Abstract
The novel antimicrobial peptide with a dimeric dendrimer scaffold, SB056, was empirically optimized by high-throughput screening. This procedure produced an intriguing primary sequence whose structure-function analysis is described here. The alternating pattern of hydrophilic and hydrophobic amino acids suggests the possibility that SB056 is a membrane-active peptide that forms amphiphilic β-strands in a lipid environment. Circular dichroism confirmed that the cationic SB056 folds as β-sheets in the presence of anionic vesicles. Lipid monolayer surface pressure experiments revealed unusual kinetics of monolayer penetration, which suggest lipid-induced aggregation as a membranolytic mechanism. NMR analyses of the linear monomer and the dendrimeric SB056 in water and in 30% trifluoroethanol, on the other hand, yielded essentially unstructured conformations, supporting the excellent solubility and storage properties of this compound. However, simulated annealing showed that many residues lie in the β-region of the Ramachandran plot, and molecular-dynamics simulations confirmed the propensity of this peptide to fold as a β-type conformation. The excellent solubility in water and the lipid-induced oligomerization characteristics of this peptide thus shed light on its mechanism of antimicrobial action, which may also be relevant for systems that can form toxic β-amyloid fibrils when in contact with cellular membranes. Functionally, SB056 showed high activity against Gram-negative bacteria and some limited activity against Gram-positive bacteria. Its potency against Gram-negative strains was comparable (on a molar basis) to that of colistin and polymyxin B, with an even broader spectrum of activity than numerous other reference compounds.
Introduction
Virtually all multicellular organisms rely on antimicrobial peptides (AMPs) to ward off pathogenic microbes (1). They are part of the innate immune system and in particular play a crucial role in organisms (the vast majority) that have not developed a more-sophisticated adaptive immune system. Even in higher vertebrates (including humans), AMPs such as defensins not only have direct microbicidal activity but also serve as signals that initiate, mobilize, and amplify adaptive immune host defenses (2). Much attention has been devoted in the last two decades or so to AMPs as a source of new anti-infective drugs, which are urgently needed to combat the growing bacterial resistance to conventional antibiotics (3). Beyond pure anti-infective activity, the potential therapeutic role of AMPs has been significantly broadened by an appreciation of their immunomodulatory properties (4). Although AMPs display a bewildering variety in their primary sequences, they generally share a cationic character and a globally amphipathic fold, with clearly distinguishable hydrophilic and hydrophobic faces. These structural features reflect their mode of action, which is primarily directed toward interacting with and damaging the target cell's plasma membrane (5). However, despite intense efforts to elucidate the mechanisms of action of AMPs, their exact roles in host defense, and possible ways of harnessing their therapeutic potential, little practical outcome has been achieved as yet. Naturally occurring AMPs have numerous drawbacks that limit their development into therapeutically applicable antibiotics, including their susceptibility to protease degradation, the high costs of manufacturing, and reduced activity in the presence of salts and divalent cations, such as those present in serum.
To overcome these problems, many researchers have developed mimics or peptidomimetics that are endowed with improved properties but retain the basic features of natural membrane-active AMPs, such as amphipathic design and cationic charge. Protein epitope mimetics, oligoacyllysines, ceragenins, synthetic lipidated peptides, peptoids, and other foldamers are only some of the routes explored so far (6,7). Among the various biomimetic approaches, multimeric peptides have recently received some attention (8–10). These so-called dendrimers are branched polymers in which peptides are attached centrally to a template or core matrix. Compared with their monomeric counterparts, dendrimeric peptides usually display increased activity, which is usually attributed to the higher local concentration of bioactive units in such polyvalent assemblies, as well as greater stability against peptidases and proteases.
Our goal in this study was to thoroughly characterize a semisynthetic peptide with a lipidated dimeric scaffold, named SB056 (Fig. 1). We investigated its antimicrobial activity against a wide range of bacteria and examined possible modes of interaction with model membranes. SB056 is a dendrimer with an entirely novel design that is meant to increase its performance against bacterial targets. To acquire detailed information about the structure-function relationship that governs its putative membrane interactions and antimicrobial activity, we compared the dendrimeric SB056 with the linear monomer throughout our studies. In this way, we expect to elucidate the exact role played by the dendrimeric scaffold and understand its significance for the peptide's mode of action. Inspection of the peptide sequence, which was previously obtained by semi-empirical optimization after high-throughput screening of a phage library (11), surprisingly revealed a striking pattern of charged and hydrophobic amino acids. With the exception of the first two residues, the sequence [WKKIRVRLSA] consists of alternating hydrophobic and cationic/polar amino acids. In this respect, it resembles the designer-made β-stranded model sequence [KIGAKI]3 (12) and several other peptides that are known to form β-sheets (see below). We thus predict that both SB056 and its linear monomer can form amphiphilic β-strands when in contact with a lipid bilayer, and we set out to verify this hypothesis as described below. Overall, the collected evidence provides a key for understanding the functional mechanism of SB056, and thus will allow further rational optimization of the dendrimeric agent.
Figure 1.
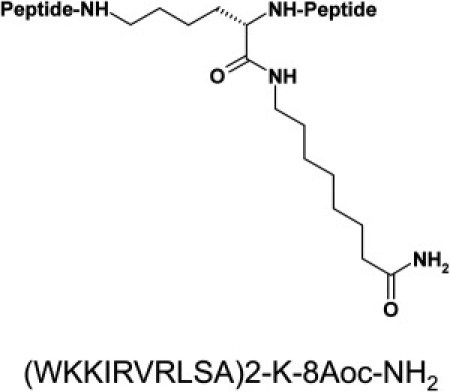
Molecular structure of SB056. Primary sequence and chemical structure of the dendrimeric (dimeric) AMP SB056. All of the amino acids have an L configuration, and 8-Aoc is 8-aminooctanoic acid, which is amidated in the SB056 construct. The monomeric peptide is amidated at the C-terminus.
