Abstract
Mitochondrial DNA (mtDNA) deletions are associated with sporadic and inherited diseases and age-associated neurodegenerative disorders. Approximately 85% of mtDNA deletions identified in humans are flanked by short directly repeated sequences; however, mechanisms by which these deletions arise are unknown. A limitation in deciphering these mechanisms is the essential nature of the mitochondrial genome in most living cells. One exception is budding yeast, which are facultative anaerobes and one of the few organisms for which directed mtDNA manipulation is possible. Using this model system, we have developed a system to simultaneously monitor spontaneous direct-repeat–mediated deletions (DRMDs) in the nuclear and mitochondrial genomes. In addition, the mitochondrial DRMD reporter contains a unique KpnI restriction endonuclease recognition site that is not present in otherwise wild-type (WT) mtDNA. We have expressed KpnI fused to a mitochondrial localization signal to induce a specific mitochondrial double-strand break (mtDSB). Here we report that loss of the MRX (Mre11p, Rad50p, Xrs2p) and Ku70/80 (Ku70p, Ku80p) complexes significantly impacts the rate of spontaneous deletion events in mtDNA, and these proteins contribute to the repair of induced mtDSBs. Furthermore, our data support homologous recombination (HR) as the predominant pathway by which mtDNA deletions arise in yeast, and suggest that the MRX and Ku70/80 complexes are partially redundant in mitochondria.
MITOCHONDRIA are essential organelles in eukaryotic organisms that generate cellular energy in the form of ATP. These organelles contain their own DNA, encoding essential electron transport chain and mitochondrial ATP synthase components. Mitochondrial DNA (mtDNA) also encodes the rRNAs and tRNAs required for mitochondrial protein synthesis. Proteins required for maintaining mtDNA are encoded in the nuclear genome, translated in the cytoplasm, and imported into mitochondria.
There is a strong correlation between the decline of oxidative phosphorylation, increased reactive oxygen species production within mitochondria, and increased accumulation of oxidative damage in aging individuals (Lee and Wei 2007). Deregulation of mitochondrial energetics and an age-associated accumulation of somatic mtDNA mutations supports the mitochondrial theory of aging (Lee and Wei 2007). The importance of mtDNA integrity is demonstrated by heritable human disease syndromes caused by mtDNA mutations (Falk and Sondheimer 2010). Accordingly, somatic mtDNA instability, and mutation accumulation over time, are associated with aging and are linked to systemic and neurodegenerative disorders including Parkinson’s, Alzheimer’s, and Huntington’s diseases and diabetes (Kajander et al. 2002; Yang et al. 2008a; Wallace 2010).
Deletions of the mitochondrial genome have been identified in yeast, plants, Drosophila, and mammalian cells (Dujon 1981; Morel et al. 2008; Bacman et al. 2009; Marechal and Brisson 2010; Oliveira et al. 2010). Interestingly, the majority of mtDNA deletions are flanked by short repetitive sequences, implicating recombination as a possible mechanism (Bianchi et al. 2001; Krishnan et al. 2008). Recombination of the mitochondrial genome has been well accepted in yeast and plants; and recombinant mtDNA molecules and homologous recombination (HR) activity have been detected in mammalian cells in vitro and in vivo (Poulton et al. 1993; Thyagarajan et al. 1996; Lakshmipathy and Campell 1999; Kajander et al. 2001; Kraytsberg et al. 2004). These studies and others provide evidence for mitochondrial double-strand break repair (mtDSBR); however, no proteins have been identified that function in these pathways.
Directly repeated sequences are hotspots for spontaneous deletion, in which one repeat and the interrepeat sequence are lost. In the nucleus, multiple pathways are implicated in the spontaneous generation of these direct-repeat–mediated deletions (DRMDs), including intrachromosomal recombination, unequal sister chromatid exchange (SCE), single-strand annealing (SSA), nonhomologous end joining (NHEJ), and polymerase slippage (Liefshitz et al. 1995). Because they share protein components and early processing steps, intrachromasomal recombination, SCE, and SSA are collectively considered HR pathways (Paques and Haber 1999). However, intrachromosomal recombination and SCE will produce reciprocal products, in addition to deletions, while SSA does not. These pathways appear to compete for substrates to some extent, since loss of one pathway can result in increased activity of other pathways. The relative contribution of each pathway to deletion formation can be altered by manipulation of the recombination substrate. For example, in yeast nuclei, when double-strand breaks (DSBs) are directed to the intervening sequence between the repeats, deletions are predominantly generated via SSA (Paques and Haber 1999).
A previous study in Drosophila utilized various promoters to target the restriction endonuclease XhoI to the mitochondrial compartment, causing a specific DSB in the cytochrome c oxidase subunit I locus. This study demonstrated the catastrophic effects of ubiquitous and selective expression of mitoXhoI and allowed the selection of mutant mtDNA genomes that were resistant to cleavage (Xu et al. 2008). Furthermore, Fukui and Moraes (2009) have generated a mouse model of mtDSB induction through mitochondrial targeting of the restriction endonuclease PstI and propose recombination and NHEJ as mechanisms of mtDSBR. Recent studies show that well-characterized nuclear DNA repair proteins also localize to mitochondria in yeast and mammals (Nakabeppu 2001; Zhang et al. 2006; Sage et al. 2010). Pertinent to this study, Rad50p and Mre11p were identified as members of the yeast mitochondrial proteome (Sickmann et al. 2003), an alternative form of Ku80 was identified in DNA end binding in mammalian mitochondria (Coffey and Campbell 2000), and Mre11 was recently found in mammalian mitochondria bound to mtDNA (Dmitrieva et al. 2011). These and other studies support the presence of mtDSBR pathways and provide the basis for the hypothesis that mtDNA repair employs the MRX and Ku70/80 complexes.
The conservation between nuclear and mitochondrial end-joining pathways has not been assessed, in part because our ability to directly compare nuclear and mitochondrial processes is limited. Although recent studies indicate that the nuclear DSBR proteins Mre11 and an isoform of Ku80 are present in mammalian mitochondria, and implicate these proteins in mtDNA maintenance (Coffey and Campbell 2000; Dmitrieva et al. 2011), extensive functional studies related to their role in recombination or DSBR have not been performed. Saccharomyces cerevisiae is a tractable model system for these studies; since these repair proteins are conserved, mtDNA is not essential for viability, and introduction of exogenous DNA is possible, allowing the generation of reporters for specific types of mtDNA mutations. We previously described the construction and use of yeast strains with reporters to simultaneously measure nuclear and mitochondrial DRMDs. Using these reporter strains we can monitor the mutagenic effect of DNA repair protein disruption on both mitochondrial and nuclear DNA in the same population of cells (Kalifa et al. 2009). In this study, we assessed mutagenic consequences of loss of the NHEJ pathway. Analysis of mutants that lack the MRX and Ku70/80 complexes reveals that recombination between repetitive sequences is the predominant mechanism to repair mtDSBs. In addition, these proteins impact repetitive DNA to a greater extent in mitochondria than in nuclei, suggesting that the interplay between the pathways leading to deletions may differ in these two compartments.
