Abstract
In this paper, we describe and evaluate a system that extracts clinical findings and body locations from radiology reports and correlates them. The system uses Medical Language Extraction and Encoding System (MedLEE) to map the reports’ free text to structured semantic representations of their content. A lightweight reasoning engine extracts the clinical findings and body locations from MedLEE’s semantic representation and correlates them. Our study is illustrative for research in which existing natural language processing software is embedded in a larger system. We manually created a standard reference based on a corpus of neuro and breast radiology reports. The standard reference was used to evaluate the precision and recall of the proposed system and its modules. Our results indicate that the precision of our system is considerably better than its recall (82.32–91.37% vs. 35.67–45.91%). We conducted an error analysis and discuss here the practical usability of the system given its recall and precision performance.
Keywords: Natural language processing, Knowledge base, Data extraction, BI-RADS
Background
It is generally believed that databases of structured medical data will help improve the diagnostic workflow, the educational and teaching process, management of care, and clinical research: see for instance the recent series of radiology reporting publications [1–3]. Despite continuing efforts to help clinicians enter data in a structured manner [4–6], a significant portion of the medical data (the vast majority of radiology and pathology reports) is still recorded in free text as it is convenient to use and allows for the expression of subtle nuances. It remains a challenge to automatically convert free text to structured database entries of the desired type.
Natural language processing (NLP) tools are available that structure free text. OpenNLP1 and GATE2 are general-purpose tools; Medical Language Extraction and Encoding System (MedLEE) [7, 8], cTAKES [9], and medKAT specialize in the medical domain. To embed these NLP tools in a system, we typically have to develop one or more post-processing modules that mine the NLP output. Depending on the nature of the overall system, the complexity of the post-processing modules ranges from simple parser modules to advanced reasoning engines.
In this paper, we describe a reasoning engine that infers the body location of findings that are extracted from free text radiology reports, as envisioned in [3]. We have divided this task into three subtasks: extracting finding and location phrases in narrative text; determining the syntactic relations between the phrases extracted; and connecting findings and locations by reasoning about the syntactic relations on a symbolic level. The three aforementioned medical NLP tools support the first subtask. In addition, as will be explained in “MedLEE section,” MedLEE’s output structure is excellently geared to solving the second subtask.
For this reason, and the fact that MedLEE [7, 8] is the state of the art in the field of medical NLP, we selected it as the preferred NLP engine for our study. In its simplest form, our reasoning engine merely filters MedLEE’s output structure, and assigns body locations to findings that were found by MedLEE to be related on syntactic grounds. Since MedLEE was not optimized for the task of correlating body locations and problem, we cannot expect superior results from the base system. Therefore, we extended this base system in two ways that reason about MedLEE’s output on a symbolic level.
MedLEE has been applied to a variety of natural language processing tasks, such as automated trend discovery in chest radiology reports [10], detection of adverse events [11], acquisition of disease drug knowledge [12], and classification of patient smoking status [13]. MedLEE can be licensed from NLP International.3
The reasoning engine can be used in a variety of applications. To support the diagnostic workflow, for instance, we can connect it to a retrieval system of previous cases to select all cases with, say, a neoplasm in the cerebellopontine angle [14]. If we have access to sufficient numbers of reports, we can use it for research purposes, e.g., to test the hypothesis that neoplasms in the lower inner breast quadrant are more likely to be malignant than neoplasms in the upper outer quadrant [10]. The reasoning engine can also be used to visualize the information in the reports by mapping the findings to a graphical representation of the imaged organ [15, 16].
The contributions of our study are twofold. First, we describe the finding–location correlation system in terms of the base system sketched above and its extensions. Second, we give an information–theoretic evaluation of the performance of the base system and its extensions on corpora of neuro and breast reports.
Methods
The reasoning engine we describe in this paper is built on MedLEE and encompasses two sub-engines. The first correlates clinical findings and body locations that are extracted from the MedLEE output. The second normalizes and completes breast locations and does not apply to non-breast reports.
