Abstract
The ability to chronicle transcription-factor binding events throughout the development of an organism would facilitate mapping of transcriptional networks that control cell-fate decisions. We describe a method for permanently recording protein–DNA interactions in mammalian cells. We endow transcription factors with the ability to deposit a transposon into the genome near to where they bind. The transposon becomes a “calling card” that the transcription factor leaves behind to record its visit to the genome. The locations of the calling cards can be determined by massively parallel DNA sequencing. We show that the transcription factor SP1 fused to the piggyBac transposase directs insertion of the piggyBac transposon near SP1 binding sites. The locations of transposon insertions are highly reproducible and agree with sites of SP1-binding determined by ChIP-seq. Genes bound by SP1 are more likely to be expressed in the HCT116 cell line we used, and SP1-bound CpG islands show a strong preference to be unmethylated. This method has the potential to trace transcription-factor binding throughout cellular and organismal development in a way that has heretofore not been possible.
MUCH of organismal development is transcriptionally regulated. Consequently, considerable effort has been expended to understand the gene expression networks that control cell division, differentiation, and migration. But mapping the transcriptional networks that control cell-fate decisions is difficult given the limitations of the tools available to measure protein–DNA interactions. Methods like ChIP-chip (Boyer et al. 2005) or ChIP-seq (Johnson et al. 2007; Robertson et al. 2007) provide a snapshot of transcription factor (TF) binding, but are unable to record transcription-factor binding events. The DamID method (Van Steensel and Henikoff 2000; Vogel et al. 2007) can measure where a transcription factor binds with targeted methylation; however, the mark is not permanent as adenine methylation is not recorded through cell division. The transient nature of both these methods makes it difficult, if not impossible, to correlate transcription-factor binding events in progenitor cells to the final fates of their progeny cells during development. To fill this experimental void, we developed transposon “calling cards.”
We attach the transposase of a transposon to a TF, thereby endowing the TF with the ability to direct insertion of the transposon into the genome near where it binds (Wang et al. 2007) (Figure 1). The transposon becomes a calling card that permanently marks the transcription factor’s visit to a particular genomic location. By harvesting the transposon calling cards along with their flanking genomic DNA, a genome-wide map of transcription factor binding can be obtained. We harnessed the piggyBac (PB) transposon as a calling card (Cary et al. 1989; Ding et al. 2005), due to its key advantages over other transposons: it is active in many different species, including yeast (Mitra et al. 2008), insects (Bossin et al. 2007), zebrafish (Lobo et al. 2006), mouse, and human (Ding et al. 2005), its transposition efficiency is high, and PB transposase is amenable to addition of proteins to its N terminus (Wu et al. 2006; Wilson et al. 2007) (Supporting Information, Figure S1).
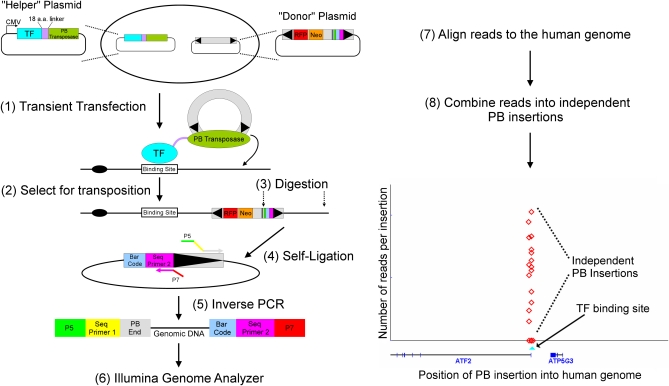
Figure 1 .
PB calling card-seq. When PB transposase is fused to a DNA-binding protein, it causes PB to integrate into the genome near the binding sites for that transcription factor (TF). After PB transposition, cells that have undergone PB transposition are selected, their genomic DNA is harvested and cleaved with restriction enzymes that cut near the end of the transposon, and the resulting fragments are ligated in dilute solution to favor their circularization. The genomic DNA flanking the end of the transposon is then amplified in an inverse PCR (PCR primers contain the Illumina sequencing primers and adaptors). The identity of the inverse-PCR products is then determined by Illumina sequencing. The Illumina sequences are then aligned to the human genome to identify the location of the PB insertions.
We describe the use of PB calling cards to accurately and reproducibly map the target genes of the transcription factor SP1 in human cells.
Methods and Materials
Cell culture
Human colon adenocarcinoma cell line HCT116 (ATCC) was maintained in McCoy's 5A Media Modified (Gibco) supplemented with 10% fetal bovine serum (Gibco). To select for PB transposition, G418 was added to the media to a final concentration 700 μg/ml. Cells were cultured at 37° in the presence of 5% CO2.
