Abstract
The Saccharomyces cerevisiae cyclin Clb5 is required for premeiotic S phase, meiotic recombination, and successful progression through meiosis. Clb5 is not essential for mitotic proliferation because Clb1–Clb4 can support DNA replication in clb5clb6 mutants. Clb1, Clb3, and Clb4 accumulate in clb5clb6 cells during meiotic differentiation yet fail to promote premeiotic DNA replication. When expressed under the regulation of the CLB5 promoter, Clb1 and Clb3 accumulate and are active in the early stages of meiotic differentiation but cannot induce premeiotic DNA replication, suggesting that they do not target Cdk1 to the necessary substrates. The Clb5 hydrophobic patch (HP) residues are important for Clb5 function but this motif alone does not provide the specificity required for Clb5 to induce premeiotic S phase. Domain exchange experiments demonstrated that the amino terminus of Clb5 when fused to Clb3 confers upon Clb3 the ability to induce premeiotic S phase. Chimeric cyclins containing smaller regions of the Clb5 amino terminus displayed reduced ability to activate premeiotic DNA replication despite being more abundant and having greater associated histone H1 kinase activity than endogenous Clb5. These observations suggest that Clb5 has a unique ability to trigger premeiotic S phase and that the amino-terminal region of Clb5 contributes to its specificity and regulates the functions performed by the cyclin–Cdk complex.
CYCLIN-dependent kinases (Cdks) in conjunction with their activating subunits, the cyclins, provide the impetus for transitions between distinct phases of the eukaryotic cell cycle (Morgan 1997). All organisms from yeast to humans express multiple cyclins that are regulated at the levels of transcription, protein stability, and subcellular localization (Pines and Hunter 1991; Lew and Reed 1992; Koepp et al. 1999). The oscillation of cyclin abundance throughout the cell cycle allows Cdk subunits to form distinct cyclin–Cdk complexes that act in a specific temporal order. The budding yeast Saccharomyces cerevisiae expresses a single Cdk (Cdk1) whose activity is required for progression through G1, S phase, and mitosis (Reed and Wittenberg 1990). Cdk1 accomplishes these functions by associating sequentially with G1 cyclins, Cln1, Cln2, and Cln3, the S-phase cyclins, Clb5 and Clb6, and finally the mitotic cyclins, Clb1–Clb4 (Nasmyth 1996).
Cyclin–Cdk activity is also required for cellular differentiation and development (Shuster and Byers 1989; Edgar and Lehner 1996; Chellappan et al. 1998). Gametogenesis is a differentiation program that employs a modified cell cycle to direct parental diploid cells through meiosis to the formation of haploid gametes (Kupiec et al. 1997; Albertini and Carabatsos 1998). S. cerevisiae initiates meiotic differentiation in response to nutrient deprivation, and the program leads to the formation of haploid spore gametes (Kupiec et al. 1997). Much of the machinery required for the mitotic cell cycle is employed to promote progression through meiotic differentiation. However, the meiotic program uses some distinct factors to achieve specific developmental aims (Simchen 1974; Orr-Weaver 1994). One example of meiosis-specific regulation is the control of premeiotic DNA replication. Clb5 is the major S-phase cyclin in S. cerevisiae but is not essential during mitotic proliferation as other cyclins can promote DNA replication in its absence (Schwob et al. 1994; Donaldson et al. 1998). Although Clb5 is not required for mitotic proliferation, it is essential for premeiotic DNA replication, the induction of meiotic recombination, some aspects of synaptonemal complex formation, and activation of the S–M checkpoint in meiosis (Dirick et al. 1998; Stuart and Wittenberg 1998; Smith et al. 2001; Henderson et al. 2006; Wan et al. 2008; Zhu et al. 2010).
A growing body of evidence supports the contention that cyclins target Cdk activity to the correct substrates and hence the cyclin provides specificity to the Cdk activity (Cross et al. 1999; Roberts 1999; Loog and Morgan 2005; Bhaduri and Pryciak 2011; Koivomagi et al. 2011; Pagliuca et al. 2011). In addition to interaction with protein substrates, the specificity of cyclins for particular functions can be imparted by the timing of accumulation, the activity of cyclin–CDK inhibitors, and by subcellular localization (Draviam et al. 2001; Miller and Cross 2001; Moore et al. 2003; Carlile and Amon 2008). This regulation can allow specific cyclins to fulfill specialized roles. For example, during meiosis the 5′-UTR of the CLB3 mRNA restricts accumulation of Clb3 to the second meiotic division, while Clb1 function is restricted to meiosis I (MI) by a specific inhibitor (Carlile and Amon 2008). The importance of these regulatory mechanisms can be masked when cyclins are overexpressed or when temporal regulation is altered. For example, overexpression of CLB1 or CLB2 can bypass the need for all other B-type cyclins in mitotically proliferating S. cerevisiae, implying that a single cyclin can perform all the S-phase and mitotic functions (Haase and Reed 1999; Hu and Aparicio 2005). Similarly a fusion of the Schizosaccharomyces pombe Cdc13 to Cdc2 can drive a relatively normal cell cycle in cells depleted of other cyclins (Coudreuse and Nurse 2010). However, the importance of substrate specificity for cyclins is implied by the observation that B-type cyclins cannot perform all the critical functions of G1 cyclins even if expressed during G1 phase (Cross et al. 1999; Donaldson 2000). Similarly, S. cerevisiae deficient in the S-phase cyclins Clb5 and Clb6 display a delay in DNA replication, and even overproduction of Clb1 or Clb2 or the expression of CLB3 under control of the CLB5 promoter does not fully suppress this defect (Hu and Aparicio 2005). Additionally, many B-type cyclin–Cdk complexes display substrate preferences in vitro (Peeper et al. 1993; Higashi et al. 1995; Loog and Morgan 2005; Joshi et al. 2009; Koivomagi et al. 2011). Thus, while there is considerable overlap between the activity and function of various cyclins, the substrate specificity of cyclin–CDK complexes remains a central mechanism that regulates cell cycle progression.
The specific functions bestowed upon Cdks by different cyclins leads to the hypothesis that distinct regions or domains of the cyclin are important for interacting with specific substrates. The most clearly defined motif in B-type cyclins that influences substrate interactions is the “hydrophobic patch” (HP), a series of residues that make hydrophobic and electrostatic contacts with the RXL and KXL motif on many high-affinity substrates (Wohlschlegel et al. 2001; Archambault et al. 2005). The hydrophobic patch is conserved among B-type cyclins, including Clb5, and mutations that change key hydrophobic residues to alanine cause loss of function in vivo and reduced activity against RXL-containing substrates in vitro (Schulman et al. 1998; Cross and Jacobson 2000). Although the hydrophobic patch residues are essential for Clb5 function, it is not clear whether the hydrophobic patch alone is sufficient to explain the differences in substrate specificity between different cyclins. The HP motif in Clb1 and Clb2 imposes regulation of the cyclin–Cdk complexes by allowing interaction with Swe1, which regulates Cdk1 activity (Hu et al. 2008). However, it is unclear whether the HP motif in these cyclins influences their substrate specificity. Additionally, the cyclin-A HP allows interaction with p27, which regulates activity of the cyclin A–Cdk2 complex (Schulman et al. 1998). Thus the HP allows modulation of cyclin–Cdk activity through interaction with both substrates and regulators.
We have investigated the role of Clb5 and its HP domain in promoting premeiotic DNA replication. Our findings show that although other B-type cyclins accumulate in clb5clb6-deficient cells undergoing meiosis, these cyclins are ineffective in activating premeiotic DNA replication. Mutational analysis revealed that the Clb5 HP motif is necessary but not sufficient to provide the function required to activate premeiotic DNA replication. Through a series of fragment exchanges, we identified an amino-terminal Clb5 region that can confer the ability to promote premeiotic DNA replication on Clb3, a cyclin that normally cannot perform this function. These results demonstrate a role for the nonconserved amino-terminal region of Clb5 in conferring specific function to the Clb5–Cdk1 complex.
Materials and Methods
Strains and growth conditions
The relevant genotypes of all strains used in the study are listed in Table 1. All strains were generated using standard genetic techniques (Rose et al. 1990). Epitope tagging and gene deletions were accomplished through a PCR-based procedure (Wach et al. 1994). Yeast strains were routinely maintained and propagated on YEP medium (1% yeast extract, 2% peptone, 10 μg/ml adenine and 10 μg/ml tryptophan) supplemented with 2% glucose (YEPD), 2% potassium acetate (YEP-KAc), or 2% galactose (YEP-GAL). Sporulation medium (SPM) was composed of 1% potassium acetate. Synthetic medium for the selection of transformants was prepared as described (Rose et al. 1990). For the selection of S. cerevisiae transformants harboring the G418 resistance gene (kanMX2 or kanMX6 cassettes), G418 sulfate was used at a final concentration of 200 μg/ml. Synchronous sporulation was achieved as previously described (Stuart and Wittenberg 1998).