Materials and Methods
Details regarding synthesis and determination of antimicrobial activity are provided in the Supporting Material.
CD spectropolarimetry
For CD spectropolarimetry, weighed amounts of the linear monomer or the dendrimeric SB056 were dissolved in 10 mM phosphate buffer to prepare stock solutions with concentrations of 50–200 μM. For further details on the sample preparation and CD spectra recording and analysis, see the Supporting Material.
NMR characterization
For liquid-state NMR structure analysis, both the dendrimeric and linear SB056 were dissolved at 1 mM concentration in 100% D2O (99.98% D; Sigma-Aldrich, St. Louis, MO), 10 vol% D2O in deionized H2O, or 30 vol% trifluoroethanol (TFE, 99.5% D; Sigma-Aldrich) in deionized H2O. In water, the final pH came to ∼6.0 at 298 K and was adjusted to 4.0 with small aliquots of HCl or NaOH 0.1 M to slow down the exchange of backbone amide protons with the solvent (13). See the Supporting Material for details on the NMR spectra recording and analysis.
Molecular-dynamics simulations
We obtained the peptide structures via a simulated annealing protocol using experimental NMR parameters as restraints, and ran molecular-dynamics (MD) simulations as described in the Supporting Material.
Surface-pressure measurements of peptide penetration in lipid monolayers
The insertion of the peptides into lipid monolayers was examined by means of a surface-pressure analysis as an indication of their ability to bind and penetrate microbial plasma membranes, as described in the Supporting Material.
Results
Determination of the minimum inhibitory concentration
The in vitro activity of SB056 and of numerous reference compounds was determined against ATCC strains and an expanded panel of recently collected clinical isolates (Table S1). All of the tested strains show resistance to several currently used antibacterial agents. SB056 has a good activity against Acinetobacter baumannii, Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia, with minimum inhibitory concentration (MIC) values in the range of 2–32 μg/ml. Against Proteus mirabilis and Serratia marcescens, SB056 exhibits higher MIC values of 32–>128 and >128 μg/ml, respectively. Polymyxin B shows an antibacterial potency similar to that of colistin, with MIC values lower than those of SB056, although the potency in terms of molarity is comparable (Table S2). The activities of SB056 against the Gram-positive bacteria Enterococcus faecalis, E. faecium, Staphylococcus epidermidis, and S. aureus are reported in Table S3. The dendrimeric peptide reveals a limited activity against enterococci with MIC values in the range of 8–128 μg/ml; against S. aureus the MIC range is 32–64 μg/ml, and against S. epidermidis the MIC value is 8 μg/ml. Colistin is not active against all Gram-positive strains tested, and polymyxin B is active only against S. epidermidis (32 μg/ml). A limited activity of SB056 against Mycobacterium smegmatis (MIC 64 μg/ml) and no activity against Candida spp. is observed (Table S4). Finally, in Table S5 we present a direct comparison between the activity of the dendrimeric SB056 and the linear monomer against standard Gram-negative and -positive reference strains. The MIC values of SB056 are decreased by several dilution steps, which clearly indicates the advantage of turning the linear monomer into the dendrimeric lipidated SB056.
CD structural analysis of the linear monomer and the dendrimeric SB056
We characterized the secondary structures of the linear monomer and the dendrimeric SB056 by circular dichroism (CD) to obtain a qualitative overview of their conformational preferences over a range of different conditions. These included unilamellar lipid vesicles of the same composition as used as in the subsequent surface-pressure experiments, as well as the widely used membrane-mimicking TFE/water mixtures to be used in the parallel NMR analysis. Fig. 2 shows the CD spectra of SB056 (linear and dendrimeric) measured in small unilamellar vesicles (SUVs) composed of DMPC or of DMPC/DMPG 1:1. In the presence of zwitterionic DMPC, the characteristic random coil lineshape suggests a mostly unordered structure for both peptides (see secondary structure estimation in Table S6), which is similar to the situation in pure phosphate buffer (see Fig. S1, Table S7, and Table S8). In anionic DMPC/DMPG liposomes, on the other hand, a pronounced conformational change is observed for the cationic peptides. The monomer shows a diminished mean residue ellipticity with two shallow minima at ∼200 and 215 nm, and a small positive Cotton effect around 190 nm. Even more pronounced is the change for the dendrimeric SB056, which exhibits a strong negative band at 195 nm, a broad positive band at ∼206 nm, and a less intense negative band below 220 nm. These spectral features clearly point to a mixture of β-sheet and β-turn, plus some unordered fraction. The quantitative secondary structure analysis of the data in Table S6 supports a substantial β-sheet content of 34% and 47%, plus β-turn fractions of 22% and 28%, for the linear monomer and the dendrimeric SB056, respectively. In agreement with the surface pressure data (see below), the excess of anionic DMPG charges in the vesicles obviously leads to electrostatic attraction of the cationic peptides. Once they are bound, the lipid environment induces a predominant β-type conformation, which is more pronounced for the dendrimeric SB056 than for the monomer. Because these fractions are taken on a mean residue ellipticity basis (and not on a total weight basis as in the MIC tests), they reflect the intrinsic tendency of the peptide chains to fold. Analogous CD measurements in the presence of 30% TFE (see Fig. S1) showed a preferential α-helical conformation, which is not unexpected because TFE is well known as a helix-inducing solvent (14). In contrast to the NMR experiments shown below, the CD measurements were carried out with low peptide concentrations (5–50 μM), where aggregation is of no concern (in the absence of a lipid bilayer surface). The CD results obtained in 30% TFE thus provide the secondary structure of monomeric (i.e., not aggregated) peptides in solution.
Figure 2.