Construction of plasmids
Plasmids pcDNA3.1Δneo- piggyBAC (PB helper), pcDNA3.1Δneo-Gal4DBD-piggyBAC (Gal4-PB helper), and PB donor (Wu et al. 2006) were obtained from Stefan Moisyadi (Hawaii University). To use “gap repair” in yeast cells (Ma et al. 1987; Wach et al. 1994) for engineering this plasmid, all three plasmids were converted into yeast vectors by inserting into their NaeI site a fragment containing CEN6, ARS, and TRP1, amplified from pRS314 (Strathern and Higgins 1991) using primers OM8191 and OM8192, to make pBM5209, and in Gal4DBD-PB helper to make pBM5210. A fragment containing CEN6, ARS, and URA3 sequences, amplified from pRS316 (Strathern and Higgins 1991) using primers OM8193 and OM8194, was cloned into the AflIII site of PB donor plasmid to make pBM5211.
All TF-PB helper constructs were built by gap repair of pBM5210 linearized by digestion with BsrGI. SP1 coding sequences were amplified using OM8747 and OM8748. Yeast cells were cotransformed with each PCR product and linearized pBM5210 selecting for Trp+ colonies. DNA extracted from yeast colonies was introduced into Escherichia coli and the plasmid was isolated. Each construct was confirmed by Sanger sequencing.
Transfection of cells and transposition of piggyBac
All plasmids used for transfection of cells were prepared using EndoFree Plasmid Maxi Kit (Qiagen) following the manufacturer’s protocol. HCT-116 cells were grown to confluency in a 25-cm flask then dispersed by adding 1 ml Trypsin-EDTA and incubating for 5 min at 37°. Media (9 ml) was added to the flask to resuspend cells thoroughly (106 cells/ml). Cell suspension, 0.5 ml, was added into one well of a six-well plate. Cells were grown in a total of 3 ml of media for 2 days until they reached 50–80% confluency. A total of 1 μg of DNA (0.33 μg helper and 0.66 μg donor) was transfected into cells with FuGENE 6 (Roche), following the manufacturer’s protocol. After 12 hr, cells were trypsinized and resuspended in 2.3 ml of media. For selection of cells in which piggyBac transposed, several 400 μl aliquots of cells were plated into one 10-cm dish with 10 ml media containing G418 (700 μg/ml), resulting in five plates for each transfection. For colony counting, 50-μl cells were plated into one 10-cm dish with 10 ml media containing G418 (700 μg/ml) for each transfection. After 7 days of selection in media containing G418, colonies from all five plates were harvested and pooled for DNA extraction. Cells were fixed for counting with PBS containing 4% paraformaldehyde for 1 hr and then stained with 0.2% methylene blue overnight.
Inverse PCR
Genomic DNA was extracted from each sample using DNeasy blood and tissue kit (Qiagen) following the manufacturer’s protocol. Each DNA sample was divided into three 2-μg aliquots, each digested by MspI, Csp6I, or TaqI individually. Digested DNA was ligated overnight at 15° in dilute solution to encourage self-ligation. After ethanol precipitation, self-ligated DNA was resuspended in 30 μl ddH2O and used as template in an inverse PCR. Primers that anneal to PB donor sequences (OM8721 and OM8722) were used to amplify the genomic regions flanking PB, and adapter sequences that allow the PCR products to be sequenced on the Illumina genome analyzer were added. The PCR products were purified using the QIAquick PCR purification kit (Qiagen) and diluted into 10 nM concentration. For each sample, the same amount of PCR product from digestion with each restriction endonuclease was pooled and submitted for Illumina sequencing.
Chromatin immunoprecipitation
HCT116 cells were grown to 50% confluency in a 20-cm dish and transfected with 0.67-μg plasmid expressing SP1 from the CMV promoter with FuGENE 6 (Roche), following the manufacturer’s protocol. After 48 hr the cells were crosslinked in 1% formaldehyde and prepared using the EZMagna ChIP A kit according to the manufacturer’s instructions (Millipore). Lysed nuclear samples were sonicated on ice to a size range of 150–700 bp. Five micrograms of SP1 (Active Motif, no. 39058) and IgG (Millipore, EZMagna ChIP A kit) were used in the IP with 50 μl of Dyna1 protein G beads overnight at 4°. The precipitated immunocomplex was treated with proteinase K for 2 hr at 65°, and the DNA was purified with the Millipore EZMagna columns. Resultant DNA was prepared according to Illumina’s protocol and sequenced on an Illumina GAIIx. Single-end reads were aligned to the human genome sequence (v. hg18) using bowtie (Langmead et al. 2009). FindPeaks (Fejes et al. 2008) was used to generate the .wig files for visualization in the UCSC genome browser (Kent et al. 2002) and MACS (Zhang et al. 2008) was used to calculate significance of each peak.