Table 1 . S. cerevisiae strains used in this study.
| Strain name | Relevant genotype |
|---|---|
| DSY 1030 | MATa SK1 |
| DSY 1031 | MATα SK1 |
| DSY 1089 | MATa/α |
| DSY 1168 | MATa clb5::URA3 clb6::TRP1 |
| DSY 1169 | MATα clb5::URA3 clb6::TRP1 |
| DSY 1649 | MATa/α clb1::KanMX2 |
| DSY 1652 | MATa/α clb3::URA3 |
| DSY 1655 | MATa/α clb1::KanMX2 clb3::URA3 |
| DSY 1658 | MATa/α clb1 clb3::C3H5-MYC |
| K3418 | MATa clb5::hisG-URA3-hisG clb6::LEU2 clb3::TRP1 clb4::HIS3 TRP1::GAL-CLB5 |
| clb5hpm | MATa/α URA3::clb5hpm-HA clb5::kanMX2 clb6::TRP1 |
| 3xclb5hpm | MATa/α URA3:: 3 x clb5hpm-HA clb5::kanMX2 clb6::TRP1 |
| clb5 clb6 | MATa/α clb5::URA3 clb6::TRP1 |
| C5 WT | MATa/α CLB5-MYC-kanMX2 |
| C3 WT | MATa/α CLB3-MYC-kanMX2 |
| C3 clb5 clb6 | MATa/α CLB3-MYC-kanMX2 clb5::URA3 clb6::TRP1 |
| C1 WT | MATa/α CLB1-MYC-kanMX2 |
| C1 clb5 clb6 | MATa/α CLB1-MYC-kanMX2 clb5::URA3 clb6::TRP1 |
| C4 WT | MATa/α CLB4-MYC-kanMX2 |
| C4 clb5 clb6 | MATa/α CLB4-MYC-kanMX2 clb5::URA3 clb6::TRP1 |
| C5 | MATa/α clb5:: PCLB5-CLB5-MYC-kanMX2 clb6::TRP1 |
| C3 | MATa/α clb5:: PCLB5-CLB3-MYC-kanMX2 clb6::TRP1 |
| C3 swe1 | MATa/α clb5:: PCLB5-CLB3-MYC-kanMX2 clb6::TRP1, swe1::LEU2 |
| C3H5 | MATa/α clb5:: PCLB5-C3H-MYC-kanMX2 clb6::TRP1 |
| C5H3 | MATa/α clb5:: PCLB5-C5H-MYC-kanMX2 clb6::TRP1 |
| C5C1 | MATa/α clb5::PCLB5-CLB1-MYC-kanMX2 clb6::TRP1 |
| C53 | MATa/α clb5:: PCLB5-C53-MYC-kanMX2 clb6::TRP1 |
| C35 | MATa/α clb5:: PCLB5-C35-MYC-kanMX2 clb6::TRP1 |
| C53A | MATa/α clb5:: PCLB5-C53A-MYC-kanMX2 clb6::TRP1 |
| C53B | MATa/α clb5:: PCLB5-C53B-MYC-kanMX2 clb6::TRP1 |
| C53C | MATa/α clb5:: PCLB5-C53C-MYC-kanMX2 clb6::TRP1 |
| C53D | MATa/α clb5:: PCLB5-C53D-MYC-kanMX2 clb6::TRP1 |
| C5T1 | MATa/α clb5:: PCLB5-C5T1-MYC-kanMX2 clb6::TRP1 |
| C53AT1 | MATa/α clb5:: PCLB5-C53AT1-MYC-kanMX2 clb6::TRP1 |
| C53AT2 | MATa/α clb5:: PCLB5-C53AT2-MYC-kanMX2 clb6::TRP1 |
| C53DT1 | MATa/α clb5:: PCLB5-C53DT1-MYC-kanMX2 clb6::TRP1 |
| C53DT2 | MATa/α clb5:: PCLB5-C53DT2-MYC-kanMX2 clb6::TRP1 |
All SK1 strains used in this study were derived from DSY1030 MATa ho::LYS2 arg4Bgl his4X leu2::hisG lys2 trp1::hisG ura3 and DSY1031 MATα ho::LYS2 arg4Nsp his4B leu2::hisG lys2 trp1::hisG ura3 (Stuart and Wittenberg 1998). The W303 strain K3418 used to test the function of chimeric cyclins in mitotic proliferation has the genotype MATα ade2-1 can1-100 leu2-2, 112 his3-11, 15 trp1-1 ura3-1 clb3::TRP1 clb4::HIS3 clb5::hisG-URA3-hisG clb6::LEU2 TRP1::GAL-CLB5 and was generously provided by Etienne Schwob (Schwob and Nasmyth 1993).
All of the mutant and chimeric alleles of CLB3 and CLB5 were constructed using a splice overlap PCR-based method (Horton et al. 1989) and integrated at the CLB5 locus in a haploid clb5clb6 strain. The fusion of the CLB5 promoter to CLB3 or CLB5–CLB3 chimera is denoted as PCLB5–CLB3 or PCLB5–CLB5–CLB3, respectively, as suggested by Fred Sherman (Sherman 1997). The fusion junctions for the chimeric cyclins were chosen in regions of local sequence conservation between Clb3 and Clb5 to avoid profoundly disrupting the protein structures. All of the open reading frames synthesized were sequenced to ensure the desired sequences had been generated with the absence of unwanted mutations. The CLB3 and CLB5 open reading frames from yeast SK1 were used for all of the constructions. The SK1 CLB3 open reading frame encodes several silent polymorphisms relative to the S288c sequence. In contrast, the CLB5 open reading frame in addition to several silent polymorphisms also encodes the following coding changes E3 D, E113 K, E114 D, M351 I, and N427 T. All of the constructs were generated to include 328 bp of the CLB5 promoter and 600 bp of sequence downstream of the CLB5 open reading frame. An epitope tag consisting of 13 copies of the C-MYC epitope was appended to each of the cyclins used. Additionally, a G418 resistance cassette was inserted immediately downstream of the CLB5 transcriptional terminator. MATa and MATα clb5::URA3clb6::TRP1 strains were transformed with each cyclin construct and transformants that had replaced clb5::URA3 with the cyclin constructs were selected for G418 and 5-fluoro-orotic acid (FOA) resistance. The correct structure of the integrants was confirmed by PCR analysis of the genomic DNA. Homozygous diploids were constructed by mating. At least three independent diploids harboring each recombinant cyclin were tested for sporulation efficiency. The C3H5 allele was installed at the CLB3 locus to create DSY1658 (clb1 C3H5) using a “loop in loop out” strategy (Rose et al. 1990). Meiotic recombination was detected by employing a “return to growth” procedure and selecting colonies that had under gone recombination to repair the heteroalleles at arg4 and his4 (Esposito and Esposito 1974) Quantitative mating assays were performed as described (Sprague 1991).
Protein extraction, Western blotting, and histone H1 kinase assays
Extracts for the analysis of protein abundance were prepared as described (Sedgwick et al. 2006). Proteins were detected with the following antibodies: α-HA acsites fluid 1:10,000 (Covance), α-MYC ascites fluid 1:10,000 (Covance), and α-PSTAIR 1:10,000 (Sigma). All primary antibodies were detected by incubation with secondary antibodies conjugated to horse radish peroxidase (HRP), followed by enhanced chemiluminescence using ECL Plus Western blotting reagents (GE Healthcare). Images were scanned and quantified with a Storm 840 phosphorimager (Molecular Dynamics) in combination with ImageQuant 1.2 software. Quantified values were normalized using positive control samples (from an asynchronous PCLB5–CLB5–MYC clb6 culture) and negative control samples (from an asynchronous clb5clb6 culture). Thus, any experimental variation in sample preparation, Western blotting, or quantitation was corrected for by using an aliquot of the same positive and negative control samples with each experiment. Additionally, the PSTAIR signal was used as a quantitative loading control to normalize the values in combination with the positive and negative controls. The values are reported in arbitrary units that reflect the fold increase relative to the positive control sample in combination with the background correction from the negative control sample and the normalization with the loading controls. Histone H1 kinase assays were performed as previously described (Raithatha and Stuart 2005).
RNA analysis and flow cytometry
Extraction of RNA and detection of RNA species by Northern blotting was performed as previously described (Raithatha and Stuart 2005). Flow cytometry to measure cellular DNA content and fluorescence microscopy of 4,6-diamino-2-phenylindole (DAPI)-stained cells was performed as previously described (Stuart and Wittenberg 1998). The proportion of cells in S phase and G2/M was determined by analysis of the flow cytometry histogram plots as described (Cha et al. 2000).
Results
Clb1, Clb3, and Clb4 accumulate in clb5 clb6 diploids but do not promote premeiotic DNA replication
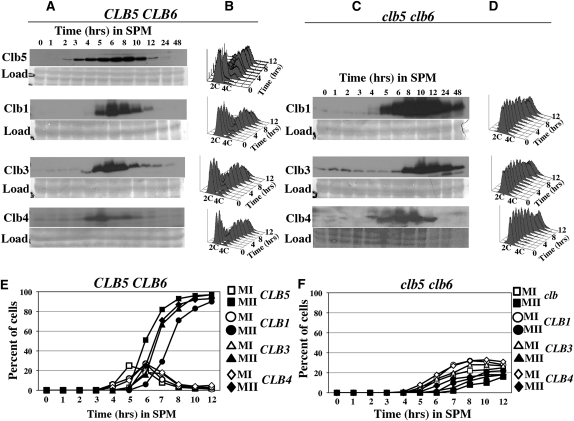
To investigate why the B-type cyclins Clb1, Clb3, and Clb4 fail to promote premeiotic DNA replication in clb5clb6 mutants, we monitored the accumulation of the Clb proteins in both CLB5CLB6 and clb5clb6 homozygous diploids that had been induced to initiate the meiotic program. During progression through the meiotic program, Clb5 began to accumulate within 2 hr and reached peak abundance between 5 and 6 hr (Figure 1A, top). The timing of Clb5 accumulation is consistent with its role in the onset of DNA replication (Figure 1B). Clb1, Clb3, and Clb4 proteins began to accumulate at ∼5 hr following the induction of sporulation in wild-type cells and reached a peak of abundance at 6–8 hr before declining to low levels at 10–12 hr (Figure 1A). In these experiments, imperfection in the synchrony of the strains was apparent when comparing progression through S phase and the meiotic chromosome divisions. In the CLB1–MYC strain, 50% of the cells had completed DNA replication after 5.25 hr (Figure 1B) and 50% of the cells had completed meiosis II (MII) at 7.5 hr (Figure 1E). In the CLB3–MYC strain, 50% of the cells had completed DNA replication after 4 hr (Figure 1B) and 50% of the cells had completed MII after 6.5 hr (Figure 1E). Although Clb1 and Clb3 both began to accumulate at about 5 hr, it is notable that at 5 hr, no cells undergoing MI were yet visible in the strain expressing Clb1, while strains expressing Clb3 were already initiating MII (Figure 1E). The different rates of progression through meiosis are due to experimental differences in the synchronization of the cultures and not a result of the epitope tags on the cyclins. Thus, in these experiments, Clb1 accumulated earlier in the meiotic program than did Clb3, consistent with the observations of Carlile and Amon (2008). In all the cultures, DNA replication initiated between 2 and 3 hr and was completed in most of the cells by 5 hr (Figure 1B). Since replication was nearly complete before Clb1, Clb3, or Clb4 accumulated, it is unlikely these cyclins make a significant contribution to initiating premeiotic DNA replication in wild-type cells.
