Abstract
We have used thiol cross-linking and electron paramagnetic resonance (EPR) to resolve structural transitions of myosin's light chain domain (LCD) and catalytic domain (CD) that are associated with force generation. Spin labels were incorporated into the LCD of muscle fibers by exchanging spin-labeled regulatory light chain for endogenous regulatory light chain, with full retention of function. To trap myosin in a structural state analogous to the elusive posthydrolysis ternary complex A.M′.D.P, we used pPDM to cross-link SH1 (Cys707) to SH2 (Cys697) on the CD. LCD orientation and dynamics were measured in three biochemical states: relaxation (A.M.T), SH1-SH2 cross-linked (A.M′.D.P analog), and rigor (A.M.D). EPR showed that the LCD of cross-linked fibers has an orientational distribution intermediate between relaxation and rigor, and saturation transfer EPR revealed slow rotational dynamics indistinguishable from that of rigor. Similar results were obtained for the CD using a bifunctional spin label to cross-link SH1-SH2, but the CD was more disordered than the LCD. We conclude that SH1-SH2 cross-linking traps a state in which both the CD and LCD are intermediate between relaxation (highly disordered and microsecond dynamics) and rigor (highly ordered and rigid), supporting the hypothesis that the cross-linked state is an A.M′D.P analog on the force generation pathway.
Introduction
Muscle contraction is driven by the actin-activated hydrolysis of ATP by myosin, resulting in the relative sliding of actin and myosin filaments. Mechanistic models propose that filament sliding is driven by a structural transition of the myosin catalytic domain (CD) from a dynamically disordered state of weak actin binding to an ordered state of strong actin binding, and a lever arm rotation of the light-chain domain (LCD) (1–4). In the lever arm model, the LCD behaves as a semirigid rod that amplifies and propagates the force-producing structural changes in the myosin CD to the thick filament core (4,5). The strongly bound actomyosin complexes (A.M or A.M.D) are quite stable and have been studied in great detail, with electron paramagnetic resonance (EPR) revealing that the orientations of the CD (6,7) and LCD (8,9) are well defined with respect to the actin filament axis. However, much less is known about the weakly bound complexes (A.M.T or A.M.D.P), which are more difficult to study due to their dynamic disorder and short lifetimes. Saturation transfer EPR (STEPR), with spin labels on the CD of myosin, in the steady state of ATP hydrolysis (10,11) or in the presence of ATPγS (12,13), has shown that weakly attached myosin heads undergo large-amplitude rotations with correlation times (τR) in the range of 1–20 μs. This dynamic disorder is consistent with the disordered appearance of myosin subfragment 1 (S1) bound to actin in electron micrographs of similar weakly bound complexes (14–16).
These spectroscopic and electron microscopic observations of weakly bound actomyosin complexes were done under conditions in which the predominant actin-attached myosin biochemical state was probably the prehydrolysis complex (A.M.T). However, the posthydrolysis ternary complex (A.M.D.P) is also of great interest, because there is evidence that force generation begins in this biochemical state, before phosphate is released but after isomerization to A.M′.D.P (17,18). This early-force complex has remained elusive, because actin: 1), greatly accelerates the rate of Pi release, thus quickly converting A.M′.D.P to A.M.D; and 2), shifts the equilibrium constant for hydrolysis toward the prehydrolysis state by a factor of 20 (19). Thus, preparation of a stable posthydrolysis ternary complex or analog is crucial for understanding myosin's force-producing structural transitions. The posthydrolysis analogs ADP.Vi and ADP.AlF4 would seem to provide an attractive approach to trapping the elusive A.M.D.P complex, but actin rapidly dissociates these phosphate analogs from myosin (20,21). The nucleotide analogs AMPPNP and pyrophosphate do remain bound in the ternary complexes with actin and myosin, but these complexes are structurally indistinguishable from the strongly bound complex A.M.D (12,22,23).
There are two proposed stable analogs of the ternary complex A.M′.D.P: the complex of actin with myosin, blebbistatin, and ADP (24) and the complex of actin with myosin that has been cross-linked with n,n′-(1,4-phenylene)dimaleimide (pPDM). In this work, pPDM cross-linking was used to trap a stable analog of the A.M′.D.P state. pPDM specifically cross-links the two most reactive cysteine residues, SH1 (Cys707) and SH2 (Cys697), on the CD of myosin (25–27). Several mechanical and biochemical studies have characterized pPDM-cross-linked myosin as a weak-binding state. Muscle fibers treated with pPDM exhibit an 85% decrease in isometric force (28) and a decrease in rigor stiffness to the level of resting untreated fibers (29). The actin-activated ATPase of pPDM-S1 is 0.2% of the rate of unmodified S1 (8), and pPDM-S1 exhibits weak actin binding, with an actin affinity at low ionic strength (Kd = 30 μM) ∼3 times stronger than S1.ATP, 100 times weaker than S1.AMPPNP, and 1000 times weaker than S1.ADP (25). However, pPDM-myosin does not trap nucleotide when bound to actin, as might be expected for a stable analog of the A.M′.D.P complex (26). Rather, pPDM appears to trap the stable A.M′.D.P complex by trapping a structural state in which the SH1 helix is disordered (see Structural Coupling Within Myosin, below).
Recent work using a bifunctional spin label, 3,4-bis-(methanethiosulfonylmethyl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxy radical (BSL) rather than pPDM to cross-link SH1 to SH2, has characterized the orientation and dynamics of the BSL-S1 actin complex (30) (Fig. 1). EPR of BSL-S1 bound to oriented actin in muscle fibers yields spectra characteristic of nearly random orientation, in contrast to the high degree of orientational order of S1 strongly bound to actin. However, STEPR of BSL-S1 reveals rotational dynamics with a correlation time (τR) ∼600 μs, which is similar to that observed for the strongly bound biochemical states A.M and A.M.D, and 30 times slower than the weakly bound ternary complex A.M.T (12). It is evident that SH1-SH2 cross-linked myosin exhibits distinct structural properties that place it intermediate between A.M.T and A.M.D, as expected for an analog of the A.M′.D.P complex (Fig. 1).
Figure 1.
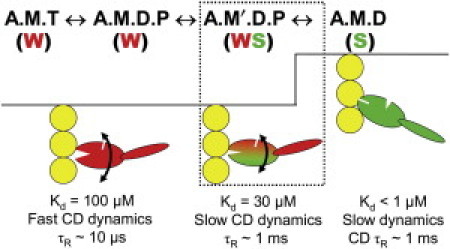
Model for coupling of actomyosin ATPase to force and movement, focusing on the coupling of biochemical transitions to orientation and dynamics of the myosin catalytic domain, adapted from Thomas et al. (4). (Red and green) Weak (preforce) and strong (force-bearing) complexes respectively. (Curved arrows) Orientational disorder. An upward step indicates the power stroke. Text at top indicates the biochemical state (defined by the active site ligand): A, actin; M, myosin; T, ATP; D, ADP; P, inorganic phosphate. Prime (′) indicates a second structural state corresponding to the same biochemical state. Text under each state indicates distinguishing properties of the catalytic domain.
