Abstract
Analyses of effects of mutants on many traits have enabled estimates to be obtained of the magnitude of pleiotropy, and in reviews of such data others have concluded that the degree of pleiotropy is highly restricted, with implications on the evolvability of complex organisms. We show that these conclusions are highly dependent on statistical assumptions, for example significance levels. We analyze models with pleiotropic effects on all traits at all loci but by variable amounts, considering distributions of numbers of traits declared significant, overall pleiotropic effects, and extent of apparent modularity of effects. We demonstrate that these highly pleiotropic models can give results similar to those obtained in analyses of experimental data and that conclusions on limits to evolvability through pleiotropy are not robust.
BIOLOGICAL organisms are complex structures, so it is not surprising that their genes often show pleiotropic effects over two or more traits. Evidence comes both from observations on substitutions at individual loci and from genetic correlations among the traits at a population level. One view is that all genes are fully pleiotropic, at least at the quantitative level, in view of the highly interdependent and interactive nature of biological systems. Another view is that such an argument is irrelevant to genetic understanding, analysis, and application in that most loci are likely to affect no more than a small minority of traits in any biologically significant way (Stearns 2010).
The general degree of pleiotropy is important in several areas, for example in understanding pathways of gene action, in assessing potential side effects of genetic manipulation of particular pathways, in assessing the impacts of selective genetic improvement programs, and in considering the opportunities for evolutionary change through new mutations that may induce both favorable and unfavorable effects on fitness.
In a recent perspectives paper Wagner and Zhang (2011) discuss the magnitude of pleiotropy for complex or quantitative traits. They argue that it is highly restricted, i.e., rather few traits are influenced by each gene, and consequently conclude there is more opportunity for evolutionary change according to Fisher’s (1930) geometric model than suggested by the analyses of Orr (2000) and for finding drugs specific for a particular genetic disease.
Different kinds of data sets were used by Wagner and Zhang, many of them collected by them and their colleagues. One is based on mapping QTL using F2 crosses of selected lines of mice (Wagner et al. 2008), but pleiotropy and linkage of nonpleiotropic QTL cannot readily be disentangled. The other is based on single mutant lines where this complication does not arise. Wang et al. (2010) analyzed four published data sets on mutants, two of which were very large: One included 253 morphological traits, each recorded on 2449 haploid lines of Saccharomyces cerevisiae, each mutant for a different gene. The mean number of traits affected by each line was 21.6 and the median, 7, was smaller because the distribution of the number per line was of exponential form. In the other data set, for 4905 genes and 308 traits in mice, the mean was 8.2 and the median 6. Overall a significant degree of pleiotropy was found for no more than 10% of traits observed.
Wagner and Zhang’s second quantitative conclusion was that genes affecting more traits have larger per-trait effects. They computed the Euclidean distance, a measure of the total of individual trait effects detected to be significant for that gene, and found that it increased more rapidly than would be expected if there were no relation between overall gene effects and number of traits affected (Wang et al. 2010, Box 4).
Their third conclusion was that there was strong modularity of effects, i.e., that genes differed in the suite of traits they affected. The association between genes and traits was represented by a bipartite network and the presence of a modular structure detected by methods developed in physics (Barber 2007). A series of random networks was generated having the same marginal properties, i.e., numbers of traits detected for each gene and correspondingly genes affecting each trait, and the association of genes and traits was found not to be randomly distributed.
Other data that indicate substantial pleiotropy have, however, been published (Stearns 2010). For example, many studies have been undertaken in Drosophila melanogaster by Mackay and colleagues. Of the P-element mutations they screened, 22% affected abdominal bristle number, 23% sternopleural bristle number, 41% starvation stress resistance, 5.6% olfactory behavior, 22% wing shape, 37% locomotor startle response, and 35% aggressive behavior (Mackay 2010). Thus many genes affect each trait and each gene affects many traits.