Figure 1 .
Clb1, Clb3, and Clb4 accumulate in clb5 clb6 mutants but do not induce premeiotic DNA replication. (A) Western blots showing the accumulation of Clb5, Clb1, Clb3, and Clb4 in CLB5 CLB6 cells progressing through the meiotic program. These strains are referred to as C5 WT, C1 WT, C3 WT, and C4 WT, respectively, in Table 1. (C) Western blots showing the accumulation of Clb1, Clb3, and Clb4 in clb5 clb6 cells progressing through the meiotic program. These strains are referred to as C1 clb5 clb6, C3 clb5 clb6, and C4 clb5 clb6 in Table 1. Samples were taken at the indicated times following the induction of sporulation. A prominent band from the amido black-stained membrane was used as a loading control (load). (B and D) Flow cytometry histograms displaying DNA content of the strains used for Western analysis in A and C. The positions representing 2C and 4C DNA content are indicated. (E) Chromatin masses in CLB5 CLB6 strains used for Western and flow cytometry analysis in A and B. (F) Chromatin masses in clb5 clb6 strains used for Western and flow cytometry analysis in C and D. Open symbols, cells with two chromatin masses; closed symbols, cells with greater than two chromatin masses.
One explanation for the dependence of premeiotic DNA replication on CLB5 is that Clb5 activity may be necessary for the expression of other B-type cyclins. Thus, the accumulation of Clb1, Clb3, and Clb4 was monitored in clb5clb6 diploids. Clb1 and Clb4 accumulated in a clb5clb6 mutant strain with timing similar to that observed in CLB5CLB6 cells (Figure 1C). Clb1 could first be detected at 5 hr; however, in contrast to wild-type cells, Clb1 continued to accumulate reaching peak abundance at 10 hr and then persisting as long as 48 hr (Figure 1C). Clb4 could first be detected at 5 hr and persisted longer than in wild-type cells, remaining detectable up to ∼12 hr (Figure 1C).
As in the CLB5CLB6 strain, the accumulation of Clb3 in clb5clb6 mutants was delayed relative to Clb1 and Clb4. Clb3 began to accumulate at ∼7 hr following induction of the meiotic program, reaching peak abundance by 8–9 hr before declining (Figure 1C). The delayed accumulation of Clb3 is likely due to the translational regulation that prevents Clb3 accumulation until the completion of the first meiotic chromosome division (Carlile and Amon 2008). No DNA replication was observed in the clb5clb6 mutant strain, indicating that although Clb1, Clb3, and Clb4 accumulated, in some cases to levels greater than observed in wild-type cells, it was insufficient to promote premeiotic DNA replication (Figure 1D). If DNA replication is not initiated on time, cells can bypass the DNA replication checkpoint and attempt to divide their unreplicated chromosomes, resulting in a dramatic loss of viability at the time that meiosis I would normally occur (Stuart and Wittenberg 1998). Clb1, Clb3, and Clb4 accumulate at about the time cells would normally initiate meiosis I; thus the critical window of opportunity during which DNA replication can initiate may be missed. The delay in Clb3 accumulation may be particularly relevant considering that Clb3 is the cyclin that is most capable of substituting for Clb5 during mitotic proliferation (Hu and Aparicio 2005). Thus, the delay in Clb3 accumulation in a clb5clb6 strain essentially results in the loss of Clb5, Clb6, and Clb3 activities during the relevant window of opportunity 2–6 hr after meiotic entry when premeiotic DNA replication is normally initiated. Both Clb3 and Clb5 accumulate in the nucleus and Clb3 can contribute to activating DNA replication in proliferating cells (Bailly et al. 2003; Hu and Aparicio 2005). Thus, we considered the possibility that Clb3 accumulation in clb5clb6 diploid strains may be particularly relevant and the potential for Clb3 to promote premeiotic DNA replication was further investigated.
Expression of either CLB1 or CLB3 under regulation of the CLB5 promoter cannot induce premeiotic S phase
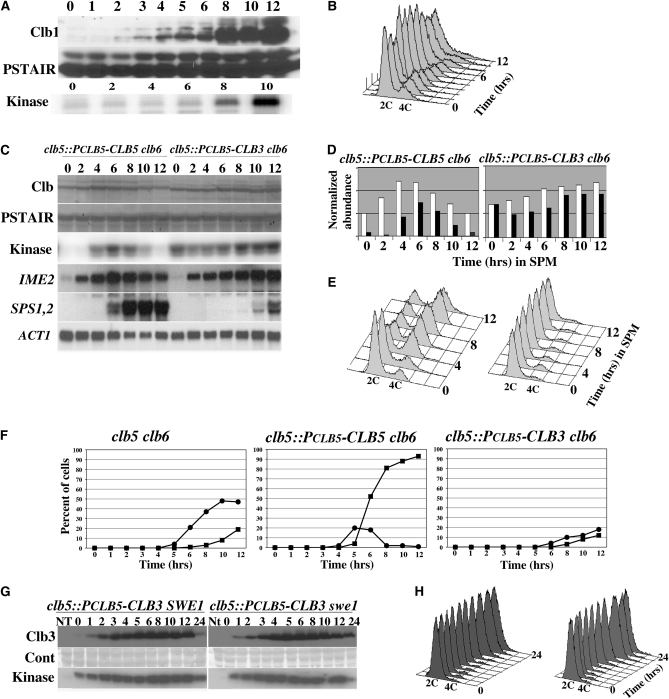
To determine whether other B-type cyclins were capable of promoting premeiotic DNA replication if expressed early in the meiotic program, we generated clb5clb6 strains that expressed either CLB1 or CLB3 under the regulation of the CLB5 promoter. In clb5clb6 cells harboring PCLB5–CLB1 the Clb1 protein accumulated poorly at early times following the induction of the meiotic program but displayed strong accumulation during the middle phase of meiosis between 8 and 12 hr (Figure 2A). Despite the accumulation of Clb1 and its associated kinase activity, the cells were unable to initiate premeiotic DNA replication (Figure 2B). In the clb5::PCLB5-CLB3clb6 strain, Clb3 began to accumulate 2 hr subsequent to the induction of sporulation (Figure 2C). The Clb3 produced in these cells displayed associated histone H1 kinase activity that was approximately proportional to the abundance of Clb3 (Figure 2C). Quantitative comparison of the kinase activity associated with Clb5 or Clb3 regulated by the CLB5 promoter demonstrated that Clb3 had relatively greater associated H1 kinase activity than Clb5 at 2 and 4 hr following the induction of sporulation (Figure 2D; shaded bars). The elevated kinase activity associated with Clb3 relative to its protein abundance at early times in the meiotic program suggested that Clb3–Cdk1 complexes might have been less susceptible to inhibition by Sic1 than were Clb5–Cdk1 complexes (Figure 2D; compare open and shaded bars). This was not investigated further but an increased sensitivity of Clb5 to Sic1 inhibition would not be surprising, considering the central role of Clb5 in regulating S-phase entry. Despite the early accumulation of Clb3 and its associated kinase activity, the clb5::PCLB5-CLB3clb6 strain displayed no premeiotic DNA replication, while the clb5::PCLB5-CLB5clb6 strain effectively replicated its chromosomal DNA with timing similar to a wild-type diploid (Figure 2E).
Figure 2 .
CLB3 expressed under the regulation of the CLB5 promoter does not induce premeiotic DNA replication. (A) Western blots displaying the accumulation of Clb1 (top), PSTAIR reactive Cdk1 (middle), and Histone H1 kinase (bottom) in clb5::PCLB5-CLB1 clb6 diploids at the indicated times following induction of sporulation. (B) Flow cytometry histogram displaying the DNA content in clb5::PCLB5-CLB1 clb6 cells at the indicated times following the induction of sporulation. (C) Western blots displaying the accumulation of Clb5, clb5::PCLB5-CLB5 clb6 (lanes 1–7) or Clb3, clb5::PCLB5-CLB3 clb6 (lanes 8–14) at the indicated times following the induction of sporulation; PSTAIR reactive Cdk1 was used as a loading control. Kinase activity associated with the indicated Clb protein was determined using histone H1 as a substrate. At each time point, samples were analyzed by Northern blot for the expression of the indicated meiotic genes. The actin transcript (ACT1) served as a loading control. (D) Protein and kinase data from A were quantified as described in Materials and Methods. Open bars represent relative protein abundance and closed bars represent relative histone H1 kinase activity. (E) Flow cytometry histograms displaying DNA content at the indicated times following the induction of sporulation. (F) MI and MII meiotic chromosome divisions based upon the appearance of two (solid circles) or greater than two (solid squares) chromatin masses. (G) Clb3 protein and associated histone HI kinase activity in SWE1 and swe1 strains detected by Western blot and autoradiography, respectively. A strip of each membrane used for the Western blots was stained with amido black as a loading control (cont). (H) Corresponding DNA content of the SWE1 and swe1 strains was determined by flow cytometry.