Materials and Methods
Media and strains
Media used in this study were previously described (Kalifa et al. 2009). S. cerevisiae strains used in this study are listed in Table 1 and are isogenic with DFS188 except NPY66, which is derived from DFS160 (Sia et al. 2000; Phadnis et al. 2005). Yeast single-deletion strains were constructed by one-step gene transplacement of the wild-type gene with a kanMX marker using standard methods (Adams et al. 1997). Strains containing two or more gene replacements were constructed by mating isogenic haploid strains, dissecting diploids, and screening for the desired genotypes by PCR. The DRMD reporter (LKY196) was constructed previously (Kalifa et al. 2009). Gene deletions in this strain were constructed by mating, followed by sporulation and dissection and screening for the appropriate genotype. These strains were cured of their mtDNA by treatment with ethidium bromide, then cytoduced with the REP96::ARG8m::cox2 mitochondrial DRMD reporter from NPY66 (Fox et al. 1991). Strains used for the analysis of nuclear point mutation accumulation were made Arg+ by mating and dissection or by standard yeast transformation with the wild-type ARG8 gene.
Table 1 . Strains used in this study.
| Strain | Relevant nuclear genotype | Mitochondrial genotype | Reference |
|---|---|---|---|
| DFS188 | MATa ura3-52 leu2-3, 112 lys2 his3 arg8::hisG | ρ+ | (Sia et al. 1997) |
| DFS160 | MATα ade2-101 leu2Δ ura3-52 arg8-Δ::URA3 kar1-1 | ρ0 | (Steele et al. 1996) |
| NPY66 | DFS160 | mit− REP96::ARG8m::cox2 | (Phadnis et al. 2005) |
| LKY196 | MATa ura3-52 leu2-3, 112 lys2 his3 arg8::hisG REP96::URA3::trp1 | mit− REP96::ARG8m::cox2 | (Kalifa et al. 2009) |
| LKY479 | LKY196 mre11-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| RCY376 | LKY196 rad50-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| RCY451 | LKY196 xrs2-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| LKY472 | LKY196 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| RCY373 | LKY196 ku70-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| RCY382 | LKY196 ku80-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| LKY477 | LKY196 ku70-Δ::kanMX ku80-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| LKY566 | LKY196 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX ku70-Δ::kanMX ku80-Δ::kanMX | mit− REP96::ARG8m::cox2 | This study |
| NPY183 | DFS188 mre11-Δ::kanMX | ρ+ | This study |
| NPY171 | DFS188 rad50-Δ::kanMX | ρ+ | This study |
| NPY179 | DFS188 xrs2-Δ::kanMX | ρ+ | This study |
| RCY416 | DFS188 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX | ρ+ | This study |
| NPY167 | DFS188 ku70-Δ::kanMX | ρ+ | This study |
| NPY169 | DFS188 ku80-Δ::kanMX | ρ+ | This study |
| RCY423 | DFS188 ku70-Δ::kanMX ku80-Δ::kanMX | ρ+ | This study |
| LKY483 | DFS188 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX ku70-Δ::kanMX ku80-Δ::kanMX | ρ+ | This study |
| NPY054 | DFS188 ARG8 | ρ+ | (Phadnis and Sia 2004) |
| LKY464 | NPY054 mre11-Δ::kanMX | ρ+ | This study |
| LKY453 | NPY054 rad50-Δ::kanMX | ρ+ | This study |
| RCY447 | NPY054 xrs2-Δ::kanMX | ρ+ | This study |
| LKY468 | NPY054 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX | ρ+ | This study |
| RCY411 | NPY054 ku70-Δ::kanMX | ρ+ | This study |
| RCY349 | NPY054 ku80-Δ::kanMX | ρ+ | This study |
| LKY455 | NPY054 ku70-Δ::kanMX ku80-Δ::kanMX | ρ+ | This study |
| LKY485 | NPY054 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX ku70-Δ::kanMX ku80-Δ::kanMX | ρ+ | This study |
| EAS748 | DFS188 | mit− REP96::ARG8m::cox2 | (Phadnis et al. 2005) |
| LKY623 | DFS188 + pYES2.1 | mit− REP96::ARG8m::cox2 | This study |
| EAS811 | DFS188 + pEAS100 | mit− REP96::ARG8m::cox2 | This study |
| LKY642 | DFS188 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX + pYES2.1 | mit− REP96::ARG8m::cox2 | This study |
| LKY648 | DFS188 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX + pEAS100 | mit− REP96::ARG8m::cox2 | This study |
| LKY644 | DFS188 ku70-Δ::kanMX ku-Δ80::kanMX + pYES2.1 | mit− REP96::ARG8m::cox2 | This study |
| LKY650 | DFS188 ku70-Δ::kanMX ku-Δ80::kanMX + pEAS100 | mit− REP96::ARG8m::cox2 | This study |
| LKY646 | DFS188 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX ku70-Δ::kanMX ku-Δ80::kanMX + pYES2.1 | mit− REP96::ARG8m::cox2 | This study |
| LKY652 | DFS188 mre11-Δ::kanMX rad50-Δ::kanMX xrs2-Δ::kanMX ku70-Δ::kanMX ku-Δ80::kanMX + pEAS100 | mit− REP96::ARG8m::cox2 | This study |
Mitochondrially localized KpnI was constructed by amplifying the bacterial KpnI gene from pETRK (Saravanan et al. 2004) using a 5′ primer containing the mitochondrial localization signal of COX4 5′-GGATCCATGCTGAGCCTGCGCCAGAGCATTCGCTTTTTTAAACC GGCGACCCGCACCCTGTGCAGCAGCCGCTATCTGGTGATGGATGTCTTTGATAAAGTTTATAG-3′ and 3′ primer 5′-AAGCTTGATATATGATAGTATTAGTGAAG-3′. The PCR product was cloned into pYES2.1 using the TOPO TA Cloning kit (Invitrogen), then transformed into Top10F′ bacterial cells containing pACMK+, a plasmid that carries the KpnI methylase (Saravanan et al. 2004). Transformants were screened by digestion with restriction endonucleases, BamHI and HindIII, and positive clones were confirmed by sequencing. pYES2.1 containing mtLS-KpnI (pEAS100) and empty pYES2.1 vector were transformed into EAS748 and mutant strains containing the mitochondrial REP96::ARG8m::cox2 reporter.
Fluctuation analysis
Rates of nuclear and mitochondrial DRMD using the 96-bp repeat reporters were determined as described previously (Kalifa et al. 2009) with minor modifications. Briefly, cells containing both intact reporters were selected on SD −Arg −Ura media, and independent colonies were isolated on YPD to release from selection. Individual colonies were resuspended in water and dilutions were plated on SD −Trp to select for nuclear deletions that restore the TRP1 gene, YPG for mitochondrial deletions restoring the COX2 gene, and on YPD for the total number of cells. All plates were incubated at 30° for 3 days. To insure that functional mtDNA was not lost in any of the DRMD reporter strains at frequencies high enough to affect the analysis, all strains were grown exactly as for the above assays and appropriate dilutions were plated on YPD. The resulting colonies were replica plated to SD −Arg and YPG. The percentages of Arg− and respiratory-deficient colonies were between 4.4 and 7.1% for all strains, with no significant differences between strains (P > 0.05 for all pairwise comparisons). For more accurate assessment of nuclear DRMD in strains, cells containing both reporters were selected on SD −Arg −Ura and independent colonies were isolated on YPD as described above. Fifteen independent colonies were then used to inoculate 5-ml YPD cultures and grown to saturation. Appropriate dilutions were plated on YPD and SD −Trp and incubated for 3 days at 30°.
