Abstract
Iron is essential for aerobic organisms. Additionally, photosynthetic organisms must maintain the iron-rich photosynthetic electron transport chain, which likely evolved in the iron-replete Proterozoic ocean. The subsequent rise in oxygen since those times has drastically decreased the levels of bioavailable iron, indicating that adaptations have been made to maintain sufficient cellular iron levels in the midst of scarcity. In combination with physiological studies, the recent sequencing of marine microorganism genomes and transcriptomes has begun to reveal the mechanisms of iron acquisition and utilization that allow marine microalgae to persist in iron limited environments.
Keywords: iron, cyanobacteria, diatoms, algae, phytoplankton, genomics, prasinophytes
Introduction
Iron is essential for all aerobic organisms, but is highly reactive and toxic via the Fenton reaction (Halliwell and Gutteridge, 1992). Consequently, organisms tightly control iron homeostasis and have highly coordinated responses to iron deficiency and iron overload. Photosynthetic organisms must also maintain the iron-rich photosynthetic electron transport chain, which likely evolved in the iron-replete reducing environments of the Proterozoic ocean (Falkowski, 2006). The levels of bioavailable iron have decreased drastically over time, concurrent with the rise in oxygen, indicating adaptations have been made to maintain sufficient iron levels in the midst of scarcity.
Still, limited iron availability impairs phytoplankton growth in as much as 40% of the ocean, notably in the Southern Ocean, equatorial Pacific Ocean, and north Pacific Ocean (Moore et al., 2001). As the levels of other nutrients are sufficient, these areas are categorized as high-nutrient, low-carbon (HNLC). This iron limitation has been evidenced by iron fertilization experiments of HNLC waters, which can produce rapidly growing algal blooms (Boyd et al., 2007; Figure 1), and ultimately, speculation that these blooms could be utilized to capture and sequester carbon from the atmosphere (Chisholm et al., 2001). At the same time, elevated atmospheric carbon will likely acidify the ocean, almost certainly altering iron bioavailability, and thus algal productivity (Shi et al., 2010). Consequently, much effort has been made to predict how climate change will affect iron availability and phytoplankton growth, and how altered phytoplankton growth will itself affect climate change.
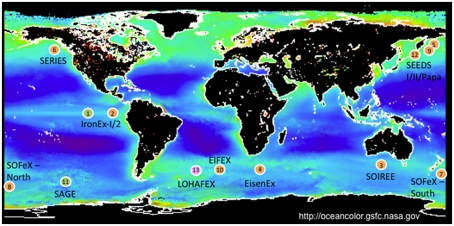
Figure 1.
Examples of iron limitation in the ocean as evidenced by rapid growth of diatoms and other plankton after iron fertilization experiments. Circle color indicates dominant plankton in resultant blooms: orange – diatoms; green – picophytoplankton; pink – zooplankton. 1 – IronEx-I, 1993; 2 – IronEx-II, 1995; 3 – SOIREE, 1999; 4 – EisenEx, 2000; 5 – SEEDS-I, 2001; 6 – SERIES, 2002; 7 – SOFeX North, 2002; 8 – SOFeX South, 2002; 9 – SEEDS-II, 2004; 10 – EIFEX, 2004; 11 – SAGE, 2004; 12 – PAPA-SEEDS, 2006; 13 – LOHAFEX, 2009. Adapted from Trick et al. (2010).
Despite the large-scale experiments related to iron and the ocean, our current understanding of the underlying mechanisms of iron homeostasis in phytoplankton remains limited. Earlier work has shown that iron quotas are often optimized in marine phytoplankton, yet the mechanisms of iron uptake in these organisms remain obscure. The sequencing of marine phytoplankton genomes and community metagenomes has revealed a plethora of genes of unknown function, many of which are species-specific (e.g., Rocap, 2003; Venter et al., 2004; Allen et al., 2008; Frias-Lopez et al., 2008; Maheswari et al., 2010). Thus it is proposed that phytoplankton survival in iron-starved waters could rely on novel adaptations encoded by these genes. Indeed, it was recently found that the newly cultured marine species Chromera velia appears to lack any of the currently characterized systems of iron uptake (Sutak et al., 2010); while the halophilic green alga Dunaliella salina was found to utilize a transferrin, an uptake system well-characterized in mammals, but otherwise unknown in plants (Paz et al., 2007). This review will focus on the recent advances in genomics in marine phytoplankton models that have begun to shed light on the adaptations that allow survival in environments where iron is vanishingly rare.
Marine Cyanobacteria
Cyanobacteria, modern examples of the oldest oxygenic phototrophs, are proposed to have begun the great oxidation event – the initial oxygenation of the earth’s atmosphere around 2.4 billion years ago (Kasting and Siefert, 2002). While metagenomic approaches have begun to reveal the diversity of bacteria in the oceans (e.g., Venter et al., 2004; Frias-Lopez et al., 2008; Zehr et al., 2008), the physiological characterization of iron homeostasis in free-living marine cyanobacteria has been primarily limited to the diazotrophs Trichodesmium and Crocosphaera watsonii, and the non-diazotrophs Synechococcus and Prochlorococcus. The genome sequencing and expression analysis in these genera, combined with physiological characterization, suggest that several mechanisms to survive iron limitation exist across marine cyanobacteria species (Table 1):
Table 1.
Iron-related genes and proteins mentioned in the review (see text for reference).
| ELECTRON TRANSPORT | |
| isiA | Novel chlorophyll-binding protein that forms chlorophyll-protein-antenna super-complexes during Fe-starvation |
| isiB (flavodoxin) | Fe-free electron transfer protein that can replace ferrodoxin during Fe-starvation. |
| petF (ferredoxin) | Fe–S cluster based electron transfer protein used in a wide variety of reactions, including electron transfer to NADP+ reductase during photosynthesis. |
| petE (plastocyanin) | Cu-based electron transfer protein that can replace cytochrome c6 during Fe-starvation; transfers electrons from cytochrome b6f complex to PSI. |
| petJ (cytochrome c6) | Cyanobacterial heme-based electron transport protein in thylakoid lumen downregulated during Fe-starvation; transfers electrons from cytochrome b6f complex to PSI. |
| cytochrome cm | Cyanobacterial heme-based electron transport protein downregulated during Fe-starvation; may function in PS and respitory electron transport chains. |
| cytochrome b6f complex | Fe-rich electron transfer and proton pumping complex in thylakoid membrane, down-regulated during Fe-starvation; mediates electron movement from PSII to PSI. |
| IRON TRANSPORT | |
| Bacterial Fe(III) transport system | |
| idiA/futA/afuA | Fe(III) binding protein |
| idiB/futB | Permease |
| idiC/futC | ATPase |
| Bacterial Fe(II) transport system | |
| feoA | Small soluble protein |
| feoB | Predicted Fe(II) permease |
| feoC | Predicted regulator |
| Divalent metal transporters | |
| ZIP | ZRT, IRT-like proteins – transports divalent transistion metals into cytoplasm, e.g., Fe(II), Zn, Mn, Cu(II), Co, Ni, Cd |
| NRAMP | Natural resistance-associated macrophage proteins – transports divalent transistion metals into cytoplasm, e.g., Fe(II), Zn, Mn, Cu(II), Co, Cd |
| Oxidase-permease based transport system | |
| FRE | Ferric chelate reductase – transfers electrons from NADH via heme to reduce Fe(III) |
| FET3 | Multicopper ferroxidase that oxidizes Fe(II) from ferric reductases and passes Fe(III) to FTR |
| FTR | High affinity iron permease – transports Fe(III) across the plasma membrane, in complex with Fet3 |
| OTHER | |
| Fur | Canonical bacterial transcriptional regulator that represses iron uptake genes |
| FER | Ferritin – sequesters and oxidizes Fe(II) in a multimer; found in the plastid and mitochondria of plants, and the cytosol and mitochondria of human; expression is induced by excess iron, thus mitigating oxidative stress. |
| Ftn | Bacterial ferritin. |
| Dps | DNA-binding proteins from starved cells – bacterial Fe-sequestering protein, that can also bind DNA. |
Iron uptake is likely mediated by the FutA/IdiA-based ABC transporter system (Figure 2), genes for which have been found in 28 unicellular cyanobacteria genomes of Prochlorococcus, Synechococcus, and Synechocystis (Rivers et al., 2009). futa/idia is predicted to encode a periplasmic iron binding protein; futb likely encodes a Fe(III) permease; and futC an ATPase binding protein. Levels of FutA/IdiA protein increase under iron starvation in Prochlorococcus (Bibby et al., 2003; Thompson et al., 2011), Trichodesmium sp. IMS 101, Crocosphaera sp. WH8501, and Synechococcus spp. WH8103 and WH7803 (Webb et al., 2001). Work in freshwater cyanobacteria found that the futA/idia ortholog futA2 encodes a periplasmic iron concentrating protein essential for Fe(III) uptake (Katoh et al., 2001; Badarau et al., 2008). Additionally, freshwater cyanobacteria possess FutA1, which also functions in iron uptake (Katoh et al., 2001) but is found to localize to the thylakoid membrane and plays an unknown role in the protection of PSII (Michel et al., 1998; Exss-Sonne et al., 2000; Tölle et al., 2002). The role, if any, of FutA1 in marine cyanobacteria remains uninvestigated.
Iron limitation remodels the machinery of photosynthesis (Barber et al., 2006). Specifically, the number of photosynthetic complexes is reduced: iron-rich PSI (12 iron atoms) decreases, in favor of PSII (three iron atoms), and the number of phycobilisomes (which are synthesized by iron-containing proteins) decreases.
Finally, genome analysis of Prochlorococcus, Synechococcus, Crocosphaera, and Trichodesmium species suggests that nickel superoxide dismutase (SOD) is utilized in place of iron SOD to remove reactive oxygen species (ROS; Dufresne et al., 2003; Palenik et al., 2003; Rocap, 2003; Eitinger, 2004).
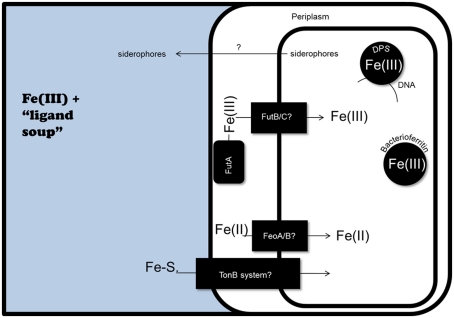
Figure 2.
