Abstract
The C. elegans distal tip cells (DTCs) are an in vivo model for the study of developmentally regulated cell migration. In this study we characterize a novel role for CCDC-55, a conserved coiled-coil domain containing protein, in DTC migration and larval development in C. elegans. Although animals homozygous for a probable null allele, ccdc-55(ok2851), display an early larval arrest, RNAi depletion experiments allow the analysis of later phenotypes and suggest that CCDC-55 is needed within the DTC for migration to cease at the end of larval morphogenesis. The ccdc-55 gene is found in an operon with rnf-121 and rnf-5, E3 ubiquitin ligases that target cell migration genes such as the β-integrin PAT-3. Genetic interaction studies using RNAi depletion and the deletion alleles rnf-121(ok848) and rnf-5(tm794) indicate that CCDC-55 and the RNF genes act at least partially in parallel to promote termination of cell migration in the adult DTC.
Keywords: coiled-coil domain containing proteins, E3 ubiquitin ligases, C. elegans, cell migration
1. Introduction
Cell migration is essential for tissue and organ morphogenesis during development. However, many questions remain about how cells integrate multiple inputs to produce coordinated, stage-appropriate cell migration (Aman and Piotrowski, 2010; Lehmann, 2001). For example, how do cells become migratory and produce the cell shape changes required to crawl to a new location? How is cell migration coordinated with other developmental events? How do cells know when to stop migrating? The translucent body, availability of cell-specific GFP markers, invariant pattern of cell migration and the relative ease of disrupting gene function using RNAi make C. elegans an excellent model system for investigating these questions. In C. elegans, studies of anchor cell invasion (Sherwood, 2006), sex myoblast migration (Chen and Stern, 1998), axon guidance (Killeen and Sybingco, 2008), and distal tip cell (DTC) migration (Kimble and Hirsh, 1979; Lehmann, 2001; Nishiwaki, 1999) have all helped to reveal the mechanisms by which migrating cells adhere to extracellular matrix (ECM) molecules, interpret guidance cues, and coordinate cell movements with embryonic and larval stages.
During the four stages of hermaphrodite larval development (L1, L2, L3, and L4), migration of the two specialized leader cells known as distal tip cells (DTCs) guides the formation of the hermaphrodite gonad (Hall and Altun, 2008). The symmetrical U-shape of each gonad arm reflects the migratory path taken by the DTCs. The two DTCs first migrate in opposite directions along the ventral basement membrane. Then, the cells take two 90° turns: toward the dorsal surface and then back towards the midline of the animal. Upon reaching the midline at the end of the L4 stage, the DTCs stop migrating and remain stationary for the rest of the life of the animal. The process is not difficult to monitor because the DTCs are clearly visible in the living worm, and if either cell fails to migrate or follows an aberrant path, malformation of the gonad arm will result (Lee and Cram, 2009).
Many well-conserved genes are required for proper DTC migration (Lehmann, 2001; Nishiwaki, 1999). DTC specification occurs early in the first larval stage, and requires the E/Daughterless transcription factor HLH-2 (Karp and Greenwald, 2004; Tamai and Nishiwaki, 2007). The DTCs migrate along the basement membrane deposited by the muscle or hypodermal cells (Kawano et al., 2009; Kubota et al., 2004; Merz et al., 2003). Metalloproteases such as GON-1 and MIG-17 modify the basement membrane matrix to permit DTC migration (Blelloch and Kimble, 1999; Nishiwaki et al., 2000). UNC-129/TGF- β (Colavita et al., 1998) and UNC-6/netrin (Hedgecock et al., 1990; Merz et al., 2001) are cues that influence the path taken by the DTCs. The β-integrin PAT-3 (Gettner et al., 1995; Lee et al., 2001) and the α-integrins INA-1 (Baum and Garriga, 1997) and PAT-2 (Meighan and Schwarzbauer, 2007), are necessary for both the mechanics and the directional guidance of DTC migration. Rac GTPases (Levy-Strumpf and Culotti, 2007; Lundquist et al., 2001) and their effectors (Lucanic and Cheng, 2008) remodel the cell cytoskeleton and allow the cells to change direction (Hall, 2005). Cessation of DTC migration is coordinated with the transition from the larval to adult stage, and requires the transcription factor VAB-3/Pax6 (Cinar and Chisholm, 2004; Meighan and Schwarzbauer, 2007; Nishiwaki, 1999). Using a genome-wide RNAi approach we have identified several novel, conserved genes that appear to play an important role in DTC migration (Cram et al., 2006). One of these genes, ccdc-55, encodes a coiled-coil domain protein, conserved throughout eukaryotes but of unknown function.
Analysis of ccdc-55(ok2851), a probable null allele, indicates that CCDC-55 is required early in larval development. Therefore, we used an RNAi-based approach to determine the role of CCDC-55 in hermaphrodite DTC migration. CCDC-55 is likely required within the DTC itself for proper DTC migration and for the cells to stop migrating at the correct position. The ccdc-55 gene is found in an operon with several other genes, including rnf-121 and rnf-5, which encode E3 ubiquitin ligase enzymes (Broday et al., 2004; Darom et al., 2010; Didier et al., 2003; Zaidel-Bar et al., 2010). Analysis of RNAi knockdowns, and the deletion alleles rnf-121(ok848) and rnf-5(tm794), indicates that the entire operon plays a role in DTC migration guidance and cessation of migration.
2. Results
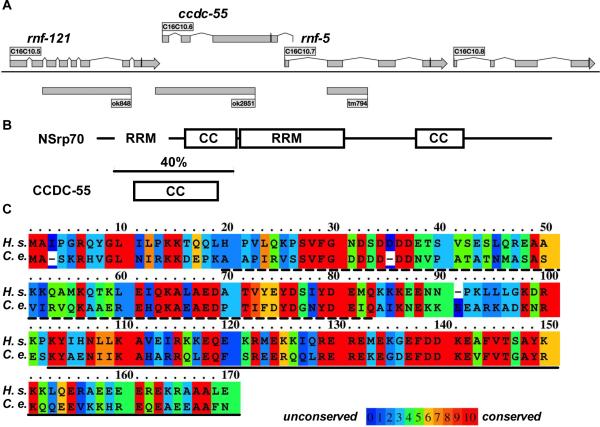
2.1 CCDC-55 is a conserved coiled-coil domain protein
The gene ccdc-55 (C16C10.6) is found in an operon (CEOP3156) with at least three other genes including two E3 ubiquitin ligases, rnf-121 (C16C10.5) and rnf-5 (C16C10.7), and the predicted nucleolar protein C16C10.8 (Garrido-Lecca and Blumenthal, 2010)(Figure 1A). ccdc-55 is predicted to encode a 392 amino acid, conserved protein with homologs in many eukaryotes (Kim et al., 2011). The gene name CCDC-55 indicates that the protein is predicted to be a coiled-coil domain containing protein. The coiled-coil domain is an interaction interface that commonly mediates homo-and hetero-oligomerization of proteins (Mason and Arndt, 2004). We used the program COILS, which calculates the probability that a given sequence will adopt a coiled-coil conformation (Lupas et al., 1991), to identify the likely coiled-coil domain in CCDC-55. The predicted coiled-coil domain (amino acids 50-150) coincides with the most conserved region of the protein (Fig. 1B). Recently, CCDC-55 has been identified as the closest C. elegans homolog to the mammalian protein, nuclear speckle splicing regulatory protein 70 (NSrp70). NSrp70 consists of two coiled-coil domains and divergent RRM-like and RS-like RNA binding domains, and has been shown to bind RNA and to regulate pre-mRNA splicing in cell culture (Kim et al., 2011). The overall homology between C. elegans CCDC-55 and human NSrp70 is 28% (Kim et al., 2011), with best conservation (40% identity, 72% similarity) in the N-terminal coiled-coil region, which partially overlaps the first RRM-like domain (Fig. 1B,C). CCDC-55 is missing the central RRM-like domain, and has low sequence homology in the C-terminal RS-like region and the second coiled-coil domain, all of which are necessary for the function of NSrp70 (Kim et al., 2011).