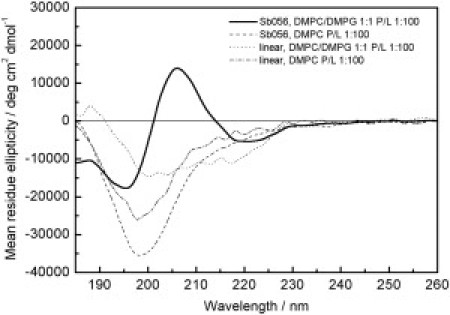
CD spectra of the linear monomer and the dendrimeric SB056 in the presence of membranes. Peptides were added to SUVs of DMPC and DMPC/DMPG (1:1) at a peptide/lipid ratio of 1:100. The lineshape of both peptides in zwitterionic DMPC is characteristic of an unordered conformation, whereas the spectra in anionic DMPC/DMPG show a combination of β-sheet, β-turn, and unordered fractions.
NMR characterization
We used 1H-NMR to analyze the structures of the dendrimeric SB056 and the linear monomer in more detail. To mimic a membrane environment, we first attempted to dissolve SB056 in SDS detergent micelles, but this led to the rapid formation of aggregates that were unsuitable for NMR. We therefore compared the peptides in pure water and in 30 vol% TFE, which was the minimum TFE concentration that induced any significantly ordered conformation in our CD analysis (see Fig. S1). Compared with the CD measurements at very low peptide concentrations (5–50 μM), however, the NMR analysis was carried out with much higher concentrations (∼1 mM). Although we observed no tendency for aggregation in TFE by CD, it is conceivable that intermolecular interactions may occur under NMR conditions due to the 100-fold higher concentration. Indeed, the first signs of peptide aggregation become apparent during these experiments: over a period of several days/weeks the TFE-containing NMR sample turned cloudy, and after prolonged storage we observed a fine precipitate at the bottom of the tube. Even though TFE is often regarded as a helix-inducing solvent (14), in the context of a peptide-peptide self-assembly equilibrium TFE can be expected to shift the system toward the more-structured conformation, regardless of whether it is α-helical or β-sheeted (15). The main effect of TFE is to enhance the formation of intra- and intermolecular hydrogen bonds, because the TFE molecules preferentially cluster around the peptide backbone, replacing the water (16–18).
On the basis of correlation spectroscopy (COSY) and total correlation spectroscopy (TOCSY) spectra, acquired both in pure water and in 30% TFE, all proton resonances from the linear monomer and the dendrimeric SB056 were assigned (Table S9). These results show neither significant differences between the linear monomer and the dendrimeric SB056 nor significant deviations from the random coil values in water (19). The values of 3JHNHα scalar coupling constants were obtained directly from the one-dimensional spectra to estimate the average values for the corresponding backbone Φ angles. In all cases, they are found to lie in the 5.5–7.1 Hz range, with a slight increase upon moving from water to 30% TFE, and hence they may reflect motional averaging of multiple conformations (13). Nevertheless, certain differences are observed in the rotational nuclear Overhauser effect spectroscopy (ROESY) and nuclear Overhauser effect spectroscopy (NOESY) spectra. Although no sequential crosspeaks were found for either peptide in pure water, some interesting 1HαN(i,i+1) signals could be assigned in 30% TFE, as shown in Fig. S2. However, no 1HNN(i,i+1) signals, which are generally expected in the case of a stable α-helical conformation (13,19), are observed. The dipolar interactions pattern turn out to be almost comparable for the two peptides, but they show slight differences at the C-terminus. In the case of the dendrimeric SB056, dαN(i,i+1) signals are observed without interruption along the entire sequence from K3 to L8 (and from K13 to L18 as defined on the other peptidic branch). On the other hand, the linear monomer shows dαN(i,i+1) from K3 to R7, as well as dαN(i,i+1) and dβN(i,i+1) between S8 and L9, which are absent in the dendrimeric SB056. Moreover, a medium-range dipolar interaction is observed between 8L-HN and 10A-Hβ. These results suggest a reduced mobility of the two peptides in 30% TFE compared with pure water. In particular, the linear monomer seems to form a kind of turn at the C-terminus, which is hindered in the dendrimeric SB056, presumably due to the attachment to the lysine linker and the resulting reduction in backbone flexibility.
As described in the Supporting Material, we applied a simulated annealing procedure to both peptides using the 3JHNHα scalar couplings and the ROE and NOE intensities as experimental parameters to restrain the Φ backbone angles and interproton distances, respectively. One hundred structures with the lowest potential energy were selected and analyzed. Fig. 3 summarizes the distribution of Φ and Ψ backbone angles on the Ramachandran plot. Of interest, for both peptides, all residues between K3 and R7 (defined as K13–R17 in the second peptidic branch) lie in the β region of the plot. Fig. 4 shows a representative structure of the linear monomer and the dendrimeric SB056, i.e., with the lowest root mean-square deviation (RMSD) from the computed average backbone conformation. A kind of coiled conformation of the N-terminus in both cases, as well as a turn at the C-terminus of the linear monomer, can be observed (Fig. 4). However, the error bars reported in Fig. 3 clearly emphasize the random-coil nature of both peptides, even in 30% TFE, in good agreement with the CD analysis (see also Supporting Material).
Figure 3.
