figure 4). This provided additional support for the idea that the Protein-A binding site is involved in aggregation, and further suggested that IgG residues that interact with P1 play a key role. There are several possible explanations for the observation that P1 inhibits aggregation to a lesser extent than Protein A: (i) conformational instability contributes to aggregation, and P1 does not stabilize the IgG structure to the same extent that Protein A does, as indicated by Tm (Figure 4); (ii) P1 has a weaker binding affinity for IgG than Protein A (Figure S7); and (iii) P1 partially blocks the aggregation-prone site, while Protein A covers the site more completely. P2 is a 14-amino acid peptide derived from the IgG-binding domain of Protein A that spans the other α-helix essential for IgG-binding (Figure S4). P2 did not prevent IgG aggregation, suggesting that P2 fails to block the aggregation-prone site and/or has a lower binding affinity for IgG than Protein A. Combining P1 and P2 did not provide greater inhibition of IgG aggregation than P1 alone, suggesting that P1 and P2 do not inhibit aggregation cooperatively. Screening a library of peptides identified only one new peptide (P3) that inhibited aggregation to an extent comparable to P1. P3 was similar to P1 in its sequence and inhibition of IgG aggregation. That P1 provides greater inhibition of aggregation than P2 may be due to the involvement of a greater number of P1 amino acids in binding to IgG. The binding of Protein A to Fc involves nine residues in the interface between the CH2 and CH3 domains of IgG (M252, I253, S254, L309, H310, Q311, N434, H435, Y436)20. Hydrophobic interactions were reported to be the dominant contribution to binding, though hydrogen bonding interactions were also indicated20. Peptide P1, derived from Protein A, contains seven amino acids that participate in this binding interaction (F124, Q128, Q129, N130, F132, Y133, L136), while P2 contains only four binding residues (N147, I150, Q151, K154) (Figure S4)20.
The involvement of the Protein A binding site in IgG aggregation is consistent with previous reports. Rispens et al. observed that a peptic fragment of intravenous immunoglobulin (IVIG), termed pFc’, blocks aggregation in this mixed IgG from pooled human plasma 24. The fragment is “almost equal to a CH3 dimer” and includes portions of the CH2 domain; therefore, it also contains the hinge region between the CH2 and CH3 domains and the Protein A binding site. The authors demonstrated that the pFc’ fragment competes with intact IgG for binding, limits Fc-Fc interactions and thus inhibits aggregation. Chennamsetty et al. predicted aggregation-prone sites on two therapeutic IgG antibodies using molecular dynamics simulations, and then modified these sites to less hydrophobic residues using site-directed mutagenesis to inhibit aggregation 8. The Protein A binding site was among the sites identified; two single point mutations (I253K and L309K) at the Protein A binding site improved stability and diminished aggregation, though mutations at other sites were also effective. DeLano et al. identified the hinge region between the CH2 and CH3 domains as a “consensus” binding site, at which at least four naturally occurring proteins bind to IgG, including Protein A and Protein G12. They stated that the site coincides with the Protein A binding site and developed peptides that competed effectively with Protein A for binding. The authors also identified other regions on the IgG surface that are solvent exposed and have low polarity, and suggested that the consensus site is preferred for binding due to its conformational adaptability.
The results presented here suggest that IgG aggregation involves this consensus binding site. While there are other solvent-exposed hydrophobic domains in IgG, this site appears to be critical for aggregation. If this were not the case, aggregation could have occurred through other aggregation-prone sites and Protein A would not have prevented aggregation so effectively. That Protein G is less effective in preventing aggregation than Protein A, despite the proximity of their binding sites on IgG, further supports the idea that the site responsible for IgG aggregation is localized. As noted above, the limited peptide library screened here does not allow the residues in the aggregation-prone site to be mapped definitively. Screening a broader library would provide better definition of the aggregation-prone site, facilitating site-directed mutagenesis or the development of designed excipients (e.g., blocking peptides, artificial chaperones) that inhibit aggregation. The latter approach may be particularly important when mutagenesis is undesirable or impossible (e.g., in IVIG). Whether the inhibition of aggregation under the stressed conditions used here is predictive of results during long-term storage is not known, and warrants further study.
