Abstract
Although bacteria are considered the simplest life forms, we are now slowly unraveling their cellular complexity. Surprisingly, not only do bacterial cells have a cytoskeleton but also the building blocks are not very different from the cytoskeleton that our own cells use to grow and divide. Nonetheless, despite important advances in our understanding of the basic physiology of certain bacterial models, little is known about Actinobacteria, an ancient group of Eubacteria. Here we review current knowledge on the cytoskeletal elements required for bacterial cell growth and cell division, focusing on actinobacterial genera such as Mycobacterium, Corynebacterium, and Streptomyces. These include some of the deadliest pathogens on earth but also some of the most prolific producers of antibiotics and antitumorals.
1. Introduction
All cells require cytoskeletal proteins for cell division and growth [1]. These structural components are essential for the maintenance of cell shape as well as for other dynamic processes critical for the cell, such as chromosomal segregation, the equal partitioning of cytosolic material, cell polarization, and motility [2].
The ubiquity of the cytoskeletal proteins reflects their early evolutionary acquisition and bacterial origin [3]. In fact, it is difficult to imagine an adaptable free-living cell without a versatile internal cytoskeleton. However, this notion is very recent since only just a decade ago it was thought that bacteria lacked a cytoskeleton. Instead, the required cell membrane support was assumed to be provided by the bacterial cell wall, which was thus considered to function as an “exoskeleton,” forming a physical barrier that contained the hydrostatic internal cell pressure and prevented the rupture of the cell membrane [4].
In fact, this exoskeleton does determine the characteristic shape of a bacterial cell, since in the absence of cell wall rod-shaped bacteria lose their morphology and become perfect spheres. But given that the chemical composition of the bacterial cell wall is essentially the same in the vast majority of Eubacteria (it is basically made of peptidoglycan or murein), it was also recognized that other factors must drive the determination of bacterial cell shape [5]. Osmotic pressure was thought to have some role in this process albeit a limited one, in view of the high morphological variability observed in different wild-type bacterial species and in strains carrying mutations in the different genes involved in cell morphology determination (morphogenes) [5]. In addition, and despite their apparent simplicity, bacterial cells undergo rapid and precise division, including chromosomal segregation and equal partitioning of the cytosolic contents, which would be impossible without the existence of an extremely dynamic but highly accurate internal organization [2, 5].
However, it is only now becoming clear that bacteria have an internal cytoskeleton made up of homologues to eukaryotic tubulin (FtsZ), actin (MreB/Mbl), and intermediate filaments (crescentin) [6]; there are even bacteria-specific cytoskeletal families of proteins, such as MinD [7] and bactofilins [8], without homologues in eukaryotes. All of these bacterial cytoskeletal proteins have pivotal roles on cell-wall synthesis and, consequently, cell-shape determination [7]. Here we review the most recent data regarding the bacterial cytoskeleton, with special emphasis on cell-shape determination by Actinobacteria, one of the three major bacterial groups.
Actinobacteria are gram positive with high GC-content genomes [9]. This ancient group of bacteria is one of the most interesting in terms of industrial and medical applications, but at the same time it is clearly understudied. Many actinobacterial species are industrially important for the bioconversion and production of antibiotics, antitumorals, amino acids, and vitamins [9]. There are also medically important species in this group, such as Mycobacterium tuberculosis, Mycobacterium leprae, and Corynebacterium diphtheriae, the causative agents of tuberculosis, leprosy, and diphtheria, respectively [9]. Yet, despite the enormous relevance for humans, little is known about the basic physiology of these and other actinobacterial species.
Progress in this field has been partially hampered by the lack of molecular tools, but mainly because of its complexity. The cell morphology of Actinobacteria varies enormously, from the almost coccoid cellular shape of Rhodococcus spp. to the fungal-like hyphae of Streptomyces spp. [10]. Furthermore, some of these bacteria have an outer lipidic layer composed of mycolic acids; this layer is essential for morphogenesis and for resistance to antimicrobials and to different stress conditions [11, 12]. In addition, some of these species sporulate, which requires two distinct molecular programs for cell-shape determination [13, 14]. Finally, recent data have demonstrated that Actinobacteria possess genus-specific morphogenes [15] that clearly differentiates them from each other but also further complicates the study of cytokinesis in this bacterial group.
2. Cytoskeletal Proteins Involved in Cell Division
Cell division in bacteria is governed by the tubulin-like protein FtsZ, a GTPase widely conserved and located within a cluster of genes involved in division and cell-wall synthesis (cluster dcw) [16]. During division, the cell membrane is constricted at the midcell, and peptidoglycan is synthesized to create a new cell wall in between the two newly formed daughter cells [17, 18]. This process starts with the polymerization of FtsZ and the assembly of the so-called Z-ring, a scaffold of cell division proteins that also generates the force needed for cell constriction [19]. No other protein is able to locate at the midcell during cell division before FtsZ; therefore, Z-ring assembly is the first step in bacterial cell division [20]. This structure recruits other proteins with different roles in cell division and cell-wall synthesis, creating a macromolecular complex called the divisome [20]. The composition of the divisome varies between different species, but a consensus has been established mainly based on Escherichia coli as the model organism.
There are positive and negative spatiotemporal regulators of FtsZ assembly that function to establish the exact location of cell division at the midcell. This results in two symmetrical daughter cells with an equal distribution of DNA and cytosolic material [17, 18]. The best studied FtsZ inhibitors are the MinCD and nucleoid occlusion systems. The MinCD system inhibits FtsZ polymerization at the cell poles to prevent asymmetrical division events [21, 22]. The nucleoid occlusion system, mediated by Noc/SlmA, is basically an inhibitor of FtsZ polymerization at those cellular locations where chromosomal DNA is present. This prohibits the unequal distribution of genetic material during cell division [22]. The combination of the two systems leaves the midcell after cell elongation and DNA replication as the only place available for FtsZ polymerization [17, 18].