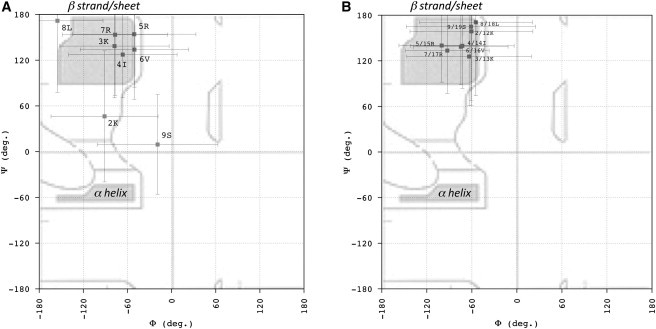
1H-NMR analysis of Φ and Ψ backbone angles on a Ramachandran plot for the linear monomer and the dendrimeric and linear SB056. A simulated annealing procedure was applied on both peptides, using 3JHNHα scalar couplings and the ROE/NOE intensities as experimental parameters to restrain the Φ backbone angles and interproton distances, respectively. In particular, dαN(I,i+1) provided the restraints for the Ψ backbone angles. One hundred structures with the lowest potential energy were selected and analyzed. The distributions of backbone Φ and Ψ angles on the Ramachandran plot were obtained for the linear peptide (A) and the dendrimeric SB056 (B). Regions typically allowed for all the standard amino acids (except for glycine) are indicated as bordered areas. Shaded regions represent the most common backbone conformations corresponding to the β-pleated and α-helical secondary structure.
Figure 4.
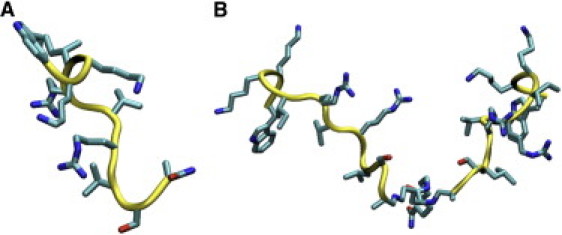
NMR-derived structures of the linear monomer (A) and the dendrimeric SB056 (B). For each peptide, the structure with the lowest RMSD from the average backbone conformation was obtained from a simulated annealing procedure with NMR-derived restraints.
MD simulations
It is important to note that the MD simulations were performed without NMR-derived restraints. Backbone RMSDs from the initial structure (Fig. S3) were higher, on average, in water than in 30% TFE for both peptides. Moreover, the residue root mean-square fluctuations (RMSFs; Fig. S4) indicate a lower conformational flexibility for both peptides in 30% TFE compared with pure water. In particular, it is interesting to note that the fluctuations of the two peptidic branches of the dendrimeric SB056 become comparable in 30% TFE, i.e., the essentially same results are obtained for homologous residues located in the two branches, in contrast to the situation observed in pure water (Fig. S4 B). The MD results for the backbone Φ and Ψ angles are consistent with these observations and with the NMR results. The Φ/Ψ distributions obtained for the linear monomer in water and in 30% TFE are shown in Fig. S5, under the same conditions as employed for the NMR experiments, and in Fig. S6 for the dendrimeric SB056.
Comprehensive Ramachandran plots are reported in Fig. 5. In water, all four quadrants are significantly populated for each of the two peptides, which is indicative of a random-coiled structure. Even though the β region is most populated, both backbone angles show considerable fluctuations, ranging from ∼−60° to −130° in Φ, and from ∼90° to 160° in Ψ. All residues exhibit a kind of bimodal distribution in these two ranges (Fig. S5 and Fig. S6) with maximum probabilities of ∼−70° and −120°, and ∼100° and 140°, respectively. This finding results in four combinations, which are almost equally populated (Fig. 5, A and B). Of interest, upon moving to 30% TFE, both positive Φ and negative Ψ contributions decrease. Moreover, backbone angles distributions in the β region change dramatically and retain only two significant maxima in the Ramachandran plot at ∼−120°/150° and −80°/90°. From residues K3–R7 (and K13–R17), both backbone Φ and Ψ angles follow a kind of alternating pattern whose maxima are already observable in pure water, whereas the distributions of K2, L8, and S9 turn out to be comparable in the two solvents investigated. This suggests that the addition of 30% TFE does indeed act in the context of a preexisting equilibrium, leading the peptide toward a preferred smaller set of conformations (15), in agreement with the NMR observations. Moreover, although the backbone angle distributions for homologous residues in the two peptidic chains of the dendrimeric SB056 are remarkably different in pure water, in 30% TFE they are almost identical to one another and comparable to the linear monomer. This finding is consistent with the RMSF values of the residues (Fig. S4) and supports the ordering role of TFE in this study.
Figure 5.
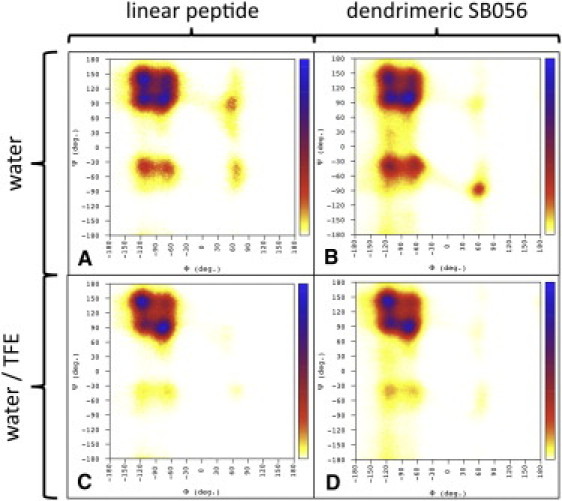
Ramachandran plots obtained from MD simulations. (A) Linear peptide in water. (B) Dendrimeric SB056 in water. (C) Linear peptide in 30% TFE. (D) Dendrimeric SB056 in 30%. The bar on the right represents the probability increasing from 0 to 1.