To determine the average frequency of respiration loss, cells were grown on YPG medium then inoculated in liquid YPD medium. Independent colonies were isolated on YPD and subsequently inoculated in YPD liquid. Dilutions were plated on YPG + 0.1% dextrose allowing for analysis of “petite” vs. respiring colonies. The rates of mitochondrial and nuclear point mutation were determined as previously described (Sia et al. 2000; Kalifa and Sia 2007).
Southern blot analysis
DNA samples for Southern blot analysis were prepared by growing cells to saturation in SD complete for DFS188, SD −Arg for EAS748, or SD −Arg −Ura for strains with the REP96::ARG8m::cox2 mitochondrial reporter and either pYES2.1 or pEAS100. These cultures were diluted into the same media to an OD600 of <0.05 and grown to an OD600 of 0.3–0.4. Cells were collected and resuspended in an equal volume of synthetic media containing 2% galactose (Sgal). Samples were collected after 0, 0.5, 1, 2, and 3 hr, and total cellular DNA was extracted using standard protocols. DNA was digested with AvaII and subjected to gel electrophoresis on a 0.8% 1× TAE agarose gel containing ethidium bromide. DNA was transferred to Amersham Hybond-XL nylon membrane (GE Healthcare) using a standard Southern blotting technique. The 583-bp COX2 DNA probe was generated by PCR with primers 5′-GACAAAAGAGTCTAAAGGTTAAG-3′ and 5′-CATCAGCAGCTGTTACAACGA-3′. The probe for the 21S rRNA sequence was constructed by amplifying ∼200 bp of the 21S rRNA mitochondrial gene using primers 5′-GGTAAATAGCAGCCTTATTATG-3′ and 5′-CGATCTATCTAATTACAGTAAAGC-3′. The nuclear 25S rRNA gene probe was generated by amplifying 193 bp with primers 5′-CCGGTTAAGATTCCGGAAC-3′ and 5-CTGTGGATTTTCACGGGCC-3′. DNA probes were isolated in low-melt agarose and labeled using the Prime-a-Gene labeling system (Promega) with α-32-dCTP. For each set of strains, DNA was obtained from three to five independent experiments; representative blots are shown in Figures 2 and 3. Quantification of Southern blots was performed using Quantity One software (Quantity One 1-D Analysis software; BioRad Laboratories, Hercules, CA). None of the bands were saturated in the exposures used for quantification. The percentage of the COX2 signal represented by deletion product and break product was calculated by measuring the intensity of the band and dividing by the total signal for intact reporter, the break product, and deletion product. The values displayed below the blot signify the average percentage of deletion and break product bands for each strain for at least three independent experiments. Statistical analyses were performed by comparing the percentages of each product obtained from three to five independent experiments using paired t tests.
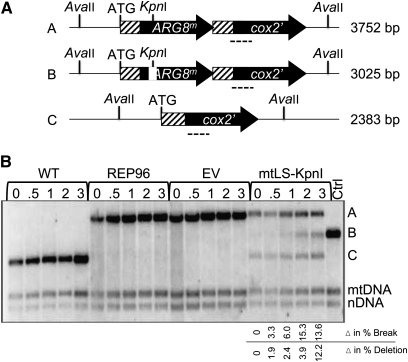
Figure 2 .
Inducing a specific mtDSB with mtLS-KpnI. (A) Schematic of probes used for detection of mtDSB. Dotted line indicates where the probe anneals. (B) Representative Southern blot of AvaII-digested DNA extracted from wild-type strains after induction of mtLS-KpnI. WT (DFS188) contains an intact COX2 gene and was grown in SD −complete media and time points were taken in Sgal −complete media. REP96 (EAS748) contains the REP96::ARG8m::cox2 reporter and was grown in SD −Arg media and time points were taken in SGal −complete media. EV (LKY623) contains the REP96::ARG8m::cox2 reporter and pYES2.1, and was grown in SD −Arg −Ura media with time points taken in SGal −Ura media. mtLS-KpnI (EAS812) contains the REP96::ARG8m::cox2 reporter and pEAS100. It was grown in SD −Arg −Ura media and time points were taken in Sgal −Ura media. Ctrl is the LKY623 T = 0 sample digested with KpnI in vitro to depict where the break product B would be relative to the other COX2 bands. The 21S rRNA gene was probed to detect total mtDNA. The nuclear 25S rRNA gene was probed for total nuclear DNA. The average increases in recombinant and break product relative to time 0 for several independent experiments are given. Data were obtained by quantification of Southern blots as described in Materials and Methods. Significance of increase by 3 hr after shift to galactose was determined using paired t tests to compare the percentage of each product before and after endonuclease induction.
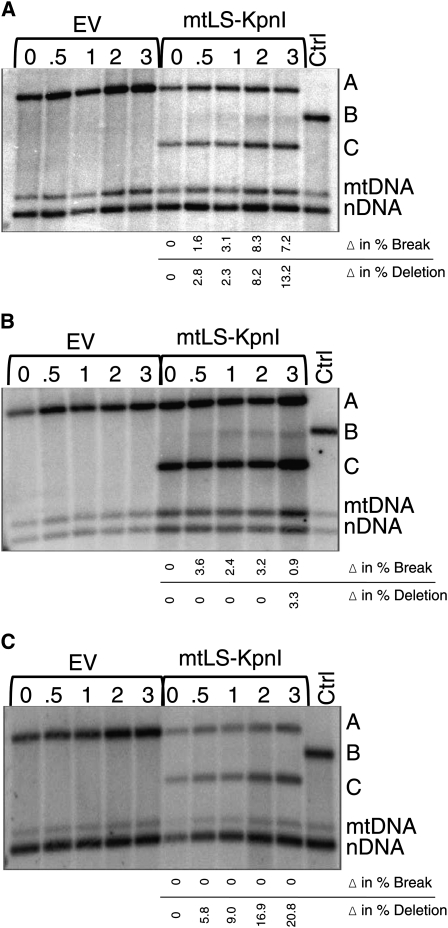
Figure 3 .
Inducing a specific mtDSB with mtLS-KpnI in MRX and KU70/80 mutant strains. (A) Induction of a specific mtDSB in mrx-Δ cells. EV (LKY642) contains the REP96::ARG8m::cox2 reporter and pYES2.1, and mtLS-KpnI (LKY648) contains the REP96::ARG8m::cox2 reporter and pEAS100. (B) Induction of a specific mtDSB in ku70/80-Δ cells. EV (LKY644) contains the REP96::ARG8m::cox2 reporter and pYES2.1, and mtLS-KpnI (LKY650) contains the REP96::ARG8m::cox2 reporter and pEAS100. A darker exposure of this blot is shown so that the break product can be seen. (C) Induction of a specific mtDSB in mrx-Δku70/80-Δ cells. EV (LKY646) contains the REP96::ARG8m::cox2 reporter and pYES2.1, and mtLS-KpnI (LKY652) contains the REP96::ARG8m::cox2 reporter and pEAS100. All strains were grown in SD −Arg −Ura media and time points were taken in SGal −Ura media. Total DNA was extracted, digested with AvaII, and subjected to gel electrophoresis on a 0.8% agarose gel prior to Southern blotting. Ctrl is the EV T = 0 sample digested with KpnI in vitro to depict where the break product B would be relative to the other COX2 bands. The 21S rRNA gene was probed to detect total mtDNA. The nuclear 25S rRNA gene was probed for total nuclear DNA. The average increases in deletion and break product relative to time 0 for several independent experiments are given. Data were obtained by quantification of Southern blots as described in Materials and Methods. Significance of increase by 3 hr after shift to galactose was determined using paired t tests to compare the percentage of each product before and after endonuclease induction.