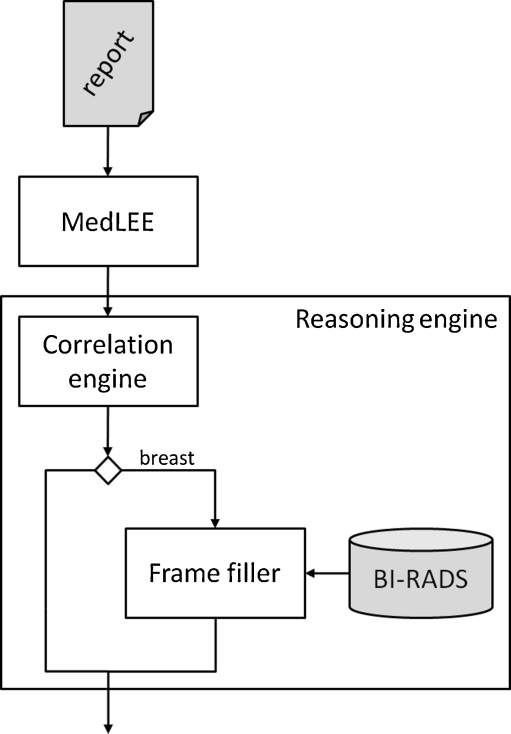
MedLEE splits the report into sentences and maps them to a normalized structure, reflecting the semantic components of the sentence (i.e., its phrases) and their interrelations. The resulting structure encompasses a labeling of the phrases such as clinical finding and body location. MedLEE’s output is transferred to the correlation engine, which assigns body locations to clinical findings. The frame filler completes partially specified breast locations based on a dedicated breast imaging vocabulary; it only considers the locations in breast reports. See Fig. 1 for an overview of the overall system.
Fig. 1.
Overview of the overall system
MedLEE
MedLEE comprises a frame-based parser that detects the grammatical structure of sentences, maps the structure found to a frame, and fills the slots in the frame with phrases. The filled frames are subjected to several normalization steps so that all phrases reside in a controlled vocabulary. In one of its settings, MedLEE returns its output in extensible markup language (XML).
Several aspects of MedLEE’s output will be of particular interest for our purposes. First, MedLEE labels the terms with their respective categories such as problem, finding, bodyloc, region, and procedure. This will allow us to extract findings and body locations from the text. Second, we can assume that the phrases that are inserted in the slots in a frame have a particular semantic relation defined by that frame.
As an example, consider the sentence This lesion is suspicious for a neoplasm such as a brainstem glioma or astrocytoma. MedLEE’s XML output comes in two parts: tagged text and structured text. In the tagged text, the sentence is split at phrase level (see Fig. 2). In the structured part, the frames are coded using XML’s nested structure (see Fig. 3). The names of the XML elements (e.g., problem and bodyloc) correspond to MedLEE’s categories. From the structured text part, using the nested structure, we can derive that the glioma and the astrocytoma are positioned in the brainstem. Note that the brainstem bodyloc is correctly applied to both the glioma and the astrocytoma based on the conjunction “or.”
Fig. 2.
Tagged text of This lesion is suspicious for a neoplasm such as a brainstem glioma or astrocytoma
Fig. 3.
Structured text of sentence from Fig. 2. We have removed some attributes and elements that are not pertinent to our discussion. We have abbreviated some elements <t… > </t > without inner text to <t…/>, as usual
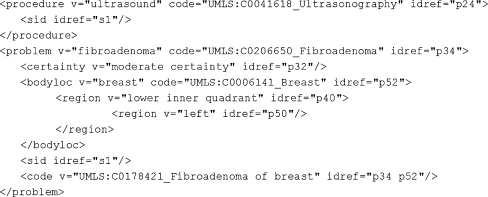
The structured text of the sentence Ultrasound evaluation demonstrates probable fibroadenoma in the left lower inner quadrant is shown in Fig. 4. The body location left lower inner quadrant is represented by a nesting of two region elements and no bodyloc element.
Fig. 4.
Structured text of Ultrasound evaluation demonstrates probable fibroadenoma in the left lower inner quadrant
Finally, consider the tagged text of the sentence There has been interval development of a 1 cm density in the far posterior medial, inferior right breast in Fig. 5. MedLEE failed to index the phrase inferior right breast. Consequently, there is no means of referring to it in the structured text (Fig. 6), and therefore, the density problem does not mention it as its body location.
Fig. 5.
Tagged text of There has been interval development of a 1 cm density in the far posterior medial, inferior right breast
Fig. 6.
Structured text of sentence from Fig. 5
Of lesser interest for the purpose of our paper are the code and certainty attributes. The first map terms to elements in the Unified Medical Language System.4 The certainty elements represent the status of the term in the report (e.g., present, possibly present, absent).
Correlation Engine
We extracted the problem and finding elements from the MedLEE output, e.g., neoplasm, glioma, and astrocytoma from Fig. 3. Similarly, we extracted the bodyloc elements and region elements. If any of them contained nested region elements, we collected them as adjectives accompanying the root element. Thus, we extracted brainstem from Fig. 3, left lower inner quadrant from Fig. 4, and posterior medial from Fig. 6.