SB056 insertion into lipid monolayers
To gain further insight into the behavior of SB056 and the linear monomer in a membrane-like environment, we explored the interactions of these peptides with lipid monolayers by surface pressure analysis (20,21). As in the CD analysis, DMPC and DMPC/DMPG (50:50, w/w) were used to mimic neutral and anionic membranes. Both SB056 and the linear monomer efficiently penetrated into both of these monolayers, as demonstrated by the increase in film surface pressure π0. A large number of measurements, starting from a wide range of different initial surface pressures, are summarized in Fig. 6, showing a general trend for both peptides and both lipid compositions. We see that the incorporation of peptide into the monolayer decreases with increasing initial values of π0, as expected in view of the greater packing density of the lipid chains at higher pressures. In the plots of Δπ as a function of π0, the monolayer exclusion pressure can be derived by extrapolating the Δπ-π0 slope to Δπ = 0. This critical surface pressure corresponds to the lipid lateral packing density at which the intercalation of a peptide into the lipid film is prevented. In DMPC it has a value of 38 mN/m and 45 mN/m for the linear monomer and the dendrimeric SB056, respectively (Fig. 6, A and B). In the case of DMPC/DMPG, these values increase to 59 mN/m and 51 mN/m, respectively (Fig. 6, A and B). By replotting the same data in Fig. 6, C and D, for the two lipid systems, we can clearly appreciate that the membrane activity of the cationic peptides is enhanced by the presence of anionic DMPG. As in the case of many other cationic AMPs, electrostatic binding plays an important role in peptide-membrane interactions. We also note that SB056 with 10 positive charges displays an overall stronger intercalation activity than the linear monomer with only five charges. This difference is most pronounced in DMPC/DMPG at low π0-values, with an interesting crossover point at a surface pressure of ∼39 mN/m (Fig. 6 D), and significantly lower penetration is observed in DMPC for all π0-values (Fig. 6 C).
Figure 6.
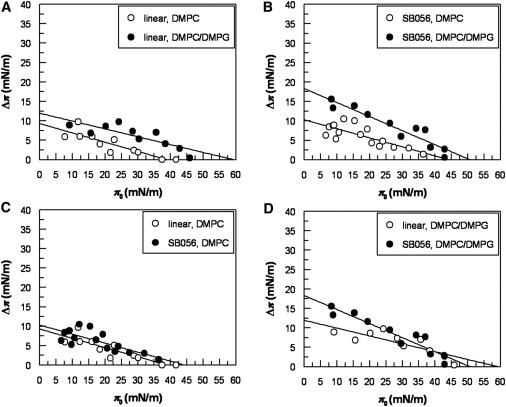
Intercalation of the linear monomer and dendrimeric SB056 into lipid monolayers. Peptide insertion into DMPC and DMPC/DMPG (1:1) was monitored from the increase in the surface pressure of lipid monolayers upon addition of 1.0 μM of the linear monomer (A) or the dendrimeric SB056 (B) into the subphase, as a function of initial surface pressure. In C and D, the same data are shown regrouped to allow a direct comparison between the two peptides in the different lipid systems.
The kinetics of the insertion of the peptides into lipid monolayers were monitored over a period of ∼30 min, and showed rather unusual time-courses for SB056 and the linear monomer. In Fig. S7 the curve of the linear monomer displays a rather constant increase in π0 with time, and within the timeframe of measurement no stabilization was reached. The curve of SB056 (Fig. S7 B) is characterized by a relatively fast peptide intercalation after injection into the subphase, followed by a gradual stabilization of π0, yet no plateau is reached. These slow, nonasymptotic kinetic patterns of intercalation differ fundamentally from those recorded previously for other cationic amphiphilic AMPs, such as the α-helical temporins and bombinins H from frog skins (21,22). Representative curves of the latter two peptides are shown in Fig. S7, C and D, respectively, for comparison. They are characterized by an explosive initial rise immediately after peptide addition, followed by an almost complete stabilization. This behavior indicates that the usual process of peptide binding and insertion into the monolayer is extremely rapid, with hardly any sign of structural relaxation or peptide reorganization. In the case of SB056 and particularly of its linear monomer, however, the whole process of peptide-membrane interaction seems to be considerably slower. Significantly more time is needed for the system to approach equilibrium, which in the case of the linear monomer is not even reached within the accessible timeframe of the experiment (i.e., before evaporation effects start to dominate).
In summary, the monolayer experiments revealed that both the linear monomer and the dendrimeric SB056 are indeed membrane-active, with SB056 displaying a stronger intercalation and faster kinetics than the linear monomer. These findings, together with the fact that monolayer penetration is enhanced by the presence of anionic DMPG, further support the idea that, just like the vast majority of AMPs studied so far, SB056 could also exert its antimicrobial activity by perturbing the plasma membrane of the target cell, with electrostatic binding playing an important role in the initial peptide-membrane interactions. The particular intercalation kinetics are rather different from and significantly slower than those recorded previously for α-helical peptides (21,22). The unusual surface-pressure curves most likely reflect some lateral peptide aggregation in the plane of the membrane, which is more efficient for SB056 than for the linear monomer. This difference makes sense, given that the two branches of the dimeric SB056 can cross-interact with one another and with neighboring peptides either before and/or during penetration of the membrane. Such behavior is expected to lead to a network of interacting β-strands with a high β-sheeted content.
Discussion
The novel AMP discussed here, SB056, belongs to the class of peptide dendrimers (branched polymers with peptides attached to a central template or core matrix). Dendrimer applications include immunogens and antigens, protein mimetics, de novo design of artificial proteins, new biopolymers, and biomaterials (23). The introduction in the late 1980s of the multiple antigenic peptide (MAP) system for the preparation of peptide immunogens was a significant advance in the field of multimeric peptides (24). With the use of MAP, multiple peptide sequences can be added to an inner core of radially branched lysine residues by standard solid-phase chemistry.