Potential iron homeostasis systems in marine cyanobacteria, as predicted by genomic analyses. At least in part, iron uptake in cyanobacteria is likely facilitated by the concentration of Fe(III) in the periplasmic space by FutA, followed by transport into the cytoplasm by the FutB/FutC ABC transporter system. The presence of FeoA/B genes in marine cyanobacteria genomes suggests Fe(II) uptake could also occur. Finally, the Synechococcus sp. PCC 7002 genome contains genes for siderophore biosynthesis, as well as Fe-siderophore (Fe-S.) uptake via a TonB dependent receptor system. Within the cell, iron could be sequestered by bacterioferritin and Dps.
Non-diazotrophs
Prochlorococcus and Synechococcus are the two most prominent genera of picoplanktonic marine cyanobacteria (Partensky et al., 1999a). Although they overlap in some ecosystems and may have participated in lateral gene transfer (Beiko et al., 2005; Zhaxybayeva et al., 2009), Synechococcus has a broader global distribution, especially in temperate latitudes and coastal regions, while Prochlorococcus is more abundant in tropical latitudes and oligotrophic environments (Zwirglmaier et al., 2008). Synechococcus and Prochlorococcus also differ in their light-harvesting apparatus: Synechococcus utilizes chlorophyll a, while Prochlorococcus utilizes divinyl chlorophylls a and b (Partensky et al., 1999b). The sequencing of these genomes is beginning to reveal the diverse genetic adaptations that allow survival in a range of nutrient environments.
Prochlorococcus
Prochlorococcus is a very small cyanobacteria (0.5–0.7 μm in diameter), ubiquitous within the latitudes 40°S to 40°N, and perhaps the most abundant photosynthetic organism on earth (Partensky et al., 1999b). Some natural populations have been shown to be somewhat iron-starved, as iron addition experiments have resulted in increased Prochlorococcus cell division, cell size, and chlorophyll levels (Cavender-Bares et al., 1999; Mann and Chisholm, 2000), and Prochlorococcus dominates Synechococcus in the iron limited equatorial Pacific (Campbell et al., 1997). Prochlorococcus populations, however, have been shown to be much less iron-starved than larger cells such as diatoms (Partensky et al., 1999b). The high surface-to-volume ratio of the small Prochlorococcus cell presumably aids nutrient uptake in these iron limited environments (Chisholm, 1992), although the MIT9313 ecotype was found to be more tolerant of iron limitation than the smaller-sized MED4 (Thompson et al., 2011). Presumably, this is because MIT9313 is from waters with 25-fold less total iron levels than MED4.
Prochlorococcus has a small genome of less than 2000 genes (Rocap et al., 2002), and the sequences of over a dozen strains have now been published. Several Prochlorococcus ecotypes possess iron homeostasis genes missing in some species of Synechococcus (Rocap, 2003), suggesting that they could be environmental adaptations. These include (Table 1):
Flavodoxin (isiB), an iron-free electron transfer protein that can replace the functionally equivalent Fe–S protein ferredoxin (petF) under iron limitation (Erdner and Anderson, 1999).
One to two ferritin genes (Figure 2). Ferritin is an iron storage protein associated with survival in low iron marine environments (Marchetti et al., 2009), and prevention of iron-induced oxidative stress in terrestrial organisms, e.g., Arabidopsis (Ravet et al., 2009) and humans (Corsi et al., 1998; Orino et al., 2001);
Two to three fur genes, the canonical bacterial transcriptional regulator that represses iron uptake genes.
Candidates for a high affinity iron scavenging system (Rocap et al., 2002).
The Prochlorococcus core genome lacks Fe–siderophore complex-related genes, but has components of a bacterial Fe(III) ABC transporter encoded by idiA/futA/afuA, futB, and futC (Rocap et al., 2003). However, it remains unclear what iron species Prochlorococcus is able to transport. MIT9313 is more sensitive to copper than MED4 and possesses putative iron transport genes that are missing in MED4, suggesting variation exists in substrate specificity of iron uptake systems between the ecotypes (Thompson et al., 2011).
Examination of the transcriptional response to iron starvation using qPCR and microarrays found that flavodoxin (isiB) is upregulated and that ferredoxin (petF) is downregulated (Bibby et al., 2003; Thompson et al., 2011), while some genes associated with the iron-rich PSI and cytochrome b6f complexes are downregulated, presumably allowing the reallocation of iron. To increase iron uptake, idiA is upregulated (although futB and futC are constitutively expressed under both iron-sufficiency and deficiency; Thompson et al., 2011). Finally, hli genes are upregulated, presumably to protect the photosystems from oxidative stress.
Comparisons of different Prochlorococcus “ecotypes” have found that there are differences in expression of iron-regulated genes, indicating there is natural variation in the iron deficiency response (Bibby et al., 2003; Thompson et al., 2011). An examination of MIT9313 and MED4 found that the iron-regulated transcriptome was enriched with genes from the genomic islands and genes outside the core genome (Thompson et al., 2011). It was previously shown that Prochlorococcus ecotypes use gene islands with nutrient transport and assimilation genes, perhaps gained by lateral gene transfer, to adapt to the phosphate and nitrogen availability of their environment (Martiny et al., 2006, 2009), so it is plausible that lateral gene transfer has also provided adaptations to iron limitation. Another potential adaptation strategy was identified in two ecotypes from low iron regions of the Eastern Equatorial Pacific upwelling and the tropical Indian Ocean through reconstruction of putative genomes of previously unidentified ecotypes from the 73 metagenomic samples of the Global Ocean Sampling expedition (Rusch et al., 2010). These two new genomes had the same assortment of iron uptake and stress genes as other ecotypes; however, six iron-containing proteins were absent. Assuming that the absence from these metagenomic data sets represents the absence from these actual genomes, this would indicate that the iron quotas are minimized via the loss of approximately 10% of the genes for iron-based proteins found in other Prochlorococcus ecotypes (Rusch et al., 2010). The missing iron-containing proteins include nitrate reductase, and several electron transfer proteins that are associated with the optimization of photosynthetic efficiency: two ferredixins, plastoquinol oxidase (PTOX), and cytochrome cm (Table 1). It was proposed that this reduces the maximum photosynthetic efficiency of these ecotypes, but allows survival in a low iron environment. At the same time, this likely limits the ability of these ecotypes to respond and grow rapidly following the appearance of iron, as iron addition experiments in this part of the ocean show only a minimal response from Prochlorococcus (Rusch et al., 2010).
Synechococcus
Iron starvation of marine Synechococcus results in accumulation of glycogen granules, decreased chlorophyll a and thylakoid leaflets, and decreased protein levels of phycocyanin, allophycocyanin, and the PSII reaction center D1 peptide PsbA (Sherman and Sherman, 1983; Webb et al., 1994; Michel et al., 2003). The iron quota is likely minimized by the use of (Table 1):
The copper-containing plastocyanin in place of the iron–protein cytochrome c6 for electron transport from cytochrome b6f complex to PSI.
A cobalt-dependent ribonucleotide reductase.
A putative nickel SOD (Rivers et al., 2009).
Flavodoxin (isiB) is present in all Prochlorococcus examined so far, but absent in nearly two-thirds of the marine Synechococcus genomes currently available. Finally, the gene isiA is present in the genomes of three out of the four marine Synechococcus species from environments that are perhaps iron limited (Bibby et al., 2009); thus it has been proposed to be an adaptation to low iron environments, although it is absent from the oligotrophic strain WH8102 (Dufresne et al., 2008). In the thermophilic freshwater species S. elongates, isiA is upregulated in response to iron starvation (Park et al., 1999; Bibby et al., 2001a), resulting in the formation of giant PSI-IsiA-chlorophyll–protein–antenna super-complexes (Bibby et al., 2001a,b; Boekema et al., 2001). Disruption of isiA in the freshwater species results in increased photoinhibition and reduced growth under iron starvation (Michel et al., 1996; Park et al., 1999), suggesting it is an important component of the iron deficiency response. Again, the relationship of freshwater IsiA to that found in marine species has not yet been investigated.
How iron moves through the outer membrane is unknown, although the Synechococcus genome is heavily enriched with genes predicted to encode transporters (Palenik et al., 2003). Both a coastal (PCC 7002) and open ocean (CCMP 1334/WH7803) species of Synechococcus can utilize a variety of siderophores (Hutchins et al., 1999), although siderophore uptake genes have not been identified in the WH8102 genome (Palenik et al., 2003). However, it was observed that the freshwater cyanobacteria Synechocystis sp. PCC 6803 utilizes Fe(III)-siderophores through reduction, and then presumably transport of Fe(II) (Kranzler et al., 2011). Some coastal marine Synechococcus species produce siderophores (Wilhelm and Trick, 1994; Ito and Alison, 2005), and the genome of one of these, PCC 7002, contains genes related to siderophore biosynthesis and uptake via putative TonB dependent receptors (Hopkinson and Morel, 2009). However, siderophore secretion has not been found in oligotrophic Synechococcus, and siderophore synthesis and uptake genes have not been identified in WH8102 (Palenik et al., 2003), nor in genomes from other open ocean strains, including CCMP 1334/WH7803 (Hopkinson and Morel, 2009).
Further sequencing has revealed variation between genomes of coastal and open ocean Synechococcus, perhaps representing environment-specific adaptations for metal homeostasis. The open ocean is a more constant environment with lower nutrient levels, while wind-driven nutrient upwellings and inputs from land result in higher total iron concentrations in coastal environments (Ryther and Kramer, 1961). Appropriately, coastal species have higher iron quotas (Sunda et al., 1991), and the genome of the coastal species CC9311 was found to possess more genes for iron-containing proteins than the open ocean WH8102 (Palenik et al., 2006). Also unique to the coastal genome was feoA/B, predicted to encode putative Fe(II) transporters absent from WH8102 (Figure 2). This is of interest because it is proposed that bioavailable Fe(II) may be more abundant in the coastal ocean through photochemical reactions with organic matter (Kuma et al., 1992). In the CC9311 genome, feoA/B was located in islands of atypical trinucleotide composition, suggesting it was acquired through horizontal gene transfer (Palenik et al., 2006). Further examination of the genomes of several coastal Synechococcus species (WH5701, RS9917, and CC9311) again found feoB, while it is absent from Prochlorococcus genomes (Rivers et al., 2009).
