Abstract
The peripheral distributions of the cardiac ryanodine receptor (RyR) and a junctional protein, junctophilin-2 (JPH2), were examined using single fluorophore localization-based super-resolution microscopy in rat ventricular myocytes. JPH2 was strongly associated with RyR clusters. Estimates of the colocalizing fraction of JPH labeling with RyR was ∼90% within 30 nm of RyR clusters. This is comparable to fractions estimated from confocal data (∼87%). Similarly, most RyRs were associated with JPH2 labeling in super-resolution images (∼81% within 30 nm of JPH2 clusters). The shape of associated RyR clusters and JPH2 clusters were very similar, but not identical, suggesting that JPH2 is dispersed throughout RyR clusters and that the packing of JPH2 into junctions and the assembly of RyR clusters are tightly linked.
Cardiac excitation-contraction coupling relies on calcium release from internal stores (the sarcoplasmic reticulum, SR) via ryanodine receptors (RyRs) (1), which are intracellular Ca2+ channels. Most RyRs are organized into clusters within narrow dyadic junctions between sarcolemmal and SR membranes (1). The RyRs are opened via the Ca2+-induced Ca2+-release mechanism (2), and the speed and fidelity of this mechanism are critically dependent on the geometry of the dyad and organization of RyRs. The dyad space itself is narrow and is formed by the close apposition of the SR and surface membrane (with ∼15 nm separation (3)) and within this space, RyRs are arranged in a quasicrystalline array (seen as “feet” (1, 4) in electron micrographs). This organization ensures a rapid response to Ca2+ influx through voltage-gated Ca2+ channels during the cardiac action-potential.
It is still unclear how junctions are formed and maintained. One protein that has been implicated in dyadic junction formation is the cardiac isoform of the junctophilin family, junctophilin-2 (JPH2). JPH2 has been suggested to promote docking of SR terminal cisterns with the sarcolemmal membrane via JPH2 cytosolic domains that have affinity for phospholipids (5). Consistent with this idea, transgenic mice lacking JPH2 showed embryonic lethality, malformed junctions, and abnormal Ca2+ transients (5). Furthermore, shRNA knockdown of JPH2 leads to a reduction in the number of junctional complexes, a greater variability in junctional geometry, and reduced efficiency of excitation-contraction (EC) coupling (6).
JPH2 has been shown to strongly colocalize with RyRs in mature cardiac myocytes (7), although, using diffraction-limited confocal microscopy, RyR and JPH clusters appear as mostly featureless puncta (∼250 nm in diameter). Recent super-resolution fluorescence microscopy has revealed a wide range of sizes and shapes for RyR clusters in peripheral junctions (8). With the increased resolution available in multicolor super-resolution microscopy (9) it is now possible to investigate the nanoscale relationship between RyRs and JPH2 to examine whether JPH2 forms “anchoring domains” within the junction.
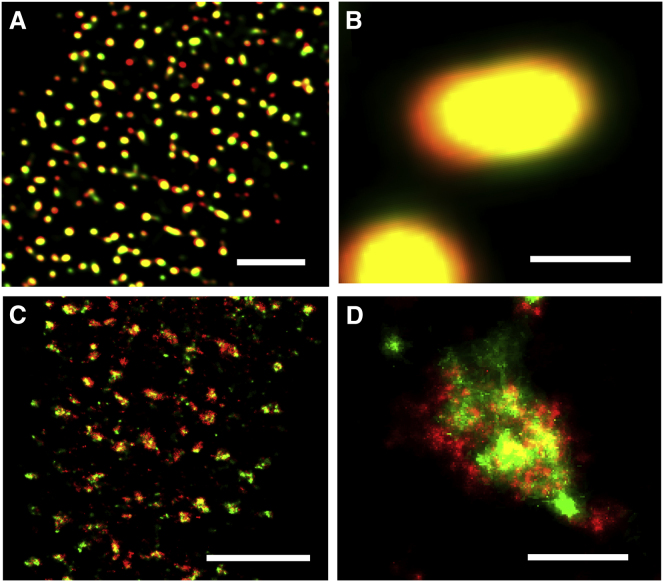
To visualize the nanoscale distribution of RyRs and JPH2 we labeled enzymatically isolated rat ventricular myocytes with specific antibodies against RyR and JPH2. The cells were imaged using both a confocal microscope and single molecule super-resolution microscopy (for detailed methods, see the Supporting Material). Imaging was restricted to the surface sarcolemma to visualize peripheral couplings, to enable comparison with previous work (8) and to allow a simpler interpretation of data from the (essentially) flat surface geometry. In agreement with previous work, both RyR and JPH2 labeling appeared as strongly overlapping diffraction-limited puncta in confocal images (Fig. 1, A and B). In contrast, super-resolution images of similar regions in RyR/JPH2 double-labeled cells revealed complex cluster shapes (Fig. 1 C). Patches of RyR labeling contained sharp edges and were incompletely filled (Fig. 1 D), similar to a previous report (8). JPH2 labeling exhibited a similar (but not identical) morphology and typically occupied most of the RyR cluster area (Fig. 1 D).