Once the timing and location of cell division have been established, FtsZ polymerizes to generate the basic scaffold of the divisome, the Z-ring. The first proteins to be recruited to the Z-ring are FtsA and ZipA, which comprise the core of the divisome in E. coli. FtsA and ZipA simultaneously bind to FtsZ and the cell membrane, thus stabilizing the Z-ring [23, 24]. Once the two proteins are located at the midcell, the remaining proteins of the divisome are sequentially recruited [18, 20]: (1) the FtsEX complex, which may facilitate constriction [25]; (2) FtsK, which is required for chromosome segregation [26]; (3) FtsQLB, a bridge protein complex between the core of the divisome and the proteins involved in peptidoglycan synthesis [27, 28], such as (4) FtsW and FtsI, a peptidoglycan precursor translocator and a penicillin-binding protein (PBP), respectively [29, 30]; finally (5) FtsN and the amidases AmiA, AmiB, and AmiC, which mediate the peptidoglycan hydrolysis required for the final separation of the two newly formed daughter cells [31–34].
Several of the divisome proteins may have partially overlapping functions [35, 36], perhaps explaining why divisome composition is relatively variable in bacteria. In general, the positive (e.g., ZapA/B/C, ZipA, or SpoIIE) and negative (e.g., Noc/SlmA, MipZ, MciZ, SulA, EzrA, or MinCDE) spatiotemporal regulators of the Z-ring assembly are poorly conserved [16, 37–41]. In fact, actinobacterial genomes do not have homologues of these regulators [12, 42], and nucleoid occlusion has not been detected; that is, FtsZ polymerization may start over nonsegregated chromosomes [43, 44]. An exception to this rule is SepF, a conserved positive regulator of FtsZ assembly [45, 46]. However, SepF was dispensable for either the growth or the cell viability of Corynebacteria [47].
Conversely, actinobacterial-specific positive and negative regulators have been recently identified. The first Z-ring positive regulator in Actinobacteria was described using M. tuberculosis as a model [48]. FtsW acts as a translocator of peptidoglycan precursors through the cell membrane during cell division [29]. This protein was also found to be a direct interaction partner of FtsZ in M. tuberculosis, suggesting that FtsW anchors the Z-ring to the cell membrane. Consequently, FtsW could be involved in the positive regulation or stabilization of the Z-ring in Actinobacteria, which lack homologues of FtsA and ZipA [48–50]. In fact, a similar role has been proposed for FtsW in Streptomyces coelicolor but only during sporulation septation [51]. Also, the Streptomyces-specific SsgA and SsgB proteins have been described as positive regulators of FtsZ assembly during spore formation [52, 53]. In general, the sporulation mechanism of S. coelicolor is better studied than its vegetative cell division process. Finally, there is evidence that the FtsZ-interacting protein A (FipA) from M. tuberculosis is a positive effector of cell division under oxidative stress conditions [54].
On the other side of the coin, there are also actinobacterial-specific inhibitors of FtsZ assembly. DivS is a cell-division suppressor that acts in response to DNA damage in Corynebacterium glutamicum [55]. PldP is a ParA-like protein that may be involved in the cell-division site selection of C. glutamicum [56]; a PldP null mutation generates minicells, a phenotype caused by the formation of septa at the cell poles and the generation of asymmetrical daughter cells with an unequal distribution of chromosomal DNA. ClpX directly interacts with FtsZ in M. tuberculosis and blocks its polymerization in response to various stress conditions, such as intramacrophage growth and antibiotic treatment [57]. Finally, the product of crgA, a small gene widely conserved in Actinobacteria, has been described as an inhibitor of Streptomyces cell division [58]. However, CrgA has been recently characterized as a facilitator of FtsI localization in M. tuberculosis [59], proving once again the complexity and variability of actinobacterial cell division.
3. Cytoskeletal Proteins Involved in Cell Elongation
In many bacillary bacteria, MreB actin-like homologues are essential for cell-wall elongation [60, 61]. The mreB gene is usually localized in the mre operon, together with mreC and mreD [62, 63]. The mreBCD cluster was identified based on the coccoid cell shape resulting from the mutation of these genes [62, 63]. MreB is an ATPase capable of polymerizing into long filaments in the presence of ATP or GTP [64–66]. During the last decade, it has been assumed that MreB forms helicoidal protofilaments that extend from pole to pole in the cell directing the synthesis of the lateral cell wall and cell elongation in many rod-shaped bacteria [61, 67, 68]. However, recent evidence suggests that MreB localizes in discrete patches that move along the cell in the company of proteins involved in peptidoglycan synthesis and translocation of cell wall precursors: Pbps and RodA, respectively [69, 70]. MreCD and also RodZ, a conserved membrane protein, are thought to act as a link between MreB and the peptidoglycan synthesis machinery [69–73]. In this new model, old peptidoglycan strands act as scaffolds of new cell wall synthesis, and the movement of the molecular machines involved in this process is powered by peptidoglycan polymerization [69, 70, 74]. MreB filaments could be required for controlling the orientation and movement of these molecular complexes and/or the recruitment of peptidoglycan precursors for their translocation across the membrane [70].
Nonetheless, MreB is essential for maintaining the cell wall synthesis and cell elongation in Bacillus subtilis, E. coli or Caulobacter crescentus [61]. In all these bacterial models the incorporation of new cell wall material occurs at the midcell during cell division (sustained by FtsZ) and at the lateral walls during cell elongation in an MreB-dependent fashion, while the polar ends of the cell are inert [68, 75, 76]. In microorganisms with a coccoidal shape and devoid of mreBCD genes like Streptococcus pneumoniae, cell wall synthesis during either cell division or cell elongation is only accomplished at the division site [68].
In contrast, all Actinobacteria studied thus far grow apically, that is, by the insertion of new peptidoglycan at the cell poles rather than at the lateral wall, which is inert [77–79]. This growth is independent of MreB; in fact, all mycobacterial and corynebacterial genomes sequenced to date lack mreB homologues, whereas Streptomyces use MreB homologues only for sporulation [80–82]. This form of cell elongation is sustained by the protein DivIVA, which localizes at the cell poles and is essential for cell viability [77–79, 83]. DivIVA polymerizes at the cell poles to generate an internal cytoskeleton that supports and recruits the cell-wall synthesis machinery in Actinobacteria. DivIVA also localizes at the division site, suggesting its role in the maturation of the newly formed cell poles [78].