The peptide sequence of SB056 was derived by rational modification and optimization of an AMP [QEKIRVRLSA] that was originally identified by selecting a random phage library against whole E. coli cells (11). As a distinctive innovative feature, SB056 carries a short lipophilic chain to increase its membrane activity. We recently exploited the MAP approach to develop other peptide dendrimers with antimicrobial properties. SB041, a tetra-branched peptide carrying four identical sequences [pyrEKKIRVRLSA] on a lysine core, was found to be particularly active against Gram-negative strains, and it strongly bound E. coli and P. aeruginosa LPS in vitro, although this binding did not translate into LPS-neutralizing activity (10). Another recent study (8) showed that peptide-derivatized dendrimers have the potential for other anti-infective applications besides targeting bacterial pathogens. The tetra-branched compound SB105 and its derivative SB105-A10 were able to inhibit replication of several strains of human cytomegalovirus in both primary fibroblasts and endothelial cells, apparently by blocking the initial attachment of the virion to heparan sulfate on the cell surface. This mode of action appears to represent a novel mechanism that could make SB105 and its derivatives attractive candidates as members of an innovative class of antiviral drugs.
The results reported here for SB056 collectively show that this novel lipodimeric dendrimer displays excellent, albeit selective, antimicrobial activity, whereas its linear counterpart is only poorly active against most of the tested strains. This study thus confirms the advantage of a dendrimeric scaffold compared with isolated sequences. As was previously demonstrated for SB041, SB056 was active against Gram-negative bacteria, and it even retained some activity against Gram-positive strains. Although the potency of SB056 against Gram-negative bacteria was comparable to that of colistin and polymyxin B (on a molar basis), its spectrum of antimicrobial activity was even broader than that displayed by these two reference compounds, which are completely inactive against Gram-positive strains. Thus, the lack of broad-spectrum activity displayed by SB056 could be due to its preferential interaction with components of the outer membrane of Gram-negative bacteria, such as LPS, for which the comparable SB041 was previously demonstrated to have good affinity (10). On the other hand, SB056's residual activity toward Gram-positive strains could be explained by its inherent tendency to bind and perturb anionic lipid membranes.
The primary sequence [WKKIRVRLSA] of the peptidic part of SB056 reveals a striking pattern of alternating hydrophilic and hydrophobic amino acids, with the exception of the first two residues. Even though this basic pattern emerged from a random phage display library, its amphiphilic features are reminiscent of the model peptide [KIGAKI]3, which was designed as an amphiphilic β-strand (12). Of interest, this artificial β-stranded model peptide exhibited a high antimicrobial activity. Its structure was experimentally confirmed to be β-pleated in the presence of membranes (25), and extensive aggregation into extended β-sheets was recently demonstrated by solid-state NMR (26). We therefore predicted that the SB056 peptide might also be able to fold in a similar way. The comparative CD structure analysis of the linear monomer and the dendrimeric SB056 showed that both peptides have a mostly unordered conformation in aqueous solution and in the presence of uncharged DMPC liposomes. In contrast, in an anionic lipid environment both peptides assume predominantly β-type conformations, which is more pronounced for the dendrimeric SB056 than for the linear monomer.
We carried out more detailed analyses by NMR and MD in water, and in 30% TFE as a membrane-mimicking solvent. At low (micromolar) peptide concentrations, CD showed that the monomeric (i.e., not aggregated) peptides had a tendency to take on an α-helical conformation. When measured at a 100-fold higher concentration by NMR, however, intermolecular aggregation started to set in. Both the linear monomer and the dendrimeric SB056 largely maintained a random-coil structure, and an inherent folding tendency was discerned in the β-region of the Ramachandran plot. Both NMR and MD point to a distinct role of the more-hydrophobic environment provided by 30% TFE compared with pure water, which reduces the conformational flexibility and pushes the preexisting random-coil↔ordered equilibrium toward its preferential conformation. Specifically in the case of the dendrimeric SB056, the two peptidic chains appear to have comparable fluctuations in 30% TFE but not in pure water, thus supporting the ordering effect of TFE. However, because TFE is an isotropic solvent, it does not trigger peptide aggregation to any extent comparable to that of the amphiphilic surface of a lipid vesicle. A genuine membrane environment leads to a very high peptide concentration in the 2D plane and induces a preordering of the amphiphilic sequences. These two effects promote intermolecular β-strand aggregation, which is something TFE cannot do in an isotropic solution.
Overall, we have collected multiple pieces of evidence showing that the linear monomer and the dendrimeric SB056 have a strong tendency to fold as β-structures, as predicted from its primary amino-acid sequence with alternating hydrophobic and hydrophilic residues. The intrinsic preference to populate the β-region of the Ramachandran plot, as revealed by NMR and MD analysis, is explicitly confirmed by the CD data in anionic membranes and by the observed rapid aggregation of SB056 induced by SDS. Obviously, the presence of a lipid bilayer or a micellar surface enhances the formation of amphiphilic β-strands and aggregated β-sheets, provided that the cationic peptides are electrostatically attracted to the structure-inducing interface. Of note, SB056 exhibits a higher content of β-type conformations than the linear monomer.
All of our observations reinforce the idea that SB056 may exert its antimicrobial action by interacting with lipid membranes, and the branched dimeric structure of the dendrimeric peptide would further facilitate the formation and stabilization of such multimolecular aggregates within the bilayer. Intriguingly, the tendency for AMPs to exist in a semifolded or prefolded state in solution (i.e., before the interaction with bacterial membranes) was previously noted for α-helical peptides and correlated with antimicrobial activity (27,28). Thus, a higher propensity to assume any kind of folded conformation in aqueous solution seems to facilitate, both thermodynamically and kinetically, complete peptide folding in the membrane environment (6).
A number of naturally occurring and synthetic AMPs are known to self-assemble into β-sheet in membranes, and these AMPs may serve as a reference for SB056 and similarly structured peptides. Natural examples are defensins and protegrins with disulfide cross-linked structures (29). Engineered sequences include some β-sheet pore-forming peptides, selected from a combinatorial library based on a 26-residue framework designed to mimic membrane-spanning β-hairpins (30). Investigators have characterized the mechanism of action of these tailored peptides on both model and biological membranes, and concluded that they form pores by the so-called carpet-like or sinking-raft models, as also observed for many natural pore-forming AMPs (30,31). According to this mechanism, the monomeric peptides bind to the outer monolayer due to hydrophobic and electrostatic interactions, and subsequently self-assemble on the membrane surface into peptide-rich domains. This leads to destabilization of the bilayer and transient leakage of vesicle contents. Much of this scenario seems to apply also to SB056.