We consider there are a number of important assumptions in the analyses by Wagner, Zhang, and colleagues that lead us to question their general conclusions on the degree and modularity of pleiotropy. Most importantly, a gene or line was declared to influence a trait if its observed effect exceeded a threshold determined by statistical significance. The power of detection of an effect then depends on its real size, the amount of replication, and the statistical significance level, typically set at a high threshold to allow for multiple comparisons. Therefore many real effects were likely to be missed. They recognize what they call “the vexing problem of detection limits,” but argue first that effects missed are likely to be smaller than those detected, which, while true overall, is not necessarily so for any particular mutant and trait.
To analyze pleiotropy, we take a radically different starting point. We assume and simulate an alternative model in which all genes affect all traits, but not necessarily by the same real amount. We find this model can give the appearance of restricted pleiotropy and modularity if data are displayed and analyzed using published methods (Wang et al. 2010).
Model and Methods
Model
The observed effects (zi) of a randomly sampled mutant on the ith of the n traits recorded are assumed to be the sum zi = ai + ei of a gene effect ai and a residual effect ei due to sampling error of the estimate. The latter may be based on many observations and its magnitude depends on the amount of replication and experimental conditions. As significance tests would be undertaken relative to the error SD, the power to detect gene effects is a function of ai/SD(ei), and so we standardize gene effects by the error SD and set SD(ei) = 1. Gene effects on each trait are assumed to vary among loci, with standard deviation σa = SD(ai)/SD(ei) being the same for all traits.
As genes influence different pathways or systems, they may have differing effects on associated traits, which we describe by a correlation ra (≥0) across traits. (We undertake two-tail significance tests, so positive correlations are not a limitation and simplify simulation.) Genes in the same pathway may have differential but correlated effects on a limited subset of traits, “modules”, within which we assume there is a correlation rma (≥0) of genetic effects. Similarly errors may be correlated across all traits; for example, if a common control is used, and these are defined by the correlations re and similarly rme within modules assuming the error deviations follow the same pathways as the gene effects.
Simulation
Normally distributed gene effects ai on trait i were sampled as the sum of effects common to all traits, N(0, raσa2), and as modular effects, independently for each module N(0, rmaσa2), and independent within module effects N(0, (1 − ra − rma)σa2). Individual trait effects ai are therefore distributed as N(0, σa2) with a block covariance structure, such that covariances of effects are (ra + rma)σa2 in the same module and raσa2 among different modules, with ra + rma ≤ 1. Components of the error effects, ei, distributed N(0,1), were sampled similarly with variances ra, rma, and (1 – re − rme), assuming the same modular structure as for the gene effects.
The “phenotypic” correlation of sampled effects within modules is given by rp = [(ra + ram)σa2 + re]/(σa2 + 1) and between modules by rmp = (raσa2 + re)/(σa2 + 1). Although these correlations and σa are sufficient to define the model, for clarity results are labeled in terms of the genetic and error correlations.
As mutant gene effects typically have a more leptokurtic distribution than the normal (Keightley and Halligan 2009), Wishart (gamma)-like gene, but not error, effects were also simulated. This was done by squaring the components of the correlated normal variates generated as above, attaching a random sign to each, and scaling such that individual trait effects had a reflected Wishart distribution with mean 0 and variance σa2. The actual correlations of these variables are similar to, but not the same as, those for the normal: for correlations of normal deviates of 0.00, 0.25, 0.50, 0.75, and 0.90, those constructed for the reflected Wishart are 0.00, 0.21, 0.44, 0.69, and 0.88, respectively.
Information is lost by comparing trait values to a threshold and constructing a binary 0/1 variable denoting significance, particularly when only a small proportion of traits are declared significant. As Wang et al. (2010) and Wagner and Zhang (2011) used only these binary results, we adopt their analyses. Sampled phenotypic effects zi were “tested” by comparison with thresholds expressed relative to the error SD, set at ±1.96, ±2.575, and ±3.09, corresponding to 5%, 1%, and 0.2% significant under a null hypothesis of no gene effect. As results for the middle threshold fell within the range of the others, most are not presented.