Expression analysis
HCT116 cells were grown to 50% confluency in a 20-cm dish and poly(A)+ RNA was extracted using DynaI magnetic beads (Invitrogen), following the manufacturer’s protocol. The purified RNA was converted to double-strand cDNA using random hexamer primers and resultant DNA was prepared using the Illumina single end library protocol and sequenced on an Illumina GAIIx. Reads were then aligned to the human genome sequence producing a .gtf file corresponding to the transcripts of RefSeq genes from the hg18 build using Tophat (Trapnell et al. 2009) and quantified with Cufflinks (Trapnell et al. 2010).
Sequence map back and gene calling
Paired reads were filtered by requiring the first bases to contain the end of the PB terminal repeat (to ensure specific amplification) on the first read, and a proper barcode and digestion site on the second read. The nongenomic sequences were trimmed and the remaining sequence from each paired-end read was aligned to hg18 using bowtie (Langmead et al. 2009) with the options –best –tryhard –minins 0 –maxins 1200. Reads that could be uniquely aligned counted as an independent insertion for every position for every unique barcode at that position.
The SP1-directed PB insertions were then clustered using a hierarchical clustering algorithm to identify other insertions within 2500 bp. The P-value of this cluster of PB insertions into that region was then modeled as a Poisson distribution, with the number of independent insertions in the “transposase-alone” experiment in the same genomic window setting the expectation. The P-value is then calculated from cumulative distribution function given the observed number of independent insertions in the TF-directed experiment. Since it is rare to find multiple PB insertions at the same TTAA site in the “transposase-alone” experiment, and this number increases dramatically in the TF-directed experiments we assigned an exponential bonus (5N) for additional insertions at the same site. Target genes were then assigned to peaks that were within 20000 bp 5′ or 5000 bp 3′ of the transcription start site for that gene.
Results
We developed a protocol for high-throughput identification of PB insertions in the human genome. Two plasmids—one encoding the PB transposase under the control of a CMV promoter, the other carrying the PB transposon with the neomycin (neo)-resistance gene and a DNA sequence “barcode,” were transfected into HCT116 cells. We used a mixture of PB transposon plasmids with different barcodes to enable the identification of multiple independent insertion events at a given site. Cells with PB transpositions were selected in medium containing G418, and their genomic DNA was isolated and cleaved with restriction endonucleases that cut near the end of PB transposon and ligated in a dilute solution to favor recircularization of the fragments. The circular DNA containing the end of the transposon and flanking genomic DNA was amplified in an “inverse PCR” using primers that include the sequences necessary for sequencing on the Illumina instrument. The DNA sequences of the products were determined by paired-end sequencing (Figure 1).
Mapping piggyBac transposition directed by the native transposase
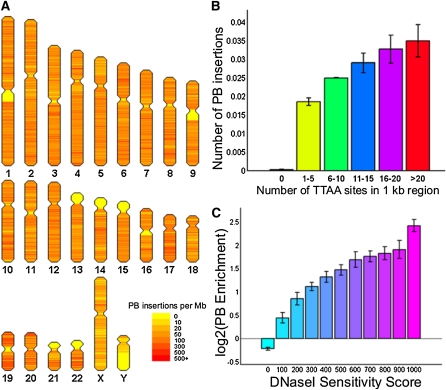
Before using our method to record the visits of DNA-binding proteins to the genome, we sought to determine the genome-wide insertion pattern of piggyBac catalyzed by its native transposase. It is known that many transposons show strong preferences for insertion into specific genomic locations (Gangadharan et al. 2010), but these preferences are not well characterized for piggyBac, since published data sets of insertions in human genomes contain fewer than 1000 mapped locations. We mapped 190,000 independent PB insertion events and found that the distribution of PB insertions was not uniform across the genome (Figure 2A). As expected, genomic regions that are enriched for piggyBac’s target sequence TTAA were more likely to host PB insertions (Figure 2B). We also observed a predilection for PB to insert into open chromatin (P < 2.2 × 10−16, χ2 test), with increasing enrichment in regions of the genome with higher DNaseI sensitivity scores (Sabo et al. 2006) (Figure 2C). There was a higher frequency of integration into RefSeq genes and into regions near transcriptional start sites (±5 kb) than into randomly selected genomic TTAA sites (Table 1), suggesting that piggyBac integrates close to actively transcribed genes, an observation that is consistent with previous studies (Wilson et al. 2007; Huang et al. 2010).