The spectroscopic results from BSL-S1, summarized above, provided insight into the structural consequences of SH1-SH2 cross-linking. However, they were acquired with probes attached only on the CD of isolated S1, leaving two questions open: 1), Are the structural dynamics of cross-linked S1 due to orientational disorder of the entire myosin head, or to disorder within the head? 2), Are these results characteristic of intact myosin in a muscle fiber? In muscle fibers, the myosin head is constrained not only by actin but also by the thick filament backbone. The resulting mechanical strain may limit the conformation of myosin as it binds actin, or dissociate a fraction of the heads due to weakened actin binding induced by cross-linking. To answer these questions, we used EPR to measure orientation and dynamics of both the LCD and CD in intact myosin in skinned skeletal muscle fibers. LCD measurements were made in three biochemical states: relaxation (A.M.T), pPDM-cross-linked (A.M′.D.P), and rigor (A.M.D). LCD orientation and dynamics in muscle fibers were measured by spin-labeling purified RLC, then exchanging it with the endogenous RLC, with retention of muscle function. The CD was studied in cross-linked fibers, using BSL to simultaneously cross-link and spin-label the myosin CD.
Methods
Muscle fiber preparation and characterization
Glycerinated rabbit psoas muscle fiber bundles were prepared and stored in a solution containing 1:1 mixture of rigor solution (60 mM KPr, 2 mM MgCl2, 1 mM EGTA, 25 mM MOPS, pH 7.0, 25°C) and glycerol at −20°C for up to six months without significant loss of function (31). Chicken gizzard RLC was prepared and labeled with 3-(5-fluoro-2,4-dinitroanilino)-2,2,5,5-tetramethyl-1-pyrrolidinyloxy (FDNASL) as described in Section S1 in the Supporting Material. Spin-labeled RLC (FDNASL-RLC) was then exchanged for endogenous RLC in fiber bundles (see Section S3 in the Supporting Material). The extent of RLC extraction and reconstitution was determined by densitometric analysis of sodium dodecyl sulfate polyacrylamide gel electrophoresis on fiber homogenates (see Section S4 in the Supporting Material). Muscle fiber function after RLC exchange was assessed by measuring the Ca dependence of myofibrillar MgATPase activity (see Section S5 in the Supporting Material).
Cross-linking with pPDM and BSL
SH1 (Cys707) and SH2 (Cys697) were cross-linked using pPDM (Sigma-Aldrich, St. Louis, MO), with the same setup used for RLC exchange (see Section S3 in the Supporting Material) except that temperature was always 4°C. The fiber bundle was washed for 15 min with cross-linking solution (190 mM KPr, 2 mM MgCl2, 1 mM EGTA, 20 mM MOPS, 5 mM MgATP), and pPDM (in dimethylformamide) was added to cross-linking solution such that the final [pPDM] was 200 μM and dimethylformamide never exceeded 1%. After 75 min, the fiber bundle was washed with RS190 (cross-linking solution without MgATP) for 15 min to remove unreacted pPDM and MgATP. K/EDTA and Ca/K ATPase activities (see Section S5 in the Supporting Material) were used to determine the extent of pPDM cross-linking. SH1 and SH2 were also cross-linked with the bifunctional spin label 3,4-bis-(methanethiosulfonylmethyl)-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-1-yloxy radical (BSL; Toronto Research Chemicals, North York, Ontario, Canada). The experimental setup and BSL cross-linking procedure was identical to that used for cross-linking with pPDM, except that the BSL concentration was 100 μM and the reaction time was 75 min.
EPR spectroscopy
EPR spectra were recorded at X-band (9.5 GHz) with a Bruker (Billerica, MA) E500 spectrometer, using either a TE102 (Bruker 4104OR-R) or TM110 (Bruker 4103TMA) cavity. The sample temperature was maintained at 23°C by flowing temperature-controlled N2 gas through a nozzle attached to the optical port on the front of the cavity (TE102) or a quartz dewar inserted into the bottom of the cavity (TM110). The sweep width was 120 G (1024 points), sweep time was typically 41.94 s (conversion time 40.96 ms), and the center field value HC was set proportionally to the microwave frequency ν (HC = ν/2.803 MHz/G, corresponding to a g value of 2.0027, the value of gz for a typical nitroxide) so that all spectra were equivalently aligned. For EPR experiments on oriented muscle fiber bundles under perfusion, the procedure was essentially the same as that used during RLC exchange (see Section S3 in the Supporting Material). The solution flow rate varied from 115 μL/min to 340 μL/min, such that a further increase in flow rate did not affect the spectra.
EPR spectra were acquired in either RS190 (190 mM KPr, 2 mM MgCl2, 1 mM EGTA, 20 mM MOPS, pH 7.0) or in relaxation solution (RS190 with 20 mM KPr rather than 190 mM KPr, plus 5 mM MgATP, 50 mM creatine phosphate, and 750 units/ml of creatine phosphokinase) (32). Ionic strength was 203 mM for both solutions. Spectra of RLC-labeled fibers in rigor and pPDM-cross-linked states were acquired while perfusing the fiber with RS190, and relaxation spectra were acquired while perfusing relaxation solution. Spectra of BSL-fibers were also acquired while perfusing the fibers with RS190, to remove free spin-label that was slowly released. For EPR experiments on randomized samples, fibers in RS190 were minced with a razor blade and placed in a quartz flat cell that contained a well of dimensions 0.5 × 1.0 × 0.05 cm (WG-806-Q; Wilmad-Labglass, Buena, NJ). Excess moisture was wicked away and a cover slip was placed over the sample well and sealed with Parafilm M (Pechiney Plastic Packaging Company, Chicago, IL) to prevent sample dehydration. The flat cell assembly was centered in the EPR cavity with its face oriented parallel or perpendicular to the applied magnetic field.
