Abstract
The classical genetic approach for exploring biological pathways typically begins by identifying mutations that cause a phenotype of interest. Overexpression or misexpression of a wild-type gene product, however, can also cause mutant phenotypes, providing geneticists with an alternative yet powerful tool to identify pathway components that might remain undetected using traditional loss-of-function analysis. This review describes the history of overexpression, the mechanisms that are responsible for overexpression phenotypes, tests that begin to distinguish between those mechanisms, the varied ways in which overexpression is used, the methods and reagents available in several organisms, and the relevance of overexpression to human disease.
Moderation is a fatal thing. Nothing succeeds like excess (Oscar Wilde).
Too much of a good thing is wonderful (Mae West).
WHEN the preceding viewpoints on the benefits of excess were conveyed by Oscar Wilde and Mae West, it is safe to assume that they were not commenting on genetic methodology after scanning the latest scientific literature, but their sentiments nonetheless ring true with geneticists who have used gene overexpression as part of their research modus operandi. An impressive variety of molecular mechanisms ensures that genes are expressed at the appropriate level and under the appropriate conditions. It is obvious that a reduction of expression below some critical threshold for any given gene will result in a mutant phenotype, since such a defect essentially mimics either a partial or complete loss of function of the target gene. It is not necessarily intuitive, however, that increased expression of a wild-type gene can also be disruptive to a cell or organism, but phenotypes caused by overexpression abound (Figure 1). Serving as dramatic examples, overexpression of HER2, MYC, REL, or AKT2 are often the driving force in a variety of human cancers (Shastry 1995), and naturally occurring overexpression due to gene amplification results in drug-, insecticide-, and heavy metal-resistance (Stark and Wahl 1984). Because overexpression of wild-type genes can cause mutant phenotypes, it has been exploited by geneticists working in tractable genetic systems as a parallel approach to loss-of-function screens. This article reviews the history, applications, methods, mechanisms, and interpretation of overexpression phenotypes, focusing on its application in genetic screens but also providing examples of the utility of targeted overexpression. Many of the principles described here arise from the numerous overexpression studies that have been reported in the yeast Saccharomyces cerevisiae, where technical advantages facilitate overexpression screens and subsequent analysis, but examples are provided from other organisms that reinforce or expand the concepts and demonstrate their validity in other systems. It is hoped that a cross-species discussion of these topics will enlighten researchers to the advantages of the approach and stimulate its greater use, especially in organisms where it has been underutilized as a genetic tool. This article builds upon a foundation established in previous reviews of overexpression in yeast (Rine 1991) and plants (Zhang 2003; Kondou et al. 2010) and its involvement in human disease (Shastry 1995).
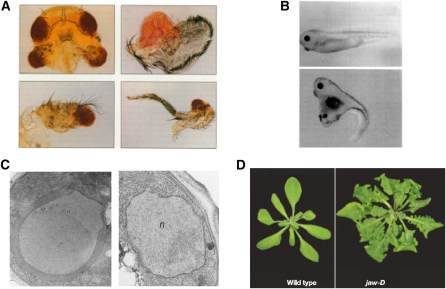
Figure 1 .
Examples of overexpression phenotypes. Overexpression has been used effectively in multiple organisms, with representative examples depicted here. (A) Overexpression of eyeless (Pax6) in the Drosophila head (top left), wing (top right), antenna (bottom left), or leg (bottom right) imaginal discs generates ectopic eye tissue (Halder et al. 1995). (B) Overexpression of Wnt-1 in Xenopus causes duplication of the dorsal body axis (Sokol et al. 1991). A dorsally injected embryo is shown on top, and an embryo injected ventrally with mouse Wnt-1 RNA is on bottom. (C) Overexpression HMG1 in yeast (left) results in the accumulation of ER stacks known as karmellae surrounding the nucleus (Wright et al. 1988). A nucleus from a cell not overexpressing HMG1 is presented on the right. (D) Overexpression of the JAW miRNA driven by a viral enhancer affects leaf development in Arabidopsis (Palatnik et al. 2003).
The Development of Overexpression as a Genetic Tool
The theoretical foundation of using overexpression as a genetic tool can be traced back to two separate lines of study that predate molecular cloning (see Box 1). Early indications that gene dosage is important for normal gene function arose from karyotype analysis showing that aneuploidies are responsible for human genetic syndromes (Lejeune et al. 1959) and for mutant phenotypes in Drosophila and plants (reviewed in Birchler and Veitia 2007). Thus, a simple increase in the copy number of entire chromosomes or portions of chromosomes, independent of discrete mutations within genes, could cause mutant phenotypes. Because the phenotypes in these cases arose from gross chromosomal duplications or translocations instead of affecting a single gene, detailed mechanistic insights were difficult to reach. Further clues emerged from classical studies on bacteriophage morphogenesis, which found that mutations that reduce expression of one gene in the bacteriophage T4 (Floor 1970; Showe and Onorato 1978) or λ (Sternberg 1976) coat proteins can be compensated for by reduced expression of other coat proteins. Although these phage studies did not involve overexpression, they provided the insight that the stoichiometry of expression is important for bacteriophage morphogenesis, and by extension, might also be important for other biological processes. Combined, these insights set the conceptual stage with the realizations that balanced gene expression is important and that even incremental changes in copy number can cause mutant phenotypes. A natural extension of this concept is that intentional overexpression of individual genes might be a useful tool for connecting genes to biological pathways.
Box 1. Overexpression vs. misexpression vs. ectopic expression.
| These terms are often used interchangeably and the distinctions between them typically are not explicitly defined. |
|---|
| The term “overexpression” predominates in studies involving unicellular organisms, implying increased expression beyond the norm, without any knowledge of the normal expression levels or pattern. |
| The terms “misexpression” and “ectopic expression” predominate in studies using metazoans, implying expression of a gene in a cell type or developmental stage or condition in which it normally is not expressed. Because the underlying genetic principles are identical, the term overexpression is used in this review, as it does not presuppose any assumptions about the basal expression of the gene. |
| The terms misexpression and ectopic expression might be more appropriate for targeted overexpression studies when details of the expression pattern are known. |
Overexpression began being exploited as a screening tool in the molecular genetics era shortly after the development of yeast transformation techniques (Beggs 1978; Hinnen et al. 1978) and the construction of genomic libraries (Nasmyth and Reed 1980; Carlson and Botstein 1982) in vectors derived from the endogenous 2μ plasmid, which are maintained at 10–30 copies per cell (Rose and Broach 1990). Transformation of a yeast strain with these libraries results in a pool of transformants, each of which contains a high-copy-number plasmid that has an average of only five or six genes, with the expression level for most genes roughly proportional to the copy number. This selective amplification of small chromosomal regions constituted a major advantage compared to investigating phenotypes caused by gross chromosomal rearrangements or aneuploidies. These yeast genomic high-copy-number plasmid libraries were used primarily to clone genes by complementation of recessive mutations until they were supplanted by the first low-copy centromeric plasmid library (Rose et al. 1987). Early studies using 2μ libraries to clone genes by complementation, however, occasionally resulted in cloning of the “wrong” gene (Hinnebusch and Fink 1983; Kuo and Campbell 1983), in which the phenotype caused by a mutation was reversed by increased copy number of a gene on the library plasmid that mapped to a different chromosomal locus than the original mutation. In effect, the genes on these plasmids were high-copy suppressors of the mutation, but initial efforts naturally focused on identifying the “correct” gene that contained the mutation, not on the high-copy suppressors.