Fig. 1.
Gene structure and homology of CCDC-55. A: Scale map of the C. elegans operon containing ccdc-55. Rectangles represent the exons and lines represent introns for each of the genes in the operon. The line through the final exon of each gene indicates the position of each stop codon. Deletion alleles (ok848,ok2851 and tm794) are indicated by gray boxes beneath the structure of the operon. B: Schematic diagram of human NSrp70 and C. elegans CCDC-55, indicating the conserved coiled-coil (CC) and the RRM-like (RRM) domains of the two proteins. The RS-like domain of NSrp70 extends from the central RRM domain to the C-terminus. The region of highest conservation (40% identity) is indicated by the bracket. The C-terminus of CCDC-55 is not well conserved. C: The gene ccdc-55 is predicted to encode a 392 aa protein. Sequence alignment of the N-terminus of human (H.s.) NSrp70 and C. elegans (C.e.) CCDC-55 is shown. The proteins are aligned at the N-terminal methionine. The RRM-like domain of NSrp70 is indicated by a dashed underscore and the coiled-coil domain of NSrp70 is indicated by a solid underscore. The rainbow color map was generated using PRALINE (Simossis and Heringa, 2005) and indicates degree of similarity for each amino acid (low homology blue, high homology red).
In order to investigate a possible role for CCDC-55 in splicing, we obtained a reporter strain (LET-2∷GFP/RFP), in which a shift from green (GFP) to red (RFP) fluorescence indicates altered splicing within a modified let-2 transcript (Kuroyanagi et al., 2010). In late-L4 animals, the ratio of red to green fluorescence in ccdc-55 RNAi treated animals (0.64 +/− 0.04, N=8) did not differ significantly from controls (0.55 +/− 0.02, N=10; p=0.08). In adult animals, the ratio of red to green fluorescence increases (Kuroyanagi et al., 2010). However, the ratio of fluorescence intensities in ccdc-55 RNAi treated adult animals (1.36 +/− 0.09, N=9) again did not differ significantly from controls (1.28 +/− 0.07, N=6; p=0.54), suggesting CCDC-55 may not play a role in the splicing of let-2 (Supplementary Fig. S1A). We next treated unc-52(e669) animals with ccdc-55 RNAi. The allele e669 results in a stop codon in an alternatively spliced exon of unc-52. Exclusion of this exon by alternative splicing results in suppression of a late larval-onset paralysis phenotype (Rogalski et al., 1995). Although positive control asd-2 RNAi strongly suppressed the unc-52(e669) paralysis phenotype (17% paralyzed, N=110) compared to the negative control (69% paralyzed, N=90), ccdc-55 RNAi did not suppress the paralysis phenotype (83% paralyzed, N=94; not significantly different from negative control at the 95% CI). (Supplementary Fig. S1 B–D). RNAi of core and alternative splicing regulators has also been shown to affect the switch from sperm to oocyte production in the C. elegans hermaphrodite (Kerins et al., 2010). Using DIC microscopy, oocyte production in young adult hermaphrodites was observed. All of the ccdc-55 RNAi treated animals (100%, N=43) and control animals (100%, N=44) produced oocytes. These data suggest oocyte production does not appear to be regulated by CCDC-55. These results are based on RNAi knockdown experiments that probably do not deplete CCDC-55 protein completely, and are limited in the tissues and genes assessed. Therefore, we cannot conclusively rule out a role for CCDC-55 in splicing. However, taken together with the weak homology data, these results suggest that CCDC-55 might not play a general role in splicing in C. elegans.
2.2 Deletion of ccdc-55 results in larval arrest
In order to investigate the role of ccdc-55 in C. elegans development, we analyzed the phenotypes resulting from a deletion allele, ok2851. The ccdc-55(ok2851) allele removes 935 bp, including the predicted start codon. There is no start codon in the remaining 244 base pairs of the open reading frame. The deletion does not disrupt the coding sequence or the two predicted poly-A signal sequences of the upstream gene, rnf-121 (Harris et al., 2010). Although the ccdc-55 genomic locus can be amplified easily in the mutant animals, no transcript is detectable by RT-PCR (data not shown). In the strain VC2493, ccdc-55(ok2851) is maintained over a balancer. Balanced heterozygous hermaphrodites produce wildtype and arrested progeny. To determine the phenotype of ccdc-55(ok2851) independent of possible balancer-related phenotypes, VC2493 hermaphrodites were crossed with wildtype males to produce heterozygous F1 progeny. The ok2851/+ hermaphrodites produced 75% fertile, wildtype appearing and 25% arrested self-progeny (14/58 arrested, Chi squared p=0.95). Larvae were genotyped using PCR, and arrested larvae were found to be homozygous for the ok2851 allele. In order to determine whether the ok2851 phenotype could be rescued with genomic sequence from the ccdc-55 locus, animals carrying a complex extrachromosomal array, including 15.5 kb of the CEOP3156 operon (from 5.9 kb upstream of rnf-121 to 500 bp downstream of C16C10.8), and the co-injection marker sur-5‷GFP, were generated. Progeny of animals homozygous for ccdc-55(ok2851), but bearing the transgenic array, were assessed 48 hours after hatching. All of the non-transgenic progeny (100%, N=39) arrested as small larvae. Although some transgenic progeny also arrested (18%, N=68), most of the progeny bearing the transgenic array developed normally (82%, N=68). These data indicate ccdc-55 genomic sequence is sufficient to rescue the larval arrest of ok2851. Taken together, these results suggest ok2851 is a recessive allele of ccdc-55 and a probable null.
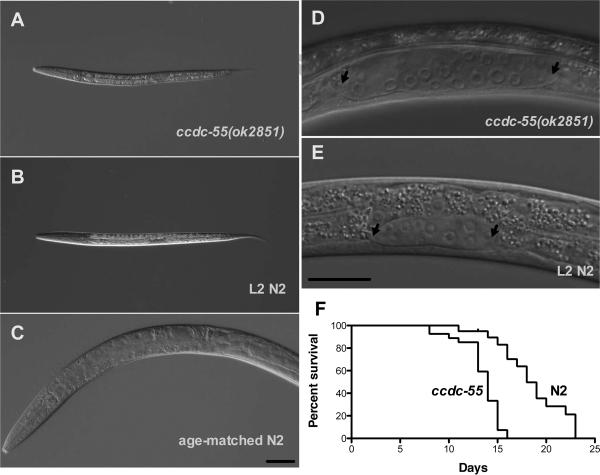
We next determined the approximate stage at which the ccdc-55(ok2851) larvae arrest. Comparison of body length in wildtype and arrested animals suggests ccdc-55(ok2851) homozygous animals arrest early in larval development (Byerly et al., 1976). By 54 hours of age, wildtype animals are fertile young adults 1038 +/− 63 μm in length (N=13). In contrast, arrested larvae (417 +/− 16 μm, N=25) are the size of wild-type mid-L2 larvae (410 +/− 7 μm, N=15; p<0.16, unpaired t-test) (Fig 2 A–C). Although the ccdc-55(ok2851) larvae remain L2-sized, no overt defects are apparent in pharyngeal morphology, rate of pharyngeal pumping, ingestion of mCherry-labeled bacteria, intestinal morphology, or overall body morphology (data not shown). The gonad in the arrested ccdc-55(ok2851) larvae at 54 hours is significantly longer (62 +/− 9 μm, N=22) than in the size-matched wildtype animals (36 +/− 3 μm, N=15; p<0.001, unpaired t-test) (Fig. 2D, E). This phenotype could be the result of precocious or too rapid expansion of the gonad in ccdc-55(ok2851) larvae, or instead could be the result of some continued gonad growth in arrested animals. We used the rescued line to determine the relationship between pre-arrest gonad and body length during early larval development, and found no statistically significant differences between rescued animals, non-rescued siblings, and N2 larvae during early development (Figure S2). These results suggest the gonad in ccdc-55(ok2851) animals continues to elongate even after the animal has stopped growing (Fig. 2 D, E). This data, in combination with the observation that ccdc-55(ok2851) DTCs are normal in general morphology suggests the DTCs are correctly specified and motile in ccdc-55(ok2851) animals. The ccdc-55(ok2851) homozygous animals remain arrested until they die at approximately 2 weeks of age (Fig. 2F).