Conventional EPR spectra (V1) of RLC-labeled fibers and BSL-fibers were acquired with the long axis of the fiber bundle oriented either parallel (using a modified TM110 cavity (9)) or perpendicular (using a TE102 cavity) to the applied magnetic field. Microwave power was set from 20 mW to 32 mW, to maximize the signal intensity without causing significant saturation. The modulation frequency was 100 kHz (first harmonic), and the peak-to-peak modulation amplitude was 2 G. The filter time constant was set equal to the conversion time (40.96 ms). Spectra of fibers were analyzed to determine the orientational distribution of the spin-label relative to the muscle fiber axis, using computational simulation and least-squares minimization (6,30,33). Briefly, the spectrum of minced fiber bundles was fit to obtain the orientation-independent parameters, including the anisotropic T and g tensors and the linewidths. Spectra of oriented fiber bundles were then fitted to determine the orientational distribution of the spin-label relative to the fiber axis, defined by the center (θ0′) and the width (Δθ′, full width at half-maximum) of the assumed Gaussian orientational distribution. Analysis of the spectrum from minced RLC-labeled fibers indicated two components with different tensor values. One component, having a wider splitting than the predominant component, composes a small fraction (≤0.15) of the total spectrum and presumably represents a small fraction of nonspecifically bound RLCs. Previously, this nonspecific component comprised 40% of the spectrum (9). This small nonspecific component was digitally subtracted before fitting.
Saturation transfer EPR spectra (V2′) of RLC-labeled fibers and BSL-fibers were acquired as described previously (30,34). Fibers were aligned perpendicular to the field to minimize the effects of orientation on the spectrum. In STEPR, the microwave power P is set such that the microwave field amplitude H1 is 0.25 G, determined from P = (H12/K)(Q0/Q), where K is determined by calibration with a sample of known saturation properties (34), Q0 is the cavity quality factor (measured by the spectrometer) during the calibration, and Q is the value measured for each experiment. For the muscle fiber setup used in this work, K was typically measured to be 1.06 G2/W and Q0 = 3000, so P was typically set at 59 mW × 3000/Q for STEPR. The following parameters were used to acquire V2′: H1 = 0.25G, modulation frequency νm = 50 kHz with the phase-sensitive detection at 100 kHz (second harmonic), peak-to-peak modulation amplitude Hm = 5.0 G, modulation phase ϕm = 90° (minimum signal at nonsaturating power), and filter time constant = 328 ms. V2′ spectra were analyzed to determine the rotational correlation time τR, based on the integrated intensity parameter ∫V2′, which is independent of the spin concentration and orientation (34).
Results
RLC exchange
Previous work using EPR to measure LCD orientation indicates a large population of RLC bound nonspecifically in skeletal muscle fibers (see Section S2 in the Supporting Material) (8,9,35). Using a modified RLC exchange method (see Section S3 in the Supporting Material), we minimized this population of nonspecific RLC to <15% of the total spin-labeled RLC (see Fig. S1 in the Supporting Material), a substantial improvement compared to previous EPR studies (9). The extent of RLC extraction and reconstitution (see Fig. S2) indicated that 54 ± 10% of the endogenous RLC was exchanged with spin-labeled RLC. Muscle function after RLC exchange was normal, as shown by the Ca dependence of myofibrillar MgATPase (Table 1). This improvement in RLC exchange technology, compared to previous EPR studies on skeletal muscle fibers (8,9,35), dramatically enhances the sensitivity and validity of LCD structural measurements.
Table 1.
ATPase assays
| Sample | MgATPase Vmax | MgATPase pKCa | K/EDTA-ATPase | Ca/K-ATPase |
|---|---|---|---|---|
| Control | 0.322 ± 0.021 | 5.87 ± 0.01 | 0.459 ± 0.021 | 0.066 ± 0.013 |
| RLC exchanged | 0.321 ± 0.024 | 5.83 ± 0.09 | — | — |
| pPDM cross-linked | — | — | 0.0508 ± 0.013 | 0.038 ± 0.024 |
All values are reported as mean ± SE (n = 4–6). Vmax, K/EDTA-ATPase, and Ca/K-ATPase values have units of μmol/mg protein/min. Control fibers underwent a mock RLC exchange or mock pPDM treatment.
Cross-linking SH1 and SH2 with pPDM
K/EDTA-ATPase and Ca/K-ATPase activities were used to determine the extent and specificity of SH1-SH2 modification with pPDM. ATPase measurements from control (no pPDM) and pPDM-treated fibers are summarized in Table 1. Both the K/EDTA- and Ca/K-ATPase are inhibited by pPDM treatment (Table 1). As shown previously (30), these values indicate that at least 90% of the myosin heads were modified at SH1 and SH2, presumably due to cross-linking by pPDM. We cannot rule out the possibility that some of the effects are due to modification of other thiols, including those on actin, tropomyosin, and troponin. However, previous studies have shown that reaction of skinned fibers with thiol-reactive probes under these conditions is remarkably specific for SH1, and that the reaction of BSL with myosin S1 is quite specific for SH1-SH2 (30), so it is likely that most of the reaction of pPDM and BSL is with SH1 and SH2.
LCD orientation measured by conventional EPR
EPR is extremely sensitive to the orientational distribution of a spin-label with respect to the applied magnetic field (H). At the frequency used here (X-band, 9.5 GHz) EPR is primarily sensitive to the angle θ between the spin-label's principal axis and H. In a well-oriented muscle fiber, aligned either parallel or perpendicular to H, the EPR spectrum detects directly the angle θ′ between the spin-label's principal axis and the fiber axis (see Fig. S3) (6). Thus, the recording of parallel and perpendicular spectra provides a clear graphical indication of the degree of orientation of the spin-label relative to the fiber axis. The parallel spectrum is then analyzed to determine the precise orientational distribution ρ(θ′).
LCD orientation, with respect to the fiber axis, was determined from conventional EPR spectra (V1) of spin-labeled RLC in skinned muscle fiber bundles. Parallel and perpendicular spectra of minced fiber bundles are nearly identical, indicating random orientation (Fig. 2, top). EPR spectra of oriented fiber bundles were acquired in three different biochemical states: relaxation (A.M.T, prepower stroke), rigor (A.M.D, postpower stroke), and SH1-SH2 cross-linked with pPDM (A.M′.D.P) (Fig. 2). In relaxation, spectra are nearly independent of fiber orientation, indicating virtually random orientation of the LCD (θ0′ = 72° ± 9°, Δθ′ = 89° ± 11°). In rigor, the spectra are quite sensitive to fiber orientation, indicating a single population with a high degree of orientational order (θ0′ = 41° ± 4°, Δθ′ = 38° ± 6°). After pPDM treatment, sensitivity to fiber orientation is intermediate between that of relaxation and rigor (θ0′ = 54° ± 6°, Δθ′ = 64° ± 16°). We conclude that in relaxation (A.M.T, prepower stroke) the LCD is randomly disordered, in rigor (A.M.D, postpower stroke) the LCD is highly ordered, and in the pPDM-cross-linked state (analog of A.M′.D.P) the LCD is partially disordered, with an orientational distribution intermediate between those of relaxation and rigor. However, this amount of disorder is enough that we cannot determine whether there is a single disordered population or a mixture of ordered and disordered populations.