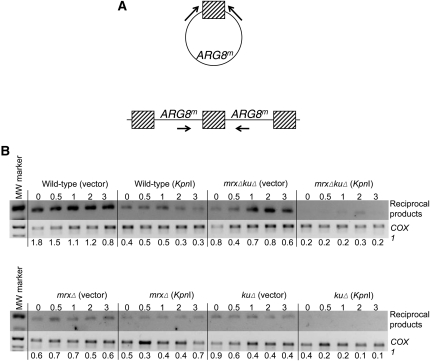
PCR analysis of reciprocal products
Yeast genomic DNA samples used for Southern blotting were used as a template for PCR amplification. The 485 bp of COX1 sequence was amplified using primers 5′-CATGTGTATT AACTTTAGCTAGTAA-3′ and 5′-TTTCCATATTGAGTATCATTAATAATA-3′, and the 489-bp fragment representing reciprocal products of recombination was amplified using primers 5′-GATGTGAATCTTCTTGATGATGTTG-3′ and 5′-TCCTTTAGCTTGTTCAGTATCAAATT-3′. To determine the appropriate template concentrations, dilutions of the templates were used for amplification at 25 cycles to ensure that amplifications were in the linear range in this cycle number. In the experiments presented in Figure 5, all samples were diluted 1:10 for PCR analysis. Equal volumes of the PCR-amplified products were electrophoresed through a 1.3% agarose gel and stained with ethidium bromide. The images were obtained using a BioRad Gel Doc XR and band intensity was determined using Quantity One software. To account for differences in intensity of ethidium bromide staining, the amount of PCR product was normalized to the corresponding 506- and 517-bp bands from the 1-kb ladder indicated in Figure 5b. After normalization, the ratio of reciprocal product to COX1 was calculated.
Figure 5 .
Analysis of KpnI resistance. Representative gel of PCR amplification and digestion with KpnI in vitro of time 0 samples grown under chronic DSB conditions. Cells were grown to saturation in SD −Arg −Ura, then subcultured twice in the same media, and total DNA was extracted for PCR. Amplification of the ARG8m fragment from LKY196 served as the control for KpnIR (R) and KpnI sensitive (S) populations. Quantification of independent experiments is present in Table 3.
Determination and sequencing of KpnI resistance under chronic mtLS-KpnI conditions
Total genomic DNA was prepared from samples that were grown to saturation, then subcultured twice and grown again to saturation, resulting in ∼16 generations. SD −Arg −Ura medium was used to select for the intact REP96::ARG8m::cox2 mitochondrial reporter and either pYES2.1 or pEAS100.
To determine whether yeast genomic DNA samples contained KpnI-resistant (KpnIR) DNA molecules, we performed PCR of a fragment of the ARG8m gene using primers 5′-CTGTAACAGCTTTAGGTCATG-3′ and 5′-CATGAGGTACTAAATCACCG-3′. PCR products were digested with KpnI to assess the percentage of KpnIR molecules in each reaction. Undigested PCR product was cloned into pCR2.1 using the TOPO TA Cloning kit and transformed into TOP10F′ cells. Transformants were screened for KpnIR by digestion with EcoRI and KpnI and analyzed on an agarose gel. KpnIR clones were sequenced using the Functional Genomics Center at the University of Rochester Medical Center, Rochester, NY.
Statistical analysis
Fluctuation analyses consisted of 10–20 independent samples each. Rates were determined using the method of the median (Lea and Coulson 1949). For strains in which the median was 0, the method developed by Luria and Delbruck (1943) was used to estimate the upper limit of mutational events and are indicated. The average rate or frequency (depending on the assay) of three or more independent experiments is shown. P-values were determined by unpaired, two-tailed t tests using either InStat 3 for Macintosh (GraphPad, San Diego) or Microsoft Excel for Macintosh 2011, unless otherwise indicated.
Results
Comparing the rates of nuclear and mitochondrial DRMD events
The conservation between nuclear and mitochondrial DSBR is largely uninvestigated. To address this, we generated genetic reporters that are integrated into the nuclear and mitochondrial genomes of the same yeast strains to measure the rate of spontaneous DRMD events in both compartments (Figure 1, a and b) (Kalifa et al. 2009). The nuclear DRMD reporter consists of the URA3 gene inserted 99 bp into the endogenous TRP1 gene on chromosome IV followed by the entire TRP1 gene lacking the start codon (Kalifa et al. 2009). The mitochondrial DRMD construct utilizes the ARG8m reporter gene, a mitochondrial derivative of the nuclear ARG8 gene, recoded to reflect a mitochondrial codon preference (Steele et al. 1996). The mitochondrial DRMD reporter consists of the ARG8m gene inserted 99 bp into the COX2 gene in the mitochondrial genome followed by the entire COX2 gene lacking the start codon (Phadnis et al. 2005). Both reporters contain 96-bp repeats flanking an intervening sequence of ∼1.4 kb. Strains with both intact reporters are phenotypically Ura+ Trp− Arg+ and respiration deficient. Nuclear DRMDs result in cells that are Ura− and Trp+ and mitochondrial DRMDs result in Arg− respiratory-proficient cells.
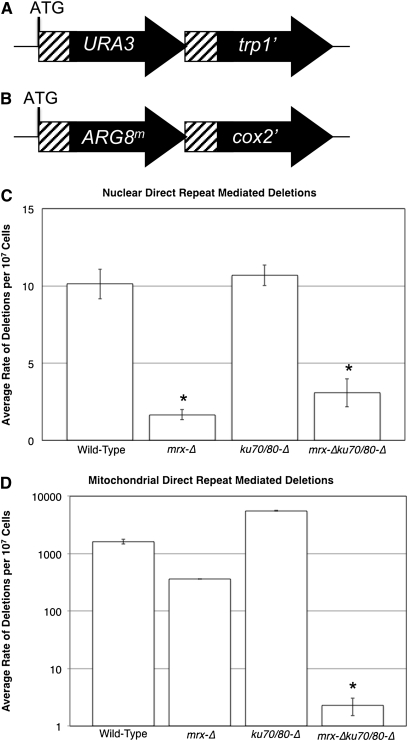
Figure 1 .
Nuclear and mitochondrial direct-repeat mediated deletions. (A) Nuclear DRMD reporter consists of the URA3 gene inserted 99 bp into the TRP1 gene followed by the entire TRP1 gene lacking the start codon (Kalifa et al. 2009). (B) Mitochondrial reporter consists of the ARG8m gene inserted 99 bp into the COX2 gene followed by the entire COX2 gene lacking the start codon (Phadnis et al. 2005). Both reporters contain 96 bp of directly repeated sequence shown in hashed boxes. Strain with intact reporter constructs are Ura+ Trp− Arg+ and respiratory deficient. (C) Average rates of nuclear DRMD. (D) Average rates of mitochondrial DRMD plotted on a log scale. Error bars indicate the SEM. *, at least one or more of the rates used to determine the average was derived using the Luria and Delbruck (1943) method due to a median value of 0.