Our correlation engine assigns at most one body location to each problem (i.e., problem or finding element), to which end it applies two rules of inference. The first rule, the in-XML rule filters the MedLEE output: a problem is assigned the body location or region that is among its children, if it has one. If it has multiple body locations and/or regions, the first body location is selected. We chose to manage multiple body locations in this way because it was not clear to us what the semantic relation is between body location/region siblings. For instance, for some problems A with body location B and region C, the concatenation of B and C was the most accurate descriptor of A’s body location, whereas in other instances B and C refer to distinct areas.
Since MedLEE was not optimized for the task of correlating body locations and problems, no superior results can be expected from the in-XML rule. We therefore define a second rule, called the in-sentence rule.
For problems that have not yet been assigned a body location by the previous rule, the in-sentence rule assigns the body location in the sentence in which the problem appears, provided that sentence contains precisely one unique body location. If neither of the two rules is applicable, the problem is assigned no body location.
For instance, the in-XML rule applies to the third and fourth problem in Fig. 3. It assigns brainstem to glioma and astrocytoma. The in-sentence rule assigns brainstem to both lesion and neoplasm since this is the only body location appearing in the sentence in which the problems appear. The in-XML rule assigns left lower inner quadrant to fibroadenoma (Fig. 4) and posterior medial to density (Fig. 6).
BI-RADS Frame Filler
We observed that the breast reports often require contextual information to determine the exact breast positions to which their body locations refer. Consider for instance the following sentences: There are several scattered smaller cysts within the left breast. A 1.8 × 2.1 cm simple cyst is present at the 12 o'clock position. The phrase 12 o’clock position is ambiguous between the 12 o’clock position in the left and right breast. We need the first sentence to resolve this ambiguity. Similarly, the phrase left lower inner quadrant is incomplete in the sentence Ultrasound evaluation demonstrates probable fibroadenoma in the left lower inner quadrant. It requires domain knowledge to infer that this refers to the lower inner quadrant of the left breast, and not, for instance, of the left axillary region.
We developed an engine that completes partial breast region locations. The engine uses a representation of breast regions that is based on Breast Imaging-Reporting and Data System (BI-RADS). This system [4] provides a standardized vocabulary for breast radiology that comprises a comprehensive terminology for describing breast regions (see Table 1).
Table 1.
BI-RADS-based location descriptors
| Laterality | Depth | Region | Breast location |
|---|---|---|---|
| Left | Anterior | Breast | Superior |
| Right | Middle | Subareolar region | Medial |
| Both | Posterior | Central region of breast | Inferior |
| Axilla region | Upper outer quadrant | ||
| Axillary tail region | Upper inner quadrant | ||
| Lower outer quadrant | |||
| Lower inner quadrant | |||
| 1–12 o’clock position |
A lexical matcher maps each body location to a BI-RADS object, accounting for morphological variations. BI-RADS objects have four attributes that can take the values as specified in Table 1. For instance, axillae are mapped to the following BI-RADS object:
laterality = both
depth = ?
region = axilla
breast location = ?
The lexical matcher yields a series of BI-RADS objects. A BI-RADS object is partial if either its laterality or its region is unknown. The frame filler completes the partial BI-RADS objects on the basis of the preceding BI-RADS objects. As for completing laterality values of BI-RADS objects, we distinguish the root XML element of the body location on which it is based. BI-RADS objects with an unknown laterality value that belong to a procedure element are assigned the value “both” since procedures (e.g., ultrasound examinations) are typically performed bilaterally unless specified otherwise. A BI-RADS object that does not belong to a procedure element simply inherits the laterality of its preceding BI-RADS object.
As for completing region values, the frame filler uses a crude rule: it sets unknown region attributes to the default value “breast.” The pseudocode in Fig. 7 implements the above rules for completing partial breast locations. Note that the BI-RADS frame filler also completes body locations that were missed by MedLEE. It assigns “breast” to the region of the BI-RADS object corresponding to the phrase posterior medial from Figs. 5 and 6.
Fig. 7.
Pseudocode for completing partial BI-RADS object
Evaluation
For the development and evaluation of the system, we used two corpora of deidentified radiology reports in the English language. The first corpus consists of 860 neuroradiology reports obtained from a US-based radiology institute. The second consists of more than 500 breast cancer reports obtained from a US-based university hospital. Six hundred sixty neuro reports and 119 breast reports were randomly selected from these respective corpora for evaluation. The remaining 200 neuro reports were used in the development phase of the correlation engine; no breast reports were used in this phase. A fraction of the remaining breast reports were used in the development phase of the BI-RADS frame filler.