The CC9311 genome also contains five copies of a bacterial ferritin, including one in an island suggestive of horizontal gene transfer (Palenik et al., 2006). Also present is dpsA (Palenik et al., 2006; Figure 2), a divergent member of the bacterioferritin superfamily found in most marine Synechococcus genomes, and absent from most Prochlorococcus genomes (Rivers et al., 2009). In bacteria and Archaea, dps genes are often expressed during periods of oxidative stress, long term nutrient deficiency, and stationary growth phase. In freshwater S. elongates species, disruption of dpsA results in death under iron starvation (Sen et al., 2000). Although its function remains unclear, DpsA from freshwater Synechococcus PCC 7942 contains heme and has weak catalase activity in vitro (Peña and Bullerjahn, 1995), is localized to the photosynthetic membranes (Durham and Bullerjahn, 2002), and can bind chromosomal DNA in vitro (Peña et al., 1995).
Diazotrophs
Nitrogen fixation by diazotrophs allows growth in nitrogen starved waters; however this process is iron intensive, as the nitrogenase protein complex is composed of the iron-rich proteins NifH (four iron atoms per homodimer) and NifDK (15 iron atoms per homodimer; Rubio and Ludden, 2008). Biological nitrogen fixation in cyanobacteria is believed to have evolved in the anoxic ocean where Fe(II) was soluble and thus more bioavailable (Falkowski, 1997). Consequently, the scarcity of readily available iron in the modern ocean limits nitrogen fixation (Berman-Frank et al., 2001; Moore et al., 2009). At the same time, oxygenic photosynthesis and nitrogen fixation must be separated due to the extreme sensitivity of the nitrogenase Fe–S clusters to oxygen (Fay, 1992). Thus, diazotrophs must balance nitrogen and iron metabolism, both in terms of iron utilization, and the spatial and temporal arrangement of these incompatible reactions. Metagenomic analysis of oligotrophic seawater identified perhaps the most extreme adaptation to this dilemma in the ostensible absence of PSII genes from the genome of UCYN-A, an uncultured nitrogen-fixing cyanobacteria (Zehr et al., 2008; Tripp et al., 2010).
Trichodesmium
Trichodesmium is a nitrogen-fixing, filamentous, non-heterocystous cyanobacteria. Abundant in tropical and subtropical surface waters, Trichodesmium forms blooms thousands of kilometers wide and completes more marine nitrogen fixation than any other organism (Capone et al., 1997). However, the combination of iron-rich photosynthetic complexes and nitrogenase results in higher intracellular iron quotas for Trichodesmium than other phytoplankton (Kustka et al., 2003a). Iron limitation in T. erythraeum IMS101 results in decreased growth, filament length, and chlorophyll levels, in addition to decreased nitrogen fixation and photosynthetic efficiency (Shi et al., 2007; Küpper et al., 2008). Similar changes were seen in four other Trichodesmium species (Chappell and Webb, 2010). A unique adaptation to this high iron quota is found in puff colonies of Trichodesmium collected from the Red Sea (although not in laboratory cultures), which actively acquire desert dust and utilize the iron (Rubin et al., 2011). Striking movies show that dust particles quickly move along the cell surface of the trichome from the colony periphery to the core, where dust and oxides are actively dissolved by an unknown mechanism. As dust inputs are correlated with Trichodesmium abundance, the ability to directly and efficiently utilize wind-blown desert dust could fuel the giant blooms.
In the iron deficiency response, a “hierarchy of iron demand” is proposed to exist in Trichodesmium, with mRNA associated with nitrogen fixation being downregulated more quickly than photosynthesis genes (Shi et al., 2007). In terms of upregulation, idiA and isiA expression were induced in response to iron starvation (Webb et al., 2001; Küpper et al., 2008), in addition to isiB (flavodoxin) and feoB (Chappell and Webb, 2010). The genomes of Trichodesmium species also contain multiple copies for the iron uptake regulator fur, and genes for bacterial ferritin and dps (Chappell and Webb, 2010). Additionally, the presence of TonB related genes suggests that Trichodesmium may have the ability to actively transport siderophores.
Crocosphaera watsonii
The unicellular diazotroph C. watsonii is found in the tropical and subtropical open ocean. Intracellular iron levels change throughout the day, increasing at night with the expression of nitrogenase (Tuit et al., 2004). Analysis of the C. watsonii transcriptomes by qPCR revealed a temporal pattern to iron demand, correlating with increased expression of flavodoxin and the iron homeostasis genes feoAB, and fur in the evening (Shi et al., 2009). This suggests coordination with nitrogenase activity. Indeed, one feoAB operon is found within the nif cluster. A similar increase in evening expression of these iron-related genes is also seen in the unicellular diazotrophic cyanobacteria Cyanothece (Stockel et al., 2008).
Absolute quantitation of the protein levels (using selected reaction monitoring mass spectrometry with isotopically labeled peptide standards) across the diel cycle found that the synthesis of photosynthesis related proteins peaks in the day, and that the proteins are degraded as evening approaches. Conversely, the nitrogenase complex proteins are absent in the day and synthesized at night (Saito et al., 2011). At the same time, bacterioferritin protein levels cycle, with peaks matching both the maximum iron utilization points for photosynthesis and nitrogen fixation. This could suggest a role in handling the iron being transferred between the two systems. Thus, it is proposed that by shifting iron from photosynthesis in the day to nitrogen fixation at night, C. watsonii minimizes its iron quota and creates temporal separation of the two incompatible systems. Indeed, C. watsonii is predicted to have half the cellular iron concentration (relative to carbon) as Trichodesmium, which fixes nitrogen during the day (Kustka et al., 2003b; Saito et al., 2011).
Eukaryotes
As described above for cyanobacteria, metagenomic approaches are uncovering a wealth of unknown marine eukaryotic phytoplankton species. In particular, alveolates and stramenopiles of great diversity and novelty are detected in almost all metagenome surveys (Massana and Pedrós-Alió, 2008). Community interactions may also be relevant to iron homeostasis. For example, a potential alga–bacteria mutualism in iron uptake between the dinoflagellate Scrippsiella trochoidea and Marinobacter could be representative of a more common iron uptake strategy among other phytoplankton (Amin et al., 2009). At the other end of the size spectrum, iron-rich whale feces also appears to serve as an important source of iron for phytoplankton in the Southern Ocean (Lavery et al., 2010). Additionally, the role of zooplankton grazing during iron fertilization experiments is of particular interest (Figure 1), as it may prevent the long term sequestration of carbon on the seafloor by diatoms (Bishop and Wood, 2009; Mazzocchi et al., 2009). The haptophyte Phaeocystis also appears to play an important role in blooms from polar iron fertilization experiments (Pollard et al., 2009). However, relatively little is known about the physiologies of many of these organisms, and the genome sequences of most of them are not yet available.
Most progress to date has been made in understanding the genetic underpinnings of iron homeostasis in green algae and diatoms. Significantly, genome and transcriptome data, combined with the development of stable transformants to overexpress and knockdown genes in diatoms (Poulsen and Kröger, 2005; Siaut et al., 2007; De Riso et al., 2009) and the green alga Ostreococcus tauri (Corellou et al., 2009) will likely accelerate our understanding of iron homeostasis in eukaryotic phytoplankton through functional genetics.
Ostreococcus
Ostreococcus are marine green algae belonging to the prasinophytes, and are described as the smallest free-living eukaryotes. Ostreococcus species possess very small, dense nuclear genomes of 12.5–13.0 Mbp. For comparison, the diatom Phaeodactylum tricornutum genome is 27.4 Mbp, and the Chlamydomonas reinhardtii genome is 120 Mbp. The two species of Ostreococcus that have been sequenced are from contrasting environments: O. lucimarinus from the open coastal waters of the Pacific Ocean (Worden et al., 2004); and O. tauri from a more nutrient-replete oyster production lagoon on the Mediterranean coast of France. TransportDB (www.membranetransport.org) predicts the presence of genes for divalent metal transporters ZIP and NRAMP (Ren et al., 2006; Table 1; Figure 3). Genes with similarity to prokaryotic siderophore uptake are present, and O. lucimarinus has genes that could represent a siderophore biosynthesis pathway (Palenik et al., 2007). Much like other marine phytoplankton, the iron quota is minimized: iron-free plastocyanin substitutes for cytochrome c6; flavodoxin is present; and Ni–SOD, Cu/Zn–SOD, and Mn–SODs appear to replace Fe–SOD (Palenik et al., 2007). Transcript levels of ferritin and a ferredoxin family protein in O. tauri are clock regulated, both peaking at dusk (Monnier et al., 2010). It would thus be worthwhile to investigate the relationship between free iron and the recently identified O. tauri redox clock (O’Neill et al., 2011).
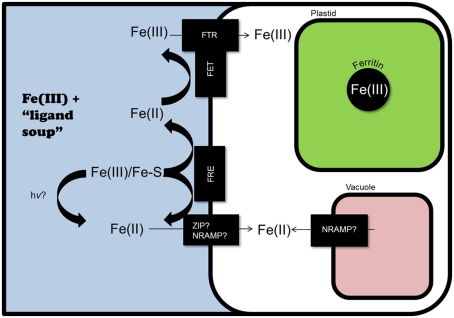
Figure 3.
Potential iron homeostasis systems in marine diatoms, as predicted by genomic analyses. Iron-regulated ferric reductase genes have been identified in T. pseudonana and P. tricornutum. These could reduce Fe(III) and Fe bound by siderophores (Fe–S), as could photoreduction (hv). Fe(II) could then enter the cytoplasm through iron-regulated transporters: ZIP in P. tricornutum, and NRAMP in T. pseudonana (although if TpNRAMP is localized to the tonoplast, it could also serve to release iron from the vacuole during iron starvation). In T. pseudonana, extracellular Fe(II) could also be reoxidized and transported through a yeast-like Fe(III) uptake system, utilizing the iron-regulated multi-copper ferroxidase (TpFET3) and Fe(III) permeases (TpFTR1 and TpFTR2). If ferritin is present (it is present in some pennate diatom genomes, but not in T. pseudonana), it can store iron, likely in the plastid.
There are species-specific differences in the repertoire of iron homeostasis genes, although it is unclear how these relate to the different nutrient profiles of their respective environments. The O. tauri genome has genes that could encode a multi-copper oxidase and two putative ferric reductases lacking in O. lucimarinus, while O. lucimarinus has two copies of ferritin genes and O. tauri has only one (Palenik et al., 2007; Jancek et al., 2008).