The potential utility of overexpression phenotypes, however, did not pass unnoticed. In 1983, a single paper attempting to identify drug targets described screens to identify overexpressed genes that reversed the growth inhibitory effects of tunicamycin, compactin/mevastatin, and ethionine (Rine et al. 1983), expanding upon previous findings that overexpression of drug targets conveys resistance to their corresponding drugs in bacteria, mouse, hamster, and human cells (Rownd et al. 1971; Alt et al. 1976; Kempe et al. 1976; Normark et al. 1977; Sandegren and Andersson 2009). This study constituted two major advances: first, library screens were used to identify the targets, instead of identifying the target by other means and then demonstrating that its overexpression reverses the drug effects and second, it showed that overexpression libraries could be used not only as functional probes to clone genes by complementation, but also can independently identify phenotypes in wild-type cells. This concept was broadened beyond the application of screening for drug resistance when it was found that disrupting the normally balanced stoichiometry of histones by directly overexpressing histone gene pairs caused a chromosome segregation defect (Meeks-Wagner and Hartwell 1986). Building upon this result, a second report from the same group described using a 2μ genomic library to screen for additional genes that cause a chromosome segregation phenotype when overexpressed (Meeks-Wagner et al. 1986). This screen yielded two genes, MIF1 and MIF2, and importantly, deletion of each gene caused phenotypes consistent with chromosome segregation defects. Thus, the link established by the screen was not an artifact of overexpression, but instead reflected the authentic biological role of these genes. Importantly, mutations in MIF1 and MIF2 had not emerged from random genomic mutant hunts for chromosome segregation defects despite significant effort, demonstrating the value of the overexpression approach. With the feasibility of overexpression screens being established by these studies, analogous successes were reported in short order for screens involving the RAS pathway (Toda et al. 1987), transcriptional regulation (Clark-Adams et al. 1988), the cell cycle (Hadwiger et al. 1989), and establishment of cell polarity (Bender and Pringle 1989), cementing its application for studying essentially any biological process.
Overexpression screens are relatively simple to perform in yeast due to the stable maintenance and facile recovery of 2μ vector-based plasmids, but they are not restricted to yeast. Conceptually analogous overexpression screens emerged from studies using sib selection of expressed cDNA library clones in Xenopus (Smith and Harland 1991, 1992), or transposon-based overexpression collections in Drosophila (Rorth 1996; Rorth et al. 1998; Staudt et al. 2005) and Arabidopsis (Kakimoto 1996). Overexpression screens thereby were established as a viable research option in multiple organisms. The major factor limiting its more widespread application was no longer theoretical, but instead was the lack of resources to facilitate routine application (see Table 1).
Table 1 . Resources for overexpression screens.
| Organism | Resource | Type | Comments | Reference |
|---|---|---|---|---|
| Saccharomyces cerevisiae | 2μ LEU2 vector with genomic fragments | Systematic | Genomic fragments containing ∼5 untagged ORFs per plasmid; endogenous promoter; ∼99% functional coverage; requires subcloning to identify responsible gene | Jones et al. (2008), Hvorecny and Prelich (2010) |
| S. cerevisiae | 2μ LEU2 vector individual ORFs, endogenous promoter | Systematic | Individual untagged ORFs expressed from endogenous promoter; barcoded; ∼80% functional coverage | Magtanong et al. (2011) |
| S. cerevisiae | 2μ GAL1p-GST-6His-ORF | Systematic | Individual tagged ORFs expressed from inducible GAL1 promoter; ∼80% functional coverage | Zhu et al. (2001) |
| S. cerevisiae | 2μ LEU2; genomic fragments | Random | One of multiple classic random genomic fragment libraries | Carlson and Botstein (1982) |
| Schizosaccharomyces pombe | SPLE-2 adh-LEU2 | Random | One of multiple size-selected cDNA libraries; driven by constitutive adh promoter | Janoo et al. (2001) |
| Drosophila melanogaster | EP transposon element insertion collection | Systematic | 3700 line collection assembled from random EP insertions; expression from GalUAS | Staudt et al. (2005) |
| Arabidopsis thaliana | FOX hunting system | Systematic | ∼10,000 full-length cDNAs expressed from CaMV 35S promoter | Ichikawa et al. (2006) |
| A. thaliana | pPCVICEn4HPTT-DNA vector | Random | Used for random insertional activation- tagging mutagenesis; integration activates adjacent gene from CaMV 35S promoter | Hayashi et al. (1992) |
| Homo sapiens | Full-length human cDNAs (OriGene) | Systematic | ∼15,000 clone cDNA collection; CMV promoter | Liu et al. (2007) |
Additional Applications of Overexpression
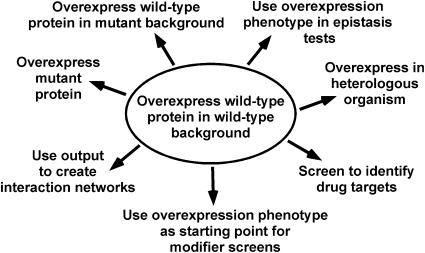
A strength of the initial screens cited above is their simplicity, overexpressing wild-type genes in a wild-type background, an approach that remains productive today. Strategies that successfully applied overexpression in additional ways soon emerged, thereby expanding the overexpression repertoire. Examples of these applications are provided here and summarized in Figure 2.
Figure 2 .
Common uses of overexpression. The prototypical strategy of overexpressing a wild-type gene in a wild-type cell is depicted in the center of the figure. The outer ring of variations on the founding core strategy is described in detail in the text.
Suppressors
A classic strategy to identify genetic interactions begins with a strain containing a mutation in a gene of interest, screening for mutations in other genes that suppress the phenotype of the original query mutation. Because suppressor hunts are used routinely and with great success (Prelich 1999), derivative selections emerged in which the phenotype caused by a mutation in one gene is suppressed by overexpression of another gene. An early example of an intentional dosage suppressor selection arose from studies of cdc2, which encodes the major regulator of cell-cycle progression in Schizosaccharomyces pombe. By selecting for high-copy plasmids that suppress the cdc2-33 temperature-sensitive phenotype, a plasmid that contained suc1 was obtained (Hayles et al. 1986b). Suc1 directly binds and regulates the Cdc2 protein kinase, and deletion of suc1 or overexpressing suc1 in an otherwise wild-type strain causes cell-cycle defects (Hayles et al. 1986a), indicating that it normally functions during cell-cycle progression. In this example, the selection identified a protein that directly contacts and regulates Cdc2, but dosage suppression can result from a wide spectrum of interesting mechanisms, including regulation by post-translational modifications, compensating for defective interactions, activating or inhibiting upstream or downstream components in the same pathway, activation of parallel pathways, and other mechanisms (Rine 1991). The variety of possible interactions that can be revealed by dosage suppressors is a clear strength of the approach, and criteria exist to distinguish them (Prelich 1999). Because dosage suppressor screens are so informative, are easy to perform, and result in the isolation of the responsible gene without any additional cloning steps, they are a routine component of the yeast genetic toolbox.
Overexpression screens were performed for more than two decades in yeast using classic random genomic libraries, but random libraries are often incomplete or biased, resulting in uncertainty about whether all possible hits are identified in any given screen. The completion of the yeast genome sequencing project in 2001 enabled development of systematic high-copy libraries that express genes from endogenous (Jones et al. 2008; Magtanong et al. 2011) or inducible (Zhu et al. 2001) promoters, opening the possibility of truly comprehensive screening. Indeed, the first two systematic overexpression screens, in which dosage suppressors were identified that reverse the transcriptional defects caused by cis-acting insertions and deletions at the HIS4 and SUC2 promoters, detected suppressors missed using random libraries (Jones et al. 2008). This past year brought the first large-scale application of systematic libraries in dosage suppressor screens. Using a combination of 51 temperature-sensitive mutations affecting varied biological processes, a systematic overexpression library containing 80% of yeast ORFs driven by the highly inducible GAL1 promoter, and barcoded microarrays to provide quantitative output, an average of five suppressors were obtained per temperature-sensitive allele tested, ranging from 0 to 24 dosage suppressors per query (Magtanong et al. 2011). Reflecting the historical tendency of 2μ library-based suppressor screens to identify functionally related genes, 80% of the suppressors analyzed in this study already were annotated to the same gene ontology category as the query gene. This study highlights the power of systematic screens and its continued use in other overexpression applications. Importantly, the first generation of conceptually analogous systematic collections of human cDNA clones (Liu et al. 2007) and transposon-mediated overexpression collections in Drosophila (Staudt et al. 2005) and plants (Kondou et al. 2010) have been assembled and are being applied in screens, although not yet at the scale of the Magtanong et al. (2011) study.