We evaluated the extent to which the proposed engine accurately extracts the problem–body location pairs from the corpus in terms of its recall and precision. In our evaluation, we regarded the reasoning engine as a module that accepts a corpus of reports and returns a list of tuples (A, B)j extracted from the corpus, where A and B are strings representing a problem and body location, respectively, and j is an index pointing at the coordinates of A in a report from the corpus. We call j a “report coordinate.” If the problem A at report coordinate j does not have a body location, we designated this by writing the empty string for B.
We defined the following parameters:
True positive (tp): number of pairs (A, B)j extracted in which A at report coordinate j is a problem with body location B.
False positive (fp): number of pairs (A, B)j extracted in which A at report coordinate j is not a problem, or B is not a body location, or A does not have body location B.
False negative (fn): number of problems A with report coordinate j and body location B for which (A, B)j is not extracted.
Recall (or sensitivity) is the fraction of problem–location pairs that are accurately extracted: tp/(tp + fn). Precision (or positive predictive value) is the fraction of extracted pairs (A, B)j that are accurate: tp/(tp + fp).
The reference standard was created by the first author whose academic background is in computer science. He has over 4 years of NLP experience in the neuro domain and over 2 years in the breast domain. The reference standard consisted of two components, one for evaluating the system’s recall and the other for evaluating its precision.
The first component was based on all occurrences of ischemia and meningioma in the evaluation part of the neuro reports and all occurrences of carcinoma, lesion, mass, and (micro)calcification in the evaluation part of the breast reports, allowing for minor morphological variations. These terms were selected for two reasons. First, we considered them typical for the respective corpora; in the neuro corpus, the two selected terms account for 9.7% of all problems extracted from the neuro reports, while the breast terms account for 34.9% of all problems extracted from the breast reports. Second, this set is a mixture of diagnoses (meningioma and carcinoma) and observations (ischemia, lesion, mass, and microcalcification). We selected four terms for the breast corpus as it was smaller in size. The number of occurrences of each term is given in Table 6; in total, the neuro set contains 812 occurrences and the breast set 194.
Table 6.
Recall results for neuro terms
| Ischemia | Meningioma | Total | |
|---|---|---|---|
| No. of occurrences | 279 | 533 | 812 |
| No. of occurrences with location | 242 | 526 | 768 |
| % Retrieved | 29.34 | 38.6 | 35.67 |
| % Retrieved by in-XML rule | 16.94 | 25.67 | 22.91 |
| % Retrieved by in-sentence rule | 12.40 | 12.93 | 12.76 |
| No. of occurrences without location | 37 | 7 | 44 |
| % Retrieved | 100 | 85.71 | 97.73 |
For each occurrence A of these terms at report coordinate j, the first author determined A’s body location B (the empty string if there is none) and whether (A, B)j was extracted. If (A, B)j was not extracted, the term A was labeled as a negative instance; otherwise, it was labeled as a positive instance. We accepted significant morphological and lexical variations between the problem and body location in the report and the strings representing them in (A, B)j.
The second component of the reference standard was based on a random selection of 1,000 pairs of (A, B)j extracted from the patient history, impressions, and conclusion sections of the neuro reports and 500 extracted from the same sections of the breast reports, accounting for 8.5% and 37.1% of all pairs extracted from the respective corpora. For each pair in which B is not the empty string, an assessment was made of whether A is mentioned at report coordinate j and whether B is a sufficient description of A’s body location. The pair was labeled negative if A was not mentioned at report coordinate j or if B was an insufficient description of A’s body location. B was regarded as insufficient if B was not a body location (e.g., in The patient left in a stable condition the word left does not refer to the left breast), if B was incorrect (e.g., left axilla vs. left breast), or if B was significantly incomplete (e.g., posterior medial breast is significantly incomplete with respect to posterior medial, inferior right breast (see Fig. 5), but posterior medial, inferior breast is significantly complete). The pair was labeled positive if it was not labeled negative.
A second annotator was invited to create a benchmark annotation for the breast reports. We decided to let him annotate all breast instances and none of the neuro instances as he indicated that he was more familiar with the breast domain than the neuro domain. The second annotator was a staff member of the first and second authors’ department, with an academic background in biomedical engineering. The second annotator had not been exposed to the reports or any aspect of this research; in fact, he was hired after the first version of this paper was submitted for review.
Like the first annotator, the second annotator labeled instances positive and negative. The “inter-rater matrix” in Table 2 compares the two annotations. The inter-rater agreement between the standard reference and the benchmark annotation is quantified by means of Cohen’s κ [17]. The purpose of this metric is to factor out the probability that the raters agree on the label of an instance by mere chance: S = (p1/t) × (p2/t) + (n1/t) × (n2/t). Using the notation of Table 2, κ is defined as (P − S)/(1 − S), where P = (a + d)/t.