Figure 2.
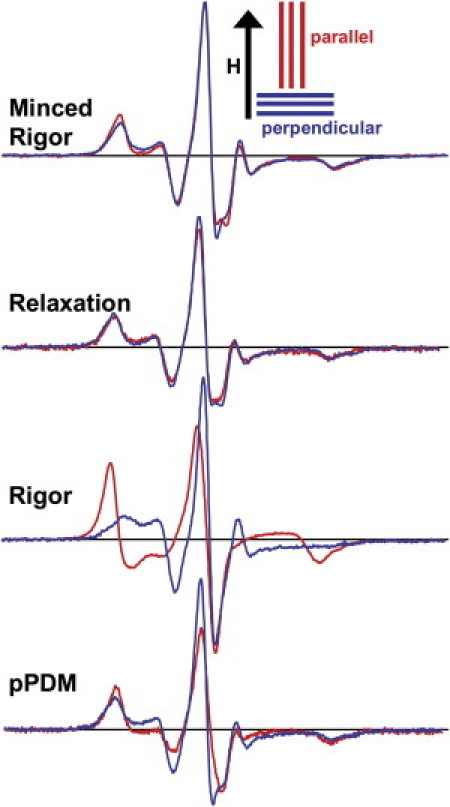
Conventional EPR spectra (V1) of spin-labeled LCD. Spectra were acquired with the flat cell (top) or the capillary containing oriented fibers (bottom three) oriented parallel (red) or perpendicular (blue) to the magnetic field H. The buffer was either RS190 (rigor, pPDM) or relaxation solution (see Methods).
Based on conventional EPR spectra (Fig. 2), the intermediate orientation of the LCD in pPDM-cross-linked fibers can be explained by two distinct models. In Model 1 (Fig. 3, left), cross-linking traps a state in which both myosin heads are bound to actin with static orientational disorder that is intermediate between relaxation (A.M.T.) and rigor (A.M.D). In Model 2 (Fig. 3, right), cross-linking traps a state in which half of the heads are dissociated from actin (and dynamically disordered as in relaxation) whereas the other half remain strongly bound as in rigor, as observed previously for the nucleotide analogs PPi and AMPPNP (22,23). It has been shown previously that both the CD (10,11,36) and LCD (37,38) of dissociated heads are dynamically disordered, with a rotational correlation time τR ∼10 μs. Thus to decide between the two models in Fig. 3, we must measure the rotational correlation time τR, to determine whether the disorder is static (Model 1) or dynamic (Model 2) on the microsecond timescale. This calls for saturation transfer EPR (STEPR).
Figure 3.
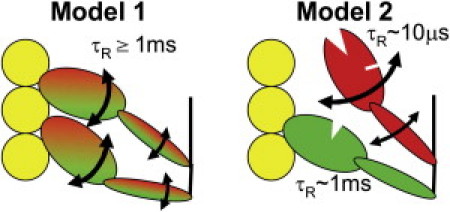
Alternative models consistent with the conventional EPR spectra of pPDM-treated fibers (see Fig. 2, bottom).
RLC dynamics measured by STEPR
In addition to being sensitive to orientation, EPR is sensitive to rotational dynamics (see Fig. S4), but conventional EPR (V1) is sensitive to rotational motion only for rotational correlation times (τR) in the picosecond-to-microsecond time range, so it is not sensitive to the slow rotational dynamics of large proteins such as myosin (4). Therefore, we must use STEPR (V2′), which is sensitive to the microsecond-to-millisecond time range (39).
STEPR was used to detect the microsecond rotational dynamics of LCD in relaxation, rigor, and the pPDM-cross-linked state (Fig. 4, bottom). Rotational correlation times were determined from STEPR spectra (V2′) using the integrated intensity parameter (∫V2′) as described in Methods (34). It is clear that the spectrum obtained in relaxation is much less intense than the other two cases (rigor and cross-linked), which are essentially identical. The values of ∫V2′ (Fig. 4, inset) in relaxation, rigor, and the cross-linked state are 0.54 ± 0.026, 1.01 ± 0.047, and 0.96 ± 0.064, respectively, corresponding to τR values of 22 ± 3.6 μs in relaxation and ≥1 ms for both rigor and pPDM (34). We conclude that the rotational dynamics of LCD in the pPDM-cross-linked state (analog of A.M′.D.P) is indistinguishable from that of rigor (A.M.D), and much slower than in relaxation (A.M.T), despite the orientational distribution being intermediate between relaxation and rigor (Fig. 4, top). These results are consistent with Model 1 but not Model 2 (Fig. 3). Cross-linking traps a state in which both heads are bound to actin with static orientational disorder.
Figure 4.
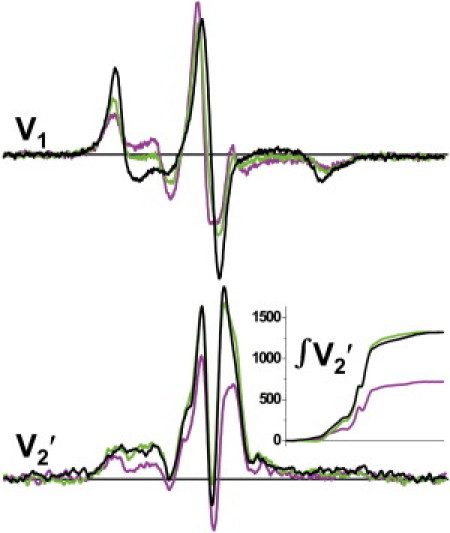
Effect of pPDM-cross-linking on EPR spectra of spin-labeled myosin LCD in skinned fiber bundles. Spectra were acquired in relaxation (magenta), rigor (black), and the pPDM-cross-linked state (green). (Top) Conventional EPR of fibers oriented parallel to the field (V1, from Fig. 2). (Bottom) STEPR spectra (V2′) of fibers oriented perpendicular to the field. (Inset) ∫V2′. The buffer was RS190 (rigor and pPDM) or relaxation solution (see Methods).
Catalytic domain orientation and dynamics of BSL-fibers
Isolated BSL-S1 bound to actin in oriented muscle fibers is almost randomly oriented and exhibits slow rotational dynamics (τR ∼ 600 μs), similar to that of rigor and 30 times slower than observed for the ternary complex A.M.T (12,30). However, it is possible that the head of endogenous myosin behaves differently due to restriction by the thick filament backbone. Thus, to gain a more complete understanding of the complex trapped by SH1-SH2 cross-linking, we measured catalytic domain orientation and dynamics in cross-linked fibers, using BSL to simultaneously cross-link and spin-label myosin.