In Actinobacteria, a change in DivIVA protein levels leads to strong morphological alterations. Specifically, the low-level expression of DivIVA produces coccoidal cells in the rod-shaped Corynebacterium and Mycobacterium; this is because the lack of DivIVA abolishes the polar synthesis of peptidoglycan [78, 79]. Presumably, cell-wall synthesis occurs uniformly along the cell, creating perfect spheres; alternatively, the cell-division apparatus is able to create a cell wall that is sufficient to allow cell enlargement [78, 79]. This latter hypothesis is more plausible since the only place where peptidoglycan synthesis has been clearly detected in DivIVA-depleted cells is the division septum. Interestingly, DivIVA is essential in Actinobacteria; that is, a knockout mutant is lethal, suggesting that this protein has another, yet unexplored role apart from the internal support of polar cell-wall synthesis [77, 78].
The overexpression of DivIVA creates large asymmetrical cells that are enlarged at one polar end, where the majority of the protein localizes [77–79, 83, 84]. DivIVA was shown to oligomerize through two coiled-coil regions [85, 86] whereas the highly conserved N-terminal domain is probably required for the protein's initial localization to the membrane at the polar end of the cell [87]. In Actinobacteria, all three domains are essential for DivIVA function; however, once DivIVA is localized at the cell's new polar end, self-interaction is probably the main force required for the protein's localization [88–90]. Overexpression of DivIVA results in greater amounts of the protein at the cell poles, which in turn become very active sites of peptidoglycan synthesis [78]. Presumably, after cell division the old polar end will have initially a larger amount of DivIVA than the recently created polar end. Therefore, the old polar end will attract a larger amount of DivIVA, which eventually will lead to an asymmetrical club-shaped cell. The balance of DivIVA levels radically changes not only the length of the cell but also its diameter [77–79, 83].
All three DivIVA domains can be exchanged without drastic consequences [88]. In fact, the only conserved region of this protein is the N-terminal domain whereas the coiled-coil regions differ greatly in size and sequence even between highly related species [91]. However, DivIVA proteins from B. subtillis or S. pneumoniae are not able to complement the lack of DivIVA in C. glutamicum, in contrast to DivIVA proteins from other Actinobacteria [78]. This suggests that actinobacterial DivIVA proteins have an unknown signature or motif in their sequences that enables their role in polar cell-wall synthesis. This is probably mediated by protein-protein interactions with class B high-molecular-weight PBPs, directly involved in cell-wall synthesis, and RodA, an essential membrane protein probably involved in the transport of peptidoglycan precursors outside the cell during cell growth [92]. RodA is in fact required for rod cell shape determination in C. glutamicum (Figure 1).
Figure 1.
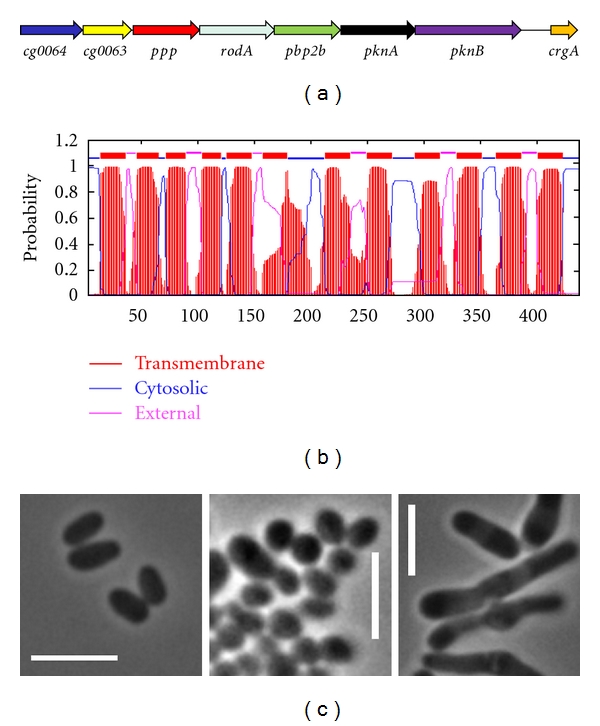
RodA is required for rod-shape maintenance of C. glutamicum. (a) In Actinobacteria, rodA is located within the conserved pkn cluster. (b) Prediction of transmembrane helices of RodA using TMHMM 2.0 software [93]. (c) Partial depletion of RodA generates coccoid cells (central) in the rod-shaped C. glutamicum (left), whereas a RodA overexpression results in club-shaped cells (right). M. Fiuza, unpublished results.
4. Other Cytoskeletal Proteins
In bacteria, tubulin-like proteins are required for cell division, whereas actin-like proteins maintain cell elongation, with the notable exception of Actinobacteria, in which, as discussed above, DivIVA directs polar cell-wall extension. Coiled-coil rich proteins such as DivIVA are common elements of the cytoskeleton of all organisms, no doubt due to their ability to oligomerize through self-interaction [94, 95]. In eukaryotes, intermediate filaments (IF) are the best example of cytoskeletal coiled-coil proteins, and in bacteria, IF-like proteins were recently identified.
The first bacterial IF-like protein was crescentin [96], which was described in C. crescentus. In this bacillary bacterium, mutations in the gene encoding crescentin, creS, straighten the curved shape of the cells. Crescentin participates in the formation of helicoidal and filamentous structures along the cell. The protein redirects cell-wall synthesis controlled by MreB to ensure a curved bacillus instead of a straight rod [96].
The main characteristics of IF-like proteins are their lack of enzymatic activity and their capacity for in vitro polymerization in the absence of any cofactors [97]. Coiled-coil self-interaction is highly resistant to mechanical stress [94, 95], making IF-like proteins perfect candidates for cytoskeletal proteins. However, the sequence homology between IFs and crescentin is very low, although all these proteins have in common a high content of coiled-coil regions [96]. Since many different amino acid sequences are able to adopt the coiled-coil structure, sequence conservation is not essential, which makes it very difficult to identify IF-like proteins in bacteria strictly by homology. Despite the low level of sequence conservation, coiled-coil regions consist of a repeated pattern of seven amino acids in which the first and fourth positions are always hydrophobic. This pattern allowed the identification of other bacterial IF-like proteins besides crescentin by in silico mining of coiled-coil regions [15, 98]. It is now becoming clear that IF-like proteins are amply distributed in bacteria; however, a cytoskeletal role has been attributed only to a few of them: CfpA in spirochaetes [99], CcrP in Helicobacter pylori [100], FilP in S. coelicolor [98], and RsmP in C. glutamicum [15].