Table 2.
Inter-rater matrix between standard reference and benchmark annotation
| Benchmark annotation | ||||
|---|---|---|---|---|
| Positive | Negative | |||
| Standard reference | Positive | a | b | p1 = a + b |
| Negative | c | d | n1 = b + d | |
| p2 = a + c | n2 = b + d | t = a + b + c + d | ||
Results
In this section, we quantify the pairs (A, B)j extracted by the system and specify the inter-rater agreements between the standard reference and the benchmark annotation. Then, we use the standard reference to measure the system’s recall and precision. We shall also specify the recall and precision of the in-XML rule and the correlation engine.
Problems and Location Pairs Extracted by System
Table 3 gives a quantitative summary of the problems, body locations, and connections between problems and body locations found by the system.
Table 3.
Quantitative summary
| Neuro | Breast | |
|---|---|---|
| No. of problems | 16,482 | 1,520 |
| No. of unique problems | 518 | 143 |
| No. of problems with location | 9,473 | 615 |
| % Derived by in-XML rule | 79.24 | 65.37 |
| % Derived by in-sentence rule | 20.76 | 34.63 |
| No. of unique locations | 2,048 | 78 |
| No. of locations completed | N/A | 236 |
Roughly 57% (neuro) and 40% (breast) of the problems were assigned a body location. In both cases, the in-XML rule derived the majority of the body locations: 79.24% (neuro) and 65.37% (breast). The breast corpus contains relatively fewer unique body locations (12.68%) than the neuro corpus (21.68%). This might be due to the BI-RADS frame filler which maps body location phrases to standardized body location objects. 38.37% (236/615) of the breast locations were completed by the BI-RADS frame filler.
Inter-rater Agreement
The distribution of positively and negatively labeled instances of the recall annotations is given in Table 4. For this distribution, S = 0.50, P = 0.85, and κ = 0.70. Similarly, Table 5 shows the distribution for the precision annotations. For this distribution, S = 0.85, P = 0.98, and κ = 0.86.
Table 4.
Inter-rater matrix of recall annotation
| Benchmark annotation | ||||
|---|---|---|---|---|
| Positive | Negative | |||
| Standard reference | Positive | 87 | 21 | 108 |
| Negative | 8 | 78 | 86 | |
| 95 | 99 | 194 | ||
Table 5.
Inter-rater matrix of precision annotation
| Benchmark annotation | ||||
|---|---|---|---|---|
| Positive | Negative | |||
| Standard reference | Positive | 179 | 3 | 182 |
| Negative | 1 | 14 | 15 | |
| 180 | 17 | 197 | ||
Recall
MedLEE failed to extract keywords only rarely: two ischemia and three meningioma occurrences. These data points are ignored in the results listed in Table 6. The vast majority of the problems considered have a body location mentioned in the report (94.58% (242 + 526/279 + 533) for neuro; 81.12% for breast). In Table 6, we see that, for instance, 242 ischemia occurrences have a body location mentioned in the report. Of these 242 ischemia body location instances, 16.94% were retrieved by the in-XML rule and 12.40% by the in-sentence rule.
In Tables 6 and 7, we see that the system retrieved 35.67% of the body locations in the neuro domain and 45.91% in the breast domain. In both domains, in roughly two thirds of the cases in which problems were assigned a body location, this was done by the in-XML rule.
Table 7.
Recall results for breast terms
| Carcinoma | Lesion | Mass | (Micro) calcification | Total | |
|---|---|---|---|---|---|
| No. of occurrences | 32 | 57 | 48 | 57 | 194 |
| No. of occurrences with location | 30 | 49 | 31 | 49 | 159 |
| % Retrieved | 36.67 | 51.02 | 48.38 | 44.89 | 45.91 |
| (% not completed/% completed) | (23.34/13.33) | (30.61/20.4) | (32.26/16.13) | (30.61/14.28) | (29.56/16.35) |
| % Retrieved by in-XML rule | 16.67 | 30.61 | 35.48 | 36.73 | 30.82 |
| (% not completed/% completed) | (6.67/10) | (18.37/12.24) | (25.81/9.68) | (26.53/10.20) | (20.13/10.69) |
| % Retrieved by in-sentence rule | 20.00 | 20.41 | 12.90 | 8.16 | 15.09 |
| (% not completed/% completed) | (16.67/3.33) | (12.24/8.16) | (6.45/6.45) | (4.08/4.08) | (9.43/5.66) |
| No. of occurrences w/o location | 2 | 8 | 17 | 8 | 35 |
| % Retrieved | 100 | 100 | 100 | 100 | 100 |
The percentage of instances that were retrieved by the respective rules is broken down into the percentage of instances that were not completed by the BI-RADS frame filler and the percentage of instances that were completed, i.e., the values x and y in (x/y), respectively
Approximately one third (16.35% of 45.91%) of all breast body locations that were assigned to a problem were completed by the BI-RADS frame filler.