Fig. 2.
Deletion of ccdc-55 results in larval arrest. DIC images of A: an arrested ccdc-55(ok2851) animal 54 hours after hatching, B: a second larval stage (L2) wildtype (N2) animal, and C: a young adult animal the same chronological age as the arrested larva in (A). Despite the larval arrest, the gonad continues to elongate. DIC images of D: the gonad of an arrested ccdc-55(ok2851) animal 54 hours after hatching, and E: the gonad of a size-matched L2 larva. Arrows indicate the positions of the DTCs. Scale bars are 20 μm. F: Lifespan analysis of wildtype (N2) and ccdc-55(ok2851) animals.
2.3 CCDC-55 is broadly expressed
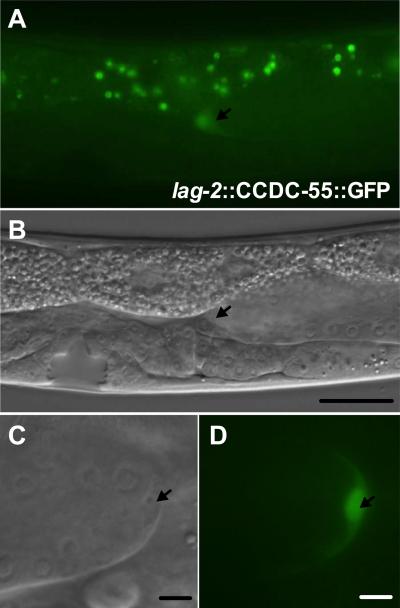
The ccdc-55 message is transcribed as part of a multi-gene operon, CEOP3156, with the predicted promoter 5' of rnf-121 (Garrido-Lecca and Blumenthal, 2010). An RNF-121 translational GFP reporter consisting of 8.1 kb of genomic sequence, including 5.7 kb 5' of rnf-121, is broadly expressed in larvae and adults, including muscle, seam, hypodermis, vulva and somatic gonad with prominent expression in the DTC (Darom et al., 2010). Because ccdc-55 closely follows rnf-121 in the operon, and because rnf-121 and ccdc-55 show similar expression levels under a variety of different conditions (Celniker et al., 2009; Spencer et al., 2011), it is likely the expression pattern seen in the rnf-121‷GFP animals is relevant for ccdc-55. In C. elegans, genes contained in operons can also be expressed independently (Blumenthal and Gleason, 2003), so the rnf-121-based expression pattern may not completely recapitulate the endogenous expression of ccdc-55. Despite the similarity of the rnf-121‷GFP construct to our rescuing fragment at the 5' end, we have been unable to generate a rescuing GFP fusion of CCDC-55 using this promoter. This suggests GFP may interfere with the function of CCDC-55 or sequences 3' of ccdc-55 may be necessary for proper expression. Therefore, in order to determine the subcellular localization of CCDC-55, we generated animals that express a CCDC-55‷GFP fusion protein in the DTC under the control of the lag-2 promoter. In these animals, we observed a nuclear and cytoplasmic distribution of CCDC-55‷GFP in the DTC (Fig. 3).
Fig. 3.
CCDC-55 is predominantly nuclear in the DTC. A: CCDC-55‷GFP expressed under the control of the DTC-specific promoter lag-2. DTC is indicated by an arrow. Green speckles in the background are autofluorescence from gut granules. B: DIC image of the same animal. DTC is indicated by an arrow and scale bar is 20 μm. C: Digital zoom DIC image of a DTC and D: corresponding CCDC-55‷GFP image. DTCs are indicated by an arrow and scale bar is 10 μm.
2.4 Depletion of ccdc-55 by RNAi results in inappropriate DTC migration
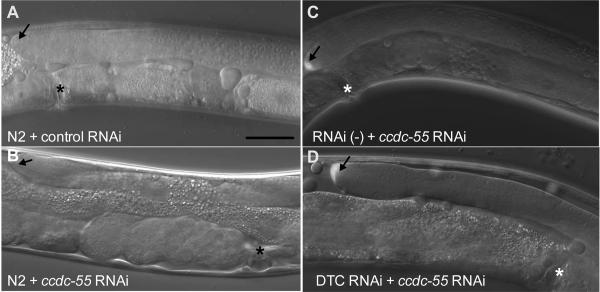
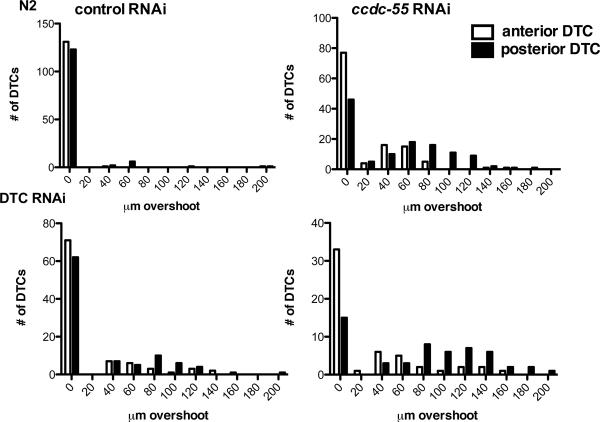
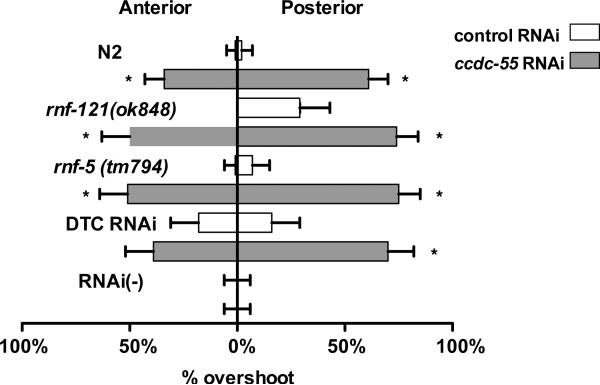
The gene ccdc-55 was isolated in a genome-wide RNAi screen as a regulator of distal tip cell (DTC) migration (Cram et al., 2006). The early larval arrest phenotype of the ok2851 allele precludes scoring of DTC migration, which occurs later in larval development. Therefore, we used an RNAi feeding approach, which does not result in larval arrest, to deplete ccdc-55 and monitored DTC migration phenotypes in young adult animals. In control animals, the DTC stops migrating by the end of L4 and is correctly positioned adjacent to the vulva (Table 1, Fig. 4A). In ccdc-55 RNAi treated animals, the DTC does not stop migrating at the correct position (Fig. 4B), but instead continues migrating during adulthood. Quantitative analysis of the distance from the vulva to the DTC indicates that when ccdc-55 is depleted, the DTCs migrate much further past the midline than DTCs in control animals (Fig. 5). The posterior DTC is more sensitive to the loss of ccdc-55 than the anterior DTC, showing increased severity (Fig. 5) and penetrance (Fig. 6) of defects. Importantly, similar results were obtained using a cDNA-based construct targeting the entire ccdc-55 open reading frame, a genomic construct targeting the middle exon, and two non-overlapping cDNA-based ccdc-55 RNAi targeting constructs (data not shown). Defects were rarely observed in animals fed control RNAi bacteria. By targeting pre-mRNA, nuclear RNAi can result in the depletion of co-transcribed genes in addition to the targeted gene (Bosher et al., 1999). Because ccdc-55 is part of an operon, we performed feeding RNAi in a nuclear RNAi defective mutant, nrde-3(gg66) (Guang et al., 2008). DTC migration defects in nrde-3(gg66) RNAi treated animals were not significantly different from those observed in wildtype animals (Table 1). These results suggest the observed DTC migration defects are specific to knockdown of ccdc-55.