CfpA is found exclusively in spirochaetes, where the protein forms helicoidal filamentous structures along the cell that are required for cell division and chromosomal segregation [99]. In H. pylori, CcrP also forms filamentous structures both in vitro and in vivo, but the protein seems to be required in the maintenance of this bacterium's helicoidal shape and its motility [100, 101]. CcrP proteins vary enormously in sequence between different strains of H. pylori; this variability may be linked to the high morphological differences encountered in clinical isolates [100].
FilP is an IF-like element of S. coelicolor [98]. Mutations in the Streptomyces filP gene cause strong morphological alterations in this bacterium and a marked deficiency in cell growth. FilP is probably required for additional support during polar cell-wall synthesis in S. coelicolor and thus contributes to the mechanical resistance of hyphae [98].
RsmP has been identified only in Corynebacteria [15]. This IF-like element is overexpressed in response to the partial depletion of DivIVA. Similarly to divIVA, rsmP is an essential gene in C. glutamicum, and its partial inhibition has been shown to result in the formation of coccoid cells while its overexpression induces a club-shaped morphology [15]. RsmP is able to produce filamentous structures in vitro and in vivo along the cell. The most interesting feature of RsmP is the change of its subcellular localization depending on its phosphorylation state. RsmP is phosphorylated at three different residues by the serine/threonine kinases PknA and PknL (see below and [15]). The cellular localization of an RsmP phosphoablative mutant is indistinguishable from the native RsmP, it still forms long filamentous structures along the cell. In contrast, a phosphomimetic mutant localizes only at the cell poles of C. glutamicum, suggesting that the phosphorylation state of RsmP is involved in the modulation of polar peptidoglycan synthesis in Corynebacteria [15]. The discovery of RsmP demonstrated that Corynebacteria have a specific molecular system for establishing their rod-shape morphology, thus distinguishing them from other Actinobacteria such as Mycobacterium and Streptomyces.
It is worth mentioning that there is also an alternative to IF-like elements, specifically in Eubacteria. Bactofilins have been recently identified in C. crescentus as a bacteria-specific cytoskeletal family of proteins that provide structural support for peptidoglycan synthesis [8]. Bactofilins also polymerize in the absence of any cofactors, forming rod-shaped filaments in vitro and polar structures in vivo during stalk morphogenesis. These proteins are widely conserved in Eubacteria (with notable exceptions like Actinobacteria), suggesting a high structural and functional versatility [8].
5. Cell Shape Control by Phosphorylation of the Bacterial Cytoskeleton
A seminal report published in 2005 demonstrated for the first time that the bacterial cytoskeleton is controlled by eukaryotic-like serine/threonine phosphorylation [102]. In that paper, Kang et al. definitively showed that two protein kinases, PknA and PknB, modulate cell shape in M. tuberculosis by changing the phosphorylation state of DivIVA [102]. Not much later, FtsZ was identified as another substrate of Pkn phosphorylation, thus linking the control of cell division with cell growth in Actinobacteria by a unique signal transduction system [103].
Both PknA and PknB are located within a highly conserved cluster in Actinobacteria [104]. This cluster includes: (1) two genes of unknown function with forkhead-associated (FHA) domains, (2) a phosphatase that antagonizes pkn kinases, (3) rodA and a pbp required for cell-wall synthesis during elongation, (4) pknAB, and (5) crgA (Figure 1(a)). Further work demonstrated that most of these genes are somehow related to cell-shape determination in Actinobacteria [59, 105–108].
Despite the high degree of conservation within pkn clusters, their functions have proven to be quite dissimilar when compared in different Actinobacteria [105]. The main differences in pkn regulation of actinobacterial cytokinesis probably involve DivIVA, the major coordinator of cell growth in these bacteria. In M. tuberculosis, PknA phosphorylates DivIVA, thus controlling cell-wall elongation [109]. In C. glutamicum, DivIVA seems to be not phosphorylated by PknA; instead, this kinase phosphorylates the Corynebacterium-specific RsmP protein, which in turn controls cell growth [15].
These distinctions are complicated by the possibility that Pkn kinases are directly involved in the modulation of the synthesis of peptidoglycan precursors, since the MurC and MurD peptidoglycan ligases are also phosphorylated by PknAB in C. glutamicum and M. tuberculosis, respectively [110, 111]. However, the biological significance of these observations has yet to be fully determined.
Finally, the pkn cluster contains two genes with FHA domains [cg0064 (fhaA) and cg0063 (fhaB) in C. glutamicum], a feature that will no doubt add another layer of complexity to our attempts to understand Pkn regulation of actinobacterial cell shape. FHA domains are phosphopeptide recognition motifs that specifically recognize phosphothreonine-containing epitopes for protein-protein interaction. In C. glutamicum, we determined that the Cg0063 (FhaB) protein is phosphorylated by PknA and PknL, another serine-threonine kinase located elsewhere in the chromosome (Figure 2). This indicates a possible role for the Cg0063 (FhaB) protein in cell-shape determination. The functions of other FhaAB proteins have been analyzed in Actinobacteria and include the maintenance of hyphal morphology in S. coelicolor [112] and the virulence of M. tuberculosis [113].
Figure 2.