Of the problems that had a body location in the report, the in-XML rule had 22.91% recall in the neuro reports. In the breast reports, assuming all body locations that were completed by the BI-RADS frame filler were significantly incomplete, and therefore incorrect, the in-XML rule had 20.13% recall.
The bottom rows in Tables 6 and 7 give the recall for the occurrences that do not have a body location in the report. For instance, in There is no evidence of meningioma, no body location is specified for the meningioma problem. We see that the reasoning engine correctly assigns no body location to such instances in all but one of the cases.
Precision
All 1,500 problems appeared in the sentence from which they were extracted. Therefore, the results in Tables 8 and 9 focus on the precision of the assigned body locations. The overall precision of the assigned body locations ranges from 82.32% (neuro) to 91.37% (breast). In both domains, the precision of the in-XML rule is higher than that of the in-sentence rule.
Table 8.
Precision results for pairs (A, B)j extracted from the breast reports
| In-XML rule | In-sentence rule | Total | |
|---|---|---|---|
| No. of problems with location (% correct) | 514 (83.66) | 114 (76.32) | 628 (82.32) |
O the 1,000 pairs, 372 have no body location (i.e., B is the empty string). Values in parentheses represent the percentage of pairs that were correctly inferred by the respective rule
Table 9.
Precision results for pairs (A, B)j extracted from the breast reports, differentiating between the rule that derived the relevant body problem and whether it was completed by the BI-RADS frame filler
| In-XML rule | In-sentence rule | Total | |
|---|---|---|---|
| Not completed | 79 | 36 | 115 |
| (% Correct) | (100) | (94.44) | (98.26) |
| Completed | 61 | 21 | 82 |
| (% Correct) | (83.61) | (76.19) | (81.71) |
| Total | 140 | 57 | 197 |
| (% Correct) | (92.86) | (87.72) | (91.37) |
Out of 500 pairs, 303 had no body location
To evaluate the influence of the BI-RADS frame filler on the precision of the overall system, we separated the problems in the breast reports with body locations that were completed by the frame filler from the problems with body locations that were not (see Table 9). The precision of the overall system is more than 16% (98.26 minus 81.71) lower for problems with partial breast locations that were completed than for breast locations that were not.
The system that only applies the in-XML rule had higher precision than the system as a whole: it had 83.66% precision in the neuro domain and 92.86% in the breast domain. If we were to discard all partial body locations instead of completing them, precision would increase to 98.26. The subsystem that only applies the in-XML rule had 100% precision.
Discussion
We conclude that there was substantial inter-rater agreement (κ = 0.70 and κ = 0.85) between the standard reference and the benchmark annotation created by the second annotator for the breast reports. We believe that the agreement is sufficiently high to draw conclusions on the performance of the modules on the basis of the standard references. This is quite beneficial for our work as it means that there is no need to create further annotations of the data and that we do not need to decide how to aggregate conflicting annotations.
Table 10 summarizes the recall and precision results of the in-XML rule, which is merely a filter on MedLEE’s output, the correlation engine, which applies the in-XML rule and the in-sentence rule, and the entire reasoning engine, which encompasses the correlation engine and the BI-RADS frame filler. To measure the overall performance of the systems, we aggregated recall (for problems with a body location) and precision in the F‐measure, i.e., their harmonic mean: (2 × recall × precision)/(recall + precision).
Table 10.
Summary of the recall and precision scores of the in-XML rule, the correlation engine, and the entire reasoning engine for the neuro and breast reports
| Recall | Precision | F‐measure | ||
|---|---|---|---|---|
| In-XML rule | Neuro | 22.91 | 83.66 | 35.97 |
| Breast | 20.13 | 100 | 33.51 | |
| Correlation engine (in-XML rule + in-sentence rule) | Neuro | 35.67 | 82.32 | 49.77 |
| Breast | 29.56 | 98.26 | 45.45 | |
| Correlation engine + frame filler | Breast | 45.91 | 91.37 | 61.11 |
We observe that the recall scores of our systems are consistently lower than their precision scores. We wish to emphasize that these recall results are not indicative of MedLEE’s performance as a medical natural language processing toolkit.