Marine diatoms
Diatoms, which carry out nearly 20% of photosynthesis on earth (Tréguer et al., 1995), are often found in the most iron limited regions of the ocean (Moore et al., 2001). Iron fertilization experiments often result in blooms dominated by diatoms, suggesting diatoms have adaptations that allow survival in iron limited waters and a subsequent rapid multiplication when iron becomes available. The recent metatranscriptomic analysis of iron fertilization bottle experiments found that diatoms possess a unique transcriptional response to the sudden appearance of iron (Marchetti et al., 2012). While other phytoplankton in the community increase gene expression for a broad array of iron–proteins like ferredoxin, cytochrome c6, and Fe–SOD, diatoms appear to prioritize expression of genes related to photosynthesis and nitrate uptake, reduction, and assimilation. Ostensibly, this strategy allows rapid diatom growth and bloom domination.
Representatives of the pennate and centric diatom lineages, Thalassiosira pseudonana and P. tricornutum, respectively, have been sequenced, and extensive molecular biology tools have been developed. Additionally, the publication of the polar pennate diatom Fragilariopsis cylindrus genome is imminent, and the bloom-forming pennate Pseudo-nitzschia multiseries is currently being sequenced. Despite these resources, predicting how diatom iron homeostasis systems function is complicated by the prevalence of unique genes of unknown function and by horizontal gene transfer. Currently, less than 50% of diatom genes have a putative function (Bowler et al., 2008; Maheswari et al., 2010), and the nuclear genome of P. tricornutum contains at least 587 genes predicted to be of bacterial origin (of which, around 60% are shared with T. pseudonana; Bowler et al., 2008).
Thalassiosira pseudonana
Thalassiosira pseudonana was the first eukaryotic marine phytoplankton sequenced. T. pseudonana has a small genome and has served as a model for marine centric diatom physiology experiments. T. pseudonana is often described as a marine coastal diatom, although recent phylogenetic analysis suggests T. pseudonana is more closely related to the freshwater and marine diatom genus Cyclotella (Alverson et al., 2011). Consequently, it was postulated that T. pseudonana may be a freshwater diatom that is adapted to salinity, rather than a marine Thalassiosira species (Alverson et al., 2011).
Iron limitation of T. pseudonana results in:
Decreased growth (Sunda and Huntsman, 1995) and photosynthetic efficiency (Bidle and Bender, 2007).
Increased cell aggregation and silica deposition on the cell wall (Mock et al., 2008), with the relative proportion of iron in the cell wall increasing (Ellwood and Hunter, 2000).
Increased oxidative stress and caspase activity, ultimately resulting in programmed cell death (Bidle and Bender, 2007; Thamatrakoln et al., 2011).
The sequencing of T. pseudonana revealed possible components of a yeast-like Fe(III) uptake system, including a multi-copper ferroxidase (TpFET3) and two iron permeases (TpFTR1 and TpFTR2; Table 1; Figure 3; Armbrust et al., 2004). Additionally, there are at least two putative ferric reductases (TpFRE1 and TpFRE2) and a putative divalent metal transporter (TpNRAMP), which could suggest a second, reduction-based uptake system, similar to those in Arabidopsis and humans. A similar combination of systems has been predicted to exist in C. reinhardtii (Merchant et al., 2006).
Transcript levels of FRE1, FTR1, FTR2, and NRAMP increase in response to iron limitation (Kustka et al., 2007), as does the transcript for flavodoxin, and those of several genes associated with oxidative stress and programmed cell death (Thamatrakoln et al., 2011). The upregulation of the ferric reductases is of additional interest, as T. pseudonana, T. weissflogii, and T. oceanica can utilize iron–siderophores via reduction (Hutchins et al., 1999; Maldonado and Price, 2001; Shaked et al., 2005). Finally, microarray analysis found that more than a third of iron-regulated genes were genes of unknown function and hypothetical proteins (Thamatrakoln et al., 2011), suggesting the existence of novel adaptations to iron starvation. Of the genes upregulated in response to iron limitation, 84 were also upregulated under silicon limitation, providing further evidence that the iron and silicon starvation pathways are interconnected, particularly at the point of cell wall synthesis (Mock et al., 2008). It was thus proposed that iron is incorporated with silicon into the cell wall, or that iron–proteins could play a role in cell wall deposition (Mock et al., 2008). X-ray fluorescence tomography of the diatom Cyclotella meneghiniana revealed distinct iron bands girding the frustules, supporting the idea that iron has a specialized function in the cell wall (de Jonge et al., 2010). Unfortunately, it is difficult to draw conclusions for T. pseudonana from this interesting result, as the sample tested was a desiccated, freshwater diatom.
Thalassiosira oceanica
Relative to other Thalassiosira species, the open ocean species T. oceanica is more adapted to iron limitation, growing faster under these conditions than the coastal species T. pseudonana (Sunda et al., 1991; Maldonado and Price, 1996) and T. weissflogii (Strzepek and Harrison, 2004). Biochemical measurements found that a novel photosynthetic architecture minimizes the iron quota of T. oceanica, decreasing the concentrations of the iron-rich PSI (12 iron atoms) and cytochrome b6f complexes (six iron atoms) by fivefold and sevenfold, respectively (Strzepek and Harrison, 2004). Additionally, the cytochrome c6 complex (one iron atom) is replaced by the copper-protein plastocyanin – a protein not found in T. weissflogii nor T. pseudonana (Peers and Price, 2006; Table 1). While photosynthetic efficiency of T. oceanica is not altered under normal light conditions, under high light it is more susceptible to photoinhibition and photosynthesis becomes nearly half as efficient as in T. weissflogii. The utilization of plastocyanin makes T. oceanica much more sensitive to copper limitation than T. weissflogii (Peers and Price, 2006). Thus, it is proposed that these adaptations to its low iron environment impair adjustment to rapid fluctuations in light intensity and copper limitation – environmental characteristics less common in the open ocean. Further adaptations to iron starvation likely exist in T. oceanica, because during iron limitation it is estimated that 100% of cellular iron is utilized by the electron transport carriers of photosynthesis, compared to only 50% in T. weissflogii (Strzepek and Harrison, 2004). The fate of the mitochondrial electron transport chain and other iron-containing proteins in iron-starved T. oceanica is worth investigating.
Finally, sequencing of the T. oceanica chloroplast genome revealed that the ferredoxin gene (petF) appears to have been transferred from the chloroplast genome to the nuclear genome (Lommer et al., 2010). Because the ferredoxin gene remains in the T. pseudonana and T. weissflogii chloroplast genomes, it is proposed that this change could alter regulation of ferredoxin expression under iron limitation, presumably contributing to the observed tolerance to iron limitation (Lommer et al., 2010); although under iron limitation the ratio of ferredoxin to flavodoxin in T. oceanica is not significantly lower than in T. weissflogii (Strzepek and Harrison, 2004).
Phaeodactylum tricornutum
Phaeodactylum tricornutum can grow at iron levels 50 times lower than T. pseudonana (Kustka et al., 2007). P. tricornutum appears to use a fundamentally different iron uptake system than T. pseudonana, raising the possibility that it could be more effective at iron uptake under limiting conditions. The ferroxidase and iron permeases indicative of a yeast-like system in T. pseudonana have not been identified in the P. tricornutum genome (Kustka et al., 2007; Bowler et al., 2008). Examination of genes upregulated in response to iron starvation using expressed sequenced tags revealed two ferric reductases, PtFRE1 and PtFRE2(Allen et al., 2008; Figure 3). The predicted PtFRE2 protein appears highly similar to the root epidermal ferric reductase required for iron uptake in Arabidopsis, AtFRO2 (Robinson et al., 1999; Bowler et al., 2008). Also highly upregulated is a putative ZIP family transporter that could serve to transport Fe(II) (Allen et al., 2008), as AtIRT1 does in Arabidopsis (Palmer and Guerinot, 2009). Finally, the presence of PtFBP, a gene orthologous to the bacterial ferrichrome binding protein FhuD, raises the possibility of iron–siderophore utilization, perhaps through the scavenging of cyanobacteria siderophores. FRE and FBP may thus play a role in the ability of P. tricornutum to utilize iron–siderophore complexes, both through reduction (Figure 3) and the apparent uptake of intact complexes (Soria-Dengg and Horstmann, 1995).
At the same time, iron limitation results in a decrease in transcripts associated with iron intensive processes like photosynthesis, mitochondrial electron transport, and nitrate assimilation (Allen et al., 2008). At the protein level, the ratio of PSII to PSI increases, cytochrome b6f and cytochrome c6 proteins decrease (Allen et al., 2008), and the activity of the iron-rich mitochondrial electron chain decreases (Kudo et al., 2000). The upregulation of transcript encoding the mitochondrial alternative oxidase (AOX) in response to iron limitation is proposed to mitigate the ROS presumably generated by iron-compromised electron transport chains (Allen et al., 2008). About 32% of genes regulated by iron in P. tricornutum have no ortholog in T. pseudonana, and iron starvation upregulates expression of several unique gene clusters in P. tricornutum (Allen et al., 2008). Of these genes, all but FRE2 are present in P. tricornutum but not T. pseudonana, and many are of unknown function. Indeed, the most highly expressed transcript under iron limitation, ISIP1, encodes a predicted protein of unknown function found in metatranscriptomic samples from iron limited waters (Marchetti et al., 2012), and with no ortholog in T. pseudonana (Allen et al., 2008). And like T. pseudonana, there is a subset of silicon starvation regulated genes that are also regulated by iron (38 out of 223 Si-sensitive genes), although there is no apparent overlap between the P. tricornutum and T. pseudonana subsets (Sapriel et al., 2009).
Pseudo-nitzschia spp.
Pseudo-nitzschia is a ubiquitous genus of pennate diatom, frequently dominating blooms in iron addition experiments (Hutchins and Bruland, 1998; de Baar, 2005; Trick et al., 2010). Among Pseudo-nitzschia species, the oceanic P. granii is more tolerant of iron limitation than the coastal species P. multiseries (Marchetti et al., 2009). P. granii is also more tolerant to iron starvation than T. oceanica, presumably because P. granii utilizes ferritin (Figure 3) to store iron during times of iron availability. Ferritin has not been detected in T. oceanica (Marchetti et al., 2009) and is absent from the T. pseudonana genome (Armbrust et al., 2004).