Table 1.
DTC migration experiment statistics.
| L4440 control RNAi | ccdc-55 RNAi | |||||
|---|---|---|---|---|---|---|
| % defective | N | 95% CI | % defective | N | 95% CI | |
| N2 | ||||||
| Anterior | 1% | 132 | 0–4% | 35% | 120 | 27–44% |
| Posterior | 7% | 132 | 3–13% | 61% | 120 | 51–69% |
| Total | 4% | 264 | 2–7% | 48% | 240 | 41–54% |
| Nuclear RNAi defective nrde-3(gg66) | ||||||
| Anterior | 0% | 46 | 0–7% | 22% | 41 | 11–38% |
| Posterior | 9% | 46 | 2–21% | 75% | 41 | 60–88% |
| Total | 4% | 92 | 1–11% | 48% | 82 | 37–60% |
| DTC-specific RNAi | ||||||
| Anterior | 24% | 95 | 16–34% | 37% | 54 | 24–51% |
| Posterior | 34% | 95 | 24–44% | 70% | 54 | 56–82% |
| Total | 29% | 190 | 23–36% | 54% | 108 | 44–63% |
| RNAi-defective rde-1(ne219) | ||||||
| Anterior | 0% | 60 | 0–6% | 0% | 56 | 0–6% |
| Posterior | 0% | 60 | 0–6% | 0% | 56 | 0–6% |
| Total | 0% | 120 | 0–3% | 0% | 112 | 0–3% |
| rnf-121(ok848) | ||||||
| Anterior | 0% | 52 | 0–7% | 50% | 62 | 37–63% |
| Posterior | 29% | 52 | 17–43% | 72% | 62 | 60–83% |
| Total | 14% | 104 | 8–23% | 61% | 124 | 52–70% |
| rnf-5(tm794) | ||||||
| Anterior | 3% | 58 | 0–11% | 51% | 55 | 37–65% |
| Posterior | 12% | 58 | 5–23% | 73% | 55 | 59–84% |
| Total | 7% | 116 | 3–13% | 62% | 110 | 52–71% |
The percentage of DTCs that have passed the correct stopping point (% defective), the number of DTCs scored (N), and the 95% confidence interval (95% CI) is given for each experiment.
Fig. 4.
CCDC-55 is likely required cell-autonomously for cessation of DTC migration. A: Wildtype N2 animals fed E. coli HT115(DE3) expressing negative control RNAi (empty L4440 vector) or B: ccdc-55 RNAi. DIC image overlaid with GFP fluorescence of C: RNAi defective (rde-1(ne219); [lag-2‷GFP]) animals treated with ccdc-55 RNAi. D: DTC-specific (rde-1(ne219); [lag-2‷GFP, lag-2‷RDE-1]) animals treated with ccdc-55 RNAi. DTC is indicated by an arrow and the vulva is indicated by an asterisk (*). The DTC has migrated A) 21μm, B) 160 μm, C) 13 μm and D) 163 μm past the vulva. Scale bar is 25 μm.
Fig. 5.
Quantification of DTC migration in ccdc-55 RNAi treated animals. Wildtype animals (top panels “N2”) or DTC-specific RNAi strain rde-1(ne219); [lag-2‷GFP, lag-2‷RDE-1] animals (bottom panels “DTC RNAi”) were fed E. coli HT115(DE3) expressing sequences from the empty RNAi vector L4440 as a negative control (left) or ccdc-55 RNAi (right). Graphs are histograms of the distribution of distances that DTCs migrated past the vulva. Quantitative data depicted in these histograms were collected from the samples described in Table 1. White bars represent the distribution of anterior DTC positions and black bars represent the distribution of posterior DTC positions in young adult animals.
Fig. 6.
Disruption of operon genes ccdc-55, rnf-121 and rnf-5 results in DTC migration defects. Animals were fed E. coli HT115(DE3) expressing sequences from the empty RNAi vector L4440 as a negative control (white bars) or ccdc-55 RNAi (gray bars). Bars indicate the proportion of animals in each treatment with DTCs that migrated past the vulva (% overshoot). Anterior DTCs are to the left and posterior are to the right. Error bars indicate the 95% confidence intervals for each set of data. Asterisks indicate significant differences between the control and ccdc-55 RNAi treated samples. Genotypes are indicated to the left of the graph. DTC RNAi animals are of the genotype (rde-1(ne219); [lag-2‷GFP, lag-2‷RDE-1]) animals and RNAi(−) animals are of the genotype (rde-1(ne219); [lag-2‷GFP]).
2.5 CCDC-55 is probably required in the DTC for cessation of migration
In order to investigate the hypothesis that CCDC-55 is required within the DTC to control cell migration, we used a cell-specific RNAi approach. Cell-specific RNAi is conducted in an rde-1(ne219) background which renders the animal resistant to RNAi (Tabara et al., 1999). The effect of RNAi is limited to a specific subset of tissues through the transgenic expression of RDE-1 with a more cell-specific promoter. DTC-specific RNAi was performed in a strain (rde-1(ne219); lag-2‷RDE-1; lag-2‷GFP) that expresses RDE-1 strongly in the DTC (DTC RNAi). The lag-2 promoter is also active in the cells of the vulva (Chen and Greenwald, 2004) and the ventral nerve cord (Siegfried et al., 2004), leaving open the possibility that ccdc-55 activity in other lag-2 expressing cell types may also play a role in DTC migration.
Using gon-1 RNAi, which produces a striking and highly reproducible DTC migration defect, we verified that genes can be silenced effectively in the DTC RNAi animals (data not shown). To further characterize the DTC RNAi animals, we treated them with talin RNAi. Talin is required for DTC migration and for the maintenance of actin in muscle cells (Cram et al., 2003). In wild type animals, depletion of talin results in DTC migration defects (74%, N=70) and paralysis (100%, N=86). In contrast, the DTC RNAi animals display fewer DTC migration defects (44%, N=68) and no evidence of paralysis (0%, N=74). These results suggest talin is required both in the DTC and in other non-rescued cell types for DTC migration, and confirms that muscle is not responsive to RNAi in this strain. The control strain (rde-1(ne219); [lag-2‷GFP]), which is not sensitive to RNAi (RNAi(−)), exhibits neither the gon-1 RNAi phenotype, the talin RNAi phenotype (data not shown), nor the ccdc-55 RNAi phenotype (Table 1, Fig. 4C). In contrast, depletion of ccdc-55 in the DTC RNAi strain results in a very similar phenotype to that seen in wildtype animals treated with ccdc-55 RNAi (Fig. 4D). Although the DTC RNAi strain exhibits background defects in DTC migration when treated with control RNAi, a significant increase in DTC migration defects is observed in ccdc-55 RNAi treated animals (Table 1, Fig. 5,6). These defects are similar in severity (Fig. 5) and penetrance (Fig. 6) to the phenotypes seen in ccdc-55 RNAi treated wildtype animals. These results suggest that CCDC-55 is likely to be required in the DTC to sense or respond to developmental and spatial cues to stop migrating.