Figure 2 .
(A) The pattern of the 190,000 mapped PB insertions catalyzed by its native transposase shows a nonuniform distribution of insertions across the genome. (B) Regions with a higher local density of TTAA sites showed an increase preference for PB to insert into that region. (C) PB insertions enrich into regions of open chromatin as assayed by DNaseI sensitivity in terms of enrichment of insertions over the number of TTAA sites available for insertion in those regions. Error bars are shown as standard deviation (SD) between three biological replicates.
Table 1 . The insertion distribution of the piggyBac transposon differs significantly from the distribution of TTAA sites relative to defined genomic loci.
| Randomized TTAA sites (%) | piggyBac distribution (%) | SP1-directed distribution (%) | |
|---|---|---|---|
| In RefSeq genes | 38.9 | 44.6 | 45.4 |
| RefSeq UTRs | 6.6 | 9.2 | 12.3 |
| RefSeq introns | 30.9 | 33.1 | 26.1 |
| RefSeq exons | 1.4 | 2.3 | 7.0 |
| First exon | 0.1 | 0.6 | 5.2 |
| First intron | 10.1 | 15.6 | 22.2 |
| ±5 kb transcription start sites | 6.1 | 13.7 | 40.1 |
| ±5 kb from CpG islands | 6.1 | 10.5 | 25.7 |
| ±1 kb from CpG islands | 1.7 | 3.7 | 13.9 |
The distribution of SP1-PBase catalyzed insertions differs from the distribution of insertions catalyzed by the native transposase with large increases in insertions near transcription start sites and CpG islands.
Since PB is thought to insert exclusively into the TTAA sequence (Wilson et al. 2007; Mitra et al. 2008), we were surprised to find that 4% of PB transposons were not flanked by this sequence. These insertion sites could be separated into two classes: in 2.5% of the cases, the transposase appears to have correctly targeted the TTAA sequence, but during the insertion of the transposon there was an addition or deletion of 1–2 bp; the remaining 1.5% of transposition events have a flanking sequence distinct from TTAA, with no TTAA site nearby. These events cannot be explained by sequencing error because the flanking sequences exactly match the reference genome. Three of these insertion sites were validated by Sanger sequencing (Figure S2). These results demonstrate that the PB transposon inserts into non-TTAA tetranucleotides at a frequency of approximately 1%.
Our data set, which is more than 300-fold larger than the previously largest available data set (Wilson et al. 2007), provides a more thorough picture of PB insertions across the human genome. The locations of all PB insertions are provided in Table S1 along with the PSWM for the sequences of all PB insertion sites in Figure S3.
Using PB calling cards to map SP1 binding sites
To demonstrate that a transcription factor can direct PB transposition, we fused the PB transposase to the C terminus of the transcription factor SP1 (Wilson et al. 2007). When this chimeric SP1 binds to its targets, it is expected to direct the insertion of the PB transposon into the genome close to SP1 binding sites (Figure 1). Using the PB calling card protocol, we mapped 40,000 insertions directed by SP1-PBase. Their distribution in the genome was dramatically different from the distribution observed with the native PB transposase (P < 2.2 × 10−16, χ2 test) (Table 1). Insertions in cells carrying the SP1-PBase fusion were significantly more clustered than insertions in cells with the native PB transposase, with a fivefold increase in the number of insertion sites with another insertion within 1 kb (P < 1 × 10−3; Student’s t-test). Furthermore, we observed a 14-fold increase in the number of TTAA sites with multiple PB insertions (P < 1 × 10−3, Student’s t-test). These clusters of insertions appear to be functionally relevant: 44.47% of sites with multiple insertions are within 5 kb of a transcription start site of a RefSeq gene, compared to only 16.53% in the cells with native PBase. The enrichment of insertions into open chromatin was also significantly increased (P < 2.2 × 10−16, χ2 test) (Figure S4), consistent with the hypothesis that these insertions are more likely to be in regulatory regions. Thus, the fusion of SP1 to the PB transposase dramatically alters the insertion pattern of PB, concentrating the calling cards near promoters of RefSeq genes.