In conclusion, SB056 is a novel lipodimeric AMP with high solubility and remarkable self-assembly properties, supporting the potential of synthetic dendrimers. The peptide shows a lipid-induced propensity to fold as an amphiphilic β-strand and thereby self-assemble into β-sheets, which may be present as aggregates as a branched network and/or as amyloid-like fibrils. The conformational properties and oligomerization dynamics are linked to the peptide's antimicrobial activity.
Acknowledgments
M. A. Scorciapino thanks the German Academic Exchange Service for support in carrying out the experiments at KIT in Karlsruhe, and CINECA and CASPUR (Italy) for providing computational time. The microbiological studies were carried out in collaboration with NeED Pharmaceuticals S.r.l (Milan, Italy). A. Giuliani and G. Pirri are minor shareholders of SpiderBiotech S.r.l.
This work was supported in part by a grant from the Italian Ministry of Education, Universities, and Research (PRIN 2008 to A.C.R.); the DFG Centre for Functional Nanostructures (TP E1.2 to A.S.U.), and Regione Autonoma della Sardegna through the Project “PO Sardegna FSE 2007-2013, L.R.7/2007 Promozione della ricerca scientifica e dell'innovazione tecnologica in Sardegna” (fellowship to A.V.V.).
Supporting Material
References
- 1.Giuliani A., Rinaldi A.C., editors. vol. 618. Humana Press; New York: 2010. (Antimicrobial peptides. Methods and Protocols. Methods in Molecular Biology). [Google Scholar]
- 2.Easton D.M., Nijnik A., Hancock R.E. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadley E.B., Hancock R.E. Strategies for the discovery and advancement of novel cationic antimicrobial peptides. Curr. Top. Med. Chem. 2010;10:1872–1881. doi: 10.2174/156802610793176648. [DOI] [PubMed] [Google Scholar]
- 4.Mayer M.L., Easton D.M., Hancock R.E.W. Fine tuning host responses in the face of infection: emerging roles and clinical applications of host defence peptides. In: Wang G., editor. Antimicrobial Peptides: Discovery, Design, and Novel Therapeutic Strategies. CABI; Wallingford, UK: 2010. pp. 195–220. [Google Scholar]
- 5.Bechinger B. Membrane association and pore formation by α-helical peptides. Adv. Exp. Med. Biol. 2010;677:24–30. doi: 10.1007/978-1-4419-6327-7_3. [DOI] [PubMed] [Google Scholar]
- 6.Giuliani A., Pirri G., Rinaldi A.C. Antimicrobial peptides: natural templates for synthetic membrane-active compounds. Cell. Mol. Life Sci. 2008;65:2450–2460. doi: 10.1007/s00018-008-8188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giuliani A., Rinaldi A.C. Beyond natural antimicrobial peptides: multimeric peptides and other peptidomimetic approaches. Cell. Mol. Life Sci. 2011;68:2255–2266. doi: 10.1007/s00018-011-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luganini A., Giuliani A., Gribaudo G. Peptide-derivatized dendrimers inhibit human cytomegalovirus infection by blocking virus binding to cell surface heparan sulfate. Antiviral Res. 2010;85:532–540. doi: 10.1016/j.antiviral.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Donalisio M., Rusnati M., Lembo D. Identification of a dendrimeric heparan sulfate-binding peptide that inhibits infectivity of genital types of human papillomaviruses. Antimicrob. Agents Chemother. 2010;54:4290–4299. doi: 10.1128/AAC.00471-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruschi M., Pirri G., Rinaldi A.C. Synthesis, characterization, antimicrobial activity and LPS-interaction properties of SB041, a novel dendrimeric peptide with antimicrobial properties. Peptides. 2010;31:1459–1467. doi: 10.1016/j.peptides.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 11.Pini A., Giuliani A., Bracci L. Antimicrobial activity of novel dendrimeric peptides obtained by phage display selection and rational modification. Antimicrob. Agents Chemother. 2005;49:2665–2672. doi: 10.1128/AAC.49.7.2665-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blazyk J., Wiegand R., Kari U.P. A novel linear amphipathic β-sheet cationic antimicrobial peptide with enhanced selectivity for bacterial lipids. J. Biol. Chem. 2001;276:27899–27906. doi: 10.1074/jbc.M102865200. [DOI] [PubMed] [Google Scholar]
- 13.Cavanagh J., Fairbrother W.J., Skelton N.J. 2nd ed. Elsevier Academic Press; Oxford: 2007. Protein NMR Spectroscopy. Principles and Practice. [Google Scholar]
- 14.Strandberg E., Ulrich A.S. NMR methods for studying membrane-active antimicrobial peptides. Concepts Magn. Reson. A. 2004;23A:89–120. [Google Scholar]
- 15.Jasanoff A., Fersht A.R. Quantitative determination of helical propensities from trifluoroethanol titration curves. Biochemistry. 1994;33:2129–2135. doi: 10.1021/bi00174a020. [DOI] [PubMed] [Google Scholar]
- 16.Roccatano D., Colombo G., Mark A.E. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proc. Natl. Acad. Sci. USA. 2002;99:12179–12184. doi: 10.1073/pnas.182199699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong D.P., Hoshino M., Goto Y. Clustering of fluorine-substituted alcohols as a factor responsible for their marked effects on proteins and peptides. J. Am. Chem. Soc. 1999;121:8427–8433. [Google Scholar]
- 18.Reiersen H., Rees A.R. Trifluoroethanol may form a solvent matrix for assisted hydrophobic interactions between peptide side chains. Protein Eng. 2000;13:739–743. doi: 10.1093/protein/13.11.739. [DOI] [PubMed] [Google Scholar]
- 19.Wüthrich K. J. Wiley & Sons; Chichester, UK: 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 20.Brockman H.L. Lipid monolayers: why use half a membrane to characterize protein-membrane interactions? Curr. Opin. Struct. Biol. 1999;9:438–443. doi: 10.1016/S0959-440X(99)80061-X. [DOI] [PubMed] [Google Scholar]