Replicate simulations with the same model were regarded as samples of different genes, each with a different array of effects, some of which were significantly different from zero. This invokes simplifications in that genes are regarded as independent and all are from the same modular distribution. Typically 10,000 replicates were used for each model, except to obtain randomized networks where much more computation was needed and we were interested only in the behavior of limited numbers of loci as in biological data.
Analysis of gene trait networks—“modularity”
The strength of a modular structure can be revealed by an increase in, for example, variation in number of traits detected per module, but requires the modular structure to be known a priori so that traits could be classified accordingly. Detection in the absence of such information can utilize a matrix in which, say, genes are rows and traits are columns and elements are equal to estimated gene effects, but we follow previous work and use binary 0/1 variables denoting significance. The gene–trait bipartite network can be represented by a modularity matrix with elements (bij − pij), where bij = 1 if trait j is significantly affected by gene i and 0 otherwise. The pij = kidi/m are probabilities in the null model network that an “edge” exists between gene i and trait j, where ki is the number of traits significantly affected by gene i, di is the number of genes that significantly affect trait j, and m is the total number of significant gene–trait pairs (edges).
Modularity (M) is defined as the extent, relative to the null model network, to which edges are formed within modules instead of between modules. The modularity of each gene–trait bipartite network was calculated from the modularity matrix with the BRIM algorithm (Barber 2007). This is an iterative algorithm for maximizing M, with the sets of gene and trait vertices each recursively drawing the other into a modular structure. Randomly wired networks that share the same marginal properties were generated with the numerical algorithm of Maslov and Sneppen (2002). First a pair of edges (A, B) and (C, D) were selected (i.e., trait A is significantly affected by locus B and C by D). The two edges were then rewired in such a way that locus B affects trait C and locus D trait A. If one or both of these new edges already existed in the gene–trait network, this step was aborted and a new pair of edges selected. Repeating this process led to a randomized version of the original network, for each of which the modularity was computed. For each network based directly on the simulated data, the average (Em) and the standard deviation (SDm) were obtained from 100 samples of randomized gene–trait networks. The scaled modularity was then obtained as (M – Em)/SDm.
Results
Distribution of number of pleiotropic traits
The mean numbers detected of 100 traits analyzed when both the gene effects and the error effects are uncorrelated are shown in Table 1 for three models of trait effect, namely constant (σa = 0; i.e., ai = a, all i, included for reference), normally distributed, and reflected Wishart-distributed gene effects. There is a high probability that real effects are missed even if ai or σa = 2, i.e., 2 SD(ei). With a leptokurtic distribution of effects, relatively more are missed than for the normal, particularly if the variance of effects is large and the threshold is low because many more genes have small effect. Absence of pleiotropy cannot be simply inferred solely from the results of tests of significance.
Table 1. Expected numbers of traits testing significant assuming 100 traits having constant effects (a) or uncorrelated normal or Wishart distributed random effects with SD = σa (by simulation).
| Threshold (P%) | ||||||
|---|---|---|---|---|---|---|
| 1.96 (5%) | 3.09 (0.2%) | |||||
| a or σa | Constant | Normal | Wishart | Constant | Normal | Wishart |
| 0 | 5.0 | 5.0 | 5.0 | 0.20 | 0.20 | 0.20 |
| 0.5 | 7.9 | 8.0 | 7.9 | 0.50 | 0.58 | 0.82 |
| 1 | 17.1 | 16.6 | 14.0 | 1.83 | 2.89 | 3.68 |
| 2 | 51.6 | 38.0 | 25.4 | 13.8 | 16.7 | 12.2 |
| 3 | 85.1 | 53.6 | 33.9 | 46.4 | 32.8 | 20.0 |
Both a and σa are expressed relative to the SD of sampling error. Error effects are uncorrelated. Two different thresholds (significant levels) are shown. Means of 10,000 replicates are shown.
If genes have correlated trait effects, whether overall or within modules, the probability an individual trait is detected and the mean number of traits detected is unchanged. The distribution of the numbers detected depends on the correlations, however (Figure 1). As the overall correlation, ra, of effects increases, so the distribution of numbers detected widens, as it becomes increasingly likely that very few or very many are detected. A correlation within modules, rma, has a qualitatively similar but quantitatively much smaller influence. Correlations among the errors have less influence than among gene effects in these examples because σa > 1.
