Abstract
In Drosophila, the Gal4-UAS system permits a transgene to be expressed in the same pattern as a gene of interest by placing the Gal4 transcription factor under control of the gene’s DNA regulatory elements. If these regulatory elements are not known, however, expression of Gal4 in the desired pattern may be difficult or impossible. To solve this problem, we have developed a method for co-expressing Gal4 with the endogenous gene by exploiting the “ribosomal skipping” mechanism of the viral T2A peptide. This method requires explicit knowledge only of the endogenous gene’s open reading frame and not its regulatory elements.
ELUCIDATING the functional role of a particular gene often requires manipulating biological processes in the cells that express it. This goal can be accomplished genetically by inducing these cells to also express transgenes that encode products that alter normal cell functions. The most versatile implementation of this strategy is found in binary systems, such as the Gal4-UAS system of Drosophila, in which the yeast transcription factor Gal4 is used to drive the expression of a broad array of effectors encoded by other transgenes (Brand and Perrimon 1993; Duffy 2002). To express Gal4 solely in cells that express an endogenous gene of interest, the Gal4-coding sequence is typically placed under the control of the endogenous gene’s regulatory elements (i.e., enhancers). When the endogenous gene’s enhancers have not been explicitly identified, transgene expression can be directly coupled to expression of the endogenous gene using either enhancer-trap techniques (Brand and Perrimon 1993) or homologous recombination (Rong and Golic 2000; Demir and Dickson 2005; Manoli et al. 2005;). In the first instance, transgene constructs randomly inserted into the genome are screened to identify insertions into regulatory regions of the gene of interest and then tested for fidelity of expression. In the second instance, the endogenous gene is simply replaced by inserting the transgene into the endogenous gene’s translation start site. Both methods are labor-intensive and prone to failure: the first often results in patterns of transgene expression that fail to fully mimic those of the endogenous gene, and the second requires positive identification of the endogenous gene’s start codon, which is often unknown and may differ between splice isoforms. An alternative strategy often used in the mouse yokes expression of the transgene to that of an endogenous gene by interposing a viral internal ribosomal entry site (i.e., IRES) between them (Douin et al. 2004). However, no IRES sequences that function robustly in Drosophila have been identified (Ye et al. 1997). A potentially promising development has been the demonstration that short viral peptides known collectively as “2A-like peptides” can be used, like IRES sequences, to couple transgene expression to the expression of an endogenous gene in mice (Madisen et al. 2010; Taniguchi et al. 2011).
Viral 2A-like peptides share an Asp-Val/Ile-Glu-X-Asn-Pro-Gly-Pro consensus motif (Donnelly et al. 2001a), which, during translation, forces the ribosome to skip from the underlined Gly to the underlined Pro codon without forming a peptide bond (Donnelly et al. 2001b). Consequently, the nascent translation product is released after the addition of the glycine residue and a new, independent protein chain is begun with the proline residue. Here, we show that, by inserting a construct consisting of the T2A- and Gal4-coding sequences in-frame into an exon of an endogenous gene, this property of 2A-like peptides can be used to co-express the Gal4 gene and the endogenous gene in Drosophila. This method is versatile and allows one to express the Gal4 gene in all cells that express a given gene or only in those that express a particular splice variant of that gene. Depending on the site of insertion of the T2A-Gal4 sequence, the translated product of the endogenous gene may be truncated or left functionally intact. This T2A-GIFF (i.e., T2A-Gal4 in-frame fusion) technique thus represents a readily adaptable technique for transgene expression in cells expressing a gene of interest.