Figure 1.
Dual-color fluorescence imaging of RyR and JPH2. (A) Overlay of a confocal section of RyR2 (red) and JPH2 (green) labeling near the surface of a fixed rat ventricular myocyte. (B) The magnified view illustrates that diffraction-limited puncta of RyR and JPH strongly overlap. (C) Overlay of super-resolution images of RyR (red) and JPH2 (green). (D) Strong, but imperfect, overlap between regions of JPH labeling and RyR cluster is seen in the magnified view of localization data. (Scale bars: A and C, 3 μm; B and D, 0.25 μm.)
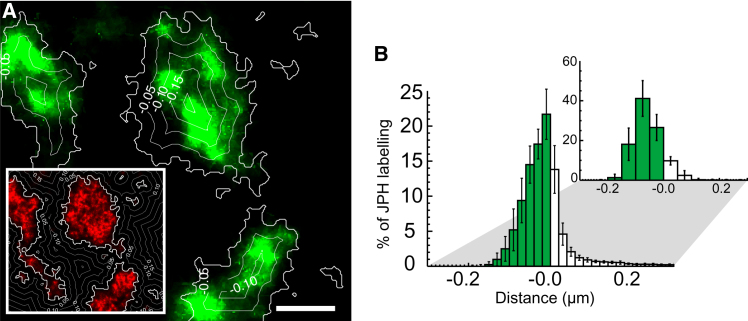
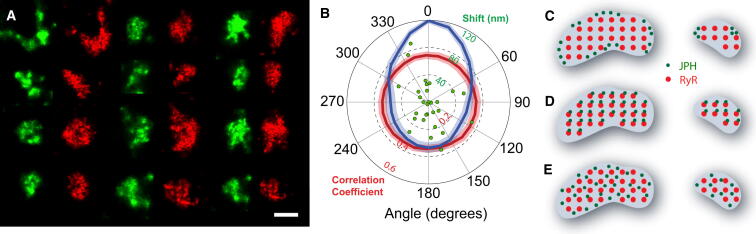
Colocalization of JPH2 and RyR was analyzed by measuring the amount of JPH2 labeling as a function of distance from the edge of RyR clusters (Fig. 2 A, and see Fig. S1 in the Supporting Material). Fig. 2 B shows histograms of the percentage of JPH2 labeling with distance to the edges of RyR clusters for both super-resolution and diffraction-limited confocal data. Estimates of this fraction were lower in super-resolution images than confocal data (72% vs. 87%, n = 5 cells), but was more comparable once a narrow area around RyR cluster edges of ∼30 nm was included (90%, n = 5 cells). Similarly, the percentage of RyR labeling colocalizing with JPH2 was 57% in super-resolution images, which increased to ∼81% by including a 30-nm band around the super-resolution cluster mask (vs. 84% in confocal data; see Fig. S2 and Table S1 in the Supporting Material). In addition, the density of JPH labeling across RyR clusters was approximately proportional to the density of RyR labeling itself (see Fig. S3). Visual inspection of the super-resolution data indicated a strong resemblance between shapes of RyR clusters and overlapping regions of JPH2 labeling. This is illustrated in Fig. 3 A, which shows a collage of randomly selected images of larger clusters. Quantification confirmed the angular and spatial alignment of corresponding RyR and JPH clusters (Fig. 3 B), in agreement with our visual impression. The correlation was ∼0.6 for aligned JPH and RyR clusters (Fig. 3 B), slightly lower than the expected correlation between identical clusters of ∼0.75 (<1 due to stochastic variations in single molecule data; see Methods in the Supporting Material). The centers of labeling intensity of large JPH and RyR clusters (mean diameter 515 nm) were mostly within ∼40 nm (Shift in Fig. 3 B). Taken together, this suggests that RyR and JPH2 distributions in clusters are similar but not identical.
Figure 2.
Analysis of the colocalization between JPH2 and RyR. (A) Euclidean distance maps (distance contours labeled in μm) were used to quantify the JPH2 labeling (green) as a function of distance from the edge of the nearest RyR cluster (red in inset). See also Fig. S1 in the Supporting Material for details. (Scale bar: 0.25 μm.) (B) A histogram showing the fraction of JPH2 labeling as a function of distance to the nearest RyR cluster edge based on the super-resolution data (front) indicates high association, broadly similar to the confocal data (back). Error bars indicate SD (n = 5 cells).
Figure 3.
JPH2 and RyR cluster shapes. (A) A collage of regions containing JPH2 labeling (green) and the corresponding RyR clusters (shown displaced to right, in red). (Scale bar: 0.25 μm.) (B) Pearson's correlation as a function of the angular alignment of the JPH2 labeling in relation to corresponding RyR clusters (blue line). JPH2 clusters correlated with randomly chosen RyR clusters exhibited no angular alignment (red line). Shaded bands indicate mean ± SE (n = 30 clusters). (Dots) Center-of-intensity distances between JPH2 and RyR clusters. (C–E) Schematic distribution of RyR and JPH2 organization. JPH2 may be confined to distinct anchoring domains (C), directly interact with RyRs (D), or randomly disperse within junctions (E).