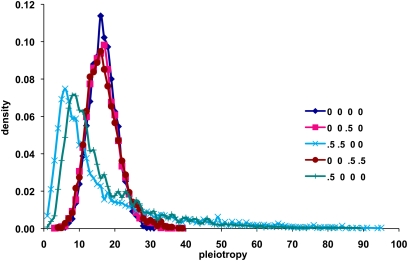
Figure 1 .
Distribution of numbers of traits declared to have a significant effect. Model: 100 traits in five modules with 5000 replicates (loci). Gene effects on traits follow a normal distribution with the threshold set at 3.09 (P = 0.2%). SD(ai)/SD(ei) = σa = 2. Correlations are ordered (ra, re, rma, rme). The mean number of traits declared significant is 17 for all the correlations displayed. The modes are 6 and 8 for (ra, re, rma, rme) = (0.5 0.5 0 0) and (0.5 0 0 0), respectively, but are ∼16 for the other examples.
The shape of the distribution and consequently its mode are most affected when there is an overall correlation of gene effects (Figure 1). With a modular structure, the degree of skew of the distribution lessens as the number of modules increases (Table A1). Skew as illustrated by the mode becomes most extreme at high thresholds and with the leptokurtic distribution.
Pleiotropic effects of detected loci
Wang et al. (2010) and Wagner and Zhang (2011) note that the Euclidean distance, TEz = √(Σi=1,mzi2), i.e., the root sum of squares of the m detected (i.e., significant) effects, would be expected to be proportional to √m if there is no relation between the magnitude of detected effects and the degree of pleiotropy (i.e., the average effect of a locus on each trait is constant). Under this assumption the coefficient of m in the power curve TEz = cmb would then be b = 0.5.
This curve was fitted to our simulated data, using both untransformed data and a weighted linear regression of log TEz on m. Both yielded very good fits as measured by R2, typically >0.95, but the actual curve can depart consistently from the simple power curve (Figure 2). At high numbers detected, the best-fitting power curve underpredicts, but these infrequent points get little weight. Similar departures are seen in some analyses of the real data (Wang et al. 2010, Figure 3; and Wagner and Zhang 2011, Box 4).
Figure 2 .
The quality of the fit of the power curve of average total effect size in Euclidean distance (TEz) vs. the degree of pleiotropy (m) TEz = cmb. The circles denote the average over samples and the red line is the best fit of the power curve for which b = 0.72. The blue line is the frequency distribution of pleiotropy. Model: 100 traits in 5 modules with 25,000 loci. Gene effects on traits follow a reflected Wishart distribution with threshold set at 1.96 (5%). SD(ai)/SD(ei) = σa = 2. Correlations (ra, re, rma, rme) = (0, 0, 0.5, 0).
The simulation results (Table 2) show that an exponent (b) of TEz in excess of 0.5 can be obtained if there is a correlation of effects or of the error, whether they are overall or within modules. Indeed, this statistic shows only that pleiotropy is present but tells little about its structure because it is insensitive to the correlations unless they are very small.
Table 2 . Mean Euclidean distance TEz for total detected phenotypic effects for each mutant expressed in terms of number of significant traits (m) for specified threshold levels.