The poor recall scores of the proposed system and its variants are due to a number of factors. First, we observed that quite a number of body locations were consistently interpreted incorrectly. For instance, in the neuro domain, MedLEE understood T1 and T2 as being the first and second thoracic vertebrae, respectively, whereas in this context they refer to T1 and T2 relaxography (MRI protocols). This happened in 19 cases. Also, the BI-RADS frame filler introduced body location errors. For instance, it assigned the default value breast to unknown region attributes. For most findings this is appropriate, but it is not in the case of lymph node-related findings, which are generally located in the axillary region.
Second, we saw that MedLEE occasionally failed to extract breast locations with multiple modifiers, such as upper inner quadrant of the left breast. We manually added the BI-RADS body location phrases to MedLEE’s vocabulary, but MedLEE would still occasionally miss these anatomy-specific terms. We concluded that MedLEE’s grammatical rules need to be updated as well to accommodate this type of body location. Our license did not allow us to modify MedLEE’s grammar, however, so we could not test this hypothesis.
The third type of missed correlation is caused by the fact that the system is incapable of making cross-sentence correlations. This is because MedLEE processes the reports in a sentence-by-sentence fashion, and the inference rules applied by the correlation engine can only connect body locations and problems that appear in the same sentence. However, radiology studies typically give a multidimensional assessment of findings (e.g., location, size, signal intensity, differential diagnoses) which are spread over multiple sentences, especially if the finding is complex. Since there is no reporting guideline that instructs radiologists to state problems and their locations in the same sentence, the correlation engine is expected to fail on complex problems in particular.
The first two error types are also discussed in [18]. They are essentially caused by the fact that MedLEE is not geared to the specifics of body location descriptors in our reports. It may not be hard to write patches for individual errors, such as extensions of the vocabulary and domain-sensitive rules that determine the meaning of abbreviations. But it remains to be seen how much work needs to be done in each new domain.
The third class of error calls for cross-sentence inference rules. These rules can be aided by anaphora resolution techniques that correlate anaphoric phrases with problems and body locations from previous sentences. To the best of our knowledge, no anaphora resolution software is currently available for medical reports [19]. General anaphora resolution software exists (e.g., OpenNLP and GATE), but it fails to account for domain-specific anaphora, such as part–whole co-references [20] or hypernym/hyponym co-references. The following sentences illustrate the latter type of co-reference: There is a spiculated mass in the left breast. The lesion measures 12 × 6 mm. The term lesion refers to the more specific phrase spiculated mass. A recent publication [21] gives a thorough analysis of the distribution of anaphora in a corpus of clinical reports, including radiology reports.
One of the reviewers suggests that the task of correlating problems and body locations considered in this paper is a type of (generalized) co-reference resolution. We find this an inspiring notion since we could think of a problem (e.g., meningioma) and a body location (e.g., anterior falx) as two linguistic devices that refer to the same discourse entity (e.g., the meningioma in the patient’s anterior falx). From this perspective, the correlation engine is an intra-sentence co-reference handler.
We saw that the heuristic in-sentence rule is less reliable than the in-XML rule. The precision of the former rule might be increased by endowing it with a table of admissible problem–location combinations, or alternatively with a table of forbidden problem–location combinations. Such a table of domain knowledge would for instance prevent the in-sentence rule from assigning left leg to headache on the basis of the sentence Numbness in left leg and headache. A table like this can be based on existing ontologies, such as Systematized Nomenclature of Medicine–Clinical Terms (SNOMED CT) or Unified Medical Language System. For instance, SNOMED CT has a finding-site relation that models exactly the relation we are interested in. An investigation should be carried out to see whether this relation is sufficiently rich for our purposes.
One of the reviewers suggests that statistical methods could also be used to model prior knowledge on the problem–location pairs. The reviewer considers that such statistics can be computed from collocation distributions of problems and body locations in sufficiently large corpora of medical texts, which could be extracted using available concept extraction tools [22]. If future research demonstrates that this approach is indeed useful, it would mean that establishing highly specific relations (i.e., relations between one particular problem descriptor and one particular body location descriptor) can be leveraged by collocation statistics, which are ipso facto not specific.
Collocation statistics can also be used to implement other rules, which might improve the recall of the system. For instance, such a rule may dictate that if there is one problem in a sentence that contains two or more body locations, we select the body location that co-occurs most frequently with the problem at hand. In its current form, the in-XML rule chooses the first body location element if a problem has several. By so doing, we ignore parts of the anatomical information that was attached to a problem. It is quite conceivable that an extension of this rule that knows when and how to aggregate multiple body locations would increase the recall of the correlation engine. Collocation statistics can also be used to this end.