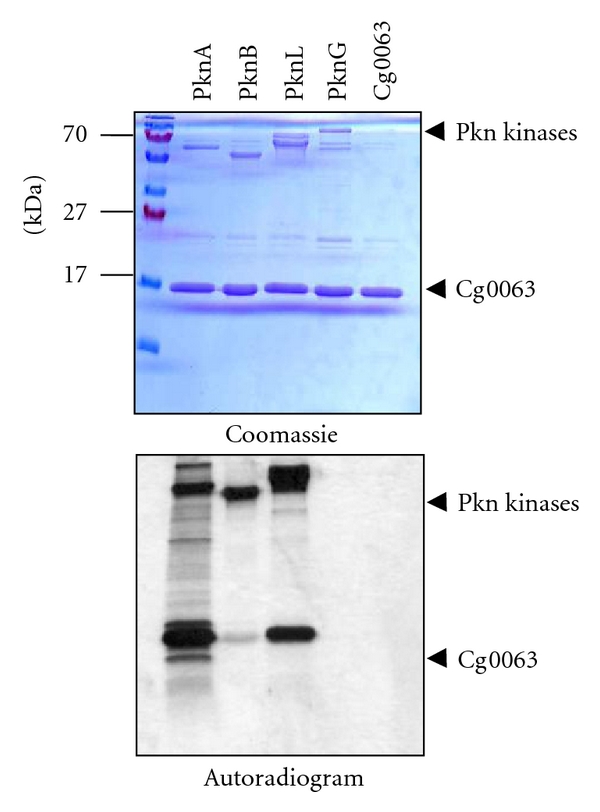
Cg0063 is phosphorylated in vitro by PknA and PknL in C. glutamicum. All four Ser/Thr Pkn kinases from C. glutamicum (PknA/B/L/G) and Cg0063 have been expressed and purified as described previously [105]. Then, Cg0063 was incubated alone or with the different Pkn kinases in the presence of [γ-33P] ATP for 30 min. Samples were separated by SDS-PAGE electrophoresis and stained with Coomassie Blue (upper panel) or visualized by autoradiography (lower panel). PknA/B/L kinases exhibit an autophosphorylation activity, whereas Cg0063 is mostly phosphorylated by PknA and PknL. M. Fiuza, unpublished results.
Further experimental work is needed before the pkn cluster is thoroughly understood; nevertheless, the evidence obtained to date strongly favors the conclusion that Pkn kinases direct actinobacterial cell division and cell elongation. Accordingly, PknA and PknB kinases have been identified as very promising targets for the development of new antituberculosis drugs [114, 115].
6. Final Remarks
During the last two decades, a good deal of progress has been made in our understanding of the basic physiology of bacteria. Once believed to be simple organisms with a low level of organization and totally unrelated to eukaryotes, bacteria are now recognized as very sophisticated forms of life that share most of the molecular tools used by our own cells to grow and replicate [1, 3, 7]. Actin-, tubulin- and IF-like proteins are being slowly identified and characterized in bacteria. Even bacterial-specific families of cytoskeletal proteins have been recently discovered [7, 8]. Most of the genes in the over-1000 bacterial genomes sequenced to date are still of unknown function; therefore many surprises probably await us. The high variability of cellular shapes in bacteria and their fantastic versatility make the study of prokaryotic cytokinesis very exciting but also extremely challenging [5]. The discovery of genus-specific molecular strategies guiding bacterial cell-shape determination equips us with unique targets for the development of new antimicrobial drugs, one of the main goals of the study of bacterial morphogenesis [116]. However, much more work is needed to completely unravel at least one model of bacterial cell division and cell growth. Yet this field of research has already yielded numerous practical applications, in the form of novel compounds that specifically inhibit FtsZ, MreB, or PknAB [114, 115, 117–119]. This progress could be vital to combating the inexorable development of new multi-drug-resistant pathogens appearing all around the world [120, 121].
Acknowledgments
M. Letek and M. Fiuza were beneficiaries of fellowships from the Ministerio de Educación y Ciencia (Spain); A. F. Villadangos from the Junta de Castilla y León. This work was funded by Grants from the Junta de Castilla y León (Ref. LE040A07), University of León (ULE 2001-08B), and Ministerio de Ciencia y Tecnología (BIO2005-02723 and BIO2008-00519).
References
- 1.Graumann PL. Cytoskeletal elements in bacteria. Annual Review of Microbiology. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- 2.Graumann PL. Dynamics of bacterial cytoskeletal elements. Cell Motility and the Cytoskeleton. 2009;66(11):909–914. doi: 10.1002/cm.20381. [DOI] [PubMed] [Google Scholar]
- 3.Erickson HP. Evolution of the cytoskeleton. BioEssays. 2007;29(7):668–677. doi: 10.1002/bies.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli . Microbiology and Molecular Biology Reviews. 1998;62(1):181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young KD. The selective value of bacterial shape. Microbiology and Molecular Biology Reviews. 2006;70(3):660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gitai Z. Diversification and specialization of the bacterial cytoskeleton. Current Opinion in Cell Biology. 2007;19(1):5–12. doi: 10.1016/j.ceb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Shih YL, Rothfield L. The bacterial cytoskeleton. Microbiology and Molecular Biology Reviews. 2006;70(3):729–754. doi: 10.1128/MMBR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kühn J, Briegel A, Mörschel E, et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus . EMBO Journal. 2010;29(2):327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventura M, Canchaya C, Tauch A, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiology and Molecular Biology Reviews. 2007;71(3):495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodfellow M. Bergey’s Manual of Systematic Bacteriology. 1989. Supragenic classification of actinomycetes; pp. 2333–2339. [Google Scholar]
- 11.Dover LG, Cerdeño-Tárraga AM, Pallen MJ, Parkhill J, Besra GS. Comparative cell wall core biosynthesis in the mycolated pathogens, Mycobacterium tuberculosis and Corynebacterium diphtheriae . FEMS Microbiology Reviews. 2004;28(2):225–250. doi: 10.1016/j.femsre.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiology and Molecular Biology Reviews. 2008;72(1):126–156. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nature Reviews Microbiology. 2009;7(1):36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh J, Larsson P, Singh B, et al. Sporulation in mycobacteria. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(26):10781–10786. doi: 10.1073/pnas.0904104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiuza M, Letek M, Leiba J, et al. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum . Journal of Biological Chemistry. 2010;285(38):29387–29397. doi: 10.1074/jbc.M110.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamames J, González-Moreno M, Mingorance J, Valencia A, Vicente M. Bringing gene order into bacterial shape. Trends in Genetics. 2001;17(3):124–126. doi: 10.1016/s0168-9525(00)02212-5. [DOI] [PubMed] [Google Scholar]
- 17.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nature Reviews Microbiology. 2009;7(9):642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 18.Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Current Biology. 2005;15(13):R514–R526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- 19.Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and Molecular Biology Reviews. 2010;74(4):504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Boer PAJ. Advances in understanding E. coli cell fission. Current Opinion in Microbiology. 2010;13(6):730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutkenhaus J. Min oscillation in bacteria. Advances in Experimental Medicine and Biology. 2008;641:49–61. doi: 10.1007/978-0-387-09794-7_4. [DOI] [PubMed] [Google Scholar]