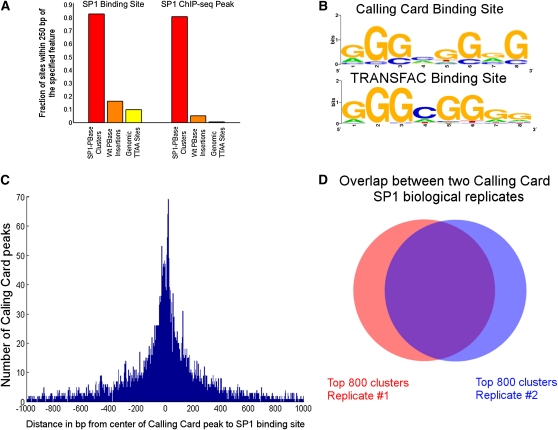
To determine if the SP1-PBase fusion protein deposited calling cards near SP1 binding sites, we created a statistical model to identify genomic loci containing more insertions than expected by chance (see Materials and Methods). We modeled the background distribution of PB insertions using our data set of transpositions catalyzed by native PBase (this is analogous to the IgG pulldown control in ChIP-seq). At a P-value threshold of 1E-3, 6373 significant “clusters” of calling cards were identified (Table S2), and 83% of these contained an SP1 binding site within 250 bp (Figure 3A). Furthermore, we were able to identify the canonical SP1 binding motif of GGGCGGGG when we searched the sequences near clusters of calling cards for a PSWM using AlignACE (Hughes et al. 2000) (Figure 3B). These results suggest that calling cards mark bona fide SP1 targets and that the SP1-PB transposase fusion is directing transposon insertion close to where the transcription factor is bound.
Figure 3 .
(A) The center of significant calling card clusters is very likely to be located within 250 bp of an SP1 binding site and an SP1 ChIP-seq peak and shows a large enrichment relative to insertions directed by the native PB transposase or random genomic TTAA sites. (B) The highest information content motif in the sequences flanking the center of the SP1-directed clusters is the canonical SP1 binding site. (C) The center of the calling card peaks shows a distribution centered on the SP1 binding site. (D) The top clusters identified by two biological replicates of SP1-directed calling cards show a 72% overlap.
Because PB prefers to insert at the tetranucleotide TTAA, we were cognizant that the resolution of the method may be limited by the local availability of TTAA sites surrounding the SP1 binding site. To investigate this possibility, we analyzed the distances between SP1 consensus sequences and centers of calling card clusters (Figure 3C). The median difference was 6 bp with a standard deviation of 293 bp. The median is on par with the same calculation for ChIP-seq data, which can range from 0 to 20 bp (Wilbanks and Facciotti 2010), although the standard deviation is somewhat higher for calling card peaks than for ChIP-seq, which is typically 20–120 bp. Thus, we conclude that the resolution of the method is not severely limited by PB’s insertion preference.
To estimate the positive predictive value of the calling card method, we compared it to the ChIP-seq method. We found that 80% of the SP1-directed calling card clusters had a nearby ChIP-seq peak (within 250 bp); 93% of the clusters had either a nearby ChIP-seq peak or a nearby SP1 site (Figure 3A). As a control we performed a similar analysis of calling cards deposited by the native PBase and found that only 5% of them had a nearby SP1 ChIP-seq peak (Figure 3A). Thus, we conclude that the calling card method has a high positive predictive value.
To estimate the sensitivity of the method, we created a list of high-confidence SP1 binding targets. To minimize the possibility of including false positives (Kharchenko et al. 2008), we required that “high-confidence” targets meet multiple criteria: they must appear in TRANSFAC (Matys et al. 2003), must contain an SP1 binding site, and must be identified by ChIP-seq in two data sets: the first being the ChIP-seq experiment, which we performed in the same cell line in which we performed the calling card experiments; the second being an SP1 ChIP-seq data set from the ENCODE project (Valouev et al. 2008) (Table S3). The sensitivity of the calling card method was then calculated as the percentage of targets on this list that have a significant calling card cluster. We found that 58% of the high-confidence targets contained significant calling card clusters. We hypothesized that some of the false negatives from these high-confidence targets may be the result of a lack of the required TTAA site near the SP1 binding site and could not allow a PB transposon to insert nearby. To test if there was a TTAA dependency we divided the high-confidence targets into two groups, on the basis of whether they had a TTAA site within 250 bp of its SP1 binding site. We observed a significant deviation between the two groups with a 79% sensitivity for targets near a TTAA site compared to a 38% sensitivity in targets without a TTAA site nearby (P = 0.0012, Fisher’s exact test), indicating that some targets may not be identified due to the stringency of the PB transposon.