The lower-resolution data obtained in previous studies (7) would be consistent with a number of different arrangements of the proteins within the junction (for example, those depicted in Fig. 3, C–E). Our data are not consistent with a junctional architecture containing specialized structural (anchoring) domains (Fig. 3 C) that are separate from Ca2+ signaling domains. The high degree of colocalization between JPH2 and RyR and the similarity in cluster shapes suggest that the ∼75 kDa JPH2 proteins are closely interspersed between the large RyR channels throughout the entire cluster (Fig. 3, D or E). In connection with this point, JPH2 has recently been shown to coimmunoprecipitate with the RyR in cardiac cell lysates, prompting the authors to suggest a direct molecular interaction between RyR and JPH2 (6). Our data show a molecular scale colocalization of RyR and JPH2 within the junction, which is compatible with this idea. We note, however, that although there is a strong shape similarity, there are discernible differences. This includes a lower correlation than would be expected between identical clusters, and colocalization only becomes substantial once a 30-nm boundary zone around clusters is included. This suggests that at least a subpopulation of RyR and JPH2 are not directly interacting, i.e., a hybrid of models in Fig. 3, D and E.
The strong molecular-scale colocalization between JPH2 and RyRs reported here supports a role for JPH2 in the formation of junctions critical for efficient EC coupling. In connection with this point, a progressive loss of JPH2 (6, 10) was linked to the development of heart failure. However, it remains unclear whether formation of JPH2 clusters precedes and acts as a possible seed site for RyR clustering (8) or if an interaction between JPH2 and RyRs helps promote formation of clusters by a form of mutual condensation. (Alternatively, JPH2 and RyR aggregation could be secondary to unidentified factors.)
These results demonstrate the utility of fluorescent multicolor super-resolution immuno-labeling to investigate protein proximity at the nanometer scale. The ability to resolve nanoscopic biological structures should give new insight into the assembly of macromolecular signaling complexes. By establishing that junctophilin is not constrained to specific domains within the junction, but broadly interspersed throughout the area occupied by RyRs, we can refine our knowledge of the molecular basis of local control (11) in cardiac EC coupling.
Acknowledgments
The Marsden Fund, the Auckland Medical Research Foundation, and the Health Research Council of New Zealand provided financial support.
Editor: Andrew McCulloch.
Footnotes
An additional Methods section, three figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(12)00148-8.
Supporting Material
References and Footnotes
- 1.Franzini-Armstrong C., Protasi F. Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions. Physiol. Rev. 1997;77:699–729. doi: 10.1152/physrev.1997.77.3.699. [DOI] [PubMed] [Google Scholar]
- 2.Fabiato A. Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am. J. Physiol. 1983;245:C1–C14. doi: 10.1152/ajpcell.1983.245.1.C1. [DOI] [PubMed] [Google Scholar]
- 3.Cannell M.B., Soeller C. Numerical analysis of ryanodine receptor activation by L-type channel activity in the cardiac muscle dyad. Biophys. J. 1997;73:112–122. doi: 10.1016/S0006-3495(97)78052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayashi T., Martone M.E., et al. Hoshijima M. Three-dimensional electron microscopy reveals new details of membrane systems for Ca2+ signaling in the heart. J. Cell Sci. 2009;122:1005–1013. doi: 10.1242/jcs.028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeshima H., Komazaki S., et al. Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 6.van Oort R.J., Garbino A., et al. Wehrens X.H. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziman A.P., Gómez-Viquez N.L., et al. Lederer W.J. Excitation-contraction coupling changes during postnatal cardiac development. J. Mol. Cell. Cardiol. 2010;48:379–386. doi: 10.1016/j.yjmcc.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baddeley D., Jayasinghe I., et al. Soeller C. Optical single-channel resolution imaging of the ryanodine receptor distribution in rat cardiac myocytes. Proc. Natl. Acad. Sci. USA. 2009;106:22275–22280. doi: 10.1073/pnas.0908971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baddeley D., Crossman D., et al. Soeller C. 4D super-resolution microscopy with conventional fluorophores and single wavelength excitation in optically thick cells and tissues. PLoS ONE. 2011;6:e20645. doi: 10.1371/journal.pone.0020645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei S., Guo A., et al. Song L.S. T-tubule remodeling during transition from hypertrophy to heart failure. Circ. Res. 2010;107:520–531. doi: 10.1161/CIRCRESAHA.109.212324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannell M.B., Kong C.H. Local control in cardiac E-C coupling. J. Mol. Cell. Cardiol. 2012;52:298–303. doi: 10.1016/j.yjmcc.2011.04.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.