2.6 ccdc-55, rnf-121, and rnf-5 all contribute to the cessation of DTC migration
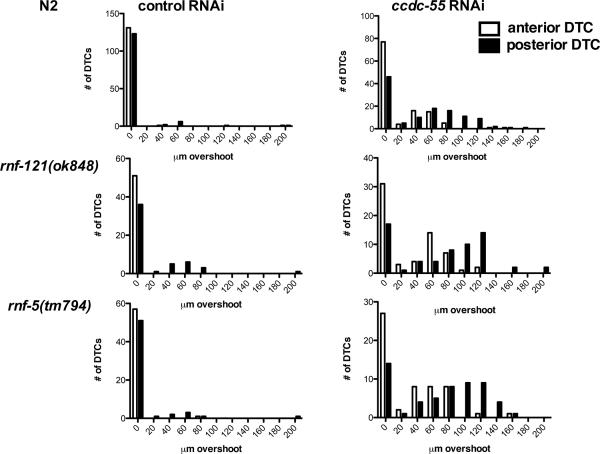
Because genes expressed in operons often regulate similar cellular processes, we next investigated the role of the ubiquitin ligases rnf-121 and rnf-5 in DTC migration. The rnf-5(tm794) probable null allele is a 647 bp deletion in exon 2 that results in a frameshift and a stop codon after 37 amino acids (our own sequencing data and (Broday et al., 2004)). The rnf-121(ok848) allele is a 1397-bp deletion that removes four of the six predicted transmembrane domains and the RING finger domain, and is also a putative null allele (Darom, 2010) (See Fig. 1). The rnf-121(ok848) and rnf-5(tm794) animals are viable and fertile, and have low penetrance, but reproducible defects in the migration of the posterior DTC (Fig. 6). When these animals are treated with ccdc-55 RNAi, the penetrance of the defects is not significantly increased compared to N2 (Table 1, Fig. 6), but the severity of the defects is enhanced (Fig. 7). The increase in severity is apparent in the rightward shift of the distribution of distances migrated past the vulva by the posterior DTCs (Fig. 7, right panels). In this subset of the data, the mean overshoot in rnf-5(tm794) ccdc-55 RNAi treated animals is 95.2 ± 5.2 μm and in rnf-121(ok848) ccdc-55 RNAi treated animals is 93 ± 5.1 μm. These means are significantly different from wildtype posterior DTCs treated with ccdc-55 RNAi, in which an average overshoot of 78 ± 3.9 μm is observed (unpaired t-test, p=0.008 and p=0.02 respectively). These results demonstrate expression of neither RNF-5 nor RNF-121 is required for ccdc-55 RNAi to produce the overshoot phenotype, and suggest rnf-5 and rnf-121 may be acting in parallel with ccdc-55 to terminate DTC migration at the end of L4.
Fig. 7.
Quantification of DTC migration in wildtype, rnf-121 and rnf-5 animals treated with ccdc-55 RNAi. Wildtype (top panels “N2”), rnf-121(ok848) (middle panels) and rnf-5(tm794) (bottom panels) animals were fed E. coli HT115(DE3) expressing sequences from the empty RNAi vector L4440 as a negative control (left) or ccdc-55 RNAi (right). Graphs are histograms of the distribution of distances that DTCs migrated past the vulva. Quantitative data depicted in these histograms were collected from the samples described in Table 1. White bars represent the distribution of anterior DTC positions and black bars represent the distribution of posterior DTC positions in young adult animals.
3. Discussion
This study presents the first characterization of C. elegans ccdc-55, a conserved novel regulator of larval development and cell migration. We identified ccdc-55 in a genome-wide in vivo RNAi screen for cell migration genes (Cram et al., 2006). Although a probable null allele results in early larval arrest, RNAi depletion experiments demonstrate that CCDC-55 has a later role in the control of DTC migration and gonad morphogenesis. Strong expression of the ccdc-55 operon in the DTCs, and data from cell-specific RNAi depletion experiments, suggest CCDC-55 is probably required within the DTC to control migration. Our data suggest that two other genes in the operon, rnf-121 and rnf-5, also play a role in determining when and where the DTC should stop migrating.
RNF-121 and RNF-5 are E3 ubiquitin ligases (Broday et al., 2004; Darom et al., 2010; Zaidel-Bar et al., 2010). RNF-5 co-localizes with UNC-95 in dense bodies, the nematode equivalent of focal adhesions (Broday et al., 2004). UNC-95 has homology to paxillin in the LIM domains and LD4 region (Broday et al., 2004). RNF-5 negatively regulates UNC-95 protein levels, most likely through ubiquitin-mediated degradation (Broday et al., 2004; Zaidel-Bar et al., 2010). Similarly, the human homolog RNF5 mediates ubiquitination of paxillin and inhibition of cell motility in mammalian cell culture (Didier et al., 2003). In C. elegans, overexpression of RNF-121 also leads to DTC migration defects, probably because of excessive degradation of the β-integrin PAT-3 and other targets (Darom et al., 2010). No effect of ccdc-55 RNAi on integrin expression levels or localization was observed (data not shown). Because deletion of either rnf-5 or rnf-121 results in inappropriate continuation of DTC migration, and exacerbates the posterior DTC migration defect caused by ccdc-55 RNAi, it seems likely that the RNF proteins and CCDC-55 are inhibiting DTC migration at least partially via parallel pathways.
DTCs in wildtype animals always stop migrating when they reach the dorsal surface adjacent to the vulva, however, little is known about how the cells make this decision. One of the few well-characterized factors known to play a role in DTC stopping is the transcription factor VAB-3/Pax6 (Cinar and Chisholm, 2004; Meighan and Schwarzbauer, 2007; Nishiwaki, 1999). VAB-3 appears to execute a stopping program at the end of larval development that involves switching between the PAT-2 and INA-1 α-integrins (Meighan and Schwarzbauer, 2007). In vab-3 mutant animals, the DTC migrates normally until the end of L4, and then continues to migrate circumferentially in the vicinity of the vulva producing “cinnamon roll” tipped gonad arms. VAB-3 is apparently not needed for the DTC to detect the correct stopping position, but is necessary to deactivate the L4 DTC migration program. This phenotype is in contrast to that seen in ccdc-55 depleted animals, in which the DTC proceeds past the vulva along the dorsal surface, producing long, straight gonad arms. Therefore, CCDC-55 is required for the DTC to detect the stopping position and for the cell to sense the developmental cue to stop migrating. Our DTC migration screen identified a small number of other genes required for DTC stopping, including cacn-1 (Cram et al., 2006). CACN-1 is also a coiled-coil domain containing protein, and is homologous to the IKB/cactus binding protein cactin (Lin et al., 2000). In cacn-1 RNAi treated animals, DTCs continue to migrate far past the correct stopping point, sometimes until they are stopped by the pharynx or the end of the tail (Tannoury et al., 2010). These phenotypes suggest there may be two different cues, a positioning cue and a developmental timing cue. VAB-3 function is apparently needed for the DTCs to interpret the timing cue, but not the positioning cue, whereas CCDC-55 and CACN-1 function may be needed for both aspects.