In addition to the utilization of ferritin, Pseudo-nitzschia is of interest because it is a eukaryotic marine phytoplankton which secretes the phytosiderophore-like compound domoic acid (DA), which is also a neurotoxin, and is causative of wildlife death and amnesic shellfish poisoning in humans. DA binds iron and copper (Rue and Bruland, 2001), and is structurally similar to the plant siderophore mugineic acid, which is secreted into soil by graminaceous plants. Because of its similarity to mugineic acid, it is proposed that DA could facilitate the extraction of iron from terrestrial sediments found in coastal waters (Rue and Bruland, 2001). The addition of exogenous DA to natural seawater samples increases iron uptake and growth of Pseudo-nitzschia (Maldonado et al., 2002; Wells et al., 2005), suggesting it is part of a Pseudo-nitzschia specific iron uptake system.
Domoic acid production has been observed to be induced by both metal availability and limitation: elevated copper (Rue and Bruland, 2001), copper limitation (Wells et al., 2005), iron limitation (Rue and Bruland, 2001), and iron fertilization (Silver et al., 2010; Trick et al., 2010). This suggests DA plays a role in both metal uptake during rapidly growing blooms and survival during limitation. DA binds iron with a low affinity, but the concentrations of DA in naturally occurring blooms are predicted to be sufficient to facilitate iron uptake (Rue and Bruland, 2001). The ability to monopolize iron availability via a species-specific phytosiderophore could thus explain the dominance of Pseudo-nitzschia in blooms.
Conclusion and Questions for Future Research
Other marine phytoplankton species have been sequenced (e.g., Emiliania huxleyi, F. cylindrus), and ever more will be. Presumably, transcriptome level analysis similar to those described above will be performed to determine which genes play a role in iron homeostasis. Additionally, metagenomic transcriptional analysis could be further applied to both classical bottle enrichment experiments (Marchetti et al., 2012), and large-scale iron fertilization experiments to elucidate the expression changes that underlie iron utilization during bloom formation.
Genome level studies have offered hints about the genes responsible for iron acquisition. This has allowed the leveraging of the extensive research done in organisms like Arabidopsis, Chlamydomonas, yeast, and humans, to identify and predict the function of marine orthologs. Nevertheless, the mechanisms of iron uptake utilized by eukaryotic marine phytoplankton ultimately remain unclear. Predicting which genes comprise the marine iron uptake systems using the well-characterized terrestrial models is complicated by the unique nature of the ocean environment, both in terms of iron–ligand chemistry (Volker and Wolf-Gladrow, 1999; Morel, 2008; Hassler et al., 2011), and in terms of the convoluted evolutionary path of organisms like diatoms (Moustafa et al., 2009).
A more daunting gap in our understanding of iron homeostasis is the abundance of genes of unknown function. These are often species-specific, and found to comprise large portions of the ever growing number of genomic and metagenomic data sets. This is further complicated by the inability to cultivate and genetically transform many of the new species that metagenomic surveys are uncovering (e.g., UCYN-A, with its apparent lack of PSII genes; Zehr et al., 2008). Additionally, the absence of canonical genes does not necessarily prove the absence of a pathway, as was recently demonstrated in freshwater cyanobacteria. Since the late 1960s, cyanobacteria were believed to lack a complete TCA cycle, as the 2-oxoglutarate dehydrogenase protein was undetected and the gene was missing from freshwater and marine cyanobacteria genomes. However, the recent functional characterization of candidate genes from Synechococcus sp. PCC 7002 identified two enzymes that perform the same role as 2-oxoglutarate dehydrogenase, completing the TCA cycle (Zhang and Bryant, 2011). It also appears that further divergence is possible, as these two genes are present in all cyanobacteria genomes except those of marine Synechococcus and Prochlorococcus.
Thus, functional characterization of putative uptake genes in model organisms is required to establish even the most basic mechanisms for iron transport in marine phytoplankton, while genetic screening (e.g., mutagenesis, expression of gene libraries derived from marine microorganisms in heterologous systems, etc.) could be utilized to identify novel iron homeostasis systems that have evolved in iron limited ocean environments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the reviewers for their helpful comments. Funding in the laboratory is supported by the Agence Nationale de la Recherche.
References
- Allen A., LaRoche J., Maheswari U., Lommer M., Schauer N., Lopez P., Finazzi G., Fernie A., Bowler C. (2008). Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443 10.1073/pnas.0711370105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson A., Beszteri B., Julius M., Theriot E. (2011). The model marine diatom Thalassiosira pseudonana likely descended from a freshwater ancestor in the genus Cyclotella. BMC Evol. Biol. 11, 125. 10.1186/1471-2148-11-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. A., Green D. H., Hart M. C., Küpper F. C., Sunda W. G., Carrano C. J. (2009). Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc. Natl. Acad. Sci. U.S.A. 106, 17071–17076 10.1073/pnas.0812469106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., Zhou S., Allen A. E., Apt K. E., Bechner M., Brzezinski M. A., Chaal B. K., Chiovitti A., Davis A. K., Demarest M. S., Detter J. C., Glavina T., Goodstein D., Hadi M. Z., Hellsten U., Hildebrand M., Jenkins B. D., Jurka J., Kapitonov V. V., Kröger N., Lau W. W. Y., Lane T. W., Larimer F. W., Lippmeier J. C., Lucas S., Medina M., Montsant A., Obornik M., Parker M. S., Palenik B., Pazour G. J., Richardson P. M., Rynearson T. A., Saito M. A., Schwartz D. C., Thamatrakoln K., Valentin K., Vardi A., Wilkerson F. P., Rokhsar D. S. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 10.1126/science.1101156 [DOI] [PubMed] [Google Scholar]
- Badarau A., Firbank S. J., Waldron K. J., Yanagisawa S., Robinson N. J., Banfield M. J., Dennison C. (2008). FutA2 is a ferric binding protein from Synechocystis PCC 6803. J. Biol. Chem. 283, 12520–12527 10.1074/jbc.M709907200 [DOI] [PubMed] [Google Scholar]
- Barber J., Nield J., Duncan J., Bibby T. S. (2006). “Accessory chlorophyll proteins in cyanobacterial photosystem I,” in Photosystem I, ed. Golbeck J. H. (Dordrecht: Springer; ), 99–117 [Google Scholar]
- Beiko R. G., Harlow T. J., Ragan M. A. (2005). Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 14332–14337 10.1073/pnas.0504068102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman-Frank I., Cullen J. T., Shaked Y., Sherrell R. M., Falkowski P. G. (2001). Iron availability, cellular iron quotas, and nitrogen fixation in Trichodesmium. Limnol. Oceanogr. 46, 1249–1260 10.4319/lo.2001.46.6.1249 [DOI] [Google Scholar]
- Bibby T., Nield J., Partensky F., Barber J. (2001a). Antenna ring around photosystem I. Nature 413, 590. 10.1038/35098153 [DOI] [PubMed] [Google Scholar]
- Bibby T. S., Nield J., Barber J. (2001b). Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743–745 10.1038/35089098 [DOI] [PubMed] [Google Scholar]
- Bibby T. S., Mary I., Nield J., Partensky F., Barber J. (2003). Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051–1054 10.1038/nature01933 [DOI] [PubMed] [Google Scholar]
- Bibby T. S., Zhang Y., Chen M. (2009). Biogeography of photosynthetic light-harvesting genes in marine phytoplankton. PLoS ONE 4, e4601. 10.1371/journal.pone.0004601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidle K. D., Bender S. J. (2007). Iron starvation and culture age activate metacaspases and programmed cell death in the marine diatom Thalassiosira pseudonana. Eukaryot. Cell 7, 223–236 10.1128/EC.00296-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. K. B., Wood T. J. (2009). Year-round observations of carbon biomass and flux variability in the Southern Ocean. Global Biogeochem. Cycles 23, GB2019. 10.1029/2008GB003206 [DOI] [Google Scholar]
- Boekema E. J., Hifney A., Yakushevska A. E., Piotrowski M., Keegstra W., Berry S., Michel K. P., Pistorius E. K., Kruip J. (2001). A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745–748 10.1038/35089104 [DOI] [PubMed] [Google Scholar]
- Bowler C., Allen A., Badger J., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 10.1038/nature07410 [DOI] [PubMed] [Google Scholar]
- Boyd P., Jickells T., Law C., Blain S., Boyle E., Buesseler K., Coale K., Cullen J., de Baar H., Follows M., Harvey M., Lancelot C., Levasseur M., Owens N., Pollard R., Rivkin R., Sarmiento J., Schoemann V., Smetacek V., Takeda S., Tsuda A., Turner S., Watson A. (2007). Mesoscale iron enrichment experiments 1993–2005: synthesis and future directions. Science 315, 612–617 10.1126/science.1131669 [DOI] [PubMed] [Google Scholar]
- Campbell L., Liu H., Nolla H. A., Vaulot D. (1997). Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at Station ALOHA during the 1991–1994 ENSO event. Deep Sea Res. Part I Oceanogr. Res. Pap. 44, 167–192 10.1016/S0967-0637(96)00102-1 [DOI] [Google Scholar]
- Capone D. G., Zehr J. P., Paerl H. W., Bergman B., Carpenter E. J. (1997). Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229 10.1126/science.276.5316.1221 [DOI] [Google Scholar]
- Cavender-Bares K. K., Mann E. L., Chisholm S. W., Ondrusek M. E., Bidigare R. R. (1999). Differential response of equatorial Pacific phytoplankton to iron fertilization. Limnol. Oceanogr. 44, 237–246 10.4319/lo.1999.44.2.0237 [DOI] [Google Scholar]