Conventional EPR spectra of BSL-fibers were acquired with the fiber axis parallel or perpendicular to the magnetic field H. Parallel and perpendicular spectra are nearly identical, indicating a highly, but not completely, disordered CD (θ0′ = 85° ± 3°, Δθ′ = 77° ± 4°) (Fig. 5, top). In contrast, previous EPR studies demonstrate that un-cross-linked fibers spin-labeled at SH1 exhibit a narrow angular distribution in rigor with Δθ′ ∼15°, and random orientation (Δθ′ ≥ 90°) in relaxation (6). Thus SH1-SH2 cross-linking produces an orientational distribution of the CD that is much more disordered than in rigor and slightly less disordered than in relaxation.
Figure 5.
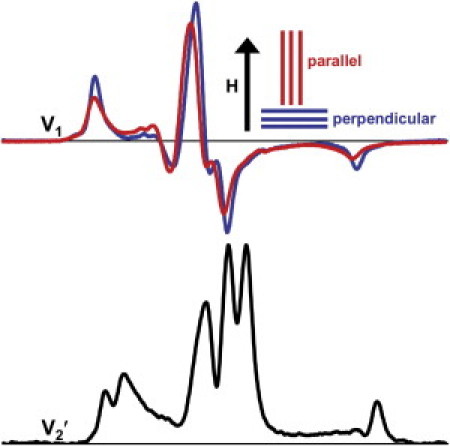
EPR spectra of skinned fiber bundles labeled on myosin CD with BSL, cross-linking SH1 and SH2. (Top) Conventional EPR (V1) of fiber bundles, with the fiber axis oriented parallel (red) and perpendicular (blue) to the magnetic field H. (Bottom) STEPR (V2′). The buffer was RS190.
The STEPR spectrum of BSL-fibers (Fig. 5, bottom) demonstrates extremely slow rotational dynamics of the CD, with ∫V2′ = 1.27 ± 0.26, corresponding to the rigid limit of STEPR (τR ≥ 1 ms). This result is similar to that of rigor, in which the CD is immobile on the microsecond timescale (36), and much slower than in relaxation (τR ∼10 μs) (10,11,36). We conclude that the CD in SH1-SH2 cross-linked fibers is much more disordered than in rigor, but not as disordered as in relaxation, and rotational motions are as slow as in rigor.
In comparing the LCD (Fig. 4) with the CD (Fig. 5), the cross-linked state in fibers has similar properties—highly disordered (as in relaxation) but static (as in rigor). However, the disorder of the LCD (Δθ′ = 64°) is significantly less than that of the CD (Δθ′ = 77°), suggesting that the source of disorder is at the actin-CD interface. In support of this hypothesis, the CD of BSL-fibers is not as disordered as the CD of BSL-S1 (30), indicating that CD disorder in intact myosin in fibers is restricted by connection to the thick filament backbone.
Discussion
Summary of results
We have used RLC exchange, myosin cross-linking, and EPR to measure the orientation and dynamics of myosin's LCD in three biochemical states, relaxation (A.M.T), SH1-SH2 cross-linked (A.M′.D.P analog), and rigor (A.M.D). Modification of a previously described RLC-exchange protocol (40) made it possible to reduce nonspecific RLC binding (see Fig. S1) while replacing 54% of the endogenous RLC with spin-labeled RLC (see Fig. S2), with retention of function as defined by the Ca-dependence of myofibrillar MgATPase (Table 1). Conventional EPR was used to measure the orientational distribution of the LCD in RLC-exchanged muscle fiber bundles (Fig. 2). In relaxation (A.M.T, prepower stroke) the LCD is highly disordered, in rigor (A.M.D, postpower stroke) the LCD is orientationally ordered, and in the pPDM-cross-linked state (A.M′.D.P) the LCD has an orientational distribution intermediate between relaxation and rigor. STEPR demonstrates that LCD dynamics in cross-linked fibers is indistinguishable from rigor (τR ≥ 1 ms) and much slower than in relaxation (τR = 22 μs) (Fig. 4). We also measured CD orientation and dynamics in fiber bundles cross-linked with BSL (Fig. 5) and found that both conventional and STEPR results are similar to those of the LCD in cross-linked fiber bundles (Fig. 4). The CD of SH1-SH2 cross-linked fibers is more disordered than in rigor, but not as disordered as in relaxation. The dynamics of the CD in SH1-SH2 cross-linked fibers is indistinguishable from rigor (τR ≥ 1 ms) and much slower than relaxation (τR ∼10 μs). Thus, SH1-SH2 cross-linking disorders both the CD and LCD, but this orientational disorder is much less dynamic than in relaxation.
Interpretation of results
During the actomyosin ATPase cycle, myosin transitions from a disordered state of weak actin binding to an ordered state of strong actin binding (Fig. 1). The weakly bound complexes (A.M.T) (10–16) and the strongly bound complexes (A.M and A.M.D) (6,7,9) have been the subject of many studies. However, relatively little is known about posthydrolysis ternary complexes, especially the force-generating A.M′.D.P. One stable equilibrium complex, proposed to be analogous to A.M′.D.P, is the complex of actin with SH1-SH2 cross-linked myosin. Recent work on BSL-S1 showed that cross-linked S1 is orientationally disordered but has very slow rotational dynamics (30). However, because that work was done with probes attached on the CD of isolated S1, two questions remained open: 1), Are the structural dynamics of cross-linked S1 due to orientational disorder of the entire myosin head, or to disorder within the head? 2), Are these results characteristic of intact myosin in a muscle fiber?
In this study, the combination of conventional and saturation transfer EPR has answered these two questions. Figs. 2 and 4 demonstrate that cross-linking stabilizes a state in which the LCD has an orientational distribution intermediate between relaxation and rigor with dynamics that is static on the microsecond timescale, as shown previously for the CD (30). Thus, cross-linking disorders the entire actin-bound myosin head. The second question is answered by EPR spectra of BSL-fibers (Fig. 5). Spectra indicate a large amount of orientational disorder (Δθ′ = 77°) (Fig. 5), but less than observed for actin-attached BSL-S1 (Δθ′ ≥ 90) (30), and slow dynamics were indistinguishable from rigor (τR ≥ 1 ms). This result is consistent with the hypothesis that the myosin head is constrained not only by actin but also by the thick filament backbone, and the resulting mechanical strain limits the conformation of myosin as it binds actin. These results indicate flexible structural-coupling between myosin's CD and LCD. That is, the structural transitions of myosin CD and LCD are similar throughout the actomyosin ATPase cycle, though not identical. It is remarkable that SH1-SH2 cross-linking on the distal CD has such a profound structural effect on the proximal LCD. However, this work does not determine whether the LCD is acting as a semirigid rod to amplify the force-producing structural changes in the CD, as proposed by the lever arm model (4,5), or if the LCD merely follows the CD throughout its force-producing structural transitions.