- 22.Bramkamp M, van Baarle S. Division site selection in rod-shaped bacteria. Current Opinion in Microbiology. 2009;12(6):683–688. doi: 10.1016/j.mib.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Molecular Microbiology. 2005;55(6):1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- 24.Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli . EMBO Journal. 2002;21(4):685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arends SJR, Kustusch RJ, Weiss DS. ATP-binding site lesions in FtsE impair cell division. Journal of Bacteriology. 2009;191(12):3772–3784. doi: 10.1128/JB.00179-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crozat E, Grainge I. FtsK DNA translocase: the fast motor that knows where it’s going. ChemBioChem. 2010;11(16):2232–2243. doi: 10.1002/cbic.201000347. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez MD, Beckwith J. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. Journal of Bacteriology. 2009;191(8):2815–2825. doi: 10.1128/JB.01597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez MD, Akbay EA, Boyd D, Beckwith J. Multiple interaction domains in FtsL, a protein component of the widely conserved bacterial FtsLBQ cell division complex. Journal of Bacteriology. 2010;192(11):2757–2768. doi: 10.1128/JB.01609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammadi T, van Dam V, Sijbrandi R, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO Journal. 2011;30(8):1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraipont C, Alexeeva S, Wolf B, et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli . Microbiology. 2011;157(1):251–259. doi: 10.1099/mic.0.040071-0. [DOI] [PubMed] [Google Scholar]
- 31.Bernhardt TG, de Boer PAJ. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Molecular Microbiology. 2003;48(5):1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters NT, Dinh T, Bernhardt TG. A Fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. Journal of Bacteriology. 2011;193(18):4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO Journal. 2010;29(8):1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerding MA, Liu B, Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. Journal of Bacteriology. 2009;191(24):7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Molecular Microbiology. 2005;58(2):596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli . Molecular Microbiology. 2007;64(5):1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter . Cell. 2006;126(1):147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 38.Handler AA, Lim JE, Losick R. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis . Molecular Microbiology. 2008;68(3):588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucet I, Feucht A, Yudkin MD, Errington J. Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE. EMBO Journal. 2000;19(7):1467–1475. doi: 10.1093/emboj/19.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K. Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Molecular Microbiology. 2008;68(3):720–735. doi: 10.1111/j.1365-2958.2008.06190.x. [DOI] [PubMed] [Google Scholar]
- 41.Durand-Heredia JM, Yu HH, de Carlo S, Lesser CF, Janakiraman A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli . Journal of Bacteriology. 2011;193(6):1405–1413. doi: 10.1128/JB.01258-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letek M, Fiuza M, Ordóñez E, et al. Cell growth and cell division in the rod-shaped actinomycete Corynebacterium glutamicum . Antonie van Leeuwenhoek. 2008;94(1):99–109. doi: 10.1007/s10482-008-9224-4. [DOI] [PubMed] [Google Scholar]
- 43.Ramos A, Letek M, Campelo AB, Vaquera J, Mateos LM, Gil JA. Altered morphology produced by ftsZ expression in Corynebacterium glutamicum ATCC 13869. Microbiology. 2005;151(8):2563–2572. doi: 10.1099/mic.0.28036-0. [DOI] [PubMed] [Google Scholar]
- 44.Flärdh K. Growth polarity and cell division in Streptomyces . Current Opinion in Microbiology. 2003;6(6):564–571. doi: 10.1016/j.mib.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Molecular Microbiology. 2006;59(3):989–999. doi: 10.1111/j.1365-2958.2005.04987.x. [DOI] [PubMed] [Google Scholar]
- 46.Gündoğdu ME, Kawai Y, Pavlendova N, et al. Large ring polymers align FtsZ polymers for normal septum formation. EMBO Journal. 2011;30(3):617–626. doi: 10.1038/emboj.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honrubia MP, Ramos A, Gil JA. The cell division genes ftsQ and ftsZ, but not the three downstream open reading frames YFIH, ORF5 and ORF6, are essential for growth and viability in Brevibacterium lactofermentum ATCC 13869. Molecular Genetics and Genomics. 2001;265(6):1022–1030. doi: 10.1007/s004380100497. [DOI] [PubMed] [Google Scholar]
- 48.Datta P, Dasgupta A, Bhakta S, Basu J. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis . Journal of Biological Chemistry. 2002;277(28):24983–24987. doi: 10.1074/jbc.M203847200. [DOI] [PubMed] [Google Scholar]
- 49.Rajagopalan M, Maloney E, Dziadek J, et al. Genetic evidence that mycobacterial FtsZ and FtsW proteins interact, and colocalize to the division site in Mycobacterium smegmatis . FEMS Microbiology Letters. 2005;250(1):9–17. doi: 10.1016/j.femsle.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 50.Datta P, Dasgupta A, Singh AK, Mukherjee P, Kundu M, Basu J. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Molecular Microbiology. 2006;62(6):1655–1673. doi: 10.1111/j.1365-2958.2006.05491.x. [DOI] [PubMed] [Google Scholar]
- 51.Mistry BV, Del SR, Wright C, Findlay K, Dyson P. FtsW is a dispensable cell division protein required for Z-ring stabilization during sporulation septation in Streptomyces coelicolor . Journal of Bacteriology. 2008;190(16):5555–5566. doi: 10.1128/JB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noens EE, Mersinias V, Willemse J, et al. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor . Molecular Microbiology. 2007;64(5):1244–1259. doi: 10.1111/j.1365-2958.2007.05732.x. [DOI] [PubMed] [Google Scholar]
- 53.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces . Genes and Development. 2011;25(1):89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sureka K, Hossain T, Mukherjee P, et al. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008590. Article ID e8590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Ogino H, Teramoto H, Inui M, Yukawa H. DivS, a novel SOS-inducible cell-division suppressor in Corynebacterium glutamicum . Molecular Microbiology. 2008;67(3):597–608. doi: 10.1111/j.1365-2958.2007.06069.x. [DOI] [PubMed] [Google Scholar]