- Chappell P. D., Webb E. A. (2010). A molecular assessment of the iron stress response in the two phylogenetic clades of Trichodesmium. Environ. Microbiol. 12, 13–27 10.1111/j.1462-2920.2009.02026.x [DOI] [PubMed] [Google Scholar]
- Chisholm S. W. (1992). “Phytoplankton size,” in Primary Productivity and Biogeochemical Cycles in the Sea, eds Falkowski P. G., Woodhead A. D. (New York, NY: Plenum Press; ), 213–237 [Google Scholar]
- Chisholm S. W., Falkowski P. G., Cullen J. J. (2001). Dis-crediting ocean fertilization. Science 294, 309–310 10.1126/science.1065349 [DOI] [PubMed] [Google Scholar]
- Corellou F., Schwartz C., Motta J.-P., Djouani-Tahri E. B., Sanchez F., Bouget F.-Y. (2009). Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote Ostreococcus. Plant Cell 21, 3436–3449 10.1105/tpc.109.068825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi B., Perrone F., Bourgeois M., Beaumont C., Panzeri M. C., Cozzi A., Sangregorio R., Santambrogio P., Albertini A., Arosio P. (1998). Transient overexpression of human H-and L-ferritin chains in COS cells. Biochem. J. 330, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Baar H. J. W. (2005). Synthesis of iron fertilization experiments: from the iron age in the age of enlightenment. J. Geophys. Res. 110, C09S16. 10.1029/2004JC002601 [DOI] [Google Scholar]
- de Jonge M. D., Holzner C., Baines S. B., Twining B. S., Ignatyev K., Diaz J., Howard D. L., Legnini D., Miceli A., McNulty I., Jacobsen C. J., Vogt S. (2010). Quantitative 3D elemental microtomography of Cyclotella meneghiniana at 400-nm resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 15676–15680 10.1073/pnas.1001469107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37, e96. 10.1093/nar/gkn913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A., Ostrowski M., Scanlan D., Garczarek L., Mazard S., Palenik B., Paulsen I., de Marsac N., Wincker P., Dossat C., Ferriera S., Johnson J., Post A., Hess W., Partensky F. (2008). Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9, R90. 10.1186/gb-2008-9-5-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A., Salanoubat M., Partensky F., Artiguenave F., Axmann I. M., Barbe V., Duprat S., Galperin M. Y., Koonin E. V., Le Gall F., Makarova K. S., Ostrowski M., Oztas S., Robert C., Rogozin I. B., Scanlan D. J., de Marsac N. T., Weissenbach J., Wincker P., Wolf Y. I., Hess W. R. (2003). Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. U.S.A. 100, 10020–10025 10.1073/pnas.1733211100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham K. A., Bullerjahn G. S. (2002). Immunocytochemical localization of the stress-induced DpsA protein in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Basic Microb. 42, 367–372 [DOI] [PubMed] [Google Scholar]
- Eitinger T. (2004). In vivo production of active nickel superoxide dismutase from Prochlorococcus marinus MIT9313 is dependent on its cognate peptidase. J. Bacteriol. 186, 7821. 10.1128/JB.186.22.7821-7825.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood M. J., Hunter K. A. (2000). The incorporation of zinc and iron into the frustule of the marine diatom Thalassiosira pseudonana. Limnol. Oceanogr. 1517–1524 10.4319/lo.2000.45.7.1517 [DOI] [Google Scholar]
- Erdner D. L., Anderson D. M. (1999). Ferredoxin and flavodoxin as biochemical indicators of iron limitation during open-ocean iron enrichment. Limnol. Oceanogr. 44, 1609–1615 10.4319/lo.1999.44.7.1609 [DOI] [Google Scholar]
- Exss-Sonne P., Toelle J., Bader K., Pistorius E., Michel K. P. (2000). The IdiA protein of Synechococcus sp. PCC 7942 functions in protecting the acceptor side of photosystem II under oxidative stress. Photosynth. Res. 63, 145–157 10.1023/A:1006322925324 [DOI] [PubMed] [Google Scholar]
- Falkowski P. G. (1997). Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387, 272–275 10.1038/387272a0 [DOI] [Google Scholar]
- Falkowski P. G. (2006). Tracing oxygen’s imprint on Earth’s metabolic evolution. Science 311, 1724. 10.1126/science.1125937 [DOI] [PubMed] [Google Scholar]
- Fay P. (1992). Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56, 340–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J., Shi Y., Tyson G. W., Coleman M. L., Schuster S. C., Chisholm S. W., DeLong E. F. (2008). Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U.S.A. 105, 3805–3810 10.1073/pnas.0708897105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B., Gutteridge J. (1992). Biologically relevant metal ion-dependent hydroxyl radical generation, an update. FEBS Lett. 307, 108–112 10.1016/0014-5793(92)80911-Y [DOI] [PubMed] [Google Scholar]
- Hassler C. S., Schoemann V., Nichols C. M., Butler E. C. V., Boyd P. W. (2011). Saccharides enhance iron bioavailability to Southern Ocean phytoplankton. Proc. Natl. Acad. Sci. U.S.A. 108, 1076–1081 10.1073/pnas.1106729108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson B., Morel F. (2009). The role of siderophores in iron acquisition by photosynthetic marine microorganisms. Biometals 22, 659–669 10.1007/s10534-009-9235-2 [DOI] [PubMed] [Google Scholar]
- Hutchins D. A., Bruland K. W. (1998). Iron-limited diatom growth and Si:N uptake ratios in a coastal upwelling regime. Nature 393, 561–564 10.1038/31203 [DOI] [Google Scholar]
- Hutchins D. A., Witter A. E., Butler A., Luther G. W. (1999). Competition among marine phytoplankton for different chelated iron species. Nature 400, 858–861 10.1038/23680 [DOI] [Google Scholar]
- Ito Y., Alison B. (2005). Structure of synechobactins, new siderophores of the marine cyanobacterium Synechococcus sp. PCC 7002. Limnol. Oceanogr. 50, 1918–1923 10.4319/lo.2005.50.6.1918 [DOI] [Google Scholar]
- Jancek S., Gourbière S., Moreau H., Piganeau G. (2008). Clues about the genetic basis of adaptation emerge from comparing the proteomes of two Ostreococcus ecotypes (Chlorophyta, Prasinophyceae). Mol. Biol. Evol. 25, 2293–2300 10.1093/molbev/msn168 [DOI] [PubMed] [Google Scholar]
- Kasting J. F., Siefert J. L. (2002). Life and the evolution of Earth’s atmosphere. Science 296, 1066–1068 10.1126/science.1071184 [DOI] [PubMed] [Google Scholar]
- Katoh H., Hagino N., Ogawa T. (2001). Iron-binding activity of FutA1 subunit of an ABC-type iron transporter in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 42, 823. 10.1093/pcp/pce106 [DOI] [PubMed] [Google Scholar]
- Kranzler C., Lis H., Shaked Y., Keren N. (2011). The role of reduction in iron uptake processes in a unicellular, planktonic cyanobacterium. Environ. Microbiol. 13, 2990–2999 10.1111/j.1462-2920.2011.02572.x [DOI] [PubMed] [Google Scholar]
- Kudo I., Miyamoto M., Noiri Y., Maita Y. (2000). Combined effects of temperature and iron on the growth and physiology of the marine diatom Phaeodactylum tricornutum (Bacillariophyceae). J. Phycol. 36, 1096–1102 10.1046/j.1529-8817.2000.99042.x [DOI] [Google Scholar]
- Kuma K., Nakabayashi S., Suzuki Y., Kudo I., Matsunaga K. (1992). Photo-reduction of Fe(III) by dissolved organic substances and existence of Fe(II) in seawater during spring blooms. Mar. Chem. 37, 15–27 10.1016/0304-4203(92)90054-E [DOI] [Google Scholar]
- Küpper H., Šetlík I., Seibert S., Prášil O., Šetlikova E., Strittmatter M., Levitan O., Lohscheider J., Adamska I., Berman-Frank I. (2008). Iron limitation in the marine cyanobacterium Trichodesmium reveals new insights into regulation of photosynthesis and nitrogen fixation. New Phytol. 179, 784–798 10.1111/j.1469-8137.2008.02497.x [DOI] [PubMed] [Google Scholar]
- Kustka A., Allen A., Morel F. (2007). Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J. Phycol. 43, 715–729 10.1111/j.1529-8817.2007.00359.x [DOI] [Google Scholar]
- Kustka A., Sañudo-Wilhelmy S., Carpenter E. J., Capone D. G., Raven J. A. (2003a). A revised estimate of the iron use efficiency of nitrogen fixation, with special reference to the marine cyanobacterium Trichodesmium spp. (Cyanophyta). J. Phycol. 39, 12–25 [Google Scholar]
- Kustka A. B., Sanudo-Wilhelmy S. A., Carpenter E. J., Capone D., Burns J., Sunda W. G. (2003b). Iron requirements for dinitrogen-and ammonium-supported growth in cultures of Trichodesmium (IMS 101): comparison with nitrogen fixation rates and iron: carbon ratios of field populations. Limnol. Oceanogr. 48, 1869–1884 10.4319/lo.2003.48.5.1869 [DOI] [Google Scholar]
- Lavery T. J., Roudnew B., Gill P., Seymour J., Seuront L., Johnson G., Mitchell J. G., Smetacek V. (2010). Iron defecation by sperm whales stimulates carbon export in the Southern Ocean. Proc. R. Soc. Lond. B Biol. Sci. 277, 3527–3531 10.1098/rspb.2010.0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommer M., Roy A.-S., Schilhabel M., Schreiber S., Rosenstiel P., LaRoche J. (2010). Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics 11, 718. 10.1186/1471-2164-11-718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswari U., Jabbari K., Petit J.-L., Porcel B., Allen A., Cadoret J.-P., De Martino A., Heijde M., Kaas R., La Roche J., Lopez P., Martin-Jezequel V., Meichenin A., Mock T., Schnitzler Parker M., Vardi A., Armbrust E. V., Weissenbach J., Katinka M., Bowler C. (2010). Digital expression profiling of novel diatom transcripts provides insight into their biological functions. Genome Biol. 11, R85. 10.1186/gb-2010-11-8-r85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado M. T., Hughes M. P., Rue E. L., Wells M. L. (2002). The effect of Fe and Cu on growth and domoic acid production by Pseudo-nitzschia multiseries and Pseudo-nitzschia australis. Limnol. Oceanogr. 47, 515–526 10.4319/lo.2002.47.2.0515 [DOI] [Google Scholar]