Previous studies in yeast have shown that the rate of deletions is positively correlated with the size of the repeat length, and this correlation is linear above a minimal threshold, called minimal efficient processed segment (MEPS) (Shen and Huang 1986; Sugawara et al. 2000; Phadnis et al. 2005). In yeast, the MEPS length is 33 bp and between 21 and 33 bp for nuclear and mitochondrial DNA, respectively (Sugawara et al. 2000; Phadnis et al. 2005). The identified repeat size flanking human mtDNA deletions is typically ≤16 bp; however, we used the 96-bp repeat reporter to facilitate accurate and sensitive measurements of both increases and decreases in the rates of nuclear and mitochondrial DRMD in yeast.
Loss of Mre11/Rad50/Xrs2 and Ku70/80 complexes significantly affects the rate of spontaneous mitochondrial deletions
The wild-type strain displays spontaneous rates of 10.1 × 10−7 deletions/cell division (nuclear) and 1629 × 10−7 deletions/cell division (mitochondrial) (Figure 1, c and d). Therefore, repeats of the same size, separated by approximately the same distance, and tested under identical extracellular and intracellular conditions appear to be 160-fold less stable in mtDNA than in nuclear DNA. It is important to note that we cannot directly compare the number of events in both compartments, as additional processes, including replication choice and segregation of mtDNA into daughter cells will also impact the observed rate of mitochondrial mutations. However, from an organismal standpoint, the end result of all of these processes is a selectable, spontaneous deletion frequency that is considerably higher than observed for similar nuclear repeats.
In the following studies, we examined whether the known NHEJ proteins are required for the high frequency of DRMDs in mitochondria. The Mre11/Rad50/Xrs2 and Ku70/80 complexes are both implicated in nuclear DRMD events (Ivanov et al. 1996; Krogh and Symington 2004). We determined that deletion of all three proteins of the MRX complex reduces the rate of spontaneous nuclear (6.1-fold; P = 6.2 × 10−4) and mitochondrial DRMD (4.5-fold; P = 1.0 × 10−3) to a similar extent relative to wild-type rates (Figure 1, c and d). This decrease in nuclear events may result from a delay in the repair of double-strand breaks (DSBs), since previous studies indicate that the MRX complex functions in the processing of breaks, and in its absence, the kinetics of SSA at direct repeats are slowed but not abolished (Sugawara and Haber 1992). The similar effect of MRX loss on nuclear and mitochondrial deletions may suggest a similar function in both compartments.
In contrast, the ku70-Δ ku80-Δ mutant did not show decreased deletion rate in either cellular compartment; its rate of nuclear DRMD was not significantly different from our wild-type strain (P > 0.7); and its mitochondrial DRMD rate was 3.4-fold higher than wild type (P = 7.6 × 10−8) (Figure 1, c and d). The lack of an effect on nuclear DRMD may reflect a minor role of Ku70/80-mediated NHEJ in this capacity within the nucleus or suggest alternate NHEJ components. These data are consistent with Ku70/80-mediated NHEJ as a mechanism for preventing spontaneous mitochondrial deletions. In the absence of the Ku70/80 proteins, mitochondrial breaks that were repaired to reconstitute the original reporter may now be repaired via recombination-dependent pathways that lead to deletions, such as SSA.
Synergistic decreases in spontaneous mitochondrial deletions suggest separate pathways of MRX and Ku complex activities
To test whether the MRX complex, which in yeast nuclei is involved in both HR and NHEJ, is important for deletions in strains lacking the Ku complex, we measured the rate of mitochondrial and nuclear DRMD in a mutant strain lacking the five genes representing both complexes. We observed a >715-fold decrease in spontaneous mitochondrial DRMD relative to wild type (P = 6.5 × 10−6) (Figure 1d). These data indicate that the mitochondrial deletions observed in the ku70-Δ ku80-Δ strain are dependent on the MRX complex. The nuclear deletion rate in the mrx mutant strain is close to the lower limit of the assay as performed. To more accurately measure the rate of nuclear mutation in these strains, we allowed independent cultures to proceed through additional divisions before measuring mutation rates, so that larger numbers of cells could be plated on selective media. We find that the average rate of nuclear DRMD in the wild-type strain (18.0 × 10−7) under these conditions is slightly higher than the rate shown in Figure 1c (10.1 × 10−7). There was a comparable increase in the rate of DRMD in the ku70/80-Δ strain (17.0 × 10−7), so that there was no significant difference in the nuclear deletion rates in these strains, consistent with our previous results. The mrx-Δ strain shows a threefold decrease relative to the wild-type (6.0 × 10−7, P = 8.6 × 10−3), similar to the mrx-Δ ku70/80-Δ strain (3.1 × 10−7). Therefore the synergistic decrease in mitochondrial DRMD in the mrx-Δ ku70/80-Δ strain contrasts with the nuclear phenotypes of these mutants, where the same strain displays a rate of DRMDs not significantly different from the mrx-Δ strain (P = 0.06) (Figure 1c). Synergistic decreases in mitochondrial deletions seen in the five-gene deletion relative to the strains lacking either MRX or Ku complexes alone implies that these proteins act in separate pathways required for the generation of mitochondrial, but not nuclear deletions. Furthermore, loss of these complexes has a greater effect on mitochondrial repetitive DNA than comparable nuclear sequences and suggests that the mechanisms of repair differ between these two compartments or that mitochondria lack an alternative repair pathway that can compensate for the loss of MRX and Ku70/80 complexes in the nucleus.
The MRX and Ku complexes affect the repair of mitochondrial double-strand breaks
Several mechanisms contribute to spontaneous DRMD events in the nucleus; it is the interplay of these recombination and end-joining pathways that gives rise to the deletion rates we observe in our wild-type strains (Prado et al. 2003). Experimentally induced DSBs between repeated sequences, however, are preferentially repaired by SSA (Sugawara and Haber 1992). Thus, the contribution of individual proteins to DRMD is influenced by how deletion events are initiated. To examine the effect of introducing a DNA break into our mitochondrial DRMD reporter, we constructed a yeast strain that allows us to introduce a mtDSB between the repeated sequence. This system takes advantage of a single KpnI site that occurs 370 bp 3′ of the ARG8m sequence start. To cleave this site, we fused an N-terminal mitochondrial localization sequence to a plasmid-borne, galactose-inducible KpnI (mtLS-KpnI). Addition of galactose to yeast growth medium induces mtLS-KpnI expression, which will be targeted to mitochondria and cleave at the mtDNA KpnI site. This cleavage results in a DSB with 4 -bp 3′ single-stranded complementary ends.
This system successfully generates the predicted deletion product (Figure 2). We find that there is no significant loss of mtDNA, as determined by the ratio of the mitochondrial 21S rRNA hybridization to the nuclear 25S rRNA control, relative to the controls strains lacking the endonuclease. Furthermore, the ratio of the total COX2 signal (A+B+C) to 21S rRNA remains constant, indicating that sequences close to the break are not preferentially lost.
At each time point, the amount of deletion product and break product was measured and is presented as a percentage of the total COX2 signal (Table 2). The values below the representative blot in Figure 2b summarize the average increase in the percentage of deletion product at 0, 0.5, 1, 2, and 3 hr after release from selection for the ARG8m reporter for several independent experiments.