Although the molecular function of CCDC-55 remains elusive, emerging evidence suggests the mammalian homolog of CCDC-55, NSrp70, regulates pre-mRNA splicing. NSrp70 has recently been proposed to contain RRM and RS-like domains in addition to the coiled-coil domain, and shown to bind RNA and to regulate splicing of specific transcripts (Kim et al., 2011). Alternative splicing of specific transcripts may be important generally for the regulation of cell migration. For example, recent work on the mechanism of epithelial-mesenchymal transition (EMT) suggests that alternative splicing of cytoskeletal, junction, and cell migration genes drives the acquisition of a migratory cell phenotype and regulates migratory behavior of human breast cancer cells (Shapiro et al., 2011). Although we have not yet been able to demonstrate a role for CCDC-55 in splicing, it seems likely that expression of specific isoforms of target genes may also be necessary for the DTC to sense and respond to temporal and spatial cues at the end of its migration program. The C. elegans DTC provides a powerful model system for understanding developmentally regulated cell migration. Probable relevance to other cell migration events, such as those underlying EMT, give these studies the potential to make an important contribution to our understanding of how cells transition between migratory and non-migratory states.
4. Experimental Procedures
4.1 C. elegans strains and culture
Nematodes were cultivated on NGM agar plates with E. coli OP50 bacteria according to standard techniques (Brenner, 1974). Nematode culture and observations were performed at 20 degrees, unless otherwise indicated. The C. elegans strain FX794 rnf-5(tm794) was obtained from the Japanese National Bioresource Project. The LET-2∷GFP/RFP line ybIs1371 [myo-3∷let-2-9G myo-3∷let-2-10R] was a gift of Hidehito Kuroyanai. NX61 rnf-121(ok848), a backcrossed version of RB953, was kindly provided by Limor Broday. DTC RNAi strains JK4135 rde-1(ne219); qIs57[lag-2∷GFP] and JK4143 rde-1(ne219); qIs57[lag-2∷GFP]; qIs140 [lag-2∷RDE-1] were used in collaboration with Judith Kimble. The wildtype reference strain N2 (Bristol), the unc-52(e669) strain CB669, the nuclear RNAi defective strain nrde-3(gg66) strain YY158 and the balanced strain VC2493 +/mT1 II; ccdc-55(ok2851)/ mT1[dpy-10(e128)] III) were obtained from the Caenorhabditis Genetics Center. VC2493 animals were backcrossed to N2 five times and maintained as ccdc-55(ok2851)/+ heterozygotes for all experiments.
4.2 RNA interference
RNA interference was performed by feeding animals dsRNA-expressing HT115 DE3 E. coli essentially as described (Cram et al., 2006). Eggs were obtained from gravid hermaphrodites using alkaline hypochlorite solution and transferred to NGM/Carbenicillin/IPTG plates seeded with RNAi bacteria. RNAi experiments were performed at 20°C. The original ccdc-55 RNAi observations were made using the Ahringer clone sjj_C16C10.6 (Cram, 2006). This construct is a genomic fragment overlapping the middle exon of the gene. The RNAi experiments described in this paper were conducted using an RNAi construct targeting the whole open reading frame (Open Biosystems C. elegans RNAi library), a clone targeting the 5' 559 bp of the sequence and a clone targeting the 3' 646 bp of the sequence. The 5' and 3' RNAi targeting constructs for ccdc-55 were constructed by RT-PCR amplification of wildtype RNA using engineered restriction sites, and subsequently cloned into pPD129.36 (Fire Vector Kit). Talin RNAi experiments were performed using the Ahringer clone sjj_Y71G12A_195.e (Cram et al., 2003). Empty pPD129.36 vector was used as a negative control in all feeding RNAi experiments. All primer sequences and cloning details are available upon request.
4.3 Analysis of larval phenotypes
Arrested ccdc-55(ok2851) homozygous animals were obtained from ccdc-55(ok2851)/+ heterozygous mothers. Heterozygotes were singled out to seeded NGM plates and allowed to lay eggs for 2 hours at room temperature, and then genotyped using PCR to confirm heterozygosity. Progeny of the heterozygous mothers were propagated for 54 hours at 20°C, at which point, arrested animals and non-arrested L4 siblings were collected. For control experiments, synchronized populations of N2 larval animals were obtained by hatching eggs prepared by alkaline lysis of gravid hermaphrodites on NGM plates with no food. Arrested L1 animals were transferred to NGM plates seeded with E. coli strain OP50, and grown for 20 hours at 20°C. Second larval stage (L2) N2 control animals, arrested ccdc-55(ok2851) animals, and age-matched controls were mounted in a drop of M9 containing 0.1M sodium azide on a slide coated with 2% agarose in water and examined using a Nikon 80i microscope with DIC optics. Body length and length of the somatic gonad were measured using Spot Advanced software version 4.6.4.6. All statistical analyses were performed using the GraphPad Prism statistical software package.
4.4 Determination of lifespan
For lifespan determination, putative ccdc-55(ok2851)/+ heterozygous animals were singled out to seeded NGM plates and allowed to lay eggs for 24 hours at 20°C. After 48 hours, arrested larvae and wildtype N2 controls were singled out to separate NGM plates and monitored each day for viability. Wildtype animals were carefully transferred to new plates every two days until egg laying stopped. Viability of non-moving animals was assessed by gentle prodding with a platinum wire. Statistical analysis of lifespan was performed using the GraphPad Prism statistical software package.
4.5 Construction of ccdc-55::GFP transgenic animals
The high-fidelity Phusion PCR polymerase (New England Biolabs, Ipswich, MA) was used to amplify 1712 bp of DNA encoding ccdc-55 from genomic DNA (forward primer 5' gcatgcctgcaggtcatggcatcaaaacggcatgtagg 3' and reverse primer 5' cctttggccaatcccaatggaggtacaacaattcc 3'). The PCR product was gel purified and inserted into the SmaI site of the GFP expression vector pPD95_77 (Fire Vector Kit) using the InFusion recombinase (Clontech) according to the manufacturer's instructions. 2848 bp of the lag-2 promoter were amplified with PstI compatible ends from the plasmid pJK590 (forward primer 5' agctgcagtccctgaactactctactccac 3', reverse primer 5'gaccttgagctttgctagaagccctgcag3'), digested with PstI, purified, and cloned into the PstI site of pPD95_77 using T4 ligase (New England Biolabs). The resulting plasmid pUN151 was used for microinjection at an approximate concentration of 25 μg/mL with the rol-6(su1006) (pRF4) plasmid at 25 μg/mL as a co-injection marker. Transgenic strains were created by standard germline transformation technique (Mello et al., 1991) of wildtype animals to create strain UN1057 xbEx1057[lag-2::ccdc-55::gfp, rol-6].
4.6 ccdc-55 genomic rescue
Complex extrachromosomal arrays were constructed and used for ccdc-55(ok2851) rescue. The high-fidelity Phusion PCR polymerase (New England Biolabs, Ipswich, MA) was used to amplify 15.5 kb of genomic DNA encompassing the CEOP3156 operon (forward primer 5'aattcccgtggtttcggatgatcagctgaagg 3' and reverse primer 5'agacttgtcgtgttctgttccctatacactgc 3'). The amplified DNA was gel purified and an injection mixture consisting of 60 ng/uL DraI digested wildtype genomic DNA, 20 ng/uL PCR-amplified ccdc-55 genomic region, 40 ng/uL pTG96 (sur-5::GFP), and 15 ng/uL pRF4 (rol-6) was injected into wildtype hermaphrodites. The rescuing genomic construct encompasses the entire operon and spans from 5.9 kb upstream of rnf-121 to 500 bp downstream of C16C10.8, for a total of 15.5 kb. The complex array was introduced into ccdc-55(ok2851) animals by genetic cross to produce the line UN1139 xbEx1139[CEOP3156(+), sur-5::GFP, rol-6(su1006)].