| Threshold | 1.96 | 3.09 | 1.96 | 3.09 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Distribution | N | W | N | W | N | W | N | W | |||
| Correlations | rma = rme= 0 | ra = r e= 0 | |||||||||
| ra | re | b | rma | rme | b | ||||||
| 0.0 | 0.0 | 0.50 | 0.50 | 0.50 | 0.51 | ||||||
| 0.0 | 0.5 | 0.61 | 0.43 | 0.54 | 0.50 | 0.0 | 0.5 | 0.54 | 0.47 | 0.52 | 0.50 |
| 0.5 | 0.0 | 0.74 | 0.81 | 0.57 | 0.61 | 0.5 | 0.0 | 0.73 | 0.85 | 0.58 | 0.65 |
| 0.5 | 0.5 | 0.73 | 0.71 | 0.56 | 0.59 | 0.5 | 0.5 | 0.74 | 0.77 | 0.58 | 0.63 |
| 0.75 | 0.0 | 0.71 | 0.74 | 0.56 | 0.60 | 0.75 | 0.0 | 0.75 | 0.87 | 0.59 | 0.65 |
| 0.9 | 0.0 | 0.67 | 0.69 | 0.55 | 0.59 | 0.9 | 0.0 | 0.74 | 0.85 | 0.58 | 0.65 |
There are 100 loci in five modules with σa = 2 for normal (N) and for Wishart (W) distributed effects. Correlations of effects of gene and error effects are, respectively, ra and re over all and rma and rme within modules. Results are expressed in terms of the parameter b of the best fit [estimated by weighted regression of log TEz = logc + blogm (R2 > 0.97)] of TEz= cmb to the simulated data. Means of 10,000 replicates are shown.
Pleiotropic effects of detected loci at the genotypic level:
Effects are detected when the phenotype (z) exceeds the threshold, but the genotypic effect (a) for a significant trait is expected to be lower than z because the expected environmental deviation of those above the threshold is also positive (equivalently, the regression baz < 1). The Euclidean distance TEa for the genotypic effects for each correlated trait declared significant was also computed. The regression of TEa on m rises more rapidly than does TEz and can exceed 1.0, whether the correlations are across or within modules (data not shown), because the regression of a on z increases as σa2 increases.
Modularity
For the model used previously with 100 traits and 100 loci in five modules of equal size, estimates of modularity and scaled modularity are given in Table 3. These show that correlations among all traits do not generate much modularity; indeed, higher correlations can lead to lower modularity. If correlations within modules are sufficiently high, in these examples >0.5, substantial scaled modularity can be generated. The modularity increases with the threshold and kurtosis of distribution of gene effects. Both the modularity and scaled modularity decrease as the correlation across modules becomes higher, but scaled modularity increases while modularity remains roughly unchanged if the correlation is within modules. As the number of traits or the number of loci is increased (Table 4), modularity falls and scaled modularity increases, with the latter reaching high values.
Table 3 . Influence on modularity (Mod) and scaled modularity (SMod) of distribution and correlations of effects for a model of 100 traits and 100 loci in five modules, each of 20 traits.
| Threshold | 1.96 | 3.09 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Distribution | Normal | Wishart | Normal | Wishart | |||||
| Mod | SMod | Mod | SMod | Mod | SMod | Mod | SMod | ||
| ra | re | rma = rme = 0 | |||||||
| 0.0 | 0.0 | 0.109 | 0.11 | 0.151 | 0.05 | 0.198 | 0.18 | 0.240 | −0.27 |
| 0.002 | 0.85 | 0.003 | 0.92 | 0.005 | 1.23 | 0.006 | 0.94 | ||
| 0.5 | 0.0 | 0.105 | −0.77 | 0.141 | −0.05 | 0.185 | −0.07 | 0.215 | −0.34 |
| 0.004 | 0.86 | 0.006 | 0.92 | 0.014 | 1.02 | 0.016 | 0.81 | ||
| 0.9 | 0.0 | 0.090 | −3.80 | 0.117 | −1.94 | 0.146 | −1.00 | 0.135 | −1.50 |
| 0.005 | 0.85 | 0.012 | 1.39 | 0.012 | 1.39 | 0.018 | 1.60 | ||
| rma | rme | ra = re = 0 | |||||||
| 0.5 | 0.0 | 0.110 | 0.58 | 0.149 | 0.04 | 0.200 | 0.29 | 0.243 | 0.51 |
| 0.003 | 1.11 | 0.003 | 0.91 | 0.006 | 1.30 | 0.007 | 1.22 | ||
| 0.5 | 0.5 | 0.109 | 0.23 | 0.150 | 0.35 | 0.200 | 0.55 | 0.242 | 0.72 |
| 0.002 | 1.06 | 0.004 | 1.04 | 0.007 | 1.17 | 0.007 | 0.86 | ||
| 0.9 | 0.0 | 0.111 | 1.80 | 0.154 | 2.55 | 0.203 | 1.17 | 0.253 | 3.12 |
| 0.003 | 1.65 | 0.004 | 1.77 | 0.005 | 1.20 | 0.006 | 2.06 | ||
σa = 2. Top rows are means and bottom rows are SD (italics), each computed from 20 replicates.