Table 2 . Effects of mitochondrial DSB induction.
| Relevent genotype | Plasmid | Time (hr) | Average % deletion product (SD) | Average % breaks (SD) | Average frequency of ρ+ |
|---|---|---|---|---|---|
| Wild type | vector | 3 | 0.35 (0.42) | — | 1.1 × 10−3 |
| Wild type | mtLS-KpnI | 0 | 26.5 (14.1) | 1.14 (1.2) | |
| 0.5 | 28.3 (10.6) | 4.42 (3.0) | |||
| 1 | 28.8 (11.0) | 7.11 (3.5) | |||
| 2 | 30.4 (13.9) | 16.4 (6.1) | |||
| 3 | 38.7 (14.7) | 4.7 (6.1) | 3.2 × 10−1 | ||
| mrx-Δ | vector | 3 | 0.15 (0.53) | — | 1.9 × 10−4 |
| mrx-Δ | mtLS-KpnI | 0 | 30.4 (10.2) | 2.62 (1.6) | |
| 0.5 | 33.2 (9.8) | 4.19 (1.4) | |||
| 1 | 32.7 (10.2) | 5.68 (1.2) | |||
| 2 | 38.6 (9.7) | 10.9 (2.1) | |||
| 3 | 43.6 (7.1) | 9.84 (2.0) | 2.2 × 10−1 | ||
| ku70/80-Δ | vector | 3 | 0.91 (0.75) | — | 1.4 × 10−3 |
| ku70/80-Δ | mtLS-KpnI | 0 | 59.6 (9.3) | 0 (2.6) | |
| 0.5 | 57.1 (9.7) | 2.51 (5.5) | |||
| 1 | 59.6 (8.9) | 1.34 (3.5) | |||
| 2 | 57.9 (7.6) | 2.10 (2.5) | |||
| 3 | 62.9 (8.7) | 0 (2.8) | 4.8 × 10−1 | ||
| mrx-Δ ku70/80-Δ | vector | 3 | 0.73 (0.29) | — | 4.0 × 10−6 |
| mrx-Δ ku70/80-Δ | mtLS-KpnI | 0 | 41.2 (4.9) | 2.92 (4.6) | |
| 0.5 | 47.0 (3.5) | 1.08 (1.0) | |||
| 1 | 50.2 (4.1) | 1.03 (1.1) | |||
| 2 | 58.2 (6.9) | 1.41 (1.6) | |||
| 3 | 62.2 (7.6) | 0.97 (1.0) | 2.2 × 10−1 |
The galactose-inducible system is not tightly controlled, and some mtLS-KpnI is expressed in dextrose, resulting in deletion product formation prior to galactose induction that comprises roughly one-fourth of the total COX2 signal. No deletion products are observed in the no-plasmid and vector-only controls, verifying that galactose does not stimulate mtDNA deletions in the absence of KpnI endonuclease expression. Furthermore, our analysis reveals that 3 hr after release from arginine selection, we continue to detect the full-length reporter. However, we see a significant increase in the percentage of deletion product and in a visible break product over the course of the experiment, suggesting that new breaks are being converted into deletions (P = 3.2 × 10−3, and 1.7 × 10−2, respectively) (Figure 2).
Examination of induced DSBs in the mrx-Δ strain reveals no significant differences from the wild-type strain with respect to either the amount of break product or the kinetics of increase in break product (Table 2, Figure 3a), in spite the 4.5-fold decrease in spontaneous events observed in the mrx-Δ strain (Figure 1d). This may indicate that the loss of MRX affects spontaneous deletions, but not deletions that are promoted by endonuclease-generated DSBs in the interrepeat sequence. Alternatively, it may reflect a greater sensitivity of the genetic assay.
In the ku70/80-Δ strain, there is a measureable increase in deletion products, representing, on average, 60% of the total COX2 signal, as compared to 26.5% in the wild-type strain (P = 9.3 × 10−3) (Table 2, Figure 3b). This is consistent with the observation that DRMD events are significantly increased in the absence of the Ku complex (Figure 1d). However, the amount of deletion product does not increase significantly after induction of KpnI in the Ku-deficient strain. Furthermore, these experiments reveal a decrease in the visible break product in strains lacking the Ku complex as a percentage of the total COX2 signal, relative to the wild-type and mrx-Δ strains.
In spite of a 715-fold reduction in spontaneous deletion events (Figure 1d), analysis of mtDNA after induction of a DSB in the mrx-Δ ku70/80-Δ mutant revealed the accumulation of the deletion product at significantly higher levels than the wild-type strain at all time points (Figure 3c). However, as in the wild-type and mrx-Δ strains, the deletion product does increase significantly in response to endonuclease induction from an average of 42.4% of the COX2 signal to 65.0% between the 0 and 3 hr time points (P ≤ 0.05), suggesting that the mechanisms of mitochondrial DRMD differ for spontaneous and break-induced events, as is also observed in the nucleus. Moreover, these data suggest that, while both increases and decreases are easily detected under spontaneous conditions, induction of a DSB between the repetitive sequences may preferentially select for deletions.
The frequency of respiring cells in these cultures following a similar 3-hr induction of mtLS-KpnI was determined (Table 2). In the wild-type strain, the frequency of cells that form colonies on glycerol medium was 3.2 × 10−1, indicating that ∼1/3 of the cells in the culture carried selectable deletions, in contrast to a frequency of 1.1 × 10−3 in the absence of breaks. Similar results were obtained for the mrx-Δ and ku70/80-Δ strains, so while there was no significant difference between the ku70/80-Δ and wild-type strains in this experiment, a statistically significant increase in deletion product was observed in the Southern blot analysis. This may indicate that there are not more ku70/80-Δ cells with deletions, but the cells with deletions carry a higher ratio of deletion product to intact reporter. In addition, we see that in the presence of breaks, the frequency of DRMD products in the five-gene deletion increases nearly five orders of magnitude.
Reciprocal products of recombination are reduced in the presence of induced mitochondrial DSBs, indicating a shift in the pathway responsible for generating deletions
In the nucleus, induction of a break between the repeats drives repair by SSA (Sugawara and Haber 1992), a recombination pathway that does not generate reciprocal products. Previously, we have demonstrated that other HR pathways such as intrachromosomal recombination or unequal sister chromatid exchange must generate some of the spontaneous mtDNA deletions, since reciprocal products of these recombination events can be detected (Phadnis et al. 2005). On the basis of our current model, we predict that DSBs induced in the interrepeat sequence will be preferentially repaired by NHEJ or SSA; thus, although deletion events are stimulated in strains expressing the KpnI endonuclease, we may see a reduction in reciprocal products.
We used a PCR-based assay to detect reciprocal products in the total cellular DNA samples used in the Southern blots shown in Figures 2 and 3. This assay detects both circular products of intramolecular recombination and duplications that would result from unequal sister chromatid exchange, using the primers shown in Figure 4a. To control for differences in template concentration, amplification of a fragment of similar size from the COX1 gene was performed. This method does not allow us to quantify the reciprocal product, since the different primer pairs show disparate amplification efficiencies, but by normalizing reciprocal product to COX1 amplification, we can compare relative amounts of reciprocal products between strains. Consistent with our model, in the wild-type strain, reciprocal products were reduced in strains containing the KpnI endonuclease plasmid (Figure 4b). This reduction was also observed in the strains lacking the Ku complex; however, no consistent reduction in reciprocal products was observed in the strain lacking the MRX complex alone. These results, taken together with our genetic data in Figure 1d, suggest that these complexes act in separate, perhaps competitive pathways for the repair of mtDSBs.