Ectopically expressed CLB3 could not induce premeiotic DNA replication; however, it did influence meiotic progression. Both clb5::PCLB5-CLB5clb6 and clb5::PCLB5-CLB3clb6 strains displayed induction of the early meiosis-specific gene IME2 at 2 hr, indicating that they had initiated the meiotic program on time (Figure 2C). However, while the strain producing Clb5 effectively induced the middle sporulation genes SPS1 and SPS2, the clb5::PCLB5-CLB3clb6 strain displayed a profound delay and reduction in the expression of SPS1 and SPS2 (Figure 2C). Delayed induction of the middle sporulation genes in this strain partly reflects the need for Cdk activity to eliminate repression of these genes by Sum1 (Shin et al. 2010). This also suggests that Clb3 is not as effective as Clb5 at eliminating Sum1 repression and is consistent with delayed progression through meiosis associated with premature accumulation of Clb3 (Carlile and Amon 2008). To determine whether the premature accumulation of Clb3 influenced other aspects of progression through the meiotic program we monitored chromosome divisions in clb5clb6, clb5::PCLB5-CLB5clb6, and clb5::PCLB5-CLB3clb6 strains. Eighteen percent of clb5clb6 cells displayed four chromatin masses after 12 hr, consistent with the completion of MII (Figure 2F). The PCLB5–CLB5 effectively drove progression through meiosis and >90% of these cells displayed four chromatin masses (Figure 2F). In contrast, PCLB5–CLB3 was ineffective in promoting progression through meiotic divisions with only 12% of the cells completing an MII division (Figure 2F). Additionally, we observed that while clb5::PCLB5-CLB5clb6 strains were effective in promoting meiotic recombination, the clb5::PCLB5-CLB3clb6 strain displayed a profound defect in recombination (supporting information, Figure S1).
The inability of Clb3 accumulating early in meiosis to promote premeiotic DNA replication in a clb5clb6 diploid suggested that there may be significant differences in specificity between Clb5 and Clb3 that affect their ability to promote DNA replication. However, B-type cyclin–Cdk complexes can be subject to inhibition by the protein kinase Swe1, and this reduces the ability of Clb3 to promote entry into S phase in proliferating cells (Hu and Aparicio 2005). To determine whether the inability of Clb3 to induce premeiotic DNA replication was due to inhibition by Swe1, we deleted SWE1 in clb5::PCLB5-CLB3clb6 diploids and induced the cells to sporulate. Clb3 accumulated with similar levels of associated histone H1 kinase activity in SWE1 and swe1 diploid strains but was unable to induce premeiotic DNA replication in either strain (Figure 2, G and H). Thus Clb3–Cdk1 inhibition by Swe1 does not prevent Clb3 from triggering DNA replication in clb5clb6 mutant strains.
The hydrophobic patch region of Clb5 is necessary for the induction of premeiotic DNA replication
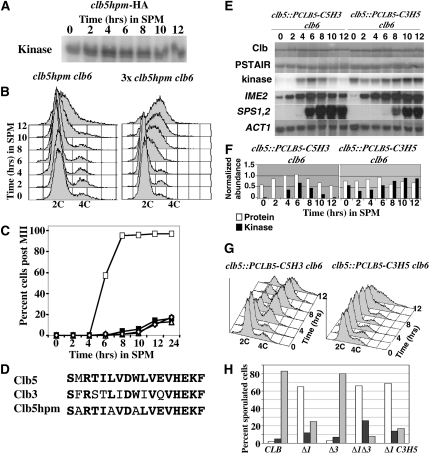
The inability of ectopically produced Clb3 to induce premeiotic DNA replication suggested that Clb5-associated kinase might phosphorylate some substrate that Clb3 recognizes poorly or not at all. Amino acid residues that make up an HP region on the surface of Clb5 confer increased affinity for substrates that possess RXL motifs and thus provide a degree of substrate specificity (Cross et al. 1999; Loog and Morgan 2005). The HP motif is required for Clb5 to promote effective DNA replication during mitotic proliferation. To determine whether the hydrophobic patch of Clb5 is essential for its ability to induce premeiotic DNA replication, the HP residues M197, L201, and W204 were changed to alanine (referred to as Clb5hpm) (Figure 3D). Although Clb5hpm displayed associated histone H1 kinase activity, the mutant was unable to promote premeiotic DNA replication (Figure 3, A and B). Modest overproduction of Clb5hpm (three copies total in a diploid clb5clb6 strain) was able to induce delayed premeiotic DNA replication, suggesting that the mutations reduced but did not completely ablate meiosis-specific functions of Clb5. Although overexpression of clbhpm could induce DNA replication, no meiotic chromosome divisions were observed in this strain (Figure 3, B and C).
Figure 3 .
Clb5 hydrophobic patch facilitates premeiotic S phase and progression through meiosis. (A) Autoradiogram displaying histone H1 kinase associated with Clb5hpm at the indicated time points during sporulation. (B) Flow cytometry histograms displaying the DNA content at the indicated times during sporulation of clb5 clb6 cells harboring clb5hpm (left) or three copies of clb5hpm (right). (C) Percentage of cells in the population that completed the M1 or MII division at the indicated time points during sporulation CLB5 CLB6 (open squares) clb5 clb6 (solid squares), clb5 clb6 URA3::clb5hpm (open diamonds), clb5 clb6 with three copies of clb5hpm (open triangles). (D) Amino acid sequences of the Clb5, Clb3, and Clb5hpm HP motifs. Conserved residues are indicated in boldface type. (E) Western blots detecting Clb5 in which the Clb5 hydrophobic patch was replaced with residues from the Clb3 HP (C5H3, top lanes 1–7), or Clb3 containing the hydrophobic patch sequence from Clb5 (C3H5, top lanes 8–14). Cdk1 (PSTAIR) was monitored as a loading control. The histone H1 kinase associated with each cyclin at the indicated times during sporulation was monitored by IP-kinase assay and autoradiography (kinase). At the indicated time points, samples of RNA from each strain were probed by Northern blot for the abundance of the meiosis-specific transcripts for IME2, SPS1, and SPS2; the ACT1 transcript served as a loading control (bottom three panels). (F) Protein and kinase data from E were quantified as described in Material and Methods. Open bars represent relative protein abundance and solid bars represent relative histone H1 kinase activity. (G) Flow cytometry histograms displaying DNA content at the indicated times following the induction of sporulation. (H) Spores per ascus produced during sporulation in wild-type diploids (CLB), clb1Δ mutants (Δ1), clb3Δ mutants (Δ3), clb1Δ clb3Δ mutants (Δ1Δ3), and clb1Δ clb3::PCLB3-C3H5 (Δ1 C3H5) strains. Open bars, monad; solid bars, dyad; shaded bars, tetrad.
The Clb5 hydrophobic patch alone is not sufficient to provide Clb5-specific functions
The loss of function imposed by the hydrophobic patch mutations indicates that the HP is necessary for Clb5’s meiotic functions but it does not confirm that the hydrophobic patch alone is sufficient to determine the specificity required to trigger premeiotic DNA replication. Clb5 can promote premeiotic S phase while Clb3 cannot, and it is possible that this could be due to differences between the HP motif sequences of these cyclins (Figure 3D). To test whether the HP motif is sufficient to provide the specificity required for premeiotic S phase, we performed a domain exchange. Residues of the Clb5 HP were replaced with the analogous residues from Clb3 (C5H3) and the HP residues of Clb3 were replaced with those of Clb5 (C3H5). If the HP residues alone confer substrate specificity to each cyclin, then we anticipated that this motif exchange would reduce or eliminate the ability of Clb5 to promote premeiotic DNA replication. Reciprocally, we hypothesized that this exchange might confer upon Clb3 the ability to promote premeiotic DNA replication. Both C3H5 and C5H3 proteins accumulated with similar timing in cells induced to initiate the meiotic program. The C5H3 protein reached a slightly higher abundance but C5H3 and C3H5 displayed nearly identical associated histone H1 kinase activities at 4 hr, indicating that they could effectively bind to and activate Cdk1 (Figure 3, E and F). The C3H5 allele displayed the ability to rescue mitotic growth of a clb3 clb4clb5clb6 strain, indicating that the changes to the HP had not inactivated the mitotic functions of CLB3 (Figure 4B). Similarly, C3H5 under the regulation of the CLB5 promoter was able to partially rescue the S-phase defect displayed by mitotically proliferating clb5clb6 mutants (Figure 4D). Very few CLB3-specific functions have been identified. However, CLB3 does display a key role in promoting the MII division in meiosis (Carlile and Amon 2008). As a result, clb1clb3 diploids accumulate dyad asci during meiosis (Grandin and Reed 1993; Dahmann and Futcher 1995). We observed that the C3H5 allele under the regulation of the CLB3 promoter was able to largely rescue the accumulation of dyad asci in a clb1 C3H5 strain relative to a clb1clb3 strain, implying that this gene retained its meiotic CLB3-specific function (Figure 3H).
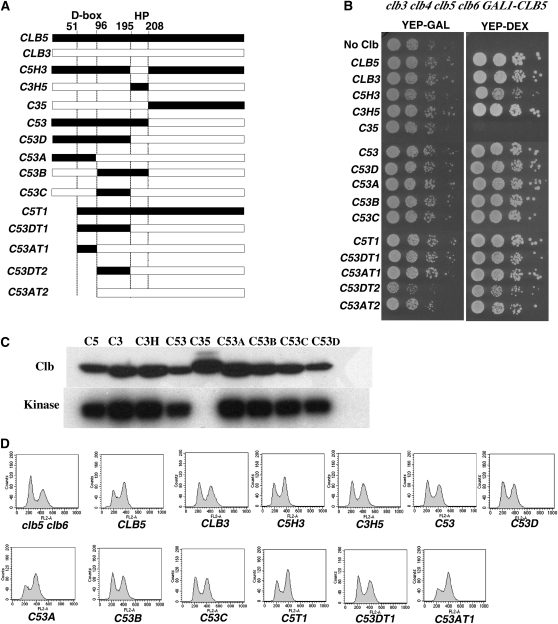
Figure 4 .
Clb5 and Clb3 chimeric cyclins are functional during mitotic proliferation. (A) Schematic representation of the chimeric and truncated CLB3 and CLB5 genes that were installed at the CLB5 locus and tested for function in a clb5 clb6 parental diploid. The relative positions of the destruction box motif (D box) and the hydrophobic patch motif (HP) are indicated. The numbers indicate the amino acid residues of Clb5. (B) Spot dilution assay testing the ability of chimeric cyclins to rescue proliferation in a clb3 clb4 clb5 clb6 GAL1-CLB5 strain, YEPGAL (GAL1–CLB5 on), and YEPDEX (GAL1–CLB5 off). Cells were spotted on the agar in 1:10 dilutions. (C, top) Western blot to determine the abundance of the indicated Clb proteins in whole cell extracts. (Bottom) Histone H1 kinase associated with the indicated Clb proteins. (D) Asynchronous flow cytometry profiles for the indicated strains grown to midlog phase in YEPD. All the strains are clb6 and the indicated cyclin replaced the endogenous CLB5 open reading frame.