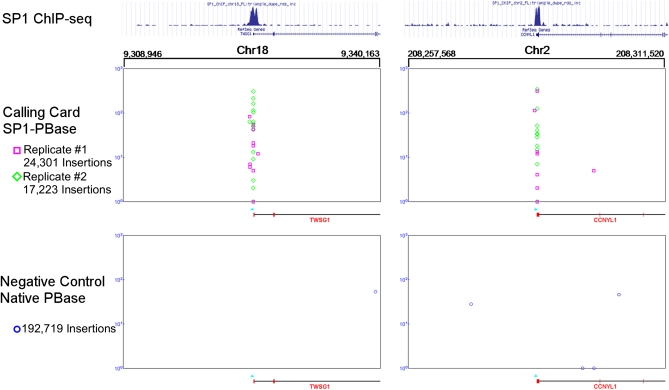
The calling card clusters displayed a high degree of concordance in two biological replicates with the top 800 clusters identified by two SP1-PBase biological replicates yielding a 72% overlap (Figure 3D). In addition, calling card clusters showed a high concordance with the ChIP-seq peaks at target genes (Figure 4). The P-values for significance in the two calling card biological replicates showed very high correlation (Pearson correlation coefficient, 0.973) (Figure 5), suggesting that the calling card method is reproducible and that the number of insertions collected is sufficient to identify most clusters.
Figure 4 .
The distribution of SP1-directed calling card insertions are enriched around SP1 binding sites (indicated by the light blue triangles) in the promoters of two genes in two biological replicates (indicated by the green diamonds and pink squares). The x-axis specifies genomic position; the y-axis represents the number of sequencing reads for each insertion (indicated by the diamond or square). The ChIP-seq distribution for each position is shown above and shows high spatial concordance with the clusters of calling cards. These clusters are not observed in the background distribution (indicated by dark blue circles) of the native PBase.
Figure 5 .
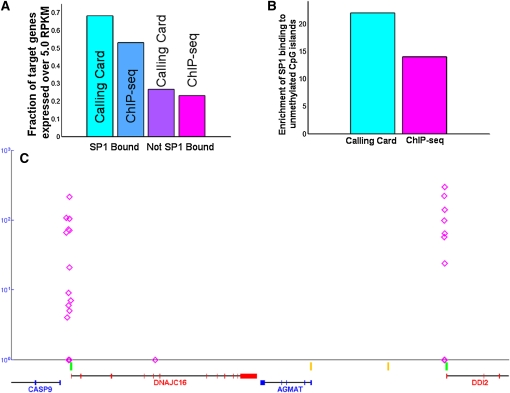
(A) The RefSeq genes identified as SP1 targets by both calling card and ChIP-seq are more likely to be expressed than genes not bound by SP1, consistent with the known function of SP1 as a general activator. (B) CpG islands bound by SP1 as assayed by both calling card and ChIP-seq are more likely to be unmethylated than methylated in the tested cell line. (C) SP1-directed PB insertions (pink diamonds) preferentially bind unmethylated CpG islands (shown in green) and over methylated CpG islands (shown in yellow).
SP1 preferentially binds unmethylated CpG islands near expressed genes
SP1 is thought to be a general transcriptional activator whose targets include TATA-less housekeeping genes. It is thought to play a role in regulating the cell cycle, the regulation of apoptosis, and the prevention of methylation at CpG islands (Philipsen and Suske 1999). A wide variety of gene ontology terms are enriched in SP1 targets identified with calling cards (Table S4), including the regulation of the cell cycle (GO:0051726, P = 1.89E-3), and the regulation of apoptosis (GO:0042981, P = 1.18E-2), so the target genes identified by calling card are consistent with the known functions of SP1.
Because SP1 is believed to behave primarily as a transcriptional activator (Philipsen and Suske 1999; Wierstra 2008), we hypothesized that SP1 target genes are more likely to be expressed than genes that are not predicted to be SP1 targets. To test this, we measured transcription levels in HCT116 cells. Both the calling card and ChIP-seq methods identified SP1 target genes whose transcripts were measured above an expression cutoff (see Materials and Methods), with genes identified by calling cards showing a 2.5-fold enrichment and genes identified by ChIP-seq showing a 2.3-fold enrichment (Figure 5A).
SP1 has been associated with the initiation and maintenance of methylation-free CpG islands (Lania et al. 1997). We observed that 49% of all significant SP1 calling card clusters are near CpG islands (within 250 bp). DNA methylation data for the same cell type (Brunner et al. 2009) revealed that SP1 preferentially binds unmethylated CpG islands. CpG islands harboring SP1 calling card clusters showed a 22-fold preference to be unmethylated vs. methylated (Figure 5, B and C). ChIP-seq revealed a similar 16-fold enrichment of SP1 binding to unmethylated CpG islands. The strong agreement between the SP1 targets identified by calling card insertions and the known functions of this transcription factor further demonstrates that the calling card method identifies functionally relevant SP1 targets.