Figure 4 .
Detection of reciprocal products of recombination. In addition to the deletion product we select, HR pathways may produce the reciprocal products shown in (A). The circular product would be produced by homologous recombination between repeats within the same mitochondrial genome, while the duplication would be produced by recombination between the first repeat on one mitochondrial genome with the second repeat on a separate mtDNA molecule. Primers indicated by the small arrows below ARG8m were used to amplify a 489 bp fragment from the reciprocal products. In (B), the amplified fragments were run on a 1.3% agarose gel and stained with ethidium bromide. Relative concentrations of mtDNA were estimated by amplifying a fragment of comparable size from the COX1 gene in the mitochondrial genome. All reactions shown in the figure for each primer pair were amplified using the same master mix. The ratio of DNA amplified from reciprocal products and from COX1 is indicated below each lane after normalizing for staining intensity as described in Materials and Methods.
Chronic DSBs result in selection for KpnI-resistant reporters in wild-type yeast
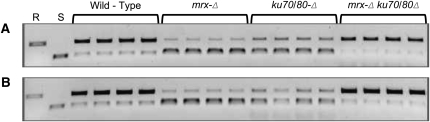
Various drivers were used to promote expression of mitochondrially targeted XhoI in flies and resulted in severe phenotypes, such as inviability or ablated tissue. Expression of endonuclease from the germline-specific nanos promoter selected for viable progeny with a homoplasmic population of mitochondrial genomes resistant to further cleavage that via mutation of the XhoI recognition site (Xu et al. 2008). Similarly, we considered the possibility that growth in dextrose in the presence of a low but chronic rate of DSB production may select for KpnIR mutants. In our experiments, the cells have two mechanisms by which to acquire resistance to further cutting: (1) maintaining the ARG8m sequence but acquiring point mutations in the KpnI restriction site, as we primarily observe in the wild-type strain, and (2) generation of the deletion product that has lost the ARG8m sequence containing the recognition site from the mitochondrial genome. Under selection for arginine prototrophy, the accumulation of these deleted genomes will be limited by the requirement for maintaining sufficient genomes that still carry the intact reporter construct.
We examined the ability of our reporter to be digested with KpnI following extended growth of the strains in dextrose medium lacking arginine. Cells were grown to saturation and subcultured twice in the same medium; we estimate that this constitutes approximately 16 generations of selective growth. Amplification of a fragment of ARG8m containing the KpnI recognition site and subsequent digestion with KpnI, revealed that ∼70% of the DNA that still contains ARG8m are KpnI resistant (KpnIR) (Figure 5, Table 3). Sequencing of cloned fragments indicates the accumulation of mutations in the KpnI recognition site blocking further cleavage (Table 4). These may be spontaneous mutations that are preexisting in the mtDNA population, but their high frequency and diversity suggest that they may result from a successful cleavage event followed by nondeletion-generating repair.
Table 3 . Percentage of KpnIR relative to total ARG8m.
| Relevant genotype | Percent KpnIR |
|---|---|
| Wild type | 71.9 (67.7–76.0) |
| mrx-Δ | 32.2 (30.7–33.7) |
| ku70/80-Δ | 44.4 (40.6–48.1) |
| mrx-Δku70/80-Δ | 78.1 (73.9–82.3) |
95% confidence limits shown in parentheses.
Table 4 . Sequencing of KpnIR clones.
| 372 | 373 | 374 | 375 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Position | T–A | T–C | A–G | C–G | C–G | C–A | C–G | C–T | Other | Total |
| Wild type | 47.6 (10) | 0 | 0 | 0 | 4.8 (1) | 0 | 42.9 (9) | 0 | 4.8 (1) | 100 (21) |
| mrx-Δ ku70/80-Δ | 34.1 (14) | 12.2 (5) | 2.4 (1) | 0 | 0 | 34.1 (14) | 0 | 4.9 (2) | 12.2 (5) | 100 (41) |
Sample size (n) shown in parentheses.
Yeast strains lacking MRX or Ku complexes show reduced KpnI-resistant mutations
Under DSB-inducing conditions, as described in Figure 3b, analysis of mtDNA in Ku-deficient strains expressing mtLS-KpnI revealed a greater accumulation of the deletion product than in the wild-type strain, at all time points. These studies show that even in medium that selects for the presence of the ARG8m gene, nearly 60% of the molecules carry a deletion of the ARG8m reporter. The maximum we have observed is 67% deletion product at the start of our induction experiments, and this may reflect the maximum that can be sustained while cells are growing under selection for arginine prototrophy. Furthermore, under chronic exposure to mtLS-KpnI, the majority (55.6%) of mtDNA molecules with the intact ARG8m reporter remain KpnI sensitive (KpnIS) in the ku70/80-Δ strain (Table 3). Consistent with the genetic data for spontaneous DRMD, deletions are preferred in the strain lacking the Ku complex. Similarly, nuclear studies have shown an increase in HR when NHEJ is defective (Clikeman et al. 2001; Mansour et al. 2008).
In the mrx-Δ strain, short-term induction of mtLS-KpnI results in deletion product accumulation at a frequency similar to the wild-type strain (Figure 3a), and after chronic exposure to endonuclease, the majority (67.8%) of intact reporters remain sensitive to KpnI digestion (Table 3). On the basis of similar nuclear studies (Boulton and Jackson 1996), we hypothesize that the KpnIS molecules in the mrx-Δ strain resulted from Ku-dependent classical NHEJ, in which the DSB is religated with high fidelity to the original sequence.
We considered the possibility that loss of the MRX or Ku complexes results in an increase in the maintenance of heteroplasmic mtDNA populations; however, in these mutant strains, the ability to maintain mixed populations was reduced (data not shown). Together, these data suggest that pathways that do not contribute to mutagenesis at the site of a DSB are preferred in the absence of MRX or Ku complexes, suggesting that the induced break is repaired by either error-free classical NHEJ or recombination.
Of the mtDNA molecules that maintain the ARG8m sequence after chronic mtLS-KpnI exposure, in the absence of both the MRX and Ku complexes, ∼80% have acquired KpnIR, suggesting a strong selective pressure for the deletion product or KpnIR molecules (Table 3). Furthermore, while all the mutations giving rise to KpnIR in the wild-type strain are transversions, only 19.5% of the mutations identified in the mrx-Δku70/80-Δ strain are the result of transversions, suggesting that the mechanism of mutagenesis may differ between the wild-type and mrx-Δku70/80-Δ strains (Table 4).
MRX and Ku complex deletion reveals distinct pathways for DRMD and petite formation
Respiration loss in yeast, scored by decreases in colony-forming units on a nonfermentable carbon source, can result from multiple types of mtDNA instability, including point mutations, large-scale deletions or rearrangements, or complete loss of the mitochondrial genome (Dujon 1981). Strains that lack respiration competence are termed petite because of their smaller colony size on solid medium. In laboratory strains, spontaneous petite formation occurs at a high frequency and most often results from large deletions and subsequent amplification of the remaining sequence. It has been proposed previously that these deletions are facilitated by direct repeats (Dujon 1981).