The early meiosis-specific gene IME2 was effectively induced in both clb5::PCLB5-C5H3 clb6 and clb5:: PCLB5-C3H5 clb6 strains, indicating that these strains entered the meiotic program on time. Expression of the middle genes SPS1 and SPS2 was greatly delayed in clb5::PCLB5-C3H5 clb6 (Figure 3E). While Clb5 with a Clb3 HP domain (C5H3) effectively triggered DNA replication, which was completed by 8 hr, no significant DNA replication could be detected in the strain producing Clb3 with a Clb5 HP (C3H5) (Figure 3G). Although some symmetric peak broadening was observed at late times, there was no significant accumulation of 4C DNA content. This symmetric peak broadening has been investigated elsewhere and may represent nonnuclear staining that occurs during late meiosis (Raithatha and Stuart 2009). Although this peak broadening may mask a limited degree DNA replication, it consistently occurred only in the late stages of meiosis.
When the clb5::PCLB5-C5H3 and clb5::PCLB5-C3H5 strains are compared to clb5::PCLB5-CLB5 and clb5::PCLB5-CLB3 strains (Figure 2C) it is apparent that the Clb5-specific HP residues have little significant impact on the overall meiosis-specific activity of the cyclins. Thus, although the Clb5 hydrophobic patch residues are necessary for the initiation of premeiotic DNA replication, they are not sufficient as independent determinants to confer the ability to induce premeiotic S phase. These observations therefore suggest that a region of Clb5 other than the HP contributes to its specific functions in meiosis.
Chimeric Clb5–Clb3 cyclins can support mitotic DNA replication
To test whether the ability to effectively induce premeiotic DNA replication could be attributed to a specific region of Clb5, we extended our domain exchange strategy. The S. cerevisiae B-type cyclins are organized into two functional domains, a carboxyl terminus that binds to and activates Cdk1 and an amino-terminal domain that harbors a protein stability determinant. In the case of Clb3 and Clb5, a destruction box sequence (D box) is present in the amino terminus that targets these proteins for regulated destruction by the anaphase promoting complex (APC). DNA fragments encoding different regions of CLB5 (which can promote premeiotic S phase) were exchanged with the analogous regions of CLB3 (a cyclin that cannot promote premeiotic S phase). The resulting recombinant open reading frames were placed under the regulation of the CLB5 promoter and installed at the endogenous CLB5 locus (Figure 4A).
The chimeric cyclins were tested for function in mitotically proliferating cells by assaying their ability to support the growth of a clb3clb4clb5clb6 PGAL1-CLB5 strain. These strains were propagated in galactose containing medium to maintain expression of the PGAL1–CLB5 but were plated on dextrose-containing medium to extinguish PGAL1–CLB5 expression and test the ability of the chimeric cyclins to support mitotic proliferation. All of the cyclins were functional in this assay with the exception of C35, a fusion of the Clb3 amino terminus to the Clb5 Cdk binding domain (Figure 4B). This chimera was found to be largely insoluble with no associated histone H1 kinase activity (Figure 4C). These chimeric cyclins are therefore biologically active and provide a physiologically relevant catalytic activity. Second, asynchronous cultures of cells expressing the chimeric cyclins were assayed by flow cytometry to determine the effectiveness of each cyclin in promoting S phase during mitotic proliferation. Relative to a clb5clb6 strain, CLB5clb6 displayed a smaller 2C DNA peak and a larger 4C DNA peak reflecting efficient entry into S phase (Figure 4D). CLB3 driven by the CLB5 promoter was incapable of fully suppressing the DNA replication defect in a clb5clb6 strain during mitotic proliferation, the asynchronous clb5::PCLB5-CLB3clb6 strain displayed a higher proportion of 4C cells than the clb5clb6 strain but less than the CLB5clb6 strain in agreement with previous studies demonstrating that CLB3 expression from the CLB5 locus is not fully effective in replacing CLB5 (Hu and Aparicio 2005). Interestingly, the majority of the strains displayed similar asynchronous DNA replication profiles to the clb5::PCLB5-CLB3clb6 strain, with slightly larger G1 peaks in comparison to their G2 peaks. Thus, the majority of the chimeric cyclins were similarly effective at promoting DNA replication during mitotic proliferation.
The amino terminus of Clb5 confers upon Clb3 the ability to induce premeiotic DNA replication
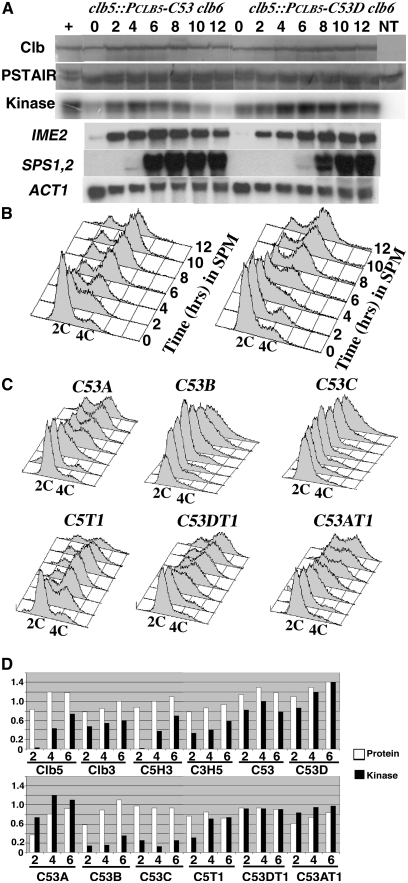
Clb5 can effectively induce premeiotic DNA replication while Clb3 cannot. If a specific region of Clb5 were responsible for its meiosis-specific function, we reasoned that replacing part of Clb3 with the appropriate region of Clb5 might confer meiosis-specific activity to the chimera. When diploid cells expressing the chimeric cyclins were induced to initiate meiotic differentiation, a subset of the strains could complete the program and form spores. Fusion of the Clb5 amino terminus to the Clb3 carboxyl terminus generated a chimera (C53) that promoted premeiotic DNA replication and sporulation with frequency and timing similar to Clb5 (Table 2, Figure 5, A and B). SPS1 and SPS2 were induced with similar timing in cells expressing CLB5 or the CLB5–CLB5 chimera, suggesting that Clb5 amino terminus conferred CLB3 with the ability to effectively eliminate Sum1-mediated repression of the middle sporulation genes. We did not directly measure DNA double strand break formation or recombination; however, the high viability of the clb5::PCLB5-CLB-CLB3clb6 spore progeny implies that the chimeric cyclin is also capable of performing the meiotic recombination functions and assembly of synaptonemal complex functions attributed to CLB5 (Table 2).
Table 2 . The ability of cyclins and chimeric cyclins to promote meiosis and sporulation.
| Cyclin | Protein (4 hr) | H1 kinase (4 hr) | % S phase (8 hr) | % MI + MII (24 hr) | % Asci (24 hr) | % Viable spores |
|---|---|---|---|---|---|---|
| CLB5 | 1.2 | 0.43 | 90 | 92.5 | 80.5 | 98.3 |
| CLB3 | 0.85 | 0.54 | 0 | 23 | 0 | 0a |
| C5H3 | 1.0 | 0.38 | 80 | 87 | 76 | 97.5 |
| C3H5 | 0.86 | 0.40 | 0 | 18.5 | 0 | 0a |
| C35 | — | — | 0 | 16.5 | 0 | ND |
| C53 | 1.3 | 1.0 | 87 | 91 | 75.5 | 87.5 |
| C53D | 1.3 | 1.2 | 85 | 75 | 48 | 83.3 |
| C53A | 0.81 | 1.2 | 60 | 40 | 18 | 49a |
| C53B | 0.89 | 0.15 | 0 | 25.5 | 0 | 0a |
| C53C | 0.94 | 0.12 | 0 | 10 | 0 | 0a |
| C5T1 | 0.85 | 0.71 | 85 | 84 | 80.5 | 89.2 |
| C53DT1 | 0.94 | 0.92 | 70 | 40 | 19 | 23.6a |
| C53AT1 | 0.74 | 0.95 | 20 | 35 | 5 | 18a |
| C53DT2 | 2.8 | 0.80 | 23 | 40 | 0 | 0a |
| C53AT2 | — | — | 30 | 28 | 5 | <1a |
All the cyclins and chimeric cyclins were installed at the clb5 locus in clb5 clb6 strains. Protein indicates the relative abundance of the cyclin protein as determined by Western blot. The protein and kinase values are expressed relative to a CLB5 clb6 strain that was used for comparison. See Material and Methods for a detailed explanation of the quantitation procedures. ND, not done. Protein and kinase data for C53AT2 are not included because of instability of the strain and lack of reproducibility.
Spore viability was determined by tetrad dissection (n = 120 spores) or random spores analysis based upon plating for 500 spores.
Figure 5 .
The Clb5 amino terminus confers the ability to induce premeiotic DNA replication on Clb3. (A, top) Accumulation of C53 in clb5::PCLB5-C53 clb6 (lanes 2–8) and C53D in clb5::PCLB5-C53D clb6 (lanes 9–16) was monitored by Western blot at the indicated times subsequent to the induction of sporulation. A CLB5 clb6 strain (+, lane 1) and an untagged CLB5 clb6 (NT, lane 16) were included for reference. The abundance of Cdk1 (PSTAIR) was monitored as a loading control. The histone H1 kinase associated with each cyclin and the control is displayed in an autoradiogram (kinase). RNA harvested at each time point was assayed by Northern blot to monitor expression of the indicated meiosis-specific genes. The ACT1 transcript served as a loading control. (B) DNA content of the strains at each time point was monitored by flow cytometry. Peaks representing 2C and 4C DNA contents are indicated. (C) Flow cytometry plots for diploid clb5 clb6 strains expressing the indicated chimeric cyclins. Samples were taken at 2-hr intervals subsequent to inducing the cells to initiate sporulation. (D) Relative cyclin protein abundance (open bars) and associated histone H1 kinase (solid bars) at the indicated times (hr) following the induction of sporulation in the indicated strains.