Discussion
The calling card method provides an accurate and reproducible way to detect transcription factor binding that is orthogonal to ChIP and may be useful for analysis of the many TFs that appear to be recalcitrant to ChIP analysis (perhaps due to poor antibody quality).
While many calling card clusters show high concordance with ChIP-seq peaks, there are a number of peaks discordant between the two methods. Other orthogonal methods for measuring genomic data can generate disparate results at certain loci; for example, DNA methylation assayed by bisulfite-based sequencing methods and by immunoprecipitation enrichment methods yield similar, but not identical, results, particularly in terms of quantification (Harris et al. 2010). Of principal concern is the constraint on piggyBac to insert almost exclusively at the sequence TTAA, which can prevent the TF-PBase from recording its visit to regions devoid of that tetranucleotide. We observed ∼1% PB insertions at non-TTAA sites, raising the prospect of engineering the PB transposase to direct insertions into a broader range of target sequences. Alternatively, it may be possible to use other, more promiscuous transposons.
A second potential limitation of the method is that it currently requires the presence of at least one of the three restriction enzyme cleavage sites to be present in the genomic sequence at an acceptable distance from the transposon (to enable amplification of the inverse PCR product on an Illumina flowcell). This limitation could be overcome by using an alternative recovery protocol, such as a single molecule sequencing platform to map calling cards locations (Harris et al. 2008; Eid et al. 2009), a strategy that would potentially eliminate other amplification biases in our recovery protocol.
Another potential limitation is that the method requires fusing a transposase to the TF, which could interfere with the binding of the TF to DNA, or preclude other cofactors from binding to it. This potential problem might be mitigated by testing N-terminal or C-terminal transposase fusions, or a different linker between the TF and the transposase could be used.
Finally, it is possible that the insertion of a transposon near the binding site of a TF may alter expression of the nearby gene. We believe this is not a common occurrence for several reasons. First, our experience applying the calling card method to the more compact Saccharomyces cerevisiae genome revealed no bias against insertion into the promoters of essential genes relative to nonessential genes (Wang et al. 2011). Second, in this study we found that many essential genes contained clusters of SP1-directed PB transposons near their transcription start sites (Table S5), indicating that cells with insertions in the promoters of essential genes remain viable. Finally, the modest transposition rate of PB (Wu et al. 2006) ensures that the probability of observing an insertion in both copies of a promoter in a diploid genome is extremely low.
The principal advantage of the calling card approach is that, unlike either ChIP or DamID, it provides a permanent record of a DNA-binding protein’s visit to the genome, thus enabling the method to approach problems that current methods are incapable of addressing. ChIP provides only a snapshot of transcription-factor binding at the moment the protein is cross-linked to DNA. ChIP also necessitates harvesting the cells at the time of cross-linking, making it impossible to connect the measured transcription factor binding events to subsequent fates of the cell. In contrast, calling cards record TF–DNA interactions that occurred at the time in the culture’s (or potentially the organism’s) development when the TF-PBase was expressed. Those binding events can be identified later, in their descendent cells, because the transposon is a permanent part of the genome. The calling card method holds great promise for understanding transcriptional regulation during cell fate decisions and the essential roles played by transcription factors during differentiation, because their binding patterns in precursor cells can be correlated to resulting cell fates.
We believe our PB calling card protocol is ready to be applied to study cell differentiation in vitro. The transiently transfected PB transposon and transposase plasmids will become diluted with each cell division, such that only the PB transposons that become integrated in the genome will be recovered after cell differentiation and selection. This approach should enable mapping TF DNA-binding events that occur throughout the differentiation of stem cells or during reprogramming of differentiated cells into iPS cells (Takahashi and Yamanaka 2006). PB calling cards offer a unique opportunity to correlate the binding of TFs in the early stage of cell differentiation or reprogramming with the final outcomes. Ultimately, we expect to be able to use calling cards in vivo to map the protein–DNA interactions that happen in early development in mice by recovering from fully grown animals the calling cards deposited during their development.
Finally, we have demonstrated deliberate delivery of transposons to a specific subset of loci in the human genome. By fusing the PB transposase to different DNA-binding proteins, PB could be delivered to genes involved in specific cellular functions or signaling pathways. By combining PB calling cards with available transposon tools (such as gene traps), one might be able to enrich for transposon insertions in genes of specific pathways for mutagenesis screens. Since piggyBac is active in many different species, including yeast (Mitra et al. 2008), insects (Bossin et al. 2007), zebrafish (Lobo et al. 2006), mouse, and human (Ding et al. 2005), the calling card method could enjoy wide use.