- 21.Zhao H.X., Rinaldi A.C., Kinnunen P.K. Interactions of the antimicrobial peptides temporins with model biomembranes. Comparison of temporins B and L. Biochemistry. 2002;41:4425–4436. doi: 10.1021/bi011929e. [DOI] [PubMed] [Google Scholar]
- 22.Coccia C., Rinaldi A.C., Mangoni M.L. Membrane interaction and antibacterial properties of two mildly cationic peptide diastereomers, bombinins H2 and H4, isolated from Bombina skin. Eur. Biophys. J. 2011;40:577–588. doi: 10.1007/s00249-011-0681-8. [DOI] [PubMed] [Google Scholar]
- 23.Tam J.P., Spetzler J.C. Synthesis and application of peptide dendrimers as protein mimetics. Curr. Protoc. Immunol. 2001 doi: 10.1002/0471142735.im0906s34. May;Chapter 18:Unit 18.5. [DOI] [PubMed] [Google Scholar]
- 24.Tam J.P. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA. 1988;85:5409–5413. doi: 10.1073/pnas.85.15.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier M., Seelig J. Length dependence of the coil <—> β-sheet transition in a membrane environment. J. Am. Chem. Soc. 2008;130:1017–1024. doi: 10.1021/ja077231r. [DOI] [PubMed] [Google Scholar]
- 26.Wadhwani P., Strandberg E., Ulrich A.S. Self-assembly of flexible β-strands into immobilized β-sheets in membranes monitored by solid-state 19F-NMR. J. Am. Chem. Soc. 2012 doi: 10.1021/ja301328f. In press. [DOI] [PubMed] [Google Scholar]
- 27.D'Abramo M., Rinaldi A.C., Aschi M. Conformational behavior of temporin A and temporin L in aqueous solution: a computational/experimental study. Biopolymers. 2006;81:215–224. doi: 10.1002/bip.20404. [DOI] [PubMed] [Google Scholar]
- 28.Bozzi A., Mangoni M.L., Aschi M. Folding propensity and biological activity of peptides: the effect of a single stereochemical isomerization on the conformational properties of bombinins in aqueous solution. Biopolymers. 2008;89:769–778. doi: 10.1002/bip.21006. [DOI] [PubMed] [Google Scholar]
- 29.Yeung A.T., Gellatly S.L., Hancock R.E. Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rausch J.M., Marks J.R., Wimley W.C. Rational combinatorial design of pore-forming β-sheet peptides. Proc. Natl. Acad. Sci. USA. 2005;102:10511–10515. doi: 10.1073/pnas.0502013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rausch J.M., Marks J.R., Wimley W.C. β-Sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes. Biochemistry. 2007;46:12124–12139. doi: 10.1021/bi700978h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 2006. Performance standards for antimicrobial susceptibility testing. Approved standard, 16th informational supplement. NCCLS document M100–S16. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 33.National Committee for Clinical Laboratory Standards. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard. 7th ed. NCCLS document M7-A7. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 34.National Committee for Clinical Laboratory Standards. 2006. Reference methods for broth dilution antifungal susceptibility testing of yeasts. Approved standard. 2nd ed. NCCLS document M27-A2. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 35.Committee for Clinical Laboratory Standards. 2006. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard. 2006. NCCLS document M24-A. National Committee for Clinical Laboratory Standards, Wayne, PA. [PubMed]
- 36.Pace C.N., Vajdos F., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 38.Sreerama N., Woody R.W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 39.Lobley A., Whitmore L., Wallace B.A. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 40.Whitmore L., Wallace B.A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32(Web Server issue) doi: 10.1093/nar/gkh371. W668-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Habeck M., Rieping W., Nilges M. Bayesian estimation of Karplus parameters and torsion angles from three-bond scalar couplings constants. J. Magn. Reson. 2005;177:160–165. doi: 10.1016/j.jmr.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 42.Hess B., Kutzner C., Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 43.Oostenbrink C., Villa A., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 44.Berendsen H.J.C., Postma J.P.M., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- 45.Schuettelkopf A.W., Van Aalten D.M.F. PRODRG—a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 46.Frisch M.J., Trucks G.W., Pople J.A. Gaussian Inc.; Pittsburgh, PA: 2003. Gaussian. [Google Scholar]
- 47.Frisch M.J., Trucks G.W., Pople J.A. Gaussian Inc.; Wallingford, CT: 2004. Gaussian 03, Revision C.02. [Google Scholar]
- 48.Barone V., Cossi M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. A. 1998;102:1995–2001. [Google Scholar]
- 49.Vargiu A.V., Ruggerone P., Carloni P. Dissociation of minor groove binders from DNA: insights from metadynamics simulations. Nucleic Acids Res. 2008;36:5910–5921. doi: 10.1093/nar/gkn561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayly C.I., Cieplak P., Kollman P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 51.Case D.A., Darden T.A., Kollman P.A. University of California; San Francisco, CA: 2006. AMBER 9. [Google Scholar]
- 52.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 53.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 54.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 55.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 56.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1976;50:1055–1076. [Google Scholar]
- 57.Perczel A., Hollósi M., Fasman G.D. Convex constraint analysis: a natural deconvolution of circular dichroism curves of proteins. Protein Eng. 1991;4:669–679. doi: 10.1093/protein/4.6.669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