To determine whether a smaller Clb5 amino-terminal domain conferring meiosis-specific activity could be localized, further domain exchanges were performed. Fusion of 198 amino acids from Clb5 to the Clb3 HP and Cdk binding domain produced a fusion protein (C53D) that was capable of promoting premeiotic DNA replication but displayed a reduced sporulation frequency (45% in contrast to 80% for C53) (Table 2, Figure 5A). Thus, although exchanging the Clb5 HP with the Clb3 HP in the native Clb5 and Clb3 proteins was not associated with any significant changes in the ability to promote premeiotic DNA replication, the Clb5 HP region appeared to play a more important role in the context of the chimeric cyclins.
Addition of an even smaller fragment of the Clb5 amino terminus (amino acids 1–95) to Clb3 resulted in a fusion protein (C53A) that still displayed the ability to promote premeiotic S phase, with 60% of the cells displaying a 4C DNA content after 8 hr (Figure 5C). However, clb5::PCLB5-C53A clb6 diploid cells displayed a reduced ability to sporulate and only 18% of the cells formed asci (Table 2). Replacing Clb3 sequences between amino acids 96 and 198 or 96 and 211 (which included the HP region) with the corresponding sequences from Clb5 yielded fusion proteins (C53C and C53B) with associated histone H1 kinase activity (Figure 4C and Table 2). However, these fusions were unable to rescue premeiotic DNA replication or sporulation in a clb5clb6 mutant (Figure 5C, Table 2).
Since the amino-terminal region of Clb5 was sufficient to confer upon Clb3 the ability to promote premeiotic S phase, it was anticipated that deleting these residues would reduce the ability of these cyclins to trigger premeiotic DNA replication. Deletion of amino acids 1–50 from the Clb5–Clb3 fusion (C53DT1) resulted in a relatively small reduction in the ability to induce premeiotic DNA replication (Figure 5C) but displayed significantly reduced asci formation, with only 19% forming asci after 24 hr (Table 2). In contrast, when amino acids 1–50 were deleted from the full-length Clb5 (C5T1), there was little effect on either premeiotic DNA replication or sporulation efficiency (Figure 5C, Table 2). Thus, the first 50 amino acids appear to make a minor contribution to the ability to promote premeiotic DNA replication within the context of the full-length Clb5 but are more significant in the context of the chimeric cyclins that carry a smaller portion of the Clb5 amino terminus.
These results collectively indicate that the Clb5 amino terminus confers the specificity to promote premeiotic DNA replication on the Clb3 protein. Although the specificity conferred by the individual regions of the Clb5 amino terminus and the Clb5 HP region appears to be dispensable when removed individually from the full-length Clb5 protein, the cumulative contributions of each of these regions becomes apparent when examined in the context of the chimeric cyclins.
The Clb5 amino-terminal region containing the D box confers a limited ability to promote premeiotic DNA replication and is important for maintaining genetic stability
Since the Clb5 amino terminus could confer meiosis-specific function on Clb3, we anticipated that deletion of the amino terminus would reduce Clb5 function. We attempted to test the meiotic function of Clb5 lacking its amino-terminal 96 amino acids (which includes the destruction box sequence); however, despite repeated attempts, we were unable to recover viable strains that had correctly installed this truncation at the CLB5 locus. Clb5 lacking amino acids 1–95 was previously found to be lethal to mitotically proliferating cells (Sullivan et al. 2008), and we interpret our inability to construct this strain to reflect the toxicity imposed by stabilized Clb5. We were able to construct strains harboring a Clb5–Clb3 fusion lacking amino acid residues 1–96 (C53DT2, see Figure 4A). Compared to the full-length Clb5–Clb3 fusion (C53), truncation of the amino terminus yielded a chimeric cyclin with a limited ability to promote premeiotic S phase, and this strain formed no asci (Table 2). Thus in the context of the Clb5–Clb3 fusion the Clb5 amino-terminal 96 residues are required to promote premeiotic DNA replication.
The observation that replacement of Clb3 amino-terminal sequences with the analogous sequences from Clb5 created a chimeric cyclin that was capable of supporting premeiotic S phase, meiosis, and sporulation suggests that the Clb5 amino terminus conferred function to Clb3 (Figure 5A). However, it was also possible that the Clb3 amino terminus prevented Clb3 from triggering premeiotic DNA replication and the chimeric cyclins were active due to loss of an inhibitory domain. To test this possibility, sequence encoding the amino-terminal 101 residues (including the destruction box) was deleted from CLB3 (C53AT2, see Figure 4A). This truncated cyclin accumulated to high levels and had associated histone H1 kinase activity (Table 2); however, its expression caused a mild growth defect as indicated by a small reduction in plating efficiency (Figure 4B). In contrast to full-length Clb3, the truncated Clb3 was capable of inducing a modest degree of premeiotic DNA replication in clb5clb6 diploids (Table 2). Three independent clb5::PCLB5-C53AT2 clb6 diploids were tested and they displayed variability in the percentage of cells that could form asci, 3–8%, but all three displayed a similar degree of DNA replication. The low frequency of sporulation may reflect chromosomal abnormalities induced by the stabilized cyclin during mitotic proliferation since clb5::PCLB5-C53AT2 clb6 diploids displayed a significant elevation in the frequency of illegal mating, an assay in which the diploid clb5::PCLB5-C53AT2 clb6 was challenged to mate with a haploid tester strain (3.4 × 10−5 ± 2.3 × 10−5 compared to 5.1 × 10−8 ± 1.9 × 10−8 for clb5clb6 diploids, P = 0.025 based on Student’s t test). The ability of the diploids to mate implies loss of or damage to one copy of chromosome III. Thus, although the Clb5 amino terminus is normally required to provide the specificity required to promote premeiotic DNA replication this requirement can be overcome when destruction-box–mediated regulation is disrupted with consequent hyperaccumulation of the cyclin. This modest gain of function comes at the cost of inducing genetic instability.
The ability of chimeric Clb5–Clb3 cyclins to promote premeiotic S phase is not related to overall levels of associated kinase activity
To investigate the possibility that the differences in effectiveness of the various chimeric cyclins was due to increased or decreased levels of associated kinase activity, the protein abundance and kinase activity were quantitated during the early stages of the meiotic program. The Clb5-associated kinase activity measured from 2 to 6 hr subsequent to the induction of sporulation likely represents the optimal level of kinase activity for activation of premeiotic DNA replication (Stuart and Wittenberg 1998). The cyclins most effective at promoting premeiotic S phase (Clb5 and C5H3) have equal or less associated kinase activity than cyclins with reduced physiological effectiveness for this function (CLB3, C53, C53D, and C53A) (Figure 5D, and Table 2). Thus, with regard to promoting progression through the meiotic program, the overall kinase activity associated with a cyclin may be less relevant than a specific function that is conferred by the Clb5 amino-terminal region.
Discussion
The results of this investigation suggest that Clb5 endows Cdk1 with the specificity required for it to promote premeiotic DNA replication and that other cyclins, at least Clb1 and Clb3, lack this ability. The idea that premeiotic DNA replication requires a highly specific cyclin activity is in contrast with suggestions that any B-type cyclin has the ability to initiate DNA replication if expressed at the correct time and if Cdk1 is not inhibited by Swe1 (Hu and Aparicio 2005). CLB1 alone, if sufficiently overexpressed, can drive complete mitotic cell cycles in clb2clb3clb4clb5clb6 cells; however, the morphology of the cells suggests that progression through the cell cycle is significantly perturbed by this manipulation (Haase et al. 2001). An even more striking demonstration of the ability of a single cyclin to drive mitotic proliferation is that a Cdc13–Cdc2 fusion protein can drive a relatively normal cell cycle in S. pombe in the absence of the other cyclins cig1+, cig2+, and puc1+ (Coudreuse and Nurse 2010). However, it is notable that in contrast to cells expressing the Cdc13–Cdc2 fusion, cig1 cig2 mutants expressing a native Cdc13 protein display a delayed S phase (Fisher and Nurse 1996). The observation that all eukaryotic cells express multiple cyclins suggests that no single cyclin is capable of promoting optimal cell cycle progression under all conditions. Multiple cyclins with different substrate preferences may be advantageous to allow responsiveness to changing environmental conditions and the demands of cellular differentiation and development. Thus, while a single cyclin can drive mitotic proliferation, the cell cycle it promotes is perturbed relative to that which is driven by the full complement of endogenous cyclins. Additionally, overexpression of either CLB1 or CLB5 or inactivation of SWE1 can promote DNA rereplication during meiosis also suggested that cyclin specificity may be a secondary issue in meiosis (Strich et al. 2004; Rice et al. 2005). However, rereplication is not a normal aspect of meiosis and may be regulated significantly differently from normal premeiotic DNA replication. Additionally, there is no evidence to suggest that CLB1 can promote any other aspects of meiosis beyond its normal function in the meiotic chromosome divisions.