We propose that cross-linking SH1 and SH2 traps an intermediate state that is minimally populated during the steady state of muscle contraction (A.M′.D.P), without trapping hydrolysis products. This is consistent with the small (often negligible) change in orientation between relaxation and contraction, as detected by spectroscopic probes on the myosin head, especially for LCD probes in skeletal muscle (8,9,35,41–43). Changes in CD orientation are usually more detectable, probably because the LCD is more flexible (38° of disorder in rigor (Fig. 2)) than the CD (15° of disorder in rigor (7)). This emphasizes the importance of measuring LCD orientation and dynamics in stable states analogous to the A.M′.D.P state, and explains why this work resolves three distinct structural states of the LCD.
Structural coupling within myosin
Function of the myosin CD requires coordinated movement of the four subdomains (upper 50 kDa, lower 50 kDa, N-terminal, and converter) coordinated with structural transitions in three flexible joints (switch II, relay helix, and SH1 helix) (44–46). Previous biochemical work has shown that the SH1 helix becomes much more flexible upon nucleotide binding (47,48), and subsequent crystal structures suggested that nucleotide-induced unfolding of the SH1 helix uncouples the LCD and converter subdomain from the rest of the CD, producing an internally uncoupled state that allows increased motion of the converter and LCD and facilitates the transition to the prepower stroke conformation (46). These results explain SH1-SH2 cross-linking studies, showing that pPDM and other cross-linkers trap a weak-binding state of myosin in a nucleotide-dependent manner (48). Spin probes at SH1 indicate that both folded and unfolded states are populated in a nucleotide-dependent equilibrium (49,50).
Taken together, these results indicate that the SH1 helix has two structural states, folded and unfolded, that are loosely coupled to the biochemical state, as defined by the bound nucleotide. Only by cross-linking SH1 to SH2 (or by crystallizing myosin under special conditions) can the unfolded state be trapped. In this study, we have exploited these principles to trap the myosin head in this weakly bound structural state on actin, and our EPR results support the proposal that the CD and LCD are partially uncoupled in this state. Measurements of myosin CD and LCD orientation and dynamics indicate that both domains exhibit increased disorder similar to relaxation (prepower stroke) but slow dynamics indistinguishable from rigor (postpower stroke), and the structural properties of the two domains are distinct—the CD is more disordered. This is consistent with the hypothesis that unwinding of the SH1 helix partially uncouples the LCD and CD (46,51), but it is notable that this uncoupling and the resulting increased freedom of the LCD results in rigor-like dynamics of the LCD (τR ≥ 1 ms), at least two-orders-of-magnitude slower than observed for weak-binding states in the presence of ATP (10–13). The slow dynamics of the RLC in the cross-linked state correlates with the actin-binding properties of this state; pPDM-S1 binds actin ∼3 times stronger than S1.ATP (25).
Though we speculate that the pPDM-cross-linked state is analogous to the A.M.D.P biochemical state, we do not propose that hydrolysis products are trapped in this state. Rather, cross-linking traps myosin in a conformation that is populated after hydrolysis and isomerization to the A.M′.D.P state, but before product release and force generation. In addition to SH1-SH2 cross-linking, myosin complexed with ADP and the myosin inhibitor blebbistatin has been proposed to stabilize an analog of the A.M.D.P state (24). However, unlike cross-linking with pPDM, 1), ADP/blebbistatin is proposed to stabilize this prepower stroke state by stabilizing the M.D.P complex and slowing Pi release (52); and 2), myosin complexed with ADP and blebbistatin retains high actin affinity (similar to M.ADP) and a primed (prepower stroke) LCD position (24).
Conclusions
These structural measurements made on skinned muscle fibers are consistent with a revised model (Fig. 6) that is distinct from the previous model (Fig. 1) for coupling of actomyosin ATPase to force and movement. We conclude that cross-linking traps myosin in a structural state that is intermediate between weak-binding states (clearly preceding the power stroke) and strong-binding states (clearly following the power stroke). Though this conclusion is speculative, it is supported by an abundance of evidence. Considered collectively, previous measurements on SH1-SH2 cross-linked myosin indicate mechanical and biochemical properties intermediate between weak- and strong-binding states. Mechanical measurements indicate that pPDM-treated fibers exhibit an 85% decrease in isometric force (28) and a decrease in rigor stiffness to the level of resting fibers (29). Biochemical studies demonstrate that pPDM-S1 exhibits actin binding 1000 times weaker than S1.ADP but ∼3 times stronger that S1.ATP (25).
Figure 6.
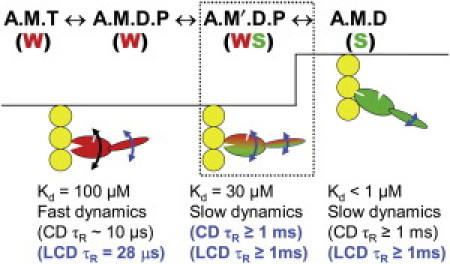
Model (updated from Fig. 1) for coupling of actomyosin biochemical transitions to orientation and dynamics of the myosin head (4). The text under each state indicates the distinguishing properties of the CD and LCD. (Blue text and curved arrows) Revisions based on this study.
This work demonstrates that the structural dynamics of this state is also intermediate between weak- and strong-binding states in a strikingly similar way as demonstrated in Fig. 2, Fig. 4, and Fig. 5, and discussed below. We measured LCD orientation and dynamics in relaxation, the pPDM-cross-linked state, and rigor (Fig. 2 and Fig. 4), demonstrating that the LCD transitions from a highly disordered state with microsecond dynamics (A.M.T) to a slightly more ordered state that is static on the microsecond timescale (A.M′.D.P analog), to a highly ordered state that is also static on the microsecond timescale (A.M.D). This sequence of structural transition of the LCD is similar to the disorder-to-order transition of the CD (6,7,36), although the CD is more dynamically disordered than the LCD in both the A.M.T and A.M′.D.P states, and the LCD is more disordered than the CD in rigor (A.M.D). An important challenge for the future is to test the model in Fig. 6 by observing the proposed intermediate not by trapping it, but by resolving it in the transient phase of a structural kinetics experiment (53).
Acknowledgments
We thank Leanne Kolb, Amanda Laden, Robert Harris, Christina Yi, Evan Smith, Doug Deitchler, Sarah Blakely, and Octavian Cornea for excellent technical support; Yuri Nesmelov, Andrew Thompson, David Kast, and Roman Agafonov for insightful discussions; Edmund Howard for EPR analysis software; and Thomas Burghardt and Katalin Ajtai for guidance regarding sodium dodecyl sulfate polyacrylamide gel electrophoresis on homogenized muscle fibers. Electron paramagnetic resonance spectroscopy was performed in the Biophysical Spectroscopy Center, and computational simulations were performed with support from the Minnesota Supercomputing Institute.