- 56.Donovan C, Schwaiger A, Krämer R, Bramkamp M. Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum . Journal of Bacteriology. 2010;192(13):3441–3451. doi: 10.1128/JB.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dziedzic R, Kiran M, Plocinski P, et al. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS ONE. 2010;5(7) doi: 10.1371/journal.pone.0011058. Article ID e11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.del Sol R, Mullins JGL, Grantcharova N, Flärdh K, Dyson P. Influence of CrgA on assembly of the cell division protein FtsZ during development of Streptomyces coelicolor . Journal of Bacteriology. 2006;188(4):1540–1550. doi: 10.1128/JB.188.4.1540-1550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plocinski P, Ziolkiewicz M, Kiran M, et al. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. Journal of Bacteriology. 2011;193(13):3246–3256. doi: 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carballido-López R. The bacterial actin-like cytoskeleton. Microbiology and Molecular Biology Reviews. 2006;70(4):888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harbor Perspectives in Biology. 2010;2(9):p. a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doi M, Wachi M, Ishino F, et al. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. Journal of Bacteriology. 1988;170(10):4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli . Journal of Bacteriology. 1987;169(11):4935–4940. doi: 10.1128/jb.169.11.4935-4940.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Esue O, Wirtz D, Tseng Y. GTPase activity, structure, and mechanical properties of filaments assembled from bacterial cytoskeleton protein MreB. Journal of Bacteriology. 2006;188(3):968–976. doi: 10.1128/JB.188.3.968-976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Popp D, Narita A, Maeda K, et al. Filament structure, organization, and dynamics in MreB sheets. Journal of Biological Chemistry. 2010;285(21):15858–15865. doi: 10.1074/jbc.M109.095901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayer JA, Amann KJ. Assembly properties of the Bacillus subtilis actin, MreB. Cell Motility and the Cytoskeleton. 2009;66(2):109–118. doi: 10.1002/cm.20332. [DOI] [PubMed] [Google Scholar]
- 67.Jones LJF, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis . Cell. 2001;104(6):913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 68.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113(6):767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 69.Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis . Science. 2011;333(6039):222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333(6039):225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 71.Alyahya SA, Alexander R, Costa T, Henriques AO, Emonet T, Jacobs-Wagner C. RodZ, a component of the bacterial core morphogenic apparatus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(4):1239–1244. doi: 10.1073/pnas.0810794106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli . EMBO Journal. 2009;28(3):193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shiomi D, Sakai M, Niki H. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO Journal. 2008;27(23):3081–3091. doi: 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Teeffelen S, Wang S, Furchtgott L, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(38):15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. Murein segregation in Escherichia coli . Journal of Bacteriology. 1997;179(9):2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Margolin W. Sculpting the bacterial cell. Current Biology. 2009;19(17):R812–R822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flärdh K. Essential role of DivlVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Molecular Microbiology. 2003;49(6):1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- 78.Letek M, Ordóñez E, Vaquera J, et al. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum . Journal of Bacteriology. 2008;190(9):3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154(3):725–735. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 80.Mazza P, Noens EE, Schirner K, et al. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Molecular Microbiology. 2006;60(4):838–852. doi: 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- 81.Kleinschnitz EM, Heichlinger A, Schirner K, et al. Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3(2) cooperate in spore wall synthesis. Molecular Microbiology. 2011;79(5):1367–1379. doi: 10.1111/j.1365-2958.2010.07529.x. [DOI] [PubMed] [Google Scholar]
- 82.Heichlinger A, Ammelburg M, Kleinschnitz EM, et al. The MreB-like protein Mbl of Streptomyces coelicolor A3(2) depends on MreB for proper localization and contributes to spore wall synthesis. Journal of Bacteriology. 2011;193(7):1533–1542. doi: 10.1128/JB.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramos A, Honrubia MP, Valbuena N, Vaquera J, Mateos LM, Gil JA. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum . Microbiology. 2003;149(12):3531–3542. doi: 10.1099/mic.0.26653-0. [DOI] [PubMed] [Google Scholar]
- 84.Letek M, Valbuena N, Ramos A, Ordóñez E, Gil JA, Mateos LM. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum . Journal of Bacteriology. 2006;188(2):409–423. doi: 10.1128/JB.188.2.409-423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muchová K, Kutejová E, Scott DJ, et al. Oligomerization of the Bacillus subtilis division protein DivIVA. Microbiology. 2002;148(3):807–813. doi: 10.1099/00221287-148-3-807. [DOI] [PubMed] [Google Scholar]
- 86.Stahlberg H, Kutejová E, Muchová K, et al. Oligomeric structure of the Bacillus subtilis cell division protien DivIVA determined by transmission microscopy. Molecular Microbiology. 2004;52(5):1281–1290. doi: 10.1111/j.1365-2958.2004.04074.x. [DOI] [PubMed] [Google Scholar]
- 87.Lenarcic R, Halbedel S, Visser L, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO Journal. 2009;28(15):2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Letek M, Fiuza M, Ordóñez E, et al. DivIVA uses an N-terminal conserved region and two coiled-coil domains to localize and sustain the polar growth in Corynebacterium glutamicum . FEMS Microbiology Letters. 2009;297(1):110–116. doi: 10.1111/j.1574-6968.2009.01679.x. [DOI] [PubMed] [Google Scholar]
- 89.Hempel AM, Wang SB, Letek M, Gil JA, Flärdh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor . Journal of Bacteriology. 2008;190(22):7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang SB, Cantlay S, Nordberg N, Letek M, Gil JA, Flärdh K. Domains involved in the in vivo function and oligomerization of apical growth determinant DivIVA in Streptomyces coelicolor . FEMS Microbiology Letters. 2009;297(1):101–109. doi: 10.1111/j.1574-6968.2009.01678.x. [DOI] [PubMed] [Google Scholar]
- 91.Letek M, Ordóñez E, Fernández-Natal I, Gil JA, Mateos LM. Identification of the emerging skin pathogen Corynebacterium amycolatum using PCR-amplification of the essential divIVA gene as a target. FEMS Microbiology Letters. 2006;265(2):256–263. doi: 10.1111/j.1574-6968.2006.00492.x. [DOI] [PubMed] [Google Scholar]