One explanation for the inability of CLB1 or CLB3 to promote premeiotic DNA replication when expressed under regulation of the CLB5 promoter, even when combined with swe1 deletion, is that they may be less effective than Clb5 at interacting with substrates required for premeiotic DNA replication. While some Cdk1 substrates are effectively phosphorylated by either Clb3–Cdk1 or Clb5–Cdk1, a subset of protein substrates displays very strong preference for either Clb3 or Clb5 (Koivomagi et al. 2011). Additionally, both two-hybrid and proteomic screens have identified distinct arrays of interacting proteins for Clb3 and Clb5 with surprisingly little overlap (Ito et al. 2001; Archambault et al. 2004). This strong substrate preference is likely determined by docking domains within both the cyclins and the substrates (Bhaduri and Pryciak 2011; Koivomagi et al. 2011). Alternatively it is possible that subcellular localization influences the effectiveness of cyclins in activating premeiotic S phase. We think this is unlikely since both Clb5 and Clb3 localize to the nucleus but we cannot formally exclude the possibility that these two cyclins experience differential localization within the nucleus that excludes Clb3 from the DNA replication machinery. Our results are consistent with the notion that the failure of Clb1 and Clb3 to promote premeiotic DNA replication can be accounted for by the different substrate specificities they confer upon Cdk1. It is possible that if sufficiently overexpressed, these cyclins might be able to activate premeiotic S phase. Indeed, it may be the combined accumulation of Clb1–Clb4 activity is what drives S phase in mitotically proliferating clb5clb6 mutants. This idea is congruent with the observation that neither Clb2 nor Clb3 is able to fully rescue the S-phase delay displayed by mitotically proliferating clb5clb6 mutants (Hu and Aparicio 2005).
Premature accumulation of Clb3–Cdk1 is not sufficient to promote premeiotic S phase; however, it does interfere with homolog disjunction leading to sister chromatid segregation in MI (Carlile and Amon 2008). Additionally, in clb5clb6 cells harboring PCLB5–CLB3, not only did CLB3 fail to induce premeiotic S phase, but meiotic recombination, chromosome divisions, and induction of middle sporulation genes were delayed and reduced beyond what was observed in clb5clb6 strains. Both meiotic recombination and many events associated with the middle phase of meiosis such as middle gene expression and the MI, MII chromosome divisions require or are regulated by B-type cyclin–Cdk1 activity (Hepworth et al. 1998; Henderson et al. 2006; Shin et al. 2010). Thus it is somewhat surprising that prematurely accumulating Clb3–Cdk1 seems to interfere with rather than accelerate these processes in clb5clb6 strains. It may be that prematurely accumulating Clb3–Cdk1 triggers the induction of a checkpoint that is usually bypassed in clb5clb6 cells owing to the lack of early Cdk activity (Stuart and Wittenberg 1998). Activation of a checkpoint such as the S/M checkpoint could potentially occur through direct phosphorylation of an inappropriate target or by Clb3–Cdk1 performing its normal function too early, leading to a premature attempt at some aspect of MII. Alternatively, it may be that progression through the meiotic program is dependent upon temporally regulated periods where Cdk1 activity is low followed by periods where it is high. Premature accumulation of Clb3–Cdk1 may disrupt that normal pattern and force the meiotic program into disarray preventing further progression and completion of the process.
The substrate specificity of some B-type cyclins is in part provided by the conserved hydrophobic patch motif (Brown et al. 1999; Cross et al. 1999; Koivomagi et al. 2011). Our results indicate that the hydrophobic patch alone is not the primary determinant of functional specificity in the context of premeiotic DNA replication. Exchanging the HP sequences between the Clb5 and Clb3 did not confer upon Clb3 the ability to effectively induce premeiotic DNA replication or reduce the ability of Clb5 to perform this function. These observations imply that Clb5 can recognize substrates through more than one specific mechanism although the HP clearly contributes to this function. This idea is further supported by the observation that overproduction of Clb5hpm, lacking an HP motif, confers the ability to promote premeiotic DNA replication, again arguing that other portions of the Clb5 protein contribute to its functional specificity.
The results we obtained from expressing chimeric cyclins suggest that the Clb5 amino terminus also plays a role in determining its functional specificity. This result is surprising as cyclin amino-terminal domains had previously been considered only in the context of conferring regulated protein instability and in some cases subcellular localization. Interestingly, the Clb5 amino-terminal domain does not independently confer an increased ability to promote DNA replication during mitotic proliferation, implying that its role is particularly important during meiotic differentiation.
Analysis of truncation mutants indicates that the entire Clb5 amino terminus is specialized for this role rather than simply containing a discrete targeting sequence. A graded loss of function in the ability to promote premeiotic S phase is observed when comparing a spectrum consisting of the native Clb5, C53, C53D, C53A, and Clb3, each of which contains a progressively smaller portion of the Clb5 amino terminus. The gradual decline in ability to promote premeiotic S phase among these cyclins is not, however, associated with a decline in kinase activity during early meiosis. In fact, the chimeric cyclins generally have associated kinase activity at least as great as native Clb5 and Clb3 during early meiosis. This suggests that overall kinase activity associated with the cyclin is less important than the specific activity conferred to the cyclin–CDK complex by the Clb5 amino terminus. This reinforces the observation that Clb5 has a higher specificity toward particular substrates, while Clb2 has a greater associated kinase activity per cyclin–CDK molecule (Loog and Morgan 2005). Thus, high, untargeted CDK activity is generally insufficient to promote premeiotic DNA replication, underscoring the importance of specific substrate targeting.
In silico structure modeling predicts that the amino-terminal regions of the B-type cyclins are disordered, which may explain their exclusion from all the reported crystal structures. Intrinsically disordered regions of proteins frequently serve functions for molecular recognition and in some cases disordered regions can assume a more stably folded structure upon binding to their targets (Tompa et al. 2009). This mechanism can allow an uncoupling of substrate recognition and binding affinity that could be important to allow proteins like cyclins to recognize and bind substrates with high affinity but then release the target once the disordered region assumes a folded structure. In addition to the apparently high degree of structural disorder, there is little sequence conservation among the amino-terminal regions of B-type cyclins. This sequence variability may support unique interactions with dramatically different arrays of substrates, thus conferring different specific functions to different cyclins.
Several distinct regions of Clb5 contribute to its meiosis-specific function. The replacement of the Clb5 HP region with that of Clb3 does not independently have any significant effect on the ability of Clb5 to promote premeiotic DNA replication nor does truncation of amino acids 1–50. However, within the context of a Clb5–Clb3 fusion protein, the chimera shows reduced function when the Clb5 HP is replaced with the Clb3 HP and when a 50-amino-acid truncation is introduced. Thus the full-length Clb5 protein has multiple specificity determinants in the amino terminus and HP region, each cumulatively contributing to the overall function of the protein without individually being essential. Thus it appears likely that cyclins may need to be extensively dissected to reveal the multiple determinants that may cumulatively contribute to their specific functions.
The importance of the Clb5 amino-terminal region as a specificity determinant may reflect a type of specialization unique to Clb5 or it may be a more widely used mechanism. Domain exchanges between human cyclins E and A2 indicate that the carboxyl-terminal regions of these two cyclins confer their substrate specificities (Horton and Templeton 1997). Nonetheless, it is intriguing to consider that there may be a higher potential for substrate specificity to develop among cyclin amino termini due to their extensive sequence variation. Since the amino-terminal regions of B-type cyclins are highly divergent and not required for interaction with the Cdk partner, any mutations occurring in this region that increased specificity for particular substrates could allow increased specialization of function. An increase in specialization has the advantage of making a cyclin more effective in its function; however, this would also demand an increase in the number of cyclins as observed in the cyclin gene duplications in budding yeast and the increased number of cyclins observed in metazoan cells.
Understanding how cyclins identify specific substrates is important to allow a complete understanding of how cell proliferation is regulated. This is of considerable interest when these concepts are extended to higher organisms. In particular, parallels have emerged between Clb5 and cyclin E, a mammalian cyclin that is involved in promoting the G1/S transition. Deletion of mouse cyclins E1 and E2 does not prevent cell cycle progression in most cell types but abnormalities are seen in spermatogenesis, trophoblast giant cells, and megakaryocytes (Geng et al. 2003). The reduced levels of spermatogenesis imply a meiosis-specific role for cyclin E, directly paralleling the meiosis-specific requirement observed with Clb5 in S. cerevisiae. Several types of amino terminally truncated cyclins have been identified as contributing factors in cancers (Van Dross et al. 2006). In many cases the effect of the mutations has been attributed to the increased stability of the cyclin. However, these mutations may also be altering the cyclin’s substrate specificity, leading to deregulation of the cyclin–Cdk complex function. Indeed, spliced variants of cyclin E with deletions in the amino terminus have been identified that alter substrate specificity without increasing stability of the protein (Porter and Keyomarsi 2000). Additionally, the inappropriate expression or activity of a single or a subset of cyclins contributes to the deregulated proliferation of some cancers, and identifying the domains used by those cyclins to direct Cdk activity to target proteins could allow the development of small molecule inhibitors to disrupt the interaction between the cyclin and the substrate.
Supplementary Material
Acknowledgments
We are grateful to Fred Cross and Etienne Schwob for the generous gift of strains used in this study and David Morgan and Michael Schultz for helpful discussions and critical reading of the manuscript. We acknowledge Xiao Dong Liu for technical assistance. This work was supported by an operating grant to D.T.S. from the Canadian Institutes for Health Research (MOP 81247).