Enhancers
In contrast with suppressors, enhancers are mutations that cause a greater-than-additive phenotype when combined with a second mutation. Mutations that display combinatorial growth defects (Bender and Pringle 1991) have proven to be extremely informative, especially when performed in large-scale systematic fashion (Tong et al. 2001; Costanzo et al. 2010), revealing networks of interactions and regulatory hubs and connecting uncharacterized genes to well-studied pathways. Analogous to the concept that an existing mutation can be suppressed by overexpression of another gene, the phenotype of a mutation can be enhanced by overexpression of a second gene. In its most severe form, when overexpression causes lethality in a mutant strain but not in a wild-type background, this phenomenon is termed synthetic dosage lethality (Kroll et al. 1996; Measday and Hieter 2002). Interestingly, in the initial proof-of-principle study (Kroll et al. 1996), synthetic lethal combinations displayed specificity and occurred more frequently than high-copy suppression, yet it has been used relatively infrequently as a screening technique. Part of the reason for its limited use is the inherent difficulty of identifying lethal combinations upon introduction of an overexpression library, but this obstacle has been circumvented by using inducible overexpression (Kroll et al. 1996), by using a systematic overexpression library (Sopko et al. 2006), or by assaying nonlethal phenotypes.
Overexpression-based enhancer screens have been performed systematically in two different ways in yeast. In one approach, a plasmid that overexpresses a single gene is introduced into the yeast deletion collection to identify deletions that have more severe growth defects when that gene is overexpressed. By applying this approach to the study of chromosome segregation, a largely nonoverlapping set of interactions was identified compared with genomic synthetic lethality screens (Measday et al. 2005). In the second approach a systematic overexpression library was introduced into a single deletion strain to identify overexpressed genes that cause a more severe phenotype in combination with that deletion (Sopko et al. 2006). In a pilot application of this strategy, 65 synthetic sick or lethal interactions were identified with deletion of PHO85, which encodes a protein kinase. Interestingly, at least five of the hits included known or novel Pho85 substrates, highlighting the potential of this approach. In an extension of this study involving 92 yeast protein kinase deletions as “queries,” known substrates were enriched, yet only accounted for 1.3% of sick or lethal interactions (B. Andrews, unpublished data). The remaining kinase interactions could include new unannotated substrates, but also likely reveal additional genetic relationships beyond substrates within those pathways. Interestingly, sick or lethal interactions in kinase deletion strains occurred at a higher frequency when assayed under conditions where the kinase was active, indicating that a basal knowledge of the initial query gene’s function could promote more effective screens. Whether using random or systematic libraries, the themes emerging from these combined studies are that overexpression-based enhancer interactions are not randomly distributed between gene ontology functional categories, but like dosage suppressors, they identify genes that are functionally related to the starting gene, and that the “interaction space” is different from those revealed by other types of genetic or physical interactions. Additional large-scale screens will be required to solidify these findings and follow-up analysis of the interactions will help to decipher the range of mechanisms underlying the enhancer phenotype.
Overexpression of mutant genes
As cloned genes became increasingly available in the 1980s, it became important to identify null phenotypes for these genes, but unfortunately it was not yet possible to create targeted deletions to infer gene function in most organisms. In this light, it was proposed that creation of point mutations or deletion derivatives that inactivate one function of a protein yet retain the ability to interact with other macromolecules, might cause mutant phenotypes by competition and that overexpression would increase the likelihood of these mutants to cause a mutant phenotype (Herskowitz 1987). The term “dominant negative” was coined to describe these interfering mutations, and this term has gained widespread usage, even though there is no substantive distinction between true dominant negatives and Muller’s previously defined “antimorphs” (see Box 2). Because overexpression of interfering mutant derivatives has the potential to selectively inactivate a given protein and the phenotype is dominant, this approach is broadly applied in diploid organisms, both to interrogate the functions of uncharacterized genes and to develop reagents for selective inactivation of characterized genes. For example, overexpression of dominant negative mutants was used to study G-protein–coupled receptors (Barren and Artemyev 2007), the stress response (Voellmy 2005), and to identify inhibitors of viral infection (Gao et al. 2002). As discussed further below (and in Herskowitz 1987), however, overexpression of both wild-type and mutant proteins can cause hypermorphic and neomorphic phenotypes, so the use of the term dominant negative or antimorphic is best reserved for when additional information on the gene’s function is available. In recent years the use of inhibitory mutants to selectively inactivate protein function has been largely superseded by RNAi methods that reduce expression of the gene of interest, yet its application at the protein level presents the advantage of being more direct, and antimorphic mutations occur naturally (Johnson et al. 1982) and remain important causes of human diseases (Veitia 2009).
Box 2. Muller’s classes of mutations.
| In 1932 Hermann J. Muller described a system for classification of mutations based on functional criteria (Muller 1932). Muller’s classification and terminology remain valid today and are applicable to both mutations and overexpression phenotypes. | |
|---|---|
| Hypermorph | A mutation that causes a gain of a wild-type function, such as hyperactivity or unregulated activity toward a normal target. |
| Antimorph | A mutant allele that antagonizes its coexpressed wild-type gene product, resulting in reduction of total activity. Over the last two decades the term dominant negative has been used synonymously with antimorph. The term antimorph was coined by Muller on the basis of formal genetic tests and in the absence of any molecular information about gene structure or function, whereas Herskowitz’s dominant negative terminology (Herskowitz 1987) arose in an era when identification of protein domains in cloned genes, construction of specific mutations or deletions, and the ability to overexpress the mutant gene came under the investigator’s control, providing mechanistic insights into Muller’s antimorphs. Overexpressed mutants are not necessarily all antimorphic, however, but can also be hypermorphic or neomorphic. |
| Neomorph | A mutation that causes a gain of an abnormal function, such as an enzyme targeting a new substrate, a DNA-binding protein obtaining altered binding specificity, or a protein localizing to an abnormal location. |
| Hypomorph | Partial loss-of-function mutation that results in reduced activity |
Overexpression in a heterologous host
For several decades recombinant proteins have been overexpressed in heterologous organisms to facilitate their purification. Heterologous expression has also been exploited to study gene functions across species barriers. A seminal example of such a strategy that surprisingly crossed prokaryotic–eukaryotic boundaries was the isolation of S. cerevisiae LEU2 (Ratzkin and Carbon 1977) and HIS3 (Struhl and Davis 1977) from a random genomic library by their ability to complement recessive mutations in Escherichia coli leuB and hisB, respectively. Similar organismal barriers were crossed while exploring functional conservation of cell cycle genes when human CDC2 was cloned by functional complementation of a Schizosaccharomyces pombe cdc2 mutation (Lee and Nurse 1987), and human and Drosophila cyclins C (Leopold and O’Farrell 1991; Lew et al. 1991) and E (Koff et al. 1991) were identified as cDNA clones that suppress S. cerevisiae cyclin mutations. Another application of heterologous expression takes advantage of deleterious effects that can arise when proteins are overexpressed in a heterologous host. For example, the pathological forms of human huntingtin or α-synuclein that cause Huntington's disease and Parkinson's disease, respectively, were overexpressed in yeast, where they formed inclusion bodies. This allowed screens for yeast deletions that enhanced their toxicity, providing insights into their pathological mechanisms (Willingham et al. 2003). In an innovative screen that led to practical application, Pseudomas aeruginosa exoenzyme S, a toxin important for Pseudomonas pathogenicity in humans, was overexpressed in S. cerevisiae, where it also caused lethality. A cell-based screen for small molecules that reversed the toxicity identified a direct inhibitor of ExoS that also displays protective effects against Pseudomonas in Chinese hamster ovary cells (Arnoldo et al. 2008). Thus overexpression can provide valuable insights when applied across species lines, either by providing functional complementation or by causing deleterious effects that can be further exploited.