This work was supported by National Institutes of Health grants to D.D.T. (No. AR32961, No. AG26160, and No. AR057220) and to R.N.M. (No. AG033495).
Supporting Material
References
- 1.Thomas D.D., Ramachandran S., Ostap E.M. The mechanism of force generation in myosin: a disorder-to-order transition, coupled to internal structural changes. Biophys. J. 1995;68:135S–141S. [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor K.A., Schmitz H., Reedy M.K. Tomographic 3D reconstruction of quick-frozen, Ca2+-activated contracting insect flight muscle. Cell. 1999;99:421–431. doi: 10.1016/s0092-8674(00)81528-7. [DOI] [PubMed] [Google Scholar]
- 3.LaConte L.E., Baker J.E., Thomas D.D. Transient kinetics and mechanics of myosin's force-generating rotation in muscle: resolution of millisecond rotational transitions in the spin-labeled myosin light-chain domain. Biochemistry. 2003;42:9797–9803. doi: 10.1021/bi034288r. [DOI] [PubMed] [Google Scholar]
- 4.Thomas D.D., Kast D., Korman V.L. Site-directed spectroscopic probes of actomyosin structural dynamics. Annu. Rev. Biophys. 2009;38:347–369. doi: 10.1146/annurev.biophys.35.040405.102118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rayment I., Rypniewski W.R., Holden H.M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 6.Thomas D.D., Cooke R. Orientation of spin-labeled myosin heads in glycerinated muscle fibers. Biophys. J. 1980;32:891–906. doi: 10.1016/S0006-3495(80)85024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke R., Crowder M.S., Thomas D.D. Orientation of spin labels attached to cross-bridges in contracting muscle fibers. Nature. 1982;300:776–778. doi: 10.1038/300776a0. [DOI] [PubMed] [Google Scholar]
- 8.Hambly B., Franks K., Cooke R. Orientation of spin-labeled light chain-2 exchanged onto myosin cross-bridges in glycerinated muscle fibers. Biophys. J. 1991;59:127–138. doi: 10.1016/S0006-3495(91)82205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker J.E., Brust-Mascher I., Thomas D.D. A large and distinct rotation of the myosin light chain domain occurs upon muscle contraction. Proc. Natl. Acad. Sci. USA. 1998;95:2944–2949. doi: 10.1073/pnas.95.6.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger C.L., Svensson E.C., Thomas D.D. Photolysis of a photolabile precursor of ATP (caged ATP) induces microsecond rotational motions of myosin heads bound to actin. Proc. Natl. Acad. Sci. USA. 1989;86:8753–8757. doi: 10.1073/pnas.86.22.8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berger C.L., Thomas D.D. Rotational dynamics of actin-bound myosin heads in active myofibrils. Biochemistry. 1993;32:3812–3821. doi: 10.1021/bi00065a038. [DOI] [PubMed] [Google Scholar]
- 12.Berger C.L., Thomas D.D. Rotational dynamics of actin-bound intermediates in the myosin ATPase cycle. Biochemistry. 1991;30:11036–11045. doi: 10.1021/bi00110a005. [DOI] [PubMed] [Google Scholar]
- 13.Berger C.L., Thomas D.D. Rotational dynamics of actin-bound intermediates of the myosin adenosine triphosphatase cycle in myofibrils. Biophys. J. 1994;67:250–261. doi: 10.1016/S0006-3495(94)80476-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craig R., Greene L.E., Eisenberg E. Structure of the actin-myosin complex in the presence of ATP. Proc. Natl. Acad. Sci. USA. 1985;82:3247–3251. doi: 10.1073/pnas.82.10.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker M., White H., Trinick J. Electron cryomicroscopy of acto-myosin-S1 during steady-state ATP hydrolysis. Biophys. J. 1994;66:1563–1572. doi: 10.1016/S0006-3495(94)80948-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker M., Trinick J., White H. Millisecond time resolution electron cryo-microscopy of the M-ATP transient kinetic state of the acto-myosin ATPase. Biophys. J. 1995;68:87S–91S. [PMC free article] [PubMed] [Google Scholar]
- 17.Smith D.A., Sleep J. Mechanokinetics of rapid tension recovery in muscle: the myosin working stroke is followed by a slower release of phosphate. Biophys. J. 2004;87:442–456. doi: 10.1529/biophysj.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagi Y., Shuman H., Goldman Y.E. Coupling between phosphate release and force generation in muscle actomyosin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:1913–1920. doi: 10.1098/rstb.2004.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White H.D., Belknap B., Webb M.R. Kinetics of nucleoside triphosphate cleavage and phosphate release steps by associated rabbit skeletal actomyosin, measured using a novel fluorescent probe for phosphate. Biochemistry. 1997;36:11828–11836. doi: 10.1021/bi970540h. [DOI] [PubMed] [Google Scholar]
- 20.Werber M.M., Peyser Y.M., Muhlrad A. Characterization of stable beryllium fluoride, aluminum fluoride, and vanadate containing myosin subfragment 1-nucleotide complexes. Biochemistry. 1992;31:7190–7197. doi: 10.1021/bi00146a023. [DOI] [PubMed] [Google Scholar]
- 21.Prochniewicz E., Walseth T.F., Thomas D.D. Structural dynamics of actin during active interaction with myosin: different effects of weakly and strongly bound myosin heads. Biochemistry. 2004;43:10642–10652. doi: 10.1021/bi049914e. [DOI] [PubMed] [Google Scholar]
- 22.Pate E., Cooke R. Energetics of the actomyosin bond in the filament array of muscle fibers. Biophys. J. 1988;53:561–573. doi: 10.1016/S0006-3495(88)83136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fajer P.G., Fajer E.A., Thomas D.D. Effects of AMPPNP on the orientation and rotational dynamics of spin-labeled muscle cross-bridges. Biophys. J. 1988;53:513–524. doi: 10.1016/S0006-3495(88)83131-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takács B., Billington N., Kovács M. Myosin complexed with ADP and blebbistatin reversibly adopts a conformation resembling the start point of the working stroke. Proc. Natl. Acad. Sci. USA. 2010;107:6799–6804. doi: 10.1073/pnas.0907585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalovich J.M., Greene L.E., Eisenberg E. Cross-linked myosin subfragment 1: a stable analogue of the subfragment-1.ATP complex. Proc. Natl. Acad. Sci. USA. 1983;80:4909–4913. doi: 10.1073/pnas.80.16.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene L.E., Chalovich J.M., Eisenberg E. Effect of nucleotide on the binding of N,N′-p-phenylenedimaleimide-modified S-1 to unregulated and regulated actin. Biochemistry. 1986;25:704–709. doi: 10.1021/bi00351a030. [DOI] [PubMed] [Google Scholar]