Supplementary Material
ACKNOWLEDGEMENTS
The authors gratefully acknowledge financial support through NIH RO1 GM085293 (PI: E. Topp) and from the College of Pharmacy at Purdue University.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Figures and tables designated “S” within the manuscript are presented in on-line supplemental information. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.ElBakri A, Nelson PN, Abu Odeh RO. The state of antibody therapy. Hum. Immunol. 2010;71(12):1243–1250. doi: 10.1016/j.humimm.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Pavlou AK, Belsey MJ. The therapeutic antibodies market to 2008. Eur. J. Pharm. Biopharm. 2005;59(3):389–396. doi: 10.1016/j.ejpb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Reichert JM. Monoclonal antibodies as innovative therapeutics. Curr. Pharm. Biotechno. 2008;9:423–430. doi: 10.2174/138920108786786358. [DOI] [PubMed] [Google Scholar]
- 4.Huang CJ, Lowe AJ, Batt CA. Recombinant immunotherapeutics: Current state and perspectives regarding the feasibility and market. Appl. Microbiol. Biot. 2010;87:401–410. doi: 10.1007/s00253-010-2590-7. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Zhu Z. Research and development of next generation of antibody-based therapeutics. Acta Pharm. Sinic. 2010;31:1198–1207. doi: 10.1038/aps.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: Mechanism and drving forces in nonnative protein aggregation. Pharm. Res. 2003;20(9):1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 7.Serno T, Carpenter JF, Randolph TW, Winter G. Inhibition of agitation-induced aggregation of an IgG-antibody by hydroxypropyl-beta-cyclodextrin. J. Pharm. Sci. 2010;99(3):1193–1206. doi: 10.1002/jps.21931. [DOI] [PubMed] [Google Scholar]
- 8.Chennamsetty N, Voynov V, Kayser V, Helk F, Trout BL. Design of therapeutic proteins with enhanced stability. P. Natl. Acad. Sci. USA. 2009;29:11937–11942. doi: 10.1073/pnas.0904191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teilum K, Olsen JG, Kragelund BB. Protein stability, flexibility and function. Biochim. Biophys. Acta. 2011;1814:969–976. doi: 10.1016/j.bbapap.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Ruiz JM. Ligand effects on protein thermodynamic stability. Biophys. Chem. 2007;126:43–49. doi: 10.1016/j.bpc.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Roque ACA, Silva CSO, Taipa MA. Affinity-based methodologies and ligands for antibody purification: Advances and perspectives. J. Chromatogr. A. 2007;1160:44–55. doi: 10.1016/j.chroma.2007.05.109. [DOI] [PubMed] [Google Scholar]
- 12.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287:1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 13.Katayama DS, Nayar R, Chou DK, Campos J, Cooper J, Vander Velde DG, Villarete L, Liu CP, Manning MC. Solution behavior of a novel type 1 interferon, interferon-tau. J. Pharm. Sci. 2005;94(12):2703–2715. doi: 10.1002/jps.20461. [DOI] [PubMed] [Google Scholar]
- 14.Hawe A, Kasper JC, Friess W, Jiskoot W. Structural properties of monoclonal antibody aggregates induced by freeze-thawing and thermal stress. Eur. J. Pharm. Sci. 2009;38(2):79–87. doi: 10.1016/j.ejps.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Wu X, Padovani D, Schubert HL, Gravel RA. Ligand-binding by catalytically inactive mutants of the cbIB complementation group defective in human ATP:cob(I)alamin adenosyltransferase. Mol. Genet. Metab. 2009;98(3):278–284. doi: 10.1016/j.ymgme.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury S, Banerjee R. Role of the dimethylbenzimidazole tail in the reaction catalyzed by coenzyme B12-dependent methylmalonyl-CoA mutase. Biochem. 1999;38(46):15287–15294. doi: 10.1021/bi9914762. [DOI] [PubMed] [Google Scholar]
- 17.He F, Woods CE, Litowski JR, Roschen LA, Gadgil HS, Razinkov VI, Kerwin BA. Effect of sugar molecules on the viscosity of high concentration monoclonal antibody solutions. Pharm. Res. 2011;28(7):1552–1560. doi: 10.1007/s11095-011-0388-7. [DOI] [PubMed] [Google Scholar]
- 18.Froese DS, Healy S, McDonald M, Kochan G, Oppermann U, Niesen FH, Gravel RA. Thermolability of mutant MMACHC protein in the vitamin B12-responsive cblC disorder. Mol. Genet. Metab. 2010;100(1):29–36. doi: 10.1016/j.ymgme.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 20.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of Protein A from Staphylococcus aureus at 2.9- and 2.8 angstrom resolution. Biochem. 1981;20(9):2361–2370. [PubMed] [Google Scholar]
- 21.Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic stability of carbonic anhydrase: Measurements of binding affinity and stoichiometry using ThermoFluor. Biochem. 2005;44(13):5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 22.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, Hui R, Marsden BD, Nordlund P, Sundstrom M, Weigelt J, Edwards AM. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. P. Natl. Acad. Sci. USA. 2006;103(43):15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer-Eriksson AE, Kleywegt GJ, Uhlen M, Jones TA. Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure. 1995;3:265–278. doi: 10.1016/s0969-2126(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 24.Rispens T, Himly M, Ooievaar-De Heer P, den Bleker TH, Aalberse RC. Traces of pFc' in IVIG interact with human IgG Fc domains and counteract aggregation. Eur. J. Pharm. Sci. 2010;40:62–68. doi: 10.1016/j.ejps.2010.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.