Table 4. Influence on mean modularity (Mod) and scaled modularity (SMod) of changing the number of loci (equivalent to replicates), number of traits, module size, and rma.
| Threshold | 1.96 | 3.09 | ||||||
|---|---|---|---|---|---|---|---|---|
| Distribution | Normal | Wishart | Normal | Wishart | ||||
| Mod | SMod | Mod | SMod | Mod | SMod | Mod | SMod | |
| Loci | 5 modules, 100 traits, rma= 0.5 | |||||||
| 100 | 0.109 | 0.15 (1.1) | 0.150 | 0.45 (1.2) | 0.199 | 0.14 (1.1) | 0.241 | 0.57 (1.0) |
| 500 | 0.076 | 0.60 (1.2) | 0.104 | 1.31 (1.1) | 0.135 | 0.84 (1.7) | 0.163 | 0.51 (1.0) |
| 1000 | 0.068 | 0.99 (1.0) | 0.094 | 1.81 (1.0) | 0.121 | 0.99 (1.1) | 0.148 | 1.67 (0.9) |
| 2 modules, 100 traits, rma= 0.5 | ||||||||
| 100 | 0.111 | 1.52 (1.7) | 0.154 | 2.59 (1.7) | 0.204 | 2.09 (1.6) | 0.247 | 3.54 (1.9) |
| 500 | 0.088 | 11.5 (2.4) | 0.125 | 14.0 (2.2) | 0.158 | 11.6 (2.7) | 0.198 | 16.1 (3.4) |
| 1000 | 0.086 | 17.6 (2.1) | 0.122 | 20.4 (2.6) | 0.155 | 19.7 (2.6) | 0.198 | 26.6 (3.5) |
| 2 modules, 200 traits, rma= 0.5 | ||||||||
| 1000 | 0.078 | 39.3 (5.2) | 0.114 | 46.2 (5.6) | 0.143 | 39.8 (5.8) | 0.184 | 41.2 (3.9) |
| 2 modules, 200 traits rma= 0.35 | ||||||||
| 1000 | 0.060 | 9.93 (3.1) | 0.087 | 17.3 (3.7) | 0.112 | 12.9 (2.2) | 0.141 | 16.0 (1.7) |
| 2 modules, 400 traits, rma= 0.35 | ||||||||
| 500 | 0.058 | 13.3 (4.5) | 0.082 | 20.7 (4.4) | 0.107 | 18.2 (4.2) | 0.136 | 25.1 (3.8) |
| 1000 | 0.054 | 27.7 (3.7) | 0.080 | 38.1 (5.7) | 0.100 | 33.4 (4.5) | 0.130 | 44.7 (4.8) |
| 2 modules, 400 traits, rma= 0.25 | ||||||||
| 500 | 0.052 | 2.35 (1.4) | 0.072 | 4.28 (2.0) | 0.095 | 3.4 (1.9) | 0.117 | 5.89 (2.4) |
| 1000 | 0.046 | 5.25 (1.9) | 0.064 | 9.45 (3.6) | 0.083 | 8.2 (2.4) | 0.107 | 15.3 (3.4) |
In each case ra= 0.0, re= 0.0, rme= 0.0, and σa = 2. Means were computed from 20 replicates. SDs are also given for scaled modularity (italics in parentheses), but SDs for modularity are very small (see Table 3) and are not listed.