- Maldonado M. T., Price N. M. (1996). Influence of N substrate on Fe requirements of marine centric diatoms. Mar. Ecol. Prog. Ser. 141, 161–172 10.3354/meps141161 [DOI] [Google Scholar]
- Maldonado M. T., Price N. M. (2001). Reduction and transport of organically bound iron by Thalassiosira oceanica (Bacillariophyceae). J. Phycol. 37, 298–310 10.1046/j.1529-8817.2001.037002298.x [DOI] [Google Scholar]
- Mann E. L., Chisholm S. W. (2000). Iron limits the cell division rate of Prochlorococcus in the eastern equatorial Pacific. Limnol. Oceanogr. 45, 1067–1076 10.4319/lo.2000.45.5.1067 [DOI] [Google Scholar]
- Marchetti A., Parker M. S., Moccia L. P., Lin E. O., Arrieta A. L., Ribalet F., Murphy M. E. P., Maldonado M. T., Armbrust E. V. (2009). Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457, 467–470 10.1038/nature07539 [DOI] [PubMed] [Google Scholar]
- Marchetti A., Schruth D. M., Durkin C. A., Parker M. S., Kodner R. B., Berthiaume C. T., Morales R., Allen A. E., Armbrust E. V. (2012). Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. U.S.A. 109, E317–E325 10.1073/pnas.1118408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny A. C., Coleman M. L., Chisholm S. W. (2006). Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U.S.A. 103, 12552–12557 10.1073/pnas.0601301103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny A. C., Kathuria S., Berube P. M. (2009). Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc. Natl. Acad. Sci. U.S.A. 106, 10787–10792 10.1073/pnas.0902532106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R., Pedrós-Alió C. (2008). Unveiling new microbial eukaryotes in the surface ocean. Curr. Opin. Microbiol. 11, 213–218 10.1016/j.mib.2008.04.004 [DOI] [PubMed] [Google Scholar]
- Mazzocchi M. G., González H. E., Vandromme P., Borrione I., DeAlcala M., Gauns M., Assmy P., Fuchs B., Klaas C., Martin P. (2009). A non-diatom plankton bloom controlled by copepod grazing and amphipod predation: preliminary results from the LOHAFEX iron-fertilisation experiment. GLOBEC International Newsletter 15, 3–6 [Google Scholar]
- Merchant S. S., Allen M. D., Kropat J., Moseley J. L., Long J. C., Tottey S., Terauchi A. M. (2006). Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763, 578–594 10.1016/j.bbamcr.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Michel K.-P., Berry S., Hifney A., Kruip J., Pistorius E. (2003). Adaptation to iron deficiency: a comparison between the cyanobacterium Synechococcus elongatus PCC 7942 wild-type and a DpsA-free mutant. Photosynth. Res. 75, 71–84 10.1023/A:1022459919040 [DOI] [PubMed] [Google Scholar]
- Michel K.-P., Exss-Sonne P., Scholten-Beck G., Kahmann U., Ruppel H. G., Pistorius E. K. (1998). Immunocytochemical localization of IdiA, a protein expressed under iron or manganese limitation in the mesophilic cyanobacterium Synechococcus PCC 6301 and the thermophilic cyanobacterium Synechococcus elongatus. Planta 205, 73–81 10.1007/s004250050298 [DOI] [PubMed] [Google Scholar]
- Michel K.-P., Thole H. H., Pistorius E. K. (1996). IdiA, a 34 kDa protein in the cyanobacteria Synechococcus sp. strains PCC 6301 and PCC 7942, is required for growth under iron and manganese limitations. Microbiology 142, 2635–2645 10.1099/00221287-142-9-2635 [DOI] [PubMed] [Google Scholar]
- Mock T., Samanta M. P., Iverson V., Berthiaume C., Robison M., Holtermann K., Durkin C., BonDurant S. S., Richmond K., Rodesch M., Kallas T., Huttlin E. L., Cerrina F., Sussman M. R., Armbrust E. V. (2008). Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc. Natl. Acad. Sci. U.S.A. 105, 1579–1584 10.1073/pnas.0707946105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier A., Liverani S., Bouvet R., Jesson B., Smith J., Mosser J., Corellou F., Bouget F.-Y. (2010). Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics 11, 192. 10.1186/1471-2164-11-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. M., Mills M. M., Achterberg E. P., Geider R. J., LaRoche J., Lucas M. I., McDonagh E. L., Pan X., Poulton A. J., Rijkenberg M. J. A., Suggett D. J., Ussher S. J., Woodward E. M. S. (2009). Large-scale distribution of Atlantic nitrogen fixation controlled by iron availability. Nat. Geosci. 2, 867–871 10.1038/ngeo667 [DOI] [Google Scholar]
- Moore J. K., Doney S. C., Glover D. M., Fung I. Y. (2001). Iron cycling and nutrient-limitation patterns in surface waters of the World Ocean. Deep Sea Res. Part I Oceanogr. Res. Pap. 49, 463–507 [Google Scholar]
- Morel F. M. M. (2008). The co evolution of phytoplankton and trace element cycles in the oceans. Geobiology 6, 318–324 10.1111/j.1472-4669.2008.00144.x [DOI] [PubMed] [Google Scholar]
- Moustafa A., Beszteri B., Maier U. G., Bowler C., Valentin K., Bhattacharya D. (2009). Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726 10.1126/science.1172983 [DOI] [PubMed] [Google Scholar]
- O’Neill J. S., van Ooijen G., Dixon L. E., Troein C., Corellou F., Bouget F.-Y., Reddy A. B., Millar A. J. (2011). Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558 10.1038/nature09654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orino K., Lehman L., Tsuji Y., Ayaki H., Torti S. V., Torti F. M. (2001). Ferritin and the response to oxidative stress. Biochem. J. 357, 241. 10.1042/0264-6021:3570241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B., Brahamsha B., Larimer F. W., Land M., Hauser L., Chain P., Lamerdin J., Regala W., Allen E. E., McCarren J., Paulsen I., Dufresne A., Partensky F., Webb E. A., Waterbury J. (2003). The genome of a motile marine Synechococcus. Nature 424, 1037–1042 10.1038/nature01943 [DOI] [PubMed] [Google Scholar]
- Palenik B., Grimwood J., Aerts A., Rouze P., Salamov A., Putnam N., Dupont C., Jorgensen R., Derelle E., Rombauts S. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 104, 7705–7710 10.1073/pnas.0611046104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B., Ren Q., Dupont C. L., Myers G. S., Heidelberg J. F., Badger J. H., Madupu R., Nelson W. C., Brinkac L. M., Dodson R. J., Durkin A. S., Daugherty S. C., Sullivan S. A., Khouri H., Mohamoud Y., Halpin R., Paulsen I. T. (2006). Genome sequence of Synechococcus CC9311: insights into adaptation to a coastal environment. Proc. Natl. Acad. Sci. U.S.A. 103, 13555–13559 10.1073/pnas.0602963103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C. M., Guerinot M. L. (2009). Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340 10.1038/nchembio.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-I., Sandström S., Gustafsson P., Öquist G. (1999). Expression of the isiA gene is essential for the survival of the cyanobacterium Synechococcus sp. PCC 7942 by protecting photosystem II from excess light under iron limitation. Mol. Microbiol. 32, 123–129 10.1046/j.1365-2958.1999.01332.x [DOI] [PubMed] [Google Scholar]
- Partensky F., Blanchot J., Vaulot D. (1999a). Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: a review. Bull. Inst. Oceanogr. Monaco Numero Spec. 19, 431–449 [Google Scholar]
- Partensky F., Hess W., Vaulot D. (1999b). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz Y., Katz A., Pick U. (2007). A multicopper ferroxidase involved in iron binding to transferrins in Dunaliella salina plasma membranes. J. Biol. Chem. 282, 8658–8666 10.1074/jbc.M606923200 [DOI] [PubMed] [Google Scholar]
- Peers G., Price N. (2006). Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature 441, 341–344 10.1038/nature04630 [DOI] [PubMed] [Google Scholar]
- Peña M. M. O., Bullerjahn G. S. (1995). The DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. J. Biol. Chem. 270, 22478–22482 10.1074/jbc.270.38.22478 [DOI] [PubMed] [Google Scholar]
- Peña M. M. O., Burkhart W., Bullerjahn G. S. (1995). Purification and characterization of a Synechococcus sp. strain PCC 7942 polypeptide structurally similar to the stress-induced Dps/PexB protein of Escherichia coli. Arch. Microbiol. 163, 337–344 10.1007/BF00404206 [DOI] [PubMed] [Google Scholar]
- Pollard R. T., Salter I., Sanders R. J., Lucas M. I., Moore C. M., Mills R. A., Statham P. J., Allen J. T., Baker A. R., Bakker D. C. E., Charette M. A., Fielding S., Fones G. R., French M., Hickman A. E., Holland R. J., Hughes J. A., Jickells T. D., Lampitt R. S., Morris P. J., Nedelec F. H., Nielsdottir M., Planquette H., Popova E. E., Poulton A. J., Read J. F., Seeyave S., Smith T., Stinchcombe M., Taylor S., Thomalla S., Venables H. J., Williamson R., Zubkov M. V. (2009). Southern Ocean deep-water carbon export enhanced by natural iron fertilization. Nature 457, 577–580 10.1038/nature07716 [DOI] [PubMed] [Google Scholar]
- Poulsen N., Kröger N. (2005). A new molecular tool for transgenic diatoms. FEBS J. 272, 3413–3423 10.1111/j.1742-4658.2005.04760.x [DOI] [PubMed] [Google Scholar]
- Ravet K., Touraine B., Boucherez J., Briat J.-F., Gaymard F., Cellier F. (2009). Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57, 400–412 10.1111/j.1365-313X.2008.03698.x [DOI] [PubMed] [Google Scholar]
- Ren Q., Chen K., Paulsen I. T. (2006). TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35, D274. 10.1093/nar/gkl925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers A. R., Jakuba R. W., Webb E. A. (2009). Iron stress genes in marine Synechococcus and the development of a flow cytometric iron stress assay. Environ. Microbiol. 11, 382–396 10.1111/j.1462-2920.2008.01778.x [DOI] [PubMed] [Google Scholar]
- Robinson N. J., Procter C. M., Connolly E. L., Guerinot M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697 10.1038/17800 [DOI] [PubMed] [Google Scholar]
- Rocap G. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 424, 1042–1047 10.1038/nature01947 [DOI] [PubMed] [Google Scholar]