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Albertini D. F., Carabatsos M. J., 1998. Comparative aspects of meiotic cell cycle control in mammals. J. Mol. Med. 76: 795–799 [DOI] [PubMed] [Google Scholar]
- Archambault V., Chang E. J., Drapkin B. J., Cross F. R., Chait B. T., et al. , 2004. Targeted proteomic study of the Cyclin-Cdk module. Mol. Cell 14: 699–711 [DOI] [PubMed] [Google Scholar]
- Archambault V., Buchler N. E., Wilmes G. M., Jacobson M. D., Cross F. R., 2005. Two-faced cyclins with eyes on the targets. Cell Cycle 4: 125–130 [DOI] [PubMed] [Google Scholar]
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M. N., 2003. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 116: 4119–4130 [DOI] [PubMed] [Google Scholar]
- Bhaduri S., Pryciak P. M., 2011. Cyclin-specific docking motifs promote phosphorylation of yeast signaling proteins by g1/s cdk complexes. Curr. Biol. 21: 1615–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. R., Noble M. E., Endicott J. A., Johnson L. N., 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1: 438–443 [DOI] [PubMed] [Google Scholar]
- Carlile T. M., Amon A., 2008. Meiosis I is established through division-specific translational control of a cyclin. Cell 133: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha R. S., Weiner B. M., Keeney S., Dekker J., Kleckner N., 2000. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11 and positively by Rec8. Genes Dev. 14: 493–503 [PMC free article] [PubMed] [Google Scholar]
- Chellappan S. P., Giordano A., Fisher P. B., 1998. Role of cyclin-dependent kinases and their inhibitors in cellular differentiation and development. Curr. Top. Microbiol. Immunol. 227: 57–103 [DOI] [PubMed] [Google Scholar]
- Coudreuse D., Nurse P., 2010. Driving the cell cycle with a minimal CDK control network. Nature 468: 1074–1079 [DOI] [PubMed] [Google Scholar]
- Cross F. R., Jacobson M., 2000. Conservation and function of a potential substrate-binding domain of the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 20: 4782–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Yuste-Rojas M., Gray S., Jacobson M., 1999. Specialization and targeting of B-type cyclins. Mol. Cell 4: 11–19 [DOI] [PubMed] [Google Scholar]
- Dahmann C., Futcher B., 1995. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics 140: 957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L., Goetsch L., Ammerer G., Byers B., 1998. Regulation of meiotic S-phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science 281: 1854–1857 [DOI] [PubMed] [Google Scholar]
- Donaldson A., 2000. Yeast mitotic cyclin CLB2 cannot substitute for S-phase cyclins in replication origin firing. EMBO Rep. 1: 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. D., Raghuraman M. K., Friedman K. L., Cross F. R., Brewer B. J., et al. , 1998. Clb5-dependent activation of late replication origins in Saccharomyces cerevisiae. Mol. Cell 2: 173–182 [DOI] [PubMed] [Google Scholar]
- Draviam V. M., Orrechia S., Lowe M., Pardi R., Pines J., 2001. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 152: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B. A., Lehner C. F., 1996. Developmental control of cell cycle regulators: a fly’s perspective. Science 274: 1646–1651 [DOI] [PubMed] [Google Scholar]
- Esposito M. S., Esposito R. E., 1974. Genetic recombination and committment to meiosis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 71: 3172–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D. L., Nurse P., 1996. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 15: 850–860 [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Yu Q., Sicinska E., Das M., Schneider J. E., et al. , 2003. Cyclin E ablation in the mouse. Cell 114: 431–443 [DOI] [PubMed] [Google Scholar]
- Grandin N., Reed S. I., 1993. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol. Cell. Biol. 13: 2113–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase S., Reed S., 1999. Evidence that a free running oscillator drives G1 events in the budding yeast cell cycle. Nature 401: 394–397 [DOI] [PubMed] [Google Scholar]
- Haase S. B., Winey M., Reed S. I., 2001. Multi-step control of spindle pole body duplication by cyclin dependent kinase. Nat. Cell Biol. 3: 38–42 [DOI] [PubMed] [Google Scholar]
- Henderson K. A., Kee K., Maleki S., Santini P. A., Keeney S., 2006. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth S., Friesen H., Segall J., 1998. NDT80 and the meiotic recombination checkpoint regulate expression of the middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 5750–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi H., Suzuki-Takahashi I., Taya Y., Segawa K., Nishimura S., et al. , 1995. Differences in substrate specificity between Cdk2-cyclin A and Cdk2-cyclin E in vitro. Biochem. Biophys. Res. Commun. 216: 520–525 [DOI] [PubMed] [Google Scholar]
- Horton L. E., Templeton D. J., 1997. The cyclin box and C-terminus of cyclins A and E specify CDK activation and substrate specificity. Oncogene 14: 491–498 [DOI] [PubMed] [Google Scholar]
- Horton R. M., Hunt M. D., Ho S. N., Pullen J. K., Pease L. R., 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Hu F., Aparicio O. M., 2005. Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102: 8910–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Gan Y., Aparicio O. M., 2008. Identification of Clb2 residues required for Swe1 regulation of Clb2-Cdc28 in Saccharomyces cerevisiae. Genetics 179: 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Yoshida M., 2001. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol. 19: S23–S27 [DOI] [PubMed] [Google Scholar]
- Joshi A. R., Jobanputra V., Lele K. M., Wolgemuth D. J., 2009. Distinct properties of cyclin-dependent kinase complexes containing cyclin A1 and cyclin A2. Biochem. Biophys. Res. Commun. 378: 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D. M., Harper J. W., Elledge S. J., 1999. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97: 431–434 [DOI] [PubMed] [Google Scholar]
- Koivomagi M., Valk E., Venta R., Iofik A., Lepiku M., et al. , 2011. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol. Cell 42: 610–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell, 1997 Meiosis and sporulation in Saccharomyces cerevisiae, pp. 889\x{2013}1036 in Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. Cell Cycle and Cell Biology, edited by J. R. Pringle, J. R. Broach, and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Lew D. J., Reed S. I., 1992. A proliferation of cyclins. Trends Cell Biol. 2: 77–81 [DOI] [PubMed] [Google Scholar]
- Loog M., Morgan D. O., 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434: 104–108 [DOI] [PubMed] [Google Scholar]
- Miller M. E., Cross F. R., 2001. Cyclin specificity: How many wheels do you need on a unicycle? J. Cell Sci. 114: 1811–1820 [DOI] [PubMed] [Google Scholar]
- Moore J. D., Kirk J. A., Hunt T., 2003. Unmasking the S-phase-promoting potential of cyclin B1. Science 300: 987–990 [DOI] [PubMed] [Google Scholar]
- Morgan D. O., 1997. Cyclin-dependent kinases: engines, clocks and microprocessors. Annu. Rev. Cell Dev. Biol. 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., 1996. At the heart of the budding yeast cell cycle. Trends Genet. 12: 405–412 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., 1994. Developmental modification of the Drosophila cell cycle. Trends Genet. 10: 321–327 [DOI] [PubMed] [Google Scholar]
- Pagliuca F. W., Collins M. O., Lichawska A., Zegerman P., Choudhary J. S., et al. , 2011. Quantitative proteomics reveals the basis for the biochemical specificity of the cell-cycle machinery. Mol. Cell 43: 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeper D. S., Parker L. L., Ewen M. E., Toebes M., Hall F. L., et al. , 1993. A- and B-type cyclins differentially modulate substrate specificity of cyclin-cdk complexes. EMBO J. 12: 1947–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T., 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 115: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. C., Keyomarsi K., 2000. Novel splice variants of cyclin E with altered substrate specificity. Nucleic Acids Res. 28: E101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raithatha S. A., Stuart D. T., 2005. Meiosis-specific regulation of the Saccharomyces cerevisiae S-phase cyclin CLB5 is dependent on MluI cell cycle box (MCB) elements in its promoter but is independent of MCB-binding factor. Genetics 169: 1329–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raithatha S. A., Stuart D. T., 2009. A comparison of fluorescent DNA binding dyes for flow cytometric analysis of sporulating Saccharomyces cerevisiae. J. Microbiol. Methods 78: 357–359 [DOI] [PubMed] [Google Scholar]
- Reed S. I., Wittenberg C., 1990. Mitotic role for the Cdc28 protein kinase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87: 5697–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L. M., Plakas C., Nickels J. T., Jr, 2005. Loss of meiotic rereplication block in Saccharomyces cerevisiae cells defective in Cdc28p regulation. Eukaryot. Cell 4: 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. M., 1999. Evolving ideas about cyclins. Cell 98: 129–132 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Heiter P., 1990. Methods in Yeast Genetics. A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schulman B. A., Lindstrom D. L., Harlow E., 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95: 10453–10458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E., Nasmyth K., 1993. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 7: 1160–1175 [DOI] [PubMed] [Google Scholar]
- Schwob E., Boehm T., Mendenhall M. D., Nasmyth K., 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1/S transition in Saccharomyces cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Sedgwick C., Rawluk M., DeCesare J., Raithatha S. A., Wohlschlegel J. A., et al. , 2006. Saccharomyces cerevisiae Ime2 phosphorylates Sic1 at multiple PXS/T sites but is insufficient to trigger Sic1 degradation. Biochem. J. 399: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., 1997. Yeast Genetics, pp. 302–325 The Encyclopedia of Molecular Biology and Molecular Medicine, edited by Meyers R. A. VCH Publishers, Weinheim, Germany [Google Scholar]
- Shin M. E., Skokotas A., Winter E., 2010. The Cdk1 and Ime2 protein kinases trigger exit from meiotic prophase in Saccharomyces cerevisiae by inhibiting the Sum1 transcriptional repressor. Mol. Cell. Biol. 30: 2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E. O., Byers B., 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., 1974. Are mitotic functions required in meiosis? Genetics 76: 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. N., Penker A., Ohta K., Klein F., Nicolas A., 2001. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 11: 88–97 [DOI] [PubMed] [Google Scholar]
- Sprague G. F., 1991. Assay of yeast mating reaction. Methods Enzymol. 194: 77–93 [DOI] [PubMed] [Google Scholar]
- Strich R., Mallory M. J., Jarnik M., Cooper K. F., 2004. Cyclin B-Cdk activity stimulates meiotic rereplication in budding yeast. Genetics 167: 1621–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D., Wittenberg C., 1998. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 12: 2698–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Holt L., Morgan D. O., 2008. Cyclin-specific control of ribosomal DNA segregation. Mol. Cell. Biol. 28: 5328–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Fuxreiter M., Oldfield C. J., Simon I., Dunker A. K., et al. , 2009. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays 31: 328–335 [DOI] [PubMed] [Google Scholar]
- Van Dross R., Browning P. J., Pelling J. C., 2006. Do truncated cyclins contribute to aberrant cyclin expression in cancer? Cell Cycle 5: 472–477 [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat R., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K. M., et al. , 2008. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 22: 386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J. A., Dwyer B. T., Takeda D. Y., Dutta A., 2001. Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol. Cell. Biol. 21: 4868–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Mori S., Oshiumi H., Matsuzaki K., Shinohara M., et al. , 2010. Cyclin-dependent kinase promotes formation of the synaptonemal complex in yeast meiosis. Genes Cells 15: 1036–1050 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.