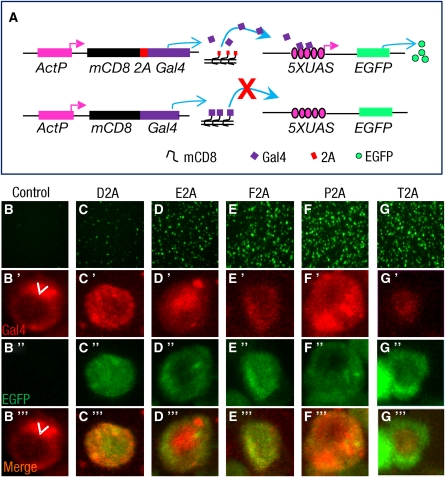
Apart from a recent study showing that a 2A peptide from the insect virus Thosea asigna known as T2A can support the translation of multiple products from a single transcript in Drosophila cell lines (Gonzalez et al. 2011), the characterization of 2A peptide activity in Drosophila has been limited. To develop the T2A-GIFF approach, we therefore first examined the efficacy of the T2A peptide in promoting ribosomal skipping compared with other candidate 2A peptides when expressed in Drosophila SL2 cells. We investigated the efficacy of five different 2A peptides (Supporting Information, File S1, Table S1) using constructs in which the coding sequence of the enhanced green fluorescent protein (i.e., EGFP) or the transcription factor, Gal4, was fused to that of a transmembrane protein, mCD8, separated by an intervening, in-frame 2A peptide sequence (Figure 1A and Figure S1A). Compared to control constructs that lacked the intervening 2A sequences, all five 2A peptides demonstrated at least some promotion of ribosomal skipping in both the EGFP (Figure S1, B–G) and the Gal4 (Figure 1, B–G′′′) assays. The T2A peptide sequence showed the highest efficiency: In SL2 cells transfected with the mCD8-T2A-EGFP construct, EGFP was dispersed throughout the cytosol and nucleus, rather than being localized to the membrane with mCD8 (Figure S1G); in mCD8-T2A-Gal4-expressing cells, Gal4 protein was quantitatively transported to the nucleus (Figure 1, G′ and G′′′) and robustly drove the expression of UAS-EGFP (Figure 1, G–G′′′).
Figure 1 .
Gal4 separated from the membrane protein mCD8 by T2A-mediated ribosomal skipping drives UAS-EGFP expression in SL2 cells. (A) Schematic of the mCD8-2A-Gal4 construct used to test the capacity of 2A peptides to support ribosomal skipping (top) and the control mCD8-Gal4 construct (bottom). Gal4 released by 2A-mediated ribosomal skipping from the mCD8 translation product of the first type of construct, but not the control construct, should enter the nucleus and drive transcription of UAS-EGFP on a cotransfected reporter plasmid. (B–G) Fluorescence images of EGFP expression in cultured SL2 cells transfected with the control and 2A fusion constructs (indicated in A). Only background EGFP fluorescence is observed in cells transfected with the control construct (B), while robust EGFP expression is observed in cells transfected with the T2A construct (G). (B′–G′′′) Fluorescence photomicrographs showing the following: (B′–G′) anti-Gal4 immunostaining, (B′′–G′′) EGFP fluorescence, and (B′′′–G′′′) merged immunostaining and fluorescence images from representative cells transfected with the control and 2A fusion constructs. In cells transfected with the control construct (B′–B′′′), little Gal4 immunoreactivity is associated with the nucleus (arrowheads) and no EGFP is expressed, while in cells transfected with the T2A construct (G′–G′′′), Gal4 immunoreactivity is strongly localized to the nucleus and accompanied by robust EGFP expression. Consistent with the small percentage of D2A-transfected cells that expressed EGFP (C), many EGFP-negative cells transfected with this construct had Gal4 immunoreactivity outside of the nucleus (data not shown).
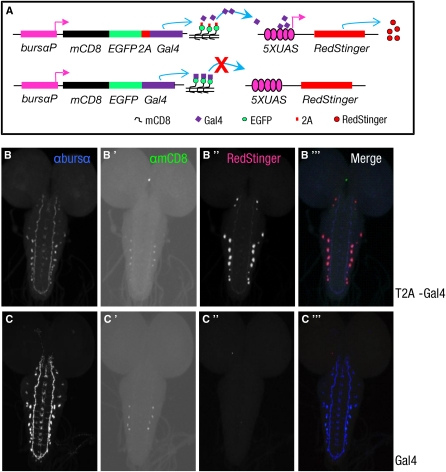
To confirm T2A’s ability to support ribosomal skipping in vivo, we generated transgenic flies that expressed a membrane-bound mCD8-EGFP-T2A-Gal4 construct only in neurons that express the hormone bursicon (Figure 2A). To detect Gal4 transcription activity, we used flies bearing a UAS-RedStinger reporter transgene. In nervous system whole mounts from these animals, confocal microscopy showed that Gal4 was not tethered to the membrane in the bursicon-expressing neurons and efficiently drove the expression of the UAS-RedStinger reporter (Figure 2, B–B′′′). The lack of tethering of Gal4 was clearly T2A mediated because animals expressing a control construct without the T2A sequence showed only faint mCD8 immunoreactivity from the membrane-associated mCD8-EGFP-Gal4 construct and no UAS-RedStinger expression (Figure 2, C–C′′′).