- Rocap G., Distel D. L., Waterbury J. B., Chisholm S. W. (2002). Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S rDNA internal transcribed spacer (ITS) sequences. Appl. Environ. Microbiol. 68, 1180–1191 10.1128/AEM.68.3.1180-1191.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G., Larimer F. W., Lamerdin J., Malfatti S., Chain P., Ahlgren N. A., Arellano A., Coleman M., Hauser L., Hess W. R., Johnson Z. I., Land M., Lindell D., Post A. F., Regala W., Shah M., Shaw S. L., Steglich C., Sullivan M. B., Ting C. S., Tolonen A., Webb E. A., Zinser E. R., Chisholm S. W. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047 10.1038/nature01947 [DOI] [PubMed] [Google Scholar]
- Rubin M., Berman-Frank I., Shaked Y. (2011). Dust- and mineral-iron utilization by the marine dinitrogen-fixer Trichodesmium. Nat. Geosci. 4, 529–534 10.1038/ngeo1181 [DOI] [Google Scholar]
- Rubio L. M., Ludden P. W. (2008). Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 62, 93–111 10.1146/annurev.micro.62.081307.162737 [DOI] [PubMed] [Google Scholar]
- Rue E., Bruland K. (2001). Domoic acid binds iron and copper: a possible role for the toxin produced by the marine diatom Pseudo-nitzschia. Mar. Chem. 76, 127–134 10.1016/S0304-4203(01)00053-6 [DOI] [Google Scholar]
- Rusch D. B., Martiny A. C., Dupont C. L., Halpern A. L., Venter J. C. (2010). Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc. Natl. Acad. Sci. U.S.A. 107, 16184–16189 10.1073/pnas.1009513107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryther J. H., Kramer D. D. (1961). Relative iron requirement of some coastal and offshore plankton algae. Ecology 42, 444–446 10.2307/1932105 [DOI] [Google Scholar]
- Saito M. A., Bertrand E. M., Dutkiewicz S., Bulygin V. V., Moran D. M., Monteiro F. M., Follows M. J., Valois F. W., Waterbury J. B. (2011). Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci. U.S.A. 108, 2184–2189 10.1073/pnas.1108376108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapriel G., Quinet M., Heijde M., Jourdren L., Tanty V., Luo G., Le Crom S., Lopez P. J. (2009). Genome-wide transcriptome analyses of silicon metabolism in Phaeodactylum tricornutum reveal the multilevel regulation of silicic acid transporters. PLoS ONE 4, e7458. 10.1371/journal.pone.0007458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A., Dwivedi K., Rice K. A., Bullerjahn G. S. (2000). Growth phase and metal-dependent regulation of the dpsA gene in Synechococcus sp. strain sp. strain PCC 7942. Arch. Microbiol. 173, 352–357 10.1007/s002030000153 [DOI] [PubMed] [Google Scholar]
- Shaked Y., Kustka A. B., Morel F. M. M. (2005). A general kinetic model for iron acquisition by eukaryotic phytoplankton. Limnol. Oceanogr. 50, 872–882 10.4319/lo.2005.50.3.0872 [DOI] [Google Scholar]
- Sherman D., Sherman L. (1983). Effect of iron deficiency and iron restoration on ultrastructure of Anacystis nidulans. J. Bacteriol. 156, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D., Xu Y., Hopkinson B. M., Morel F. M. M. (2010). Effect of ocean acidification on iron availability to marine phytoplankton. Science 327, 676–679 10.1126/science.1183517 [DOI] [PubMed] [Google Scholar]
- Shi T., Sun Y., Falkowski P. G. (2007). Effects of iron limitation on the expression of metabolic genes in the marine cyanobacterium Trichodesmium erythraeum IMS101. Environ. Microbiol. 9, 2945–2956 10.1111/j.1462-2920.2007.01406.x [DOI] [PubMed] [Google Scholar]
- Shi Y., Tyson G. W., DeLong E. F. (2009). Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature 459, 266–272 10.1038/nature08055 [DOI] [PubMed] [Google Scholar]
- Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., Manfredonia A., Falciatore A., Bowler C. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406, 23–35 10.1016/j.gene.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Silver M. W., Bargu S., Coale S. L., Benitez-Nelson C. R., Garcia A. C., Roberts K. J., Sekula-Wood E., Bruland K. W., Coale K. H. (2010). Toxic diatoms and domoic acid in natural and iron enriched waters of the oceanic Pacific. Proc. Natl. Acad. Sci. U.S.A. 107, 20762–20767 10.1073/pnas.1006968107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria-Dengg S., Horstmann U. (1995). Ferrioxamines B and E as iron sources for the marine diatom Phaeodactylum tricornutum. Mar. Ecol. Prog. Ser. 127, 269–277 10.3354/meps127269 [DOI] [Google Scholar]
- Stockel J., Welsh E. A., Liberton M., Kunnvakkam R., Aurora R., Pakrasi H. B. (2008). Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic processes. Proc. Natl. Acad. Sci. U.S.A. 105, 6156–6161 10.1073/pnas.0711068105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzepek R., Harrison P. (2004). Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692 10.1038/nature02954 [DOI] [PubMed] [Google Scholar]
- Sunda W., Huntsman S. A. (1995). Iron uptake and growth limitation in oceanic and coastal phytoplankton. Mar. Chem. 50, 189–206 10.1016/0304-4203(95)00035-P [DOI] [Google Scholar]
- Sunda W. G., Swift D. G., Huntsman S. A. (1991). Low iron requirement for growth in oceanic phytoplankton. Nature 351, 55–57 10.1038/351055a0 [DOI] [Google Scholar]
- Sutak R., Šlapeta J., San Roman M., Camadro J.-M., Lesuisse E. (2010). Nonreductive iron uptake mechanism in the marine alveolate Chromera velia. Plant Physiol. 154, 991–1000 10.1104/pp.110.159947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamatrakoln K., Korenovska O., Niheu A. K., Bidle K. D. (2011). Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ. Microbiol. 14, 67–81 10.1111/j.1462-2920.2011.02468.x [DOI] [PubMed] [Google Scholar]
- Thompson A. W., Huang K., Saito M. A., Chisholm S. W. (2011). Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability. ISME J. 5, 1580–1594 10.1038/ismej.2011.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tölle J., Michel K.-P., Kruip J., Kahmann U., Preisfeld A., Pistorius E. K. (2002). Localization and function of the IdiA homologue Slr1295 in the cyanobacterium Synechocystis sp. strain PCC 6803. Microbiology 148, 3293–3305 [DOI] [PubMed] [Google Scholar]
- Tréguer P., Nelson D. M., Van Bennekom A. J., DeMaster D. J., Leynaert A., Quéguiner B. (1995). The silica balance in the world ocean: a reestimate. Science 268, 375. 10.1126/science.268.5209.375 [DOI] [PubMed] [Google Scholar]
- Trick C. G., Bill B. D., Cochlan W. P., Wells M. L., Trainer V. L., Pickell L. D. (2010). Iron enrichment stimulates toxic diatom production in high-nitrate, low-chlorophyll areas. Proc. Natl. Acad. Sci. U.S.A. 107, 5887–5892 10.1073/pnas.0910579107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp H. J., Bench S. R., Turk K. A., Foster R. A., Desany B. A., Niazi F., Affourtit J. P., Zehr J. P. (2010). Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 464, 90–94 10.1038/nature08786 [DOI] [PubMed] [Google Scholar]
- Tuit C., Waterbury J., Ravizza G. (2004). Diel variation of molybdenum and iron in marine diazotrophic cyanobacteria. Limnol. Oceanogr. 49, 978–990 10.4319/lo.2004.49.4.0978 [DOI] [Google Scholar]
- Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D., Paulsen I., Nelson K. E., Nelson W., Fouts D. E., Levy S., Knap A. H., Lomas M. W., Nealson K., White O., Peterson J., Hoffman J., Parsons R., Baden-Tillson H., Pfannkoch C., Rogers Y.-H., Smith H. O. (2004). Environmental genome shotgun sequencing of the Sargasso sea. Science 304, 66–74 10.1126/science.1093857 [DOI] [PubMed] [Google Scholar]
- Volker C., Wolf-Gladrow D. A. (1999). Physical limits on iron uptake mediated by siderophores or surface reductases. Mar. Chem. 65, 227–244 10.1016/S0304-4203(99)00004-3 [DOI] [Google Scholar]
- Webb E. A., Moffett J. W., Waterbury J. B. (2001). Iron stress in open-ocean cyanobacteria (Synechococcus, Trichodesmium, and Crocosphaera spp.): identification of the IdiA protein. Appl. Environ. Microbiol. 67, 5444–5452 10.1128/AEM.67.12.5444-5452.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb R., Troyan T., Sherman D., Sherman L. A. (1994). MapA, an iron-regulated, cytoplasmic membrane protein in the cyanobacterium Synechococcus sp. strain PCC7942. J. Bacteriol. 176, 4906–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells M. L., Trick C. G., Cochlan W. P., Hughes M. P., Trainer V. L. (2005). Domoic acid: the synergy of iron, copper, and the toxicity of diatoms. Limnol. Oceanogr. 50, 1908–1917 10.4319/lo.2005.50.6.1908 [DOI] [Google Scholar]
- Wilhelm S. W., Trick C. G. (1994). Iron-limited growth of cyanobacteria: multiple siderophore production is a common response. Limnol. Oceanogr. 39, 1979–1984 10.4319/lo.1994.39.8.1979 [DOI] [Google Scholar]
- Worden A. Z., Nolan J. K., Palenik B. (2004). Assessing the dynamics and ecology of marine picophytoplankton: the importance of the eukaryotic component. Limnol. Oceanogr. 49, 168–179 10.4319/lo.2004.49.1.0168 [DOI] [Google Scholar]
- Zehr J. P., Bench S. R., Carter B. J., Hewson I., Niazi F., Shi T., Tripp H. J., Affourtit J. P. (2008). Globally distributed uncultivated oceanic N2-fixing cyanobacteria lack oxygenic photosystem II. Science 322, 1110–1112 10.1126/science.1165340 [DOI] [PubMed] [Google Scholar]
- Zhang S., Bryant D. A. (2011). The tricarboxylic acid cycle in cyanobacteria. Science 334, 1551–1553 10.1126/science.1210369 [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O., Doolittle W. F., Papke R. T., Gogarten J. P. (2009). Intertwined evolutionary histories of marine Synechococcus and Prochlorococcus marinus. Genome Biol. Evol. 1, 325–339 10.1093/gbe/evp032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwirglmaier K., Jardillier L., Ostrowski M., Mazard S., Garczarek L., Vaulot D. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161 [DOI] [PubMed] [Google Scholar]