Combinatorial overexpression
Although most of the examples provided above entail overexpression of a single gene, informative phenotypes sometimes require overexpression of multiple genes. For example, combinatorial overexpression has been applied dramatically in the creation and differentiation of stem cells. When Oct4, Sox2, c-Myc, and Klf4 were co-overexpressed in mouse fibroblasts, induced pluripotent stem cells (iPS cells) were generated, while expression of the individual genes or pairs of genes was ineffective (Takahashi and Yamanaka 2006). In another application of overexpression to stem cell technology, combinatorial overexpression of Brn2, Ascl1, and Myt1l induces the generation of neuronal cells from fibroblasts or from human pluripotent stem cells (Pang et al. 2011), a phenomenon that is conceptually similar to the pioneering work using overexpression to demonstrate roles for MyoD (Davis et al. 1987) and Wnt (Smith and Harland 1991) in differentiation. In these examples from the stem cell literature, the genes chosen for combinatorial overexpression emerged from extensive knowledge of the relevant pathways, not from an unbiased screen. The application of a combinatorial screening protocol for a desired phenotype might prove difficult, but would be a welcome addition to the overexpression arsenal.
Using an overexpression phenotype as a starting point for finding genetic modifiers
Mirroring the concept that overexpression can suppress or enhance the phenotype of preexisting mutations, phenotypes caused by overexpression can be used as the starting point for modifier screens. One example of this approach arose from overexpression of the strong Gal4-VP16 transcriptional activator, which caused toxicity in yeast, presumably by titrating general transcription factors. Mutations were identified that reversed the Gal4-VP16 overexpression-mediated cytotoxicity, and those genes (ADA1–ADA5) encode subunits of the SAGA complex, an important transcriptional regulator (Berger et al. 1992; Marcus et al. 1994). This strategy of generating a phenotype by targeted overexpression, which then becomes the starting point for a classic genomic modifier screen, is used effectively in Caenorhabditis elegans and Drosophila (see Bulow et al. 2002; Secombe et al. 2007 for examples), organisms where overexpression screens are more challenging, although in screens such as these it is important to ensure that suppression does not simply reduce expression of the overexpressed query gene. Because overexpression phenotypes often result from competition-based mechanisms (see below), they can also be reversed by co-overexpression of a target protein. For example, when GAL3 is overexpressed in yeast, it causes the inappropriate expression of galactose-induced genes in glucose-containing medium, which is reversed by co-overexpression of its target, Gal80 (Suzuki-Fujimoto et al. 1996). Similarly, the cytotoxicity and increased amount of ubiquitin conjugates caused by overexpression of the ubiquitin-binding protein Dsk2 are suppressed by co-overexpression of the proteosomal subunit Rpn10, which binds directly to Dsk2 (Matiuhin et al. 2008). In addition to directed tests, dosage suppressors of overexpression phenotypes can also be uncovered by screening; expression of mutant forms of human FUS that cause aggregation and familial amyotrophic lateral sclerosis (ALS) in human patients triggers similar aggregation properties in yeast, which allowed screens for overexpressed yeast and human genes that reverse the FUS-induced toxicity (Ju et al. 2011; Sun et al. 2011). Phenotypes caused by targeted overexpression of a given gene therefore can be suppressed by mutations in a second gene or by overexpression of another gene, with the potential to identify direct physical interactors.
Application in epistasis tests
Mutations that cause opposite phenotypes can be used in epistasis tests to infer the order of action of those gene products within a pathway. Epistasis tests also can be performed when one of the phenotypes is caused by overexpression. An example of this application arose from ordering a portion of the yeast MAP kinase signaling pathway involved in mating; overexpression of STE12 causes constitutive activation of pheromone-responsive genes required for mating, mutations in STE7 or STE11 result in inability to induce those pheromone-responsive genes, and overexpression of STE12 in ste7Δ or ste11Δ strains results in constitutive expression (Dolan and Fields 1990). Thus, the STE12 overexpression phenotype is independent of the STE7 or STE11 genotype, implying that STE12 functions downstream. Analogous logic allowed ordering of components involved in meiosis (Smith and Mitchell 1989) in yeast, and the cell death pathway in C. elegans (Shaham and Horvitz 1996) using overexpression phenotypes. The relative ease of ordering pathways using this approach provides strong incentive for determining whether a gene of interest causes a phenotype upon targeted overexpression.
Mechanisms that result in an overexpression phenotype
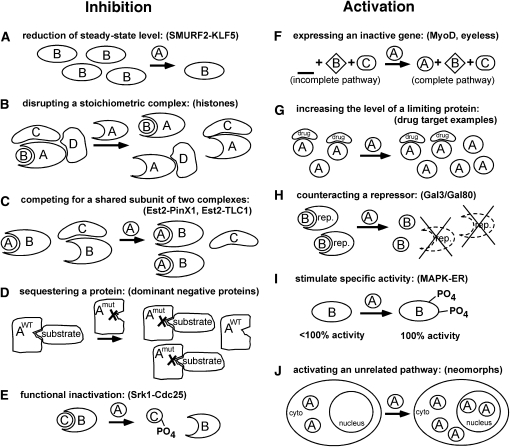
With this rich history of success, mechanistic insights have emerged to explain how overexpression can cause mutant phenotypes. As with any mutations, overexpression phenotypes can be categorized on an abstract genetic level as being either hypermorphic, hypomorphic, antimorphic, or neomorphic according to Muller’s classic criteria (Muller 1932) (see Table 1). In addition, we now have a more concrete understanding of how overexpression can cause inhibition or activation of a protein, a complex, or a pathway by different molecular mechanisms (Figure 3). In this section the mechanisms and their variations are described, followed by tests to distinguish which of these mechanisms is responsible.
Figure 3 .
Mechanisms responsible for overexpression phenotypes. Overexpression mechanisms can be placed into two broad categories that can inhibit (left column) or activate (right column) proteins, with specific mechanisms depicted below. For each variation, a representative example that is discussed more fully in the text is provided in parentheses. (A) Overexpression can simply reduce the steady-state levels of other proteins, by affecting their transcription, translation, or their rate of degradation. (B) Overexpression of subunit A that make multiple contacts within a multi-protein complex can result in partial A-B, A-D, or A-C subassemblies, thereby reducing the amount of the intact functional A-B-C-D complex. (C) If protein B participates in two separate A-B and A-C complexes, overexpression of nonshared subunit A could effectively compete for limiting amounts of protein B, reducing the amount of functional B-C complexes. (D) Overexpression of a mutant enzyme A that can still bind to its substrate competes that substrate from wild-type enzyme A by a classical dominant negative or antimorphic mechanism. (E) Overexpression can functionally inactivate proteins independent of competition-based mechanisms. Depicted here, post-translational modification of one subunit disrupts a protein–protein interaction. (F) Expressing a normally silent gene under a condition where the rest of a pathway is intact can activate its pathway. (G) Overexpressing a gene under a condition where it is expressed, but limiting, could increase total activity and stimulate output. (H) Counteracting a repressor by any number of mechanisms, including degradation of the repressor, inactivating it by post-translational modification or by direct competition could activate a pathway. In this example, overexpression of gene A results in degradation of a repressor (rep.), releasing active protein B. (I) Overexpression can increase the specific activity of other proteins. The most common mechanism is likely via post-translational modifications. (J) Overexpression can activate new pathways via neomorphic effects. Here, overexpression of the normally cytoplasmic protein A results in accumulation of a subpopulation in the nucleus, which causes a novel phenotype. See text for details.