Figure 2 .
Bicistronic constructs containing the Gal4 transgene can be expressed in targeted cells using the T2A peptide. (A) Schematics of the two constructs used to test the efficacy of the T2A peptide in transgenic flies. As indicated, T2A-mediated ribosomal skipping is expected to cause the Gal4-coding sequence in the mCD8-EGFP-T2A-Gal4 construct (top) to be translated independently of mCD8-EGFP and thus produce transcriptionally active Gal4, whereas the mCD8-EGFP-Gal4 control construct (bottom) is expected to be translated as a single, membrane-bound fusion protein that includes (transcriptionally inactive) Gal4. Both constructs are under control of a promoter that selectively drives their expression in neurons expressing the α-subunit of the hormone bursicon (bursα), and Gal4 activity is monitored by expression of a UAS-RedStinger reporter. (B–B′′′and C–C′′′) Representative confocal images of two CNS whole mounts from third instar larvae expressing the mCD8-EGFP-T2A-Gal4 construct (B–B′′′) or the mCD8-EGFP-Gal4 control (C–C′′′). (B and C) Bursα immunoreactivity in each whole mount shows the bursα-expressing neurons and their processes. (B′ and C′) mCD8-EGFP labeling, as detected by anti-mCD8 immunostaining. (B′′ and C′′) UAS-RedStinger-associated fluorescence. (B′′′ and C′′′) Merged images. Although mCD8-EGFP labeling is observed in preparations expressing both the control and the T2A-containing constructs, only the latter shows RedStinger immunofluorescence, indicating Gal4 transcriptional activity. All images were collected under identical conditions, but contrast and brightness were adjusted in B′ and C′ to enhance mCD8-GFP expression, the low level of which reflects the weakness of the burs α promoter. Even with enhancement, expression in some cells remained below the detection threshold of the anti-mCD8 antibody.
The results of both our in vitro and in vivo studies thus confirm the effectiveness of the T2A peptide in promoting the simultaneous expression of two distinct protein products from a single transcript in Drosophila and indicate that the efficacy of T2A in fly neurons is similar to what has been previously reported for mammalian neurons (Tang et al. 2009). Importantly for the development of the T2A-GIFF technique, our results also demonstrate that T2A promotes ribosomal skipping during membrane protein translation, when the nascent protein strand is likely to be threaded into the endoplasmic reticulum. Furthermore, they show that the residual proline left at the N terminus of Gal4 upon its separation from the remainder of the T2A peptide does not impair Gal4 transcriptional activity.
To directly validate the T2A-GIFF approach, we implemented it in cells that express the Drosophila rickets (rk) gene. rickets is the fruit-fly ortholog of mammalian stem-cell markers encoded by the LGR5 and LGR6 genes (Barker and Clevers 2010). The role of the Rickets protein (RK) in fly stem-cell biology is unknown, but it does have a well-characterized role in promoting adult wing expansion (Baker and Truman 2002). Characterizing RK and the cells that express it has been impeded in the fly by the complexity of the rk gene locus. Two splice variants, rk-RA and rk-RF, with different 5′ start sites, are predicted from the rk genomic sequence, and other transcripts have been identified (Eriksen et al. 2000; Nishi et al. 2000). The longest variant, rk-RA, has highly conserved, putative regulatory elements throughout its first, large intron and multiple possible translation start sites, the first of which is preceded by six out-of-frame ATG triplets in the putative 5′ UTR (Eriksen et al. 2000). Direct knock-in of the Gal4 transgene into the predicted start site by homologous recombination resulted in no detectable expression of Gal4 (F. Diao and B. H. White, unpublished observations). Gaining genetic access to RK-expressing cells has thus proved challenging, making this an excellent candidate for the T2A-GIFF approach described here.