4.7 Analysis of gonad phenotypes
Partially synchronized populations of young adult animals were obtained by rearing embryos isolated using alkaline hypochlorite solution and propagating the animals at 23°C for 54 (N2) or 60 hours (JK4135 and JK4143). In order to document gonad morphology, animals were mounted in a drop of M9 containing 0.1M sodium azide on a slide coated with 2% agarose in water and examined using a Nikon 80i microscope with DIC optics. DTC migration defects were inferred from the resulting shape of the gonad arms. Defects such as insufficient distance migrated along the ventral surface, inappropriate or extra turns, and failure to cease migrating at the vulva were counted as DTC migration defects. The most common defect was failure to stop migrating at the correct position. Distance traveled by the DTC past the vulva was quantified using Spot Advanced software version 4.6.4.6. All proportions were compared for statistical significance by calculating binomial 95% confidence intervals with JavaStat. Non-overlapping intervals indicate significance at the p<0.05 level. Means and standard deviations are compared using Student's t-test. All statistical analyses were performed using GraphPad Prism statistical software package.
Supplementary Material
let-2 and unc-52 do not require CCDC-55 for normal alternative splicing. A: In order to investigate a possible role for CCDC-55 in splicing, we obtained a reporter strain (LET-2∷GFP/RFP ybIs1371 [myo-3∷let-2-9G myo-3∷let-2-10R]), in which a shift from green (GFP) to red (RFP) fluorescence indicates alternative splicing within a modified let-2 transcript (Kuroyanagi et al., 2010). Stop codons have been introduced into exon 9 (E9) or 10 (E10) of let-2 DNA such that inclusion of exon 9 leads to expression of GFP and inclusion of E10 results in the expression of RFP. The fluorescence gradually shifts from green to red as the animals develop, reflecting the splicing patterns of endogenous let-2 (Kuroyanagi et al., 2010). The LET-2∷GFP/RFP animals were treated with ccdc-55 or empty RNAi vector L4440 as a negative control, and L4 (48 hour) and adult (72 hour) individuals were imaged to quantify the level of green and red fluorescence. RFP and GFP fluorescence intensity was determined by imaging at 20× magnification using standardized exposure conditions. Amount of fluorescence in each channel was determined using ImageJ. ccdc-55 RNAi produced no significant difference in the overall splicing pattern. B–D: The allele e669 results in a stop codon in an alternatively spliced exon of unc-52. Exclusion of this exon by alternative splicing results in suppression of the late larval-onset paralysis phenotype (Rogalski et al., 1995). Movement of animals was assessed on plates after 48 hours of treatment. Randomly selected fields of view were imaged on plates using a Nikon SMZ800 dissecting microscope equipped with a Spot Insight 2 camera. Paralyzed animals were identified by their stick-straight body posture and curved animals were scored as normal. B: Negative control empty vector RNAi, C: positive control asd-2 RNAi and D: ccdc-55 RNAi.
Relationship between gonad size and body size in pre-arrest ccdc-55 larvae is normal. Eggs laid by rescued UN1139 or wildtype (N2) hermaphrodites were transferred to a new seeded plate, and larvae were analyzed after 18, 23, 28, 32, or 40 hours of incubation at 20°. Gonad length and body length were assessed at each time point using DIC imaging and Spot Advanced software. N2 larvae are represented by black triangles, rescued UN1139 by green circles, and non-rescued ccdc-55 larvae by orange squares. Linear regression of the data is indicated by the black, green, and orange lines. For nonrescued ccdc-55 animals, the 95% confidence interval of the regression is shown (orange wedge).
Highlights
The conserved coiled-coil domain protein CCDC-55 is a novel regulator of cell migration
CCDC-55 is necessary for larval development in C. elegans
CCDC-55 likely functions in the distal tip cells
ccdc-55 is found in an operon with the ubiquitin ligases rnf-121 and rnf-5
CCDC-55, RNF-121 and RNF-5 regulate the final position of the distal tip cells
Acknowledgments
C. elegans strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Center for Research Resources, National Institutes of Health. We thank Judith Kimble and Dana Byrd for the use of the DTC-specific RNAi strains. We thank Melissa LaBonty and Vivek Krishnan for help with C. elegans maintenance and experimental protocols, Limor Broday for helpful discussions and Hiba Tannoury for critical reading of the manuscript. This work was supported by grant GM085077 from the National Institutes of Health to E.J.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Baum PD, Garriga G. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants. Neuron. 1997;19:51–62. doi: 10.1016/s0896-6273(00)80347-5. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 1999;399:586–590. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Gleason KS. Caenorhabditis elegans operons: form and function. Nat Rev Genet. 2003;4:112–120. doi: 10.1038/nrg995. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Dufourcq P, Sookhareea S, Labouesse M. RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics. 1999;153:1245–1256. doi: 10.1093/genetics/153.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday L, Kolotuev I, Didier C, Bhoumik A, Podbilewicz B, Ronai Z. The LIM domain protein UNC-95 is required for the assembly of muscle attachment structures and is regulated by the RING finger protein RNF-5 in C. elegans. J Cell Biol. 2004;165:857–867. doi: 10.1083/jcb.200401133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly L, Cassada RC, Russell RL. The life cycle of the nematode Caenorhabditis elegans. I. Wild-type growth and reproduction. Dev Biol. 1976;51:23–33. doi: 10.1016/0012-1606(76)90119-6. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EB, Stern MJ. Understanding cell migration guidance: lessons from sex myoblast migration in C. elegans. Trends Genet. 1998;14:322–327. doi: 10.1016/s0168-9525(98)01507-8. [DOI] [PubMed] [Google Scholar]
- Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell. 2004;6:183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- Cinar HN, Chisholm AD. Genetic analysis of the Caenorhabditis elegans pax-6 locus: roles of paired domain-containing and nonpaired domain-containing isoforms. Genetics. 2004;168:1307–1322. doi: 10.1534/genetics.104.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 1998;281:706–709. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Clark SG, Schwarzbauer JE. Talin loss-of-function uncovers roles in cell contractility and migration in C. elegans. J Cell Sci. 2003;116:3871–3878. doi: 10.1242/jcs.00705. [DOI] [PubMed] [Google Scholar]
- Cram EJ, Shang H, Schwarzbauer JE. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci. 2006;119:4811–4818. doi: 10.1242/jcs.03274. [DOI] [PubMed] [Google Scholar]
- Darom A, Bening-Abu-Shach U, Broday L. RNF-121 is an endoplasmic reticulum-membrane E3 ubiquitin ligase involved in the regulation of beta-integrin. Mol Biol Cell. 2010;21:1788–1798. doi: 10.1091/mbc.E09-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier C, Broday L, Bhoumik A, Israeli S, Takahashi S, Nakayama K, Thomas SM, Turner CE, Henderson S, Sabe H, et al. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol Cell Biol. 2003;23:5331–5345. doi: 10.1128/MCB.23.15.5331-5345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Lecca A, Blumenthal T. RNA polymerase II C-terminal domain phosphorylation patterns in Caenorhabditis elegans operons, polycistronic gene clusters with only one promoter. Mol Cell Biol. 2010;30:3887–3893. doi: 10.1128/MCB.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettner SN, Kenyon C, Reichardt LF. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- Hall DH, Altun ZF. C. elegans Atlas. Cold Spring Harbor Laboratory Press; New York: 2008. [Google Scholar]
- Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- Karp X, Greenwald I. Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev Biol. 2004;272:460–469. doi: 10.1016/j.ydbio.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Kawano T, Zheng H, Merz DC, Kohara Y, Tamai KK, Nishiwaki K, Culotti JG. C. elegans mig-6 encodes papilin isoforms that affect distinct aspects of DTC migration, and interacts genetically with mig-17 and collagen IV. Development. 2009;136:1433–1442. doi: 10.1242/dev.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerins JA, Hanazawa M, Dorsett M, Schedl T. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev Dyn. 2010;239:1555–1572. doi: 10.1002/dvdy.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen MT, Sybingco SS. Netrin, Slit and Wnt receptors allow axons to choose the axis of migration. Dev Biol. 2008;323:143–151. doi: 10.1016/j.ydbio.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Kim YD, Lee JY, Oh KM, Araki M, Araki K, Yamamura K, Jun CD. NSrp70 is a novel nuclear speckle-related protein that modulates alternative pre-premRNA splicing in vivo. Nucleic Acids Res. 2011;39:4300–4314. doi: 10.1093/nar/gkq1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Kuroki R, Nishiwaki K. A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr Biol. 2004;14:2011–2018. doi: 10.1016/j.cub.2004.10.047. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc. 2010;5:1495–1517. doi: 10.1038/nprot.2010.107. [DOI] [PubMed] [Google Scholar]
- Lee M, Cram EJ. Quantitative analysis of distal tip cell migration in C. elegans. Methods Mol Biol. 2009;571:125–136. doi: 10.1007/978-1-60761-198-1_8. [DOI] [PubMed] [Google Scholar]
- Lee M, Cram EJ, Shen B, Schwarzbauer JE. Roles for beta(pat-3) integrins in development and function of Caenorhabditis elegans muscles and gonads. J Biol Chem. 2001;276:36404–36410. doi: 10.1074/jbc.M105795200. [DOI] [PubMed] [Google Scholar]
- Lehmann R. Cell migration in invertebrates: clues from border and distal tip cells. Curr Opin Genet Dev. 2001;11:457–463. doi: 10.1016/s0959-437x(00)00217-3. [DOI] [PubMed] [Google Scholar]
- Levy-Strumpf N, Culotti JG. VAB-8, UNC-73 and MIG-2 regulate axon polarity and cell migration functions of UNC-40 in C. elegans. Nat Neurosci. 2007;10:161–168. doi: 10.1038/nn1835. [DOI] [PubMed] [Google Scholar]
- Lin P, Huang LH, Steward R. Cactin, a conserved protein that interacts with the Drosophila IkappaB protein cactus and modulates its function. Mech Dev. 2000;94:57–65. doi: 10.1016/s0925-4773(00)00314-2. [DOI] [PubMed] [Google Scholar]
- Lucanic M, Cheng HJ. A RAC/CDC-42-independent GIT/PIX/PAK signaling pathway mediates cell migration in C. elegans. PLoS Genet. 2008;4:e1000269. doi: 10.1371/journal.pgen.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- Meighan CM, Schwarzbauer JE. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors. Genes Dev. 2007;21:1615–1620. doi: 10.1101/gad.1534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz DC, Alves G, Kawano T, Zheng H, Culotti JG. UNC-52/perlecan affects gonadal leader cell migrations in C. elegans hermaphrodites through alterations in growth factor signaling. Dev Biol. 2003;256:173–186. doi: 10.1016/s0012-1606(03)00014-9. [DOI] [PubMed] [Google Scholar]
- Merz DC, Zheng H, Killeen MT, Krizus A, Culotti JG. Multiple signaling mechanisms of the UNC-6/netrin receptors UNC-5 and UNC-40/DCC in vivo. Genetics. 2001;158:1071–1080. doi: 10.1093/genetics/158.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K. Mutations affecting symmetrical migration of distal tip cells in Caenorhabditis elegans. Genetics. 1999;152:985–997. doi: 10.1093/genetics/152.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki K, Hisamoto N, Matsumoto K. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science. 2000;288:2205–2208. doi: 10.1126/science.288.5474.2205. [DOI] [PubMed] [Google Scholar]
- Rogalski TM, Gilchrist EJ, Mullen GP, Moerman DG. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics. 1995;139:159–169. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Blasamo M, Condeelis J, Oktay MH, Burge CB, Gertler FB. An EMT-Driven Alternative Splicing Program Occurs in Human Breast Cancer and Modulates Cellular Phenotype. PLoS Genet. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood DR. Cell invasion through basement membranes: an anchor of understanding. Trends Cell Biol. 2006;16:250–256. doi: 10.1016/j.tcb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Kidd AR, 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004;166:171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tamai KK, Nishiwaki K. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev Biol. 2007;308:562–571. doi: 10.1016/j.ydbio.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Tannoury H, Rodriguez V, Kovacevic I, Ibourk M, Lee M, Cram EJ. CACN-1/Cactin interacts genetically with MIG-2 GTPase signaling to control distal tip cell migration in C. elegans. Dev Biol. 2010;341:176–185. doi: 10.1016/j.ydbio.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Miller S, Kaminsky R, Broday L. Molting-specific downregulation of C. elegans body-wall muscle attachment sites: the role of RNF-5 E3 ligase. Biochem Biophys Res Commun. 2010;395:509–514. doi: 10.1016/j.bbrc.2010.04.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
let-2 and unc-52 do not require CCDC-55 for normal alternative splicing. A: In order to investigate a possible role for CCDC-55 in splicing, we obtained a reporter strain (LET-2∷GFP/RFP ybIs1371 [myo-3∷let-2-9G myo-3∷let-2-10R]), in which a shift from green (GFP) to red (RFP) fluorescence indicates alternative splicing within a modified let-2 transcript (Kuroyanagi et al., 2010). Stop codons have been introduced into exon 9 (E9) or 10 (E10) of let-2 DNA such that inclusion of exon 9 leads to expression of GFP and inclusion of E10 results in the expression of RFP. The fluorescence gradually shifts from green to red as the animals develop, reflecting the splicing patterns of endogenous let-2 (Kuroyanagi et al., 2010). The LET-2∷GFP/RFP animals were treated with ccdc-55 or empty RNAi vector L4440 as a negative control, and L4 (48 hour) and adult (72 hour) individuals were imaged to quantify the level of green and red fluorescence. RFP and GFP fluorescence intensity was determined by imaging at 20× magnification using standardized exposure conditions. Amount of fluorescence in each channel was determined using ImageJ. ccdc-55 RNAi produced no significant difference in the overall splicing pattern. B–D: The allele e669 results in a stop codon in an alternatively spliced exon of unc-52. Exclusion of this exon by alternative splicing results in suppression of the late larval-onset paralysis phenotype (Rogalski et al., 1995). Movement of animals was assessed on plates after 48 hours of treatment. Randomly selected fields of view were imaged on plates using a Nikon SMZ800 dissecting microscope equipped with a Spot Insight 2 camera. Paralyzed animals were identified by their stick-straight body posture and curved animals were scored as normal. B: Negative control empty vector RNAi, C: positive control asd-2 RNAi and D: ccdc-55 RNAi.
Relationship between gonad size and body size in pre-arrest ccdc-55 larvae is normal. Eggs laid by rescued UN1139 or wildtype (N2) hermaphrodites were transferred to a new seeded plate, and larvae were analyzed after 18, 23, 28, 32, or 40 hours of incubation at 20°. Gonad length and body length were assessed at each time point using DIC imaging and Spot Advanced software. N2 larvae are represented by black triangles, rescued UN1139 by green circles, and non-rescued ccdc-55 larvae by orange squares. Linear regression of the data is indicated by the black, green, and orange lines. For nonrescued ccdc-55 animals, the 95% confidence interval of the regression is shown (orange wedge).