- 92.Valbuena N, Letek M, Ordóñez E, et al. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Molecular Microbiology. 2007;66(3):643–657. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- 93.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 94.Lupas A. Coiled coils: new structures and new functions. Trends in Biochemical Sciences. 1996;21(10):375–382. [PubMed] [Google Scholar]
- 95.Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem. 2004;5(2):170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- 96.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115(6):705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 97.Ausmees N. Intermediate filament-like cytoskeleton of Caulobacter crescentus . Journal of Molecular Microbiology and Biotechnology. 2006;11(3–5):152–158. doi: 10.1159/000094051. [DOI] [PubMed] [Google Scholar]
- 98.Bagchi S, Tomenius H, Belova LM, Ausmees N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces . Molecular Microbiology. 2008;70(4):1037–1050. doi: 10.1111/j.1365-2958.2008.06473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Izard J. Cytoskeletal cytoplasmic filament ribbon of Treponema: a member of an intermediate-like filament protein family. Journal of Molecular Microbiology and Biotechnology. 2006;11(3–5):159–166. doi: 10.1159/000094052. [DOI] [PubMed] [Google Scholar]
- 100.Waidner B, Specht M, Dempwolff F, et al. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori . PLoS Pathogens. 2009;5(11) doi: 10.1371/journal.ppat.1000669. Article ID e1000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Specht M, Schätzle S, Graumann PL, Waidner B. Helicobacter pylori possesses four coiled-coil-rich proteins (Ccrp) that form extended filamentous structures and control cell shape and motility. Journal of Bacteriology. 2011;193(17):4523–4530. doi: 10.1128/JB.00231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kang CM, Abbott DW, Sang TP, Dascher CC, Cantley LC, Husson RN. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes and Development. 2005;19(14):1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thakur M, Chakraborti PK. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. Journal of Biological Chemistry. 2006;281(52):40107–40113. doi: 10.1074/jbc.M607216200. [DOI] [PubMed] [Google Scholar]
- 104.Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Molecular Microbiology. 2010;75(5):1064–1077. doi: 10.1111/j.1365-2958.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 105.Fiuza M, Canova MJ, Zanella-Cléon I, et al. From the characterization of the four serine/threonine protein kinases (PknA/B/G/L) of Corynebacterium glutamicum toward the role of PknA and PknB in cell division. Journal of Biological Chemistry. 2008;283(26):18099–18112. doi: 10.1074/jbc.M802615200. [DOI] [PubMed] [Google Scholar]
- 106.Dasgupta A, Datta P, Kundu M, Basu J. The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology. 2006;152(2):493–504. doi: 10.1099/mic.0.28630-0. [DOI] [PubMed] [Google Scholar]
- 107.Schultz C, Niebisch A, Schwaiger A, et al. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Molecular Microbiology. 2009;74(3):724–741. doi: 10.1111/j.1365-2958.2009.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sajid A, Arora G, Gupta M, Upadhyay S, Nandicoori VK, Singh Y. Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB. PLoS ONE. 2011;6(3) doi: 10.1371/journal.pone.0017871. Article ID e17871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jani C, Eoh H, Lee JJ, et al. Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiology. 2010;10:p. 327. doi: 10.1186/1471-2180-10-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fiuza M, Canova MJ, Patin D, et al. The MurC ligase essential for peptidoglycan biosynthesis is regulated by the serine/threonine protein kinase PknA in Corynebacterium glutamicum . Journal of Biological Chemistry. 2008;283(52):36553–36563. doi: 10.1074/jbc.M807175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thakur M, Chakraborti PK. Ability of PknA, a mycobacterial eukaryotic-type serine/threonine kinase, to transphosphorylate MurD, a ligase involved in the process of peptidoglycan biosynthesis. Biochemical Journal. 2008;415(1):27–33. doi: 10.1042/BJ20080234. [DOI] [PubMed] [Google Scholar]
- 112.Jones G, Del SR, Dudley E, Dyson P. Forkhead-associated proteins genetically linked to the serine/threonine kinase PknB regulate carbon flux towards antibiotic biosynthesis in Streptomyces coelicolor . Microbial Biotechnology. 2011;4(2):263–274. doi: 10.1111/j.1751-7915.2010.00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gupta M, Sajid A, Arora G, Tandon V, Singh Y. Forkhead-associated domain-containing protein Rv0019c and polyketide-associated protein PapA5, from substrates of serine/threonine protein kinase PknB to interacting proteins of Mycobacterium tuberculosis . Journal of Biological Chemistry. 2009;284(50):34723–34734. doi: 10.1074/jbc.M109.058834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Székely R, Wáczek F, Szabadkai I, et al. A novel drug discovery concept for tuberculosis: inhibition of bacterial and host cell signalling. Immunology Letters. 2008;116(2):225–231. doi: 10.1016/j.imlet.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 115.Magnet S, Hartkoorn RC, Székely R, et al. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis. 2010;90(6):354–360. doi: 10.1016/j.tube.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 116.Vicente M, Hodgson J, Massidda O, Tonjum T, Henriques-Normark B, Ron EZ. The fallacies of hope: will we discover new antibiotics to combat pathogenic bacteria in time? FEMS Microbiology Reviews. 2006;30(6):841–852. doi: 10.1111/j.1574-6976.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- 117.Kumar K, Awasthi D, Berger WT, Tonge PJ, Slayden RA, Ojima I. Discovery of anti-TB agents that target the cell-division protein FtsZ. Future Medicinal Chemistry. 2010;2(8):1305–1323. doi: 10.4155/fmc.10.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Singh P, Panda D. FtsZ inhibition: a promising approach for anti-staphylococcal therapy. Drug News and Perspectives. 2010;23(5):295–304. doi: 10.1358/dnp.2010.23.5.1429489. [DOI] [PubMed] [Google Scholar]
- 119.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120(3):329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 120.Caminero JA, Sotgiu G, Zumla A, Migliori GB. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. The Lancet Infectious Diseases. 2010;10(9):621–629. doi: 10.1016/S1473-3099(10)70139-0. [DOI] [PubMed] [Google Scholar]
- 121.Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. Journal of Antimicrobial Chemotherapy. 2011;66(supplement 4):iv43–iv48. doi: 10.1093/jac/dkr076. [DOI] [PubMed] [Google Scholar]