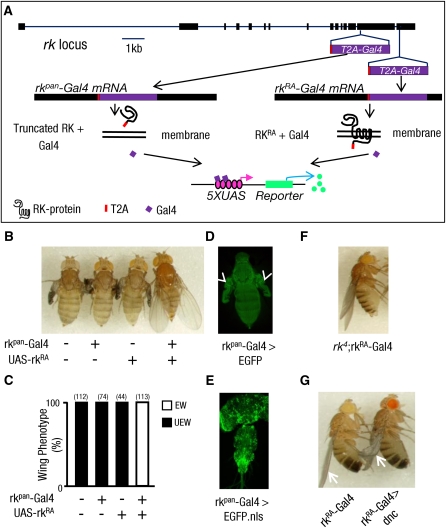
To target Gal4 expression to rk-expressing cells, we inserted the T2A-Gal4 sequence in-frame into the P[acman] genomic clone CH322-119A8 (Venken et al. 2006, 2009), which contains the entire predicted coding sequence of the rk gene and all the necessary enhancer elements required for rk expression (Figure S2). To ensure Gal4 expression in all rk-expressing cells, we inserted the T2A-Gal4 sequence into exon 14 of the rk gene, which is shared by all predicted rk splice variants, at a highly conserved site just prior to the first transmembrane-spanning region (Figure 3A, rkpan-Gal4). The parent CH322-119A8 clone rescues the wing-expansion deficits of rk mutants, but because the T2A-Gal4 insertion should produce a nonfunctional, truncated RK product, we expected the rkpan-Gal4 construct to lack this ability. Consistent with this prediction, wing-expansion deficits were not rescued in flies bearing the rkpan-Gal4 transgene (Figure 3, B and C). However, the rkpan-Gal4 transgene fully rescued wing-expansion deficits when combined with a UAS-rk-RA construct encoding the full-length RA isoform of the rk gene. This result confirms both the successful release of the Gal4 activity from the upstream RK fragment and the faithful expression of the rkpan-Gal4 driver in the pattern of the endogenous rk gene. We further confirmed the fidelity of the rkpan-Gal4 expression pattern by examining the expression of a UAS-EGFP reporter. EGFP was readily detected in tissues known to have RK activity, such as the epidermis and the unfolded wings of newly emerged flies (Davis et al. 2007) (Figure 3D), as well as in diverse cells of the central nervous system (Peabody et al. 2008) (Figure 3E). The observed tissue distribution of EGFP also closely matched the distribution of rk mRNA reported at FlyAtlas (Chintapalli et al. 2007), with all tissues having high message levels and showing robust EGFP signals, except the adult fat body (Table 1).
Figure 3 .
In-frame fusions of T2A-Gal4 can couple Gal4 expression to that of an endogenous gene. (A) Schematic of the intron–exon structure of the rickets gene locus. The in-frame insertion sites of the T2A-Gal4 sequence are indicated. Two insertions were made in the rk sequence of the genomic P[acman] clone CH322-119A8 (see Figure S2), one within exon 14, which is common to all rk transcripts, and one just prior to the stop codon of the rk-RA transcript located in exon 15. Transgenic flies bearing the first of these constructs, rkpan-Gal4, produce a truncated, presumably secreted RK protein without transmembrane domains, while flies bearing the second construct produce a full-length RK protein with a C-terminal T2A peptide fusion. (B) Pictures of rk4 mutant flies (−/−) carrying either the rkpan-Gal4 transgene, a UAS-rkRA rescue transgene, or both. The wing-expansion deficits of rk4 mutants were rescued only when the UAS-rkRA expression was driven by rkpan-Gal4. (C) Bar graph summarizing the frequency of flies of the various genotypes with unexpanded wings (UEW) vs. expanded wings (EW). Total numbers of flies scored is in parentheses above each bar. (D) Fluorescence image of epidermal UAS-EGFP expression driven by rkpan-Gal4 in a newly eclosed fly. Arrowheads indicate prominent labeling of the wing epidermis prior to expansion. (E) Confocal micrograph of UAS-EGFP.nls labeling in the central nervous system driven by rkpan-Gal4 in the pharate adult. (F) Wing-expansion phenotype of an rk4 mutant fly expressing the rkRA-Gal4 transgene. Rescue of the wing-expansion deficits shows that the RK protein produced by the rkRA-Gal4 construct is functional. (G) A representative fly (right) in which the rkRA-Gal4 transgene drives expression of the dunce gene, which encodes a cAMP-phosphodiesterase, to disrupt the second messenger pathway activated by RK protein. Wings (arrows) in such flies remained incompletely expanded, compared with flies not expressing UAS-dunce (left), indicating that cells within the rkRA-Gal4 expression pattern contribute to wing expansion.