We find that deletion of the MRX complex causes a 2.4-fold increase in respiration-deficient cells compared to wild type (P = 3.2 × 10−4), while deletion of KU70 and KU80 did not affect the frequency of respiration loss (Table 5). However, deletion of all five genes increased respiration loss relative to both wild type (3.5-fold; P = 2.6 × 10−3) and the mrx-Δ mutant alone (1.5-fold; P = 0.04). The increase in respiration loss concurrent with a dramatic decrease in DRMD in the mrx-Δku70/80-Δ mutant suggests that respiration loss in yeast is mechanistically distinct from these DRMD pathways.
Table 5 . Analysis of mutational events.
| Average median frequency of respiration loss | Average rate of erythromycin resistance | Average rate of canavanine resistance | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Strains | % | Fold change | P-value | Rate (× 108) | Fold change | P-value | Rate (× 108) | Fold change | P-value |
| Wild type | 2.3 | — | — | 10.6 | — | — | 22.2 | — | — |
| mrx-Δ | 5.4 | 2.4 | 3.2 × 10−4 | 9.4 | 1.1 | 0.3 | 52.1 | 2.3 | 1.1 × 10−3 |
| ku70/80-Δ | 1.8 | 1.2 | 0.5 | 10.4 | 1.0 | 0.9 | 23.1 | 1.0 | 0.8 |
| mrx-Δ ku70/80-Δ | 8.0 | 3.5 | 2.6 × 10−3 | 0.9a | 12.3 | 0.01 | 47.8 | 2.2 | 2.1 × 10−4 |
At least one rate was calculated using the method of Luria and Delbruck (1943).
Nuclear point mutation accumulation does not correlate with mitochondrial phenotypes
We considered the possibility that the dramatic effects on mitochondrial DRMD events in the strains lacking the MRX and Ku complexes result from increased nuclear mutagenesis. We observe that nuclear DRMDs are not increased in the mutant strains; however, it is possible that point mutations may increase. We used the spontaneous acquisition of canavanine resistance (CanR) to estimate the rate of nuclear DNA point mutation. Deletion of KU70/80 had no effect on CanR rate (P = 0.8), while loss of the MRX complex or the combined five-gene deletion resulted in a modest twofold increase (P = 1.1 × 10−3 and P = 2.1 × 10−4, respectively) (Table 5).
To estimate mtDNA point mutation accumulation, we measured spontaneous resistance to erythromycin (EryR). Specific changes to the mitochondrial 21S rRNA gene confer EryR; therefore, we can estimate the genome-wide rate of spontaneous mtDNA point mutation by measuring the rate of resistance to the drug. We found that in the five-gene deletion mutant there is a 12.3-fold decrease in EryR rate relative to wild type (P = 0.01), while loss of each separate complex has no effect (P ≥ 0.3, in both cases) (Table 5).
Nuclear point mutation accumulation cannot account for the mitochondrial phenotypes we observe, because the strain lacking the MRX complex alone, and the five-gene deletion strain, display the same nuclear point mutation rates, but different mitochondrial DRMD and point mutation rates. These data demonstrate the relatively modest effect of MRX and Ku70/80 on nuclear mutation avoidance, while supporting a hypothesis in which these complexes contribute directly to mitochondrial mutagenesis, possibly via error-prone joining of DSBs. DSB repair-dependent mutagenesis has been observed in organisms, including Escherichia coli, yeast, and humans (Strathern et al. 1995; Bentley et al. 2004; Yang et al. 2008b; Shee et al. 2011). In E. coli the same mechanisms have been found to be responsible for mutagenesis at the sites of DSBs as well as spontaneous mutations (Shee et al. 2011).
Discussion
Genes essential for electron transport and oxidative phosphorylation are encoded within the mitochondrial genome, making the maintenance of mtDNA essential for most eukaryotic cells. In spite of the requirement for functional mitochondria, for many years the prevailing theories held that DNA repair mechanisms were limited relative to nuclear repair pathways. The multicopy nature of the mitochondrial genome made it possible to posit that damaged genomes were degraded, or simply not replicated, and early experiments seemed to support this hypothesis (Liu and Demple 2010). Although mitochondrial base excision repair (BER) pathways were well studied in the repair of oxidative damage, and isoforms of the nuclear BER proteins were found in mitochondria in diverse organisms (Boesch et al. 2011), recombination and other forms of DSBR remained controversial, in part due to the difficulty in directly detecting recombination in a uniparentally inherited genome. As a result, many of the tests performed to assess recombination have been indirect, and the results have been contradictory (Jorde and Bamshad 2000; Elson et al. 2001; Wiuf 2001; Innan and Nordborg 2002; Piganeau and Eyre-Walker 2004). A recent study suggests that these indirect tests may not be sufficient to detect recombination in even in a dataset where recombination has been demonstrated experimentally (White and Gemmell 2009).
However, more direct experimental evidence for recombination and end joining in mammalian mitochondria, derived from in vivo and in vitro studies, has accumulated in recent years (Poulton et al. 1993; Thyagarajan et al. 1996; Coffey et al. 1999; Lakshmipathy and Campell 1999; Kajander et al. 2001; Kraytsberg et al. 2004). The finding that some of the known nuclear DSBR proteins are localized to the mitochondria has suggested that, like BER, some of these pathways may be operational in both nuclear and mitochondrial compartments.
Initial studies suggest that Mre11 and Ku80 are localized to mammalian mitochondria and function to maintain mtDNA (Coffey and Campbell 2000; Dmitrieva et al. 2011); however, detailed mechanistic studies of recombination and DSB repair remain difficult in this system. In addition, it remains possible that although the rate of recombination may be lower in mammalian mtDNA than that of yeast, the mechanisms may be conserved.
We, therefore, employed the S. cerevisiae model system, in which specific types of mutagenic events can be efficiently assessed. Taken together, our data provide evidence of a role for the MRX and Ku70/80 complexes in at least partially redundant pathways in mitochondrial genome maintenance. In addition, the significant changes we observe in spontaneous DRMD in mtDNA in the absence of these complexes are more substantial than the nuclear DRMD phenotype conferred by their loss. Pathways dependent on either Ku or MRX complexes are responsible for the bulk of repeat-mediated deletions observed in our system.
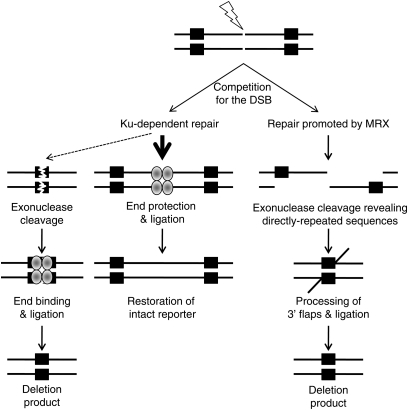
These results support a model in which there are at least two mechanisms for mtDSBR: one that results in deletion of one homologous repeat and the intervening sequence via HR and another in which the break is processed and ligated, resulting in point mutations surrounding the break point (Figure 6). Our construct cannot distinguish between uncleaved DNA and DNA that has been cleaved and ligated with fidelity. We interpret the increase in deletions that we see in the absence of the Ku complex as support for such error-free repair, occurring through classical NHEJ, and compensated for by alternative pathways when NHEJ is compromised. This observation is consistent with a competition for substrates between HR and NHEJ and suggests that, as in the nucleus, multiple related HR and NHEJ pathways exist in mitochondria.
Figure 6 .
Model for repair of DSB in the mitochondrial compartment. A proposed model is shown for mtDSBR when the DSB occurs between directly repeated sequences. There is competition between MRX and Ku70/80 complexes for the break substrate. When repair is promoted by