Inhibition
A conceptually straightforward way that overexpression can inhibit another protein is simply to reduce the amount of that protein. Steady-state levels can be reduced by affecting any level of gene expression including inhibiting its transcription or translation, or by increasing its rate of degradation. Serving as clear examples of the latter, overexpression of the mammalian ubiquitin E3 ligase MKRN1 results in the degradation of the hTERT telomerase subunit (Kim et al. 2005) and overexpression of the SMURF2 ubiquitin E3 destabilizes the KLF5 DNA-binding protein (Du et al. 2011). Consistent with its identification as a specific regulator, knockdown of the SMURF2 E3 ligase increases the level of KLF5. In contrast with this first mechanism, many examples have been identified in which inhibition occurs at a functional level, frequently involving competition with other macromolecules. In principle, competition could disrupt a multiprotein complex into nonfunctional subassemblies, compete shared factors away from participation in other complexes, or sequester individual proteins. A classic example of the first mechanism arose from the studies on histone overexpression discussed previously. Overexpression of either histone H2A-H2B or histone H3-H4 gene pairs causes aberrant chromosome segregation (Meeks-Wagner and Hartwell 1986) and gene expression defects (Clark-Adams et al. 1988), yet co-overexpression of all four core histones together abolishes these effects due to restoration of the normal histone stoichiometry. Disruption of stoichiometry is reported to be relatively common; on the basis of a systematic overexpression study, stoichiometry issues were inferred to cause ∼23% of observed overexpression phenotypes on cell morphology (Sopko et al. 2006). Competition for shared subunits of two distinct complexes is exemplified by overexpression of yeast PinX1, which results in telomeric shortening due to its binding the telomerase subunit Est2 into an inactive PinX1–Est2 complex instead of an alternative active Est2–TLC1 complex (Lin and Blackburn 2004). Supporting the existence of alternative complexes, levels of the PinX1–Est2 complex increase when TLC1 is deleted and decrease when TLC1 is overexpressed. The most efficient class of proteins that produces an overexpression phenotype by competition is likely to be dominant negative mutants. Although wild-type proteins are capable of competing with their binding partners, the underlying logic of dominant negative proteins is that they more effectively sequester proteins due to the loss of a second function (Herskowitz 1987). Mutations in the active site of enzymes, for example, might result in inactive enzyme-substrate complexes when the catalytically inactive enzyme is overproduced. Inhibitory proteins typically are isolated by creation of directed deletions or point mutations and are not expected to emerge often from systematic screens that express full-length wild-type proteins. One of the advantages of random library screens relative to systematic screens is that dominant negatives should emerge more frequently due to production of truncated proteins.
Although most examples of competition involve protein–protein interactions, competition can also result from increased level of RNAs or cis-acting DNA sequences. In perhaps the best example of this phenomenon, the first Drosophila and Arabidopsis microRNAs that were identified emerged from overexpression screens, not from loss-of-function mutations; overexpression of either the Drosophila bantam miRNA (Hipfner et al. 2002; Brennecke et al. 2003; Xu et al. 2003) or mir-14 (Xu et al. 2003) causes tissue-specific defects in apoptosis, while overexpression of Arabidopsis miR-JAW regulates plant leaf development (Palatnik et al. 2003). Increased copy number of regulatory DNA elements also can cause mutant phenotypes; the HMR locus that binds silencing factors in yeast was obtained in a screen for high-copy-number plasmids that disrupt repression of a reporter gene under the control of a synthetic silencer (Zhang and Buchman 1997). Increased copy number of the regulatory sites presumably titrated a limiting silencing factor, resulting in expression of the reporter.
A final inhibitory mechanism that operates independently of competition is functional inactivation, in which the specific activity of a target protein, but not its level, is reduced. Overexpression of the stress-activated kinase Srk1 in S. pombe (Lopez-Aviles et al. 2005), for example, results in phosphorylation and inactivation of the Cdc25 protein phosphatase, thereby causing G2 cell-cycle arrest. Thus overexpression can be a productive approach for identifying inhibitory post-translational modifiers.
Activation
The second broad category of overexpression mechanisms generates phenotypes by activating a step in a pathway (Figure 3, F–J). The simplest examples to envision consist of signaling pathways in which the expression of a key regulatory step can trigger the pathway. This phenomenon is not uncommon; overexpression of MyoD causes fibroblasts to differentiate into muscle (Davis et al. 1987), overexpression of eyeless causes the development of ectopic eyes (Halder et al. 1995), and overexpression of Neurogenin (Lee et al. 1995; Ma et al. 1996) or NeuroD (Lee et al. 1995) causes ectopic neuronal differentiation in Drosophila and Xenopus. For each of these genes, the unifying principle is that their expression is normally limited to specific conditions or to specific cell types while other parts of the pathway are intact, and their overexpression completes or triggers the pathway. Although it remains possible that overexpression of only a single “master regulator” can elicit the response, in some cases overexpression of several genes that constitute a regulatory cascade can have the same effect. As an example, overexpression of any one of four sequentially acting bHLH transcription factors (MyoD, myogenin, Myf5, or MRF4) causes myogenic gene expression when expressed in nonmuscle cells (Olson and Klein 1994). A similar regulatory cascade operates in the eyeless pathway, where overexpression of eyeless, twin of eyeless, sine oculus, eyes absent, and dachshund causes formation of ectopic eyes to varying extents, whereas deletion of those genes results in defective eye formation (Wawersik and Maas 2000). In these cases a combination of epistasis tests and investigation of the temporal expression patterns of the relevant genes was required to uncover the regulatory relationships among these factors. The ability to identify rate-limiting steps, consisting of genes that are both required for a pathway and whose increased expression is sufficient to trigger a response, is a distinct advantage of overexpression studies.
Activation can occur by other mechanisms beyond the expression of a completely inactive gene. In one scenario, overexpression can increase the total activity of a protein beyond a critical threshold, causing a mutant phenotype. This situation has been applied to identifying drug targets by adding the drug at a suboptimal dose, such that its target’s activity becomes limiting, which then can be overcome by overexpression of its target protein (Rine et al. 1983). In a second scenario, some pathways are completely intact in vivo, yet kept in an inactive state by an inhibitor. Overexpression can activate such pathways by overcoming or counteracting the inhibitor at several levels, including blocking its expression or causing its degradation, resulting in net activation of the pathway. For example, the well-studied transcriptional activator Gal4 is expressed under repressing conditions and binds to DNA, but is maintained in an inactive state by the Gal80 repressor. This repressed state can be overcome by overexpression of Gal4, which simply titrates out Gal80 (Nogi et al. 1984) or by overexpression of Gal3, which binds directly to Gal80 (Suzuki-Fujimoto et al. 1996). An additional way that overexpression can increase the total activity of a protein is by post-translational modification, resulting in stimulation of its specific activity. A classic example of such an effect is the stimulation of estrogen receptor transcriptional activity by overexpression of the Ras–MAPK cascade (Kato et al. 1995). Finally, similar to dominantly acting neomorphic mutations (see Table 1) that arise from a gain of an abnormal function, overexpression is also likely to occasionally generate neomorphic phenotypes. As one example, overexpression of Drosophila hairy interferes with sex determination even though apparently it normally has no role in the process. Overexpression of hairy still was informative, because it interferes with sex determination by competing with helix-loop-helix proteins that are important players in the pathway (Parkhurst et al. 1990; Erickson and Cline 1991). On the basis of results from a systematic overexpression study in yeast, neomorphic effects are relatively rare (Sopko et al. 2006). Thus, although neomorphic effects remain possible both for dominant mutations and for overexpression effects, they can remain informative, and tests can be performed to focus on the other classes.
Distinguishing the mechanisms
In light of the variety of mechanisms summarized above, how can one discern which mechanism is responsible for generating a mutant phenotype, especially for uncharacterized genes where binding partners or involvement in a specific pathway are unknown? Fortunately, experience provides a framework that can begin distinguishing the possibilities.
Determining the loss-of-function phenotype
The primary test to distinguish the mechanism responsible for an overexpression phenotype is determining the loss-of-function phenotype of the gene of interest. Three outcomes can be envisioned: loss-of-function could cause either the opposite phenotype of overexpression, the same phenotype, or no phenotype. The simplest scenario to interpret is when overexpression and deletion cause opposite phenotypes. Examples of this phenomenon are common; for example, overexpression of eyeless in Drosophila causes formation of ectopic eyes, while an eyeless deletion blocks eye formation (Halder et al. 1995), and deletion of WOR1 blocks white-opaque switching in Candida while overexpression triggers switching (Zordan et al. 2006). The interpretation is that overexpression results in an unregulated or hyperactive protein. This hypermorphic effect is indicative of an authentic stimulatory role in the pathway, either due to expression of a rate-limiting factor or modifying protein that is also required for that pathway (a la MyoD or eyeless) or by overexpression counteracting an inhibitor.