Table 1 . Comparison of expression patterns between rkpan-Gal4 and FlyAtlas in adults.
| Tissue | rkpan-Gal4 | FlyAtlas |
|---|---|---|
| Brain | ++++ | +++ |
| Ventral nerve cord | ++++ | +++ |
| Midgut | ++ | — |
| Tubule | + | — |
| Fat body | — | + |
| Virgin spermatheca | ++++ | ++++ |
| Mated spermatheca | ++++ | ++++ |
The number of “+” signs indicate the message levels or relative levels of EGFP fluorescence in different tissues. The expression level of rk in FlyAtlas is based on the present call and enrichment of rk expression in adults reported at http://flyatlas.org/ (Chintapalli et al. 2007).
To further demonstrate the utility of the T2A-GIFF approach, we selectively targeted Gal4 expression to cells that express only the rk-RA splice variant, by inserting T2A-Gal4 just prior to its stop codon in exon 15 (Figure 3A). Unlike rkpan-Gal4, this 3′ terminal insertion construct (i.e., rkRA-Gal4) should yield full-length RK-RF and RK-RA proteins in addition to Gal4. Consistent with this, we found that the rkRA-Gal4 transgene restored wing expansion in rk mutants even without driving expression of the UAS-rk-RA rescue construct (Figure 3F). However, partial wing-expansion deficits could be induced, along with a variety of other developmental impairments, by rkRA-Gal4-directed expression of a UAS-dunce transgene (Figure 3G, right fly). dunce (i.e. dnc) encodes a cAMP-phosphodiesterase that is expected to block second-messenger signaling induced by RK activation. The partial effects of dnc overexpression on wing expansion may reflect incomplete blockage of the cAMP pathway. Alternatively, the rkRA-Gal4 expression pattern may include many, but not all, cells required for complete wing expansion. Comparison of the rkRA-Gal4 and rkpan-Gal4 expression patterns showed overlap in the wing epidermis and many other tissues (Table 1), but differences at the cellular level may exist and would require more refined analysis to detect.
Taken together, our results show that the T2A-GIFF technique can be used to gain genetic access to otherwise inaccessible cells in Drosophila using only the coding sequence of a gene of interest. T2A-GIFF readily allows the full range of Gal4-mediated manipulations of cellular function and can be implemented not only by using recombineered genomic clones as illustrated here, but also by introducing a T2A transgene into the endogenous gene’s genomic sequence by homologous recombination (Rong and Golic 2000) or by recombinase-mediated cassette exchange, using, for example, the recently described MiMIC insertions (Venken et al. 2011). Finally, we have demonstrated how T2A-GIFF can be used to selectively mark or manipulate cells expressing individual splice variants of a gene of interest and, if desired, to mutate the gene to investigate its role in cellular function. Our approach complements similar 2A-based methods recently described for the cre-lox system in transgenic mice (Madisen et al. 2010; Taniguchi et al. 2011), and it is readily adaptable for use with other binary expression systems. We thus anticipate that T2A-GIFF and related methods based on in-frame fusions of 2A transgenes will be broadly applicable to the investigation of cellular functions in Drosophila as well as other genetic model organisms.