Discussion
We are not arguing here that there may not be restricted pleiotropy or modularity of gene action, but that the statistical evidence has to be considered critically. In particular we point out that a simple appearance of restricted numbers of traits shown to be significant can simply be a consequence of sampling error and high threshold on variable effects among traits, and the threshold P-values used here are much lower than if corrections were made for multiple testing. Thus Wagner and Zhang (2011) give a theoretical example for a study with 30 traits examined with a power of 80%. If significant effects were found for 3 of them, the probability that 27 effects of the same or larger size are missed is <(1 − 0.8)27 ∼ 10−19. So they conclude that, if the power is 80% and few effects are detected, it is unlikely the true number is much higher; i.e., the detection bias is likely to be small in well-designed studies. We think this conclusion is overstated and the potential bias is severe. In their example, if the effects for all traits are actually the same and those missed are due only to sampling error, the probability that no others are missed is 0.827 ∼ 0.0024, and the expected number of traits missed is 27 × 0.2 ∼ 5, i.e., more than the number detected.
Hence the magnitude of actual pleiotropy is inevitably underestimated unless some alternative steps are taken. The use of basic trait data would considerably improve any analysis and we recommend this be done rather than merely judging significance, especially when thresholds are high so the binary information is a small fraction of that in the actual records. Information on sign can also be lost. We consider it would be better to assume a joint distribution of effects of loci on the traits and attempt to estimate parameters of it. This would not be easy, but there is opportunity to set prior distributions and test fit in some kind of iterative structure. Any further analysis is well beyond the scope of this article, however.
We also consider the evidence that the power curve of effects as measured by Euclidean distance that shows the magnitude of effects increases with the degree of pleiotropy is not well founded. A gene that has a large effect correlated across traits is likely to be detected for more traits, so there is a nonrandom association between rate of detection and mean effect. It does not imply that ones of larger effect show more real pleiotropy as the degree of pleiotropy is the same for all pairs of loci in the model used here. The relationship between mean and mode of numbers detected is indicative of widespread pleiotropy if the mode is much smaller than the mean. This relationship is more extreme in the absence of modularity. Thus the more extreme distributions noted by Wagner and Zhang (2011) (e.g., their Figure 3B) are indicative more of correlations of gene effects across all traits than of correlations within modules.
The strongest evidence for modularity of gene action is undoubtedly provided by the specific analysis of networks. Correlation of effects as in a modular structure does indeed generate such evidence, albeit simulated assuming a model of universal pleiotropy with variable correlated effects. We do not doubt that the results cited by Wagner and Zhang (2011) show modularity of gene action, but we can simulate such results merely by assuming there is a moderate correlation of effects among loci, even though there is universal pleiotropy. To assess the sampling properties of the modularity and scaled modularity, sets of data were sampled using the same parameter values (number of genes, traits, and correlations). The results showed that the variation in the modularity is rather low relative to its mean, whereas that in the scaled modularity is relatively much larger. This indicates that, though scaled modularity has high sampling error when the system is small, the modularity is more robust. Therefore our results indicate that large values of scaled modularity (e.g., >10) as obtained by Wang et al. can arise with large data sets with a fully pleiotropic model, even if the correlation within modules is ≤0.5. The simulations show that the high values of modularity they obtained (>0.2 say) are more likely in our model if there are large correlations within modules, high threshold values, high kurtosis of distribution of gene effects, and low variance of gene effects (i.e., low σa).
Taken together with the lack of evidence for a relationship between the degree of pleiotropy and significance of effects, it implies that we have to be wary of drawing conclusions about lack of evolutionary constraints. A deeper analysis is required.
Appendix
Table A1. Influence of correlations across and within modules on the mode of numbers detected.