In contrast with the previous examples, overexpression of the wild-type gene can also cause identical phenotypes as loss-of-function mutations. Because overexpression mimics a loss of function, it presumably interferes at some level with the function of the protein or its complex, acting as an antimorph. For example, overexpression of histone pairs, SPT5, SPT6, or SPT16 each causes the same transcription-related phenotypes as loss-of-function mutations in those genes (Clark-Adams and Winston 1987; Clark-Adams et al. 1988; Malone et al. 1991; Swanson et al. 1991). These genes all function as part of multiprotein complexes, suggesting that this phenomenon is due to disrupting stoichiometry or otherwise interfering with the function of their respective complexes. In the case of histones, co-overexpressing the other histone pair restores the wild-type phenotype, confirming that disruption of the complex is the cause of the defect.
A final scenario is when a gene that causes an overexpression phenotype has no obvious deletion phenotype. An informative example of this phenomenon is suppression of the cdc28-4 mutation by overexpression of CLN2 or CLN3 (Hadwiger et al. 1989). Deletion of CLN2 or CLN3 individually has no detectable phenotype, but deletion of both genes results in cell-cycle defects that mirror the original cdc28 mutant phenotype, indicating that CLN2 and CLN3 are redundant. This is an informative case, as it accentuates the importance of saturated selections. If the selection was not saturated and only CLN2 had been isolated, the lack of phenotype caused by cln2Δ would have been interpreted as possible redundancy with an unknown gene. The additional isolation of CLN3 as a high-copy suppressor allowed a direct test and confirmation of the redundancy model. Although the lack of a knockout phenotype can be disappointing to an investigator, this category highlights the major incentive for initiating overexpression studies, namely that it generates insights into function even when knockouts are uninformative.
Overexpressing a catalytically defective mutant
A second criterion for understanding an overexpression phenotype is to assess the effect of overexpressing a catalytically inactive derivative. Three outcomes are possible, with the first possibility being that overexpression of the wild-type but not the mutant gene causes the phenotype. Here the inference is that catalytic activity is required, indicative of either a hypermorphic or neomorphic effect. Examining the null phenotype should distinguish between these two possibilities, as an opposite phenotype is expected when overexpression causes a hypermorphic effect, whereas an unrelated phenotype is expected when overexpression is neomorphic. Examples where catalytic activity are required are abundant; serving as two examples, overexpression of the S. pombe histone demethylase Jmj2 but not a catalytically dead version reduces effects caused by histone methylation (Huarte et al. 2007), and overexpression of the catalytically inactive cathepsin D protease does not cause the apoptotic effects observed when wild-type cathepsin D is overexpressed (Beaujouin et al. 2006). The second possible outcome, where phenotypes are caused by overexpression of the catalytically inactive protein but not by the wild-type protein, is characteristic of a dominant negative (antimorphic) mechanism. A clear example of this phenomenon is the dominant negative effects on transcription exhibited when an ATPase-defective form of yeast Swi2 or its human ortholog is overexpressed (Khavari et al. 1993). The final outcome, where overexpression of either the wild-type protein or the catalytically inactive mutant causes the phenotype, is exemplified by overexpression of HMGCoA reductase (HMG1), which causes hyperproliferation of membrane stacks surrounding the nucleus (karmellae) in yeast (Wright et al. 1988), and by overexpression of DNA ligase, which causes a genome instability phenotype (Subramanian et al. 2005). Because overexpression of catalytically inactive versions of these proteins results in the same karmellae hyperproliferation and genome instability phenotypes, the effect cannot be due to increased activity of the protein, but instead must arise from an alternative mechanism.
Determining the regions needed for the overexpression phenotype
The strategy of overexpressing catalytically inactive derivatives can provide mechanistic insights when well-characterized catalytic site mutations are available. A parallel strategy that can be effective when the protein has not been characterized extensively or when no obvious domains are present is to express deletion derivatives with the goal of determining the regions that are required for the overexpression phenotype. More specifically, do the regions needed for the overexpression phenotype correlate with regions required for complementation or function in vivo, and do they correlate with binding sites for other macromolecules? This type of analysis was informative for the HMG1 and DNA ligase examples cited above that did not require catalytic activity; the HMG1 overexpression phenotype required a region of HMGCoA reductase that lies within the ER lumen (Parrish et al. 1995), and the region of DNA ligase required for the genome stability phenotype corresponded to a region that binds to PCNA (Subramanian et al. 2005).
Insights into the relative frequency that these mechanisms occur emerged from the first applications of nearly complete systematic libraries in yeast overexpression screens (Sopko et al. 2006; Magtanong et al. 2011). Sopko overexpressed ∼80% of the genome as GAL1p–GST–ORF fusions, finding that 184 transformants caused aberrant morphology. Forty-two of the 184 colonies (23%) caused the same phenotype as annotated loss-of-function phenotype in that gene, suggesting that overexpression interferes with their function at some level. The other 142 (77%) transformants did not resemble the null phenotype and were assumed to be due to a gain of function (hypermorphic). Examples where overexpression had antimorphic effects were rare in this study.
Finally, it is worth remembering that these genetic criteria typically are only one aspect of a multipronged investigation into the overexpression phenotype. Biochemical analysis of binding partners, investigation of any known biochemical activities, knowledge of cellular localization under normal and overexpressed conditions, information about gene expression patterns, and genetic interactions gleaned from other approaches all have the potential to provide insights into interpreting the phenotype and direction for further investigation, especially when considered in combination.
Relevance of overexpression to human health
The lessons learned from overexpression studies have several implications for human health, impacting our understanding of the causes and treatment of diseases. First, there are numerous examples in which human diseases are directly caused by increased gene expression (Shastry 1995; Santarius et al. 2010), sometimes accompanied by gene amplification, highlighting the importance of understanding at least in broad terms the mechanisms by which overexpression can cause mutant phenotypes. Second, even when overexpression does not cause overt diseases, changes in gene expression patterns or levels can contribute to phenotypic variation, diversity, and evolution (Carroll 2008). For example, human copy number variants (CNVs) can cause human familial diseases and are likely to contribute to more complex disease phenotypes (Zhang et al. 2009). The contributions to phenotypic variability by CNVs and noncoding polymorphisms that increase expression levels are only beginning to be explored but will be an important area of future investigation. Third, the successful application of systematic overexpression studies in organisms such as yeasts, Drosophila, and Arabidopsis strongly suggests that analogous systematic overexpression collections of human genes will be valuable basic research tools in cell culture systems to reveal additional therapeutic applications of gene overexpression. The generation of iPS cells (Takahashi and Yamanaka 2006) and the induction of neurons from fibroblasts (Pang et al. 2011) by combinatorial targeted overexpression highlight the application of overexpression to potential therapeutic use. Finally, the realization that overexpression can cause phenotypes, including diseases in humans, accentuates the importance of establishing correct levels of expression in gene therapy strategies. Concerns about integration of gene therapy vectors inadvertently increasing expression of adjacent genes has triggered extensive research into development of retroviral vectors that block increased expression of genes adjacent to the viral integration site (Maier et al. 2010).
Summary and Future Directions
A lesson emerging from systemic knockout studies is that loss-of-function mutations alone are insufficient to deduce gene functions. If additional genetic approaches are needed, where then are we to turn? It is difficult to argue with success, and overexpression studies certainly have a rich history of establishing functional links for essentially any cellular process in several species. As summarized here, overexpression studies provide several advantages: (1) it is a versatile tool that can be applied in several ways in wild-type and mutant backgrounds; (2) it can identify regulatory rate-limiting steps; (3) it has dominant effects, so it can be performed readily in diploid organisms; (4) it provides functional links even for redundant genes; and (5) it identifies complementary interactions from loss-of-function screens.
At least two barriers, on the other hand, have hampered the wider use of overexpression. Targeted overexpression of an individual gene can be performed in essentially any organism, but technical limitations and the lack of appropriate resources have inhibited routine genome-wide overexpression screens. Biological limitations, such as the inability to maintain introduced DNA as plasmids, will remain in some species, but the lack of resources is not an insurmountable challenge. A barrier that is more difficult to assess is the misinformed opinion that it is difficult to glean meaningful biological insights when genes are expressed at nonphysiological levels. No experimental method is without its caveats, but the concerns of studying an overexpression phenotype are no different from those associated with any other mutant background; cells are perturbed regardless of whether a pathway is disrupted by a knockout, by a dominant gain-of-function mutation, or by overexpression. Although potentially confounding neomorphic effects remain a possibility, experience and results from the first large-scale systematic screens (Sopko et al. 2006) suggest that they are infrequent and overwhelmingly balanced by the abundant benefits provided. Most importantly, the examples provided here are merely the tip of the iceberg, demonstrating that when used appropriately with reasonable secondary screening criteria, overexpression can be as effective, informative, and as versatile as any other screening technique.