Supplementary Material
Acknowledgments
We thank Bruce Paterson and Chi-hon Lee for the pPacPL and pC-attB plasmids, respectively; Randall Hewes for UAS-dnc flies; and the National Institute of Neurological Disorders and Stroke DNA sequencing facility for sequencing services. We also thank Grace Gray for comments on the manuscript. This research was supported by the Intramural Research Program of the National Institute of Mental Health (Project 1ZIA-MH-002800-07).
Footnotes
Communicating editor: P. K. Geyer
Literature Cited
- Baker J. D., Truman J. W., 2002. Mutations in the Drosophila glycoprotein hormone receptor, rickets, eliminate neuropeptide-induced tanning and selectively block a stereotyped behavioral program. J. Exp. Biol. 205: 2555–2565 [DOI] [PubMed] [Google Scholar]
- Barker N., Clevers H., 2010. Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology 138: 1681–1696 [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. T., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720 [DOI] [PubMed] [Google Scholar]
- Davis M. M., O’Keefe S. L., Primrose D. A., Hodgetts R. B., 2007. A neuropeptide hormone cascade controls the precise onset of post-eclosion cuticular tanning in Drosophila melanogaster. Development 134: 4395–4404 [DOI] [PubMed] [Google Scholar]
- Demir E., Dickson B. J., 2005. fruitless splicing specifies male courtship behavior in Drosophila. Cell 121: 785–794 [DOI] [PubMed] [Google Scholar]
- Donnelly M. L., Hughes L. E., Luke G., Mendoza H., ten Dam E., et al. , 2001a The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 82: 1027–1041 [DOI] [PubMed] [Google Scholar]
- Donnelly M. L., Luke G., Mehrotra A., Li X., Hughes L. E., et al. , 2001b Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol. 82: 1013–1025 [DOI] [PubMed] [Google Scholar]
- Douin V., Bornes S., Creancier L., Rochaix P., Favre G., et al. , 2004. Use and comparison of different internal ribosomal entry sites (IRES) in tricistronic retroviral vectors. BMC Biotechnol. 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15 [DOI] [PubMed] [Google Scholar]
- Eriksen K. K., Hauser F., Schiott M., Pedersen K. M., Sondergaard L., et al. , 2000. Molecular cloning, genomic organization, developmental regulation, and a knock-out mutant of a novel Leu-rich repeats-containing G protein-coupled receptor (DLGR-2) from Drosophila melanogaster. Genome Res. 10: 924–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M., Martin-Ruiz I., Jimenez S., Pirone L., Barrio R., et al. , 2011. Generation of stable Drosophila cell lines using multicistronic vectors. Sci. Rep. 1: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., et al. , 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli D. S., Foss M., Villella A., Taylor B. J., Hall J. C., et al. , 2005. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436: 395–400 [DOI] [PubMed] [Google Scholar]
- Nishi S., Hsu S. Y., Zell K., Hsueh A. J. W., 2000. Characterization of two fly LGR (leucine-rich repeat-containing, G protein-coupled receptor) proteins homologous to vertebrate glycoprotein hormone receptors: constitutive activation of wild-type fly LGR1 but not LGR2 in transfected mammalian cells. Endocrinology 141: 4081–4090 [DOI] [PubMed] [Google Scholar]
- Peabody N. C., Diao F., Luan H., Wang H., Dewey E. M., et al. , 2008. Bursicon functions within the Drosophila CNS to modulate wing expansion behavior, hormone secretion, and cell death. J. Neurosci. 28: 14379–14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Tang W., Ehrlich I., Wolff S. B., Michalski A. M., Wolfl S., et al. , 2009. Faithful expression of multiple proteins via 2A-peptide self-processing: a versatile and reliable method for manipulating brain circuits. J. Neurosci. 29: 8621–8629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H., He M., Wu P., Kim S., Paik R., et al. , 2011. A resource of cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71: 995–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., He Y. C., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J. T., Schulze K. L., Haelterman N. A., Pan H. L., He Y. C., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Fong P., Iizuka N., Choate D., Cavener D. R., 1997. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell. Biol. 17: 1714–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.