Supplementary Material
Acknowledgments
We are especially grateful to Stefan Moisyadi (Hawaii University) for generously providing reagents and advice and to Jessica Hoisington-Lopez, Laura Langton, Brian Koebbe, and Jim Dover (Washington University) and the Washington University Genome Technology Access Center for their expert assistance with the use of Illumina Genome Analyzer. This work was supported by National Institutes of Health grants R21RR023960, 5R01DA025744-02, 5P50HG003170-03, and T32 HG000045 and by funds provided by the James S. McDonnell Foundation.
Sequence data from this article have been deposited with the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE34791.
Communicating editor: O. Hobert
Literature Cited
- Bossin H., Furlong R. B., Gillett J. L., Bergoin M., Shirk P. D., 2007. Somatic transformation efficiencies and expression patterns using the JcDNV and piggyBac transposon gene vectors in insect. Insect Mol. Biol. 16: 37–47 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., et al. , 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A. L., Johnson D. S., Kim S. W., Valouev A., Reddy T. E., et al. , 2009. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 19: 1044–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary L. C., Goebel M., Corsaro B. G., Wang H. G., Rosen E., et al. , 1989. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia in transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172: 156–169 [DOI] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., Han M., Zhuang Y., et al. , 2005. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122: 473–483 [DOI] [PubMed] [Google Scholar]
- Eid J., Fehr A., Gray J., Luong K., Lyle J., et al. , 2009. Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138 [DOI] [PubMed] [Google Scholar]
- Fejes A. P., Robertson G., Bilenky M., Varhol R., Bainbridge M., et al. , 2008. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics 24: 1729–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan S., Mularoni L., Fain-Thornton J., Wheelan S. J., Craig N. L., 2010. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci. USA 107: 21966–21972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. A., Wang T., Coarfa C., Nagarajan R. P., Hong C., et al. , 2010. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 28: 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. D., Buzby P. R., Babcock H., Beer E., Bowers J., et al. , 2008. Single-molecule DNA sequencing of a viral genome. Science 320: 106–109 [DOI] [PubMed] [Google Scholar]
- Huang X., Guo H., Tammana S., Jung Y. C., Mellgren E., et al. , 2010. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and PiggyBac transposons in human primary T cells. Mol. Ther. 18: 1803–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. D., Estep P. W., Tavazoie S., Church G. M., 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B., 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P. V., Tolsotorukov M. Y., Park P. J., 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lania L., Majello B., De Luca P., 1997. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell Biol. 29: 1313–1323 [DOI] [PubMed] [Google Scholar]
- Lobo N. F., Fraser T. S., Adams J. A., Fraser M. J., 2006. Interplasmid transposition demonstrates piggyBac mobility in vertebrate species. Genetica 128: 347–357 [DOI] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D., 1987. Plasmid construction by homologous recombination in yeast. Gene 58: 201–216 [DOI] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., et al. , 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R., Fain-Thornton J., Craig N. L., 2008. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 27: 1097–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen S., Suske G., 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27: 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., et al. , 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4: 651–657 [DOI] [PubMed] [Google Scholar]
- Sabo P. J., Kuehn M. S., Thurman R., Johnson B. E., Johnson E. M., et al. , 2006. Genome-scale mapping of DNaseI sensitivity in vivo using tiling DNA microarrays. Nat. Methods 3: 511–518 [DOI] [PubMed] [Google Scholar]
- Strathern J. N., Higgins D. R., 1991. Recovery of plasmids from yeast into Escherichia coli: shuttle vectors. Methods Enzymol. 194: 319–329 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S., 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A., Johnson D. S., Sundquist A., Medina C., Anton E., et al. , 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steensel B., Henikoff S., 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18: 424–428 [DOI] [PubMed] [Google Scholar]
- Vogel M. J., Peric-Hupkes D., van Steensel B., 2007. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2: 1467–1478 [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wang H., Johnston M., Mitra R. D., 2007. Calling cards for DNA-binding proteins. Genome Res. 17: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Mayhew D., Chen X., Johnston M., Mitra R. D., 2011. Calling Cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome Res. 21: 748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I., 2008. SP1: emerging roles: beyond constituitive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 372: 1–13 [DOI] [PubMed] [Google Scholar]
- Wilbanks E. G., Facciotti M. T., 2010. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE 5: e11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. H., Coates C. J., George A. L., 2007. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15: 139–145 [DOI] [PubMed] [Google Scholar]
- Wu S. C., Meir Y. J., Coates C. J., Handler A. M., Pelczar P., et al. , 2006. PiggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl. Acad. Sci. USA 103: 15008–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.