Paralleling trends occurring in other areas of genetics, overexpression studies have entered a new phase. Although directed overexpression of single genes provides valuable information, and random screens will continue to be a powerful tool with distinct advantages, systematic approaches to querying the genome are likely to dominate the coming decade. Pilot systematic screens have been performed in yeast, flies, plants, and human tissue culture systems using large yet incomplete overexpression resources. A challenge for the future will be the completion of the resource collections and developing high-throughput screening technologies to facilitate their use. On the basis of the initial systematic overexpression screening studies in yeast, we can expect overexpression interaction networks to contribute new genetic links as the results are incorporated with other large datasets such as physical interactions and deletion collection results.
Acknowledgments
Work from the author’s lab was supported primarily by National Institutes of Health grant GM52486. The author gratefully acknowledges Fred Winston, Scott Hawley, Brenda Andrews, and Kenneth Robzyk for examples, comments, and unpublished results.
Footnotes
Communicating editor: J. Rine
Literature Cited
- Alt F. W., Kellems R. E., Schimke R. T., 1976. Synthesis and degradation of folate reductase in sensitive and methotrexate-resistant lines of S-180 cells. J. Biol. Chem. 251: 3063–3074 [PubMed] [Google Scholar]
- Arnoldo A., Curak J., Kittanakom S., Chevelev I., Lee V. T., et al. , 2008. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS Genet. 4: e1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barren B., Artemyev N. O., 2007. Mechanisms of dominant negative G-protein alpha subunits. J. Neurosci. Res. 85: 3505–3514 [DOI] [PubMed] [Google Scholar]
- Beaujouin M., Baghdiguian S., Glondu-Lassis M., Berchem G., Liaudet-Coopman E., 2006. Overexpression of both catalytically active and -inactive cathepsin D by cancer cells enhances apoptosis-dependent chemo-sensitivity. Oncogene 25: 1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs J. D., 1978. Transformation of yeast by a replicating hybrid plasmid. Nature 275: 104–109 [DOI] [PubMed] [Google Scholar]
- Bender A., Pringle J. R., 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA 86: 9976–9980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Pringle J. R., 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L., Pina B., Silverman N., Marcus G. A., Agapite J., et al. , 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70: 251–265 [DOI] [PubMed] [Google Scholar]
- Birchler J. A., Veitia R. A., 2007. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M., 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Bulow H. E., Berry K. L., Topper L. H., Peles E., Hobert O., 2002. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc. Natl. Acad. Sci. USA 99: 6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D., 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28: 145–154 [DOI] [PubMed] [Google Scholar]
- Carroll S. B., 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Winston F., 1987. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F., 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2: 150–159 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B., 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Fields S., 1990. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 4: 492–502 [DOI] [PubMed] [Google Scholar]
- Du, J. X., E. G. Hagos, M. O. Nandan, A. B. Bialkowska, B. Yu et al., 2011 The E3 ubiquitin ligase, SMAD ubiquitination regulatory factor 2, negatively regulates Kruppel-like factor 5. J. Biol. Chem. 286: 40354–40364. [DOI] [PMC free article] [PubMed]
- Erickson J. W., Cline T. W., 1991. Molecular nature of the Drosophila sex determination signal and its link to neurogenesis. Science 251: 1071–1074 [DOI] [PubMed] [Google Scholar]
- Floor E., 1970. Interaction of morphogenetic genes of bacteriophage T4. J. Mol. Biol. 47: 293–306 [DOI] [PubMed] [Google Scholar]
- Gao G., Guo X., Goff S. P., 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297: 1703–1706 [DOI] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Richardson H. E., de Barros Lopes M., Reed S. I., 1989. A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. USA 86: 6255–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G., Callaerts P., Gehring W. J., 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267: 1788–1792 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Czaja I., Lubenow H., Schell J., Walden R., 1992. Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Science 258: 1350–1353 [DOI] [PubMed] [Google Scholar]
- Hayles J., Aves S., Nurse P., 1986a suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. EMBO J. 5: 3373–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Beach D., Durkacz B., Nurse P., 1986b The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol. Gen. Genet. 202: 291–293 [DOI] [PubMed] [Google Scholar]
- Herskowitz I., 1987. Functional inactivation of genes by dominant negative mutations. Nature 329: 219–222 [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R., 1983. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80: 5374–5378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R., 1978. Transformation of yeast. Proc. Natl. Acad. Sci. USA 75: 1929–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner D. R., Weigmann K., Cohen S. M., 2002. The bantam gene regulates Drosophila growth. Genetics 161: 1527–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M., Lan F., Kim T., Vaughn M. W., Zaratiegui M., et al. , 2007. The fission yeast Jmj2 reverses histone H3 Lysine 4 trimethylation. J. Biol. Chem. 282: 21662–21670 [DOI] [PubMed] [Google Scholar]
- Hvorecny K. L., Prelich G., 2010. A systematic CEN library of the Saccharomyces cerevisiae genome. Yeast 27: 861–865 [DOI] [PubMed] [Google Scholar]
- Ichikawa T., Nakazawa M., Kawashima M., Iizumi H., Kuroda H., et al. , 2006. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 48: 974–985 [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S., 1982. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell 30: 873–882 [DOI] [PubMed] [Google Scholar]
- Jones G. M., Stalker J., Humphray S., West A., Cox T., et al. , 2008. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 5: 239–241 [DOI] [PubMed] [Google Scholar]
- Ju S., Tardiff D. F., Han H., Divya K., Zhong Q., et al. , 2011. A yeast model of FUS/TLS-dependent cytotoxicity. PLoS Biol. 9: e1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T., 1996. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274: 982–985 [DOI] [PubMed] [Google Scholar]
- Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., et al. , 1995. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270: 1491–1494 [DOI] [PubMed] [Google Scholar]
- Kempe T. D., Swyryd E. A., Bruist M., Stark G. R., 1976. Stable mutants of mammalian cells that overproduce the first three enzymes of pyrimidine nucleotide biosynthesis. Cell 9: 541–550 [DOI] [PubMed] [Google Scholar]
- Khavari P. A., Peterson C. L., Tamkun J. W., Mendel D. B., Crabtree G. R., 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366: 170–174 [DOI] [PubMed] [Google Scholar]
- Kim J. H., Park S. M., Kang M. R., Oh S. Y., Lee T. H., et al. , 2005. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 19: 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A., Cross F., Fisher A., Schumacher J., Leguellec K., et al. , 1991. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 66: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Kondou Y., Higuchi M., Matsui M., 2010. High-throughput characterization of plant gene functions by using gain-of-function technology. Annu. Rev. Plant Biol. 61: 373–393 [DOI] [PubMed] [Google Scholar]
- Kroll E. S., Hyland K. M., Hieter P., Li J. J., 1996. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics 143: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L., 1983. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8–1 mutation. Mol. Cell. Biol. 3: 1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Hollenberg S. M., Snider L., Turner D. L., Lipnick N., et al. , 1995. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science 268: 836–844 [DOI] [PubMed] [Google Scholar]
- Lee M. G., Nurse P., 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327: 31–35 [DOI] [PubMed] [Google Scholar]
- Lejeune J., Turpin R., Gautier M., 1959. Mongolism; a chromosomal disease (trisomy) Bull. Acad. Natl. Med. 143: 256–265 (in French) [PubMed] [Google Scholar]
- Leopold P., O’Farrell P. H., 1991. An evolutionarily conserved cyclin homolog from Drosophila rescues yeast deficient in G1 cyclins. Cell 66: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Dulic V., Reed S. I., 1991. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66: 1197–1206 [DOI] [PubMed] [Google Scholar]
- Lin J., Blackburn E. H., 2004. Nucleolar protein PinX1p regulates telomerase by sequestering its protein catalytic subunit in an inactive complex lacking telomerase RNA. Genes Dev. 18: 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kern J. T., Walker J. R., Johnson J. A., Schultz P. G., et al. , 2007. A genomic screen for activators of the antioxidant response element. Proc. Natl. Acad. Sci. USA 104: 5205–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Aviles S., Grande M., Gonzalez M., Helgesen A. L., Alemany V., et al. , 2005. Inactivation of the Cdc25 phosphatase by the stress-activated Srk1 kinase in fission yeast. Mol. Cell 17: 49–59 [DOI] [PubMed] [Google Scholar]
- Ma Q., Kintner C., Anderson D. J., 1996. Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87: 43–52 [DOI] [PubMed] [Google Scholar]
- Magtanong L., Ho C. H., Barker S. L., Jiao W., Baryshnikova A., et al. , 2011. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat. Biotechnol. 29: 505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier P., von Kalle C., Laufs S., 2010. Retroviral vectors for gene therapy. Future Microbiol. 5: 1507–1523 [DOI] [PubMed] [Google Scholar]
- Malone E. A., Clark C. D., Chiang A., Winston F., 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus G. A., Silverman N., Berger S. L., Horiuchi J., Guarente L., 1994. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 13: 4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matiuhin Y., Kirkpatrick D. S., Ziv I., Kim W., Dakshinamurthy A., et al. , 2008. Extraproteasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol. Cell 32: 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Baetz K., Guzzo J., Yuen K., Kwok T., et al. , 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA 102: 13956–13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Hieter P., 2002. Synthetic dosage lethality. Methods Enzymol. 350: 316–326 [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner D., Hartwell L. H., 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44: 43–52 [DOI] [PubMed] [Google Scholar]
- Meeks-Wagner D., Wood J. S., Garvik B., Hartwell L. H., 1986. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell 44: 53–63 [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1932 Further studies on the nature and causes of gene mutations. Proceedings of the 6th International Congress on Genetics, Ithaca, NY, pp. 213–255.