In its current form, the overall system is particularly useful for applications that require high precision, such as tools for researching trends in sizeable report repositories. MedLEE was used in such a way in [23]. On the other hand, the system is less suitable for applications that require high recall, such as tools that summarize and/or visualize the contents of one particular report. Such an application is described in [15, 16], but no performance analysis is given of the NLP system used in these publications with respect to the problem–location correlation task, nor were we able to reproduce it from the description.
As discussed above, we only consulted neuro reports in the development phase of the correlation engine. In Table 10, we see that the aggregated performance of the correlation engine in the breast and neuro domains is comparable (F‐measure 45.45 vs. 49.77). These figures seem to indicate that the correlation engine is sufficiently domain-independent and may be applicable to other anatomical regions. Different domains may have different reporting styles, though, which may pose different NLP challenges. Dedicated engines, such as our BI-RADS frame filler, can be developed to face these challenges, resulting in an increased F‐measure of the overall system.
Conclusion
In this paper, we have described a system that automatically correlates body locations and clinical problems. The system was fully specified and relatively easy, and it should therefore be straightforward to reproduce. Our evaluation on the basis of two corpora shows that the system’s precision is satisfactory, but that its recall lags behind. We have outlined the repercussions of the usability of the system in real applications. We have sketched several ways to improve the results, one of which touches on anaphora resolution in medical data. This seems to us a new and exciting problem area for natural language processing. This system can be used in various applications, such as anatomy-based retrieval of cases, researching problem-finding site trends, and visualization of report data.
Acknowledgments
The authors gratefully acknowledge Marco Janssen for creating the benchmark annotation and the reviewers for their excellent comments and suggestions, which improved the paper considerably.
Footnotes
References
- 1.Reiner BI. The challenges, opportunities, and imperative of structured reporting in medical imaging. J Digit Imaging. 2009;22:562–568. doi: 10.1007/s10278-009-9239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiner B. Uncovering and improving upon the inherent deficiencies of radiology reporting through data mining. J Digit Imaging. 2010;23:109–118. doi: 10.1007/s10278-010-9279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reiner BI. Customization of medical report data. J Digit Imaging. 2010;23:363–373. doi: 10.1007/s10278-010-9307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breast Imaging Reporting and Data System Atlas. Reston: American College of Radiology; 2003. [Google Scholar]
- 5.Dunnick NR, Langlotz CP. The radiology report of the future: a summary of the 2007 Intersociety Conference. J Am Coll Radiol. 2008;5:626–629. doi: 10.1016/j.jacr.2007.12.015. [DOI] [PubMed] [Google Scholar]
- 6.Weiss DL, Langlotz CP. Structured reporting: patient care enhancement or productivity nightmare? Radiology. 2008;249:739–747. doi: 10.1148/radiol.2493080988. [DOI] [PubMed] [Google Scholar]
- 7.Friedman C, Alderson PO, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman C, Hripcsak G, et al. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc. 1999;6:76–87. doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savova GK, Masanz JJ, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hripcsak G, Austin JHM, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224:157–163. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 11.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. J Am Med Inform Assoc. 2005;12:448–457. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen ES, Hripcsak G, et al. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc. 2008;15:87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick PJ, Elhadad N, et al: Use of semantic features to classify patient smoking status. AMIA Annual Symposium, pp. 450–454, 2008 [PMC free article] [PubMed]
- 14.Sevenster M, van Ommering R, Qian Y: Bridging the text-image gap: a decision support tool for real-time PACS browsing. J Digit Imaging doi:10.1007/s10278-011-9414-x, in press [DOI] [PMC free article] [PubMed]
- 15.Sinha U, Dai B, et al. Interactive software for generation and visualization of structured findings in radiology reports. AJR Am J Roentgenol. 2000;175:609–612. doi: 10.2214/ajr.175.3.1750609. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DB, Taira RK, et al. Hyperad: augmenting and visualizing free text radiology reports. Radiographics. 1998;18:507–515. doi: 10.1148/radiographics.18.2.9536493. [DOI] [PubMed] [Google Scholar]
- 17.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20:37–46. doi: 10.1177/001316446002000104. [DOI] [Google Scholar]
- 18.Jain NL, Knirsch CA, et al: Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. AMIA Annual Symposium, pp. 542–6, 1996 [PMC free article] [PubMed]
- 19.Chapman WW, Savova GK, et al: Characteristics of Anaphoric Reference in Clinical Reports. AMIA Annual Symposium, pp. 1007, 2010
- 20.Feldman R, Sanger J. The Text Mining Handbook: Advanced Approaches in Analyzing Unstructured Data. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 21.Savova GK, Chapman WW, Zheng J, et al. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc. 2011;18:459–465. doi: 10.1136/amiajnl-2011-000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hripcsak G, Friedman C, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122:681–688. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]