- 27.Bobkov A.A., Reisler E. Is SH1-SH2-cross-linked myosin subfragment 1 a structural analog of the weakly-bound state of myosin? Biophys. J. 2000;79:460–467. doi: 10.1016/S0006-3495(00)76307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaen S., Shimada M., Sugi H. Evidence for cooperative interactions of myosin heads with thin filament in the force generation of vertebrate skeletal muscle fibers. J. Biol. Chem. 1986;261:13632–13636. [PubMed] [Google Scholar]
- 29.Barnett V.A., Ehrlich A., Schoenberg M. Formation of ATP-insensitive weakly-binding crossbridges in single rabbit psoas fibers by treatment with phenylmaleimide or para-phenylenedimaleimide. Biophys. J. 1992;61:358–367. doi: 10.1016/S0006-3495(92)81842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson A.R., Naber N., Thomas D.D. Structural dynamics of the actomyosin complex probed by a bifunctional spin label that cross-links SH1 and SH2. Biophys. J. 2008;95:5238–5246. doi: 10.1529/biophysj.108.138982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prochniewicz E., Lowe D.A., Thomas D.D. Functional, structural, and chemical changes in myosin associated with hydrogen peroxide treatment of skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 2008;294:C613–C626. doi: 10.1152/ajpcell.00232.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roopnarine O., Thomas D.D. Orientational dynamics of indanedione spin-labeled myosin heads in relaxed and contracting skeletal muscle fibers. Biophys. J. 1995;68:1461–1471. doi: 10.1016/S0006-3495(95)80319-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fajer P.G., Bennett R.L.H., Thomas D.D. General method for multiparameter fitting of high-resolution EPR spectra using a simplex algorithm. J. Magn. Reson. 1990;88:111–125. [Google Scholar]
- 34.Squier T.C., Thomas D.D. Methodology for increased precision in saturation transfer electron paramagnetic resonance studies of rotational dynamics. Biophys. J. 1986;49:921–935. doi: 10.1016/S0006-3495(86)83720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hambly B., Franks K., Cooke R. Paramagnetic probes attached to a light chain on the myosin head are highly disordered in active muscle fibers. Biophys. J. 1992;63:1306–1313. doi: 10.1016/S0006-3495(92)81717-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnett V.A., Thomas D.D. Microsecond rotational motion of spin-labeled myosin heads during isometric muscle contraction. Saturation transfer electron paramagnetic resonance. Biophys. J. 1989;56:517–523. doi: 10.1016/S0006-3495(89)82698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roopnarine O., Szent-Györgyi A.G., Thomas D.D. Microsecond rotational dynamics of spin-labeled myosin regulatory light chain induced by relaxation and contraction of scallop muscle. Biochemistry. 1998;37:14428–14436. doi: 10.1021/bi9808363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandran S., Thomas D.D. Rotational dynamics of the regulatory light chain in scallop muscle detected by time-resolved phosphorescence anisotropy. Biochemistry. 1999;38:9097–9104. doi: 10.1021/bi9902945. [DOI] [PubMed] [Google Scholar]
- 39.Thomas D.D., Dalton L.R., Hyde J.S. Rotational diffusion studied by passage saturation transfer electron paramagnetic resonance. J. Chem. Phys. 1976;65:3006–3024. [Google Scholar]
- 40.Roopnarine O. Mechanical defects of muscle fibers with myosin light chain mutants that cause cardiomyopathy. Biophys. J. 2003;84:2440–2449. doi: 10.1016/S0006-3495(03)75048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostap E.M., Barnett V.A., Thomas D.D. Resolution of three structural states of spin-labeled myosin in contracting muscle. Biophys. J. 1995;69:177–188. doi: 10.1016/S0006-3495(95)79888-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corrie J.E., Brandmeier B.D., Irving M. Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature. 1999;400:425–430. doi: 10.1038/22704. [DOI] [PubMed] [Google Scholar]
- 43.Burghardt T.P., Li J., Ajtai K. Single myosin lever arm orientation in a muscle fiber detected with photoactivatable GFP. Biochemistry. 2009;48:754–765. doi: 10.1021/bi8017703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houdusse A., Kalabokis V.N., Cohen C. Atomic structure of scallop myosin subfragment S1 complexed with MgADP: a novel conformation of the myosin head. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 45.Houdusse A., Szent-Gyorgyi A.G., Cohen C. Three conformational states of scallop myosin S1. Proc. Natl. Acad. Sci. USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Himmel D.M., Gourinath S., Cohen C. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc. Natl. Acad. Sci. USA. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huston E.E., Grammer J.C., Yount R.G. Flexibility of the myosin heavy chain: direct evidence that the region containing SH1 and SH2 can move 10 Å under the influence of nucleotide binding. Biochemistry. 1988;27:8945–8952. doi: 10.1021/bi00425a011. [DOI] [PubMed] [Google Scholar]
- 48.Nitao L.K., Reisler E. Probing the conformational states of the SH1-SH2 helix in myosin: a cross-linking approach. Biochemistry. 1998;37:16704–16710. doi: 10.1021/bi9817212. [DOI] [PubMed] [Google Scholar]
- 49.Barnett V.A., Thomas D.D. Resolution of conformational states of spin-labeled myosin during steady-state ATP hydrolysis. Biochemistry. 1987;26:314–323. doi: 10.1021/bi00375a044. [DOI] [PubMed] [Google Scholar]
- 50.Agafonov R.V., Nesmelov Y.E., Thomas D.D. Muscle and nonmuscle myosins probed by a spin label at equivalent sites in the force-generating domain. Proc. Natl. Acad. Sci. USA. 2008;105:13397–13402. doi: 10.1073/pnas.0801342105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gourinath S., Himmel D.M., Cohen C. Crystal structure of scallop myosin s1 in the pre-power stroke state to 2.6 Å resolution: flexibility and function in the head. Structure. 2003;11:1621–1627. doi: 10.1016/j.str.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Allingham J.S., Smith R., Rayment I. The structural basis of blebbistatin inhibition and specificity for myosin II. Nat. Struct. Mol. Biol. 2005;12:378–379. doi: 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- 53.Nesmelov Y.E., Agafonov R.V., Thomas D.D. Structural kinetics of myosin by transient time-resolved FRET. Proc. Natl. Acad. Sci. USA. 2011;108:1891–1896. doi: 10.1073/pnas.1012320108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