| Distribution of gene effects | Normal | Wishart | ||||||
|---|---|---|---|---|---|---|---|---|
| Threshold | 1.96 | 2.575 | 3.09 | 1.96 | 2.575 | 3.09 | ||
| σa ; 2 | ||||||||
| Mean no. detecteda | 38.0 | 24.9 | 16.7 | 25.4 | 16.7 | 12.2 | ||
| ra | re | rp | rma ; 0, rme ; 0 | |||||
| 0.0 | 0.0 | 0.0 | 37 | 25 | 17 | 26 | 17 | 12 |
| 0.0 | 0.5 | 0.1 | 38 | 24 | 15 | 23 | 15 | 12 |
| 0.5 | 0.0 | 0.4 | 28 | 15 | 8 | 16 | 8 | 4 |
| 0.5 | 0.5 | 0.5 | 23 | 12 | 6 | 12 | 7 | 3 |
| 0.6 | 0.1 | 0.5 | 24 | 12 | 6 | 12 | 5 | 2 |
| 0.4 | 0.9 | 0.5 | 25 | 12 | 6 | 12 | 7 | 4 |
| 0.75 | 0.0 | 0.6 | 19 | 8 | 3 | 9 | 3 | 1 |
| 0.9 | 0.0 | 0.72 | 12 | 3 | 1 | 7 | 1 | 1 |
| No. modules | ra ; 0, re ; 0, rma ; 0.5, rme ; 0.5 | |||||||
| 20 | 39 | 24 | 16 | 24 | 16 | 11 | ||
| 10 | 37 | 23 | 16 | 26 | 15 | 11 | ||
| 5 | 37 | 22 | 12 | 22 | 13 | 9 | ||
| 2 | 28 | 14 | 7 | 14 | 8 | 5 | ||
| σa ; 1, rma ; 0, rme ; 0 | ||||||||
| Mean no. detecteda | 16.6 | 6.9 | 2.9 | 14.0 | 6.6 | 3.7 | ||
| ra | re | rp | ||||||
| 0.0 | 0.0 | 0.0 | 17 | 6 | 2 | 14 | 6 | 3 |
| 0.5 | 0.0 | 0.25 | 12 | 4 | 1 | 9 | 3 | 1 |
| 0.75 | 0.25 | 0.5 | 6 | 1 | 0b | 4 | 0b | 0b |
| σa ; 3, rma ; 0, rme ; 0 | ||||||||
| Mean no. detecteda | 53.6 | 41.6 | 32.8 | 33.4 | 25.1 | 20.0 | ||
| ra | re | rp | ||||||
| 0.0 | 0.0 | 0.0 | 53 | 42 | 34 | 35 | 25 | 20 |
| 0.5 | 0.0 | 0.45 | 43 | 29 | 21 | 23 | 14 | 9 |
| 0.75 | 0.25 | 0.7 | 30 | 16 | 9 | 10 | 4 | 2 |
There are 100 traits. ra, re, and rp ; (raσa2 + re)/(σa2 + 1) are correlations across modules of gene, error, and “phenotypic” effects, and similarly rma, rme, and rmp are correlations within modules.
The mean number detected (numbers in italics) does not depend on the correlations. If, however, the correlations across module (ra, re) are high, the probability that no traits are detected to be significant rises, and the mean conditional on at least one trait detected may be much higher than shown.
Mode of number detected is 0. Mode conditional on at least 1 trait detected is >0.
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Barber M. J., 2007. Modularity and community detection in bipartite networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 76: 066102. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection, Oxford University Press, Oxford [Google Scholar]
- Keightley P. D., Halligan D. L., 2009. Analysis and implications of mutational variation. Genetica 136: 359–369 [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., 2010. Mutations and quantitative genetic variation: lessons from Drosophila. Philos. Trans. R. Soc. B 365: 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S., Sneppen K., 2002. Specificity and stability in topology of protein networks. Science 296: 910–913 [DOI] [PubMed] [Google Scholar]
- Orr H. A., 2000. Adaptation and the cost of complexity. Evolution 54: 13–20 [DOI] [PubMed] [Google Scholar]
- Stearns F. W., 2010. One hundred years of pleiotropy: a retrospective. Genetics 186: 767–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J., 2011. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 12: 206–213 [DOI] [PubMed] [Google Scholar]
- Wagner G. P., Kenney-Hunt J. P., Pavlicev M., Peck J. R., Waxman D., et al. , 2008. Pleiotropic scaling of effects and the ‘cost of complexity’. Nature 452: 470–473 [DOI] [PubMed] [Google Scholar]
- Wang Z., Liao B., Zhang J., 2010. Genomic patterns of pleiotropy and the evolution of complexity. Proc. Natl. Acad. Sci. USA 107: 18034–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]