- Nasmyth K. A., Reed S. I., 1980. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc. Natl. Acad. Sci. USA 77: 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Shimada H., Matsuzaki Y., Hashimoto H., Fukasawa T., 1984. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. II. The isolation and dosage effect of the regulatory gene GAL80. Mol. Gen. Genet. 195: 29–34 [DOI] [PubMed] [Google Scholar]
- Normark S., Edlund T., Grundstrom T., Bergstrom S., Wolf-Watz H., 1977. Escherichia coli K-12 mutants hyperproducing chromosomal beta-lactamase by gene repetitions. J. Bacteriol. 132: 912–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. N., Klein W. H., 1994. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8: 1–8 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., et al. , 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., et al. , 2011. Induction of human neuronal cells by defined transcription factors. Nature 476: 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Bopp D., Ish-Horowicz D., 1990. X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell 63: 1179–1191 [DOI] [PubMed] [Google Scholar]
- Parrish M. L., Sengstag C., Rine J. D., Wright R. L., 1995. Identification of the sequences in HMG-CoA reductase required for karmellae assembly. Mol. Biol. Cell 6: 1535–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G., 1999. Suppression mechanisms: themes from variations. Trends Genet. 15: 261–266 [DOI] [PubMed] [Google Scholar]
- Ratzkin B., Carbon J., 1977. Functional expression of cloned yeast DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 74: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., 1991. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 194: 239–251 [DOI] [PubMed] [Google Scholar]
- Rine J., Hansen W., Hardeman E., Davis R. W., 1983. Targeted selection of recombinant clones through gene dosage effects. Proc. Natl. Acad. Sci. USA 80: 6750–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth P., Szabo K., Bailey A., Laverty T., Rehm J., et al. , 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057 [DOI] [PubMed] [Google Scholar]
- Rose A. B., Broach J. R., 1990. Propagation and expression of cloned genes in yeast: 2-micron circle-based vectors. Methods Enzymol. 185: 234–279 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R., 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60: 237–243 [DOI] [PubMed] [Google Scholar]
- Rownd R., Kasamatsu H., Mickel S., 1971. The molecular nature and replication of drug resistance factors of the Enterobacteriaceae. Ann. N. Y. Acad. Sci. 182: 188–206 [DOI] [PubMed] [Google Scholar]
- Sandegren L., Andersson D. I., 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7: 578–588 [DOI] [PubMed] [Google Scholar]
- Santarius T., Shipley J., Brewer D., Stratton M. R., Cooper C. S., 2010. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10: 59–64 [DOI] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L., Eisenman R. N., 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21: 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S., Horvitz H. R., 1996. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 10: 578–591 [DOI] [PubMed] [Google Scholar]
- Shastry B. S., 1995. Overexpression of genes in health and sickness. A bird’s eye view. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 112: 1–13 [DOI] [PubMed] [Google Scholar]
- Showe M. K., Onorato L., 1978. Kinetic factors and form determination of the head of bacteriophage T4. Proc. Natl. Acad. Sci. USA 75: 4165–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. E., Mitchell A. P., 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W. C., Harland R. M., 1991. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell 67: 753–765 [DOI] [PubMed] [Google Scholar]
- Smith W. C., Harland R. M., 1992. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 70: 829–840 [DOI] [PubMed] [Google Scholar]
- Sokol S., Christian J. L., Moon R. T., Melton D. A., 1991. Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell 67: 741–752 [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., et al. , 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 21: 319–330 [DOI] [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M., 1984. Gene amplification. Annu. Rev. Biochem. 53: 447–491 [DOI] [PubMed] [Google Scholar]
- Staudt N., Molitor A., Somogyi K., Mata J., Curado S., et al. , 2005. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 1: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N., 1976. A genetic analysis of bacteriophage lambda head assembly. Virology 71: 568–582 [DOI] [PubMed] [Google Scholar]
- Struhl K., Davis R. W., 1977. Production of a functional eukaryotic enzyme in Escherichia coli: cloning and expression of the yeast structural gene for imidazole-glycerolphosphate dehydratase (his3). Proc. Natl. Acad. Sci. USA 74: 5255–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian J., Vijayakumar S., Tomkinson A. E., Arnheim N., 2005. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics 171: 427–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Diaz Z., Fang X., Hart M. P., Chesi A., et al. , 2011. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9: e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Fujimoto T., Fukuma M., Yano K. I., Sakurai H., Vonika A., et al. , 1996. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol. Cell. Biol. 16: 2504–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. S., Malone E. A., Winston F., 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11: 4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S., 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Toda T., Cameron S., Sass P., Zoller M., Wigler M., 1987. Three different genes in S. cerevisiae encode the catalytic subunits of the cAMP-dependent protein kinase. Cell 50: 277–287 [DOI] [PubMed] [Google Scholar]
- Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., et al. , 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Veitia R. A., 2009. Dominant negative factors in health and disease. J. Pathol. 218: 409–418 [DOI] [PubMed] [Google Scholar]
- Voellmy R., 2005. Dominant-positive and dominant-negative heat shock factors. Methods 35: 199–207 [DOI] [PubMed] [Google Scholar]
- Wawersik S., Maas R. L., 2000. Vertebrate eye development as modeled in Drosophila. Hum. Mol. Genet. 9: 917–925 [DOI] [PubMed] [Google Scholar]
- Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., Muchowski P. J., 2003. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science 302: 1769–1772 [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D’Ari L., Rine J., 1988. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Vernooy S. Y., Guo M., Hay B. A., 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 13: 790–795 [DOI] [PubMed] [Google Scholar]
- Zhang F., Gu W., Hurles M. E., Lupski J. R., 2009. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10: 451–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Z., 2003. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 6: 430–440 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Buchman A. R., 1997. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol. Cell. Biol. 17: 5461–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., et al. , 2001. Global analysis of protein activities using proteome chips. Science 293: 2101–2105 [DOI] [PubMed] [Google Scholar]
- Zordan R. E., Galgoczy D. J., Johnson A. D., 2006. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc. Natl. Acad. Sci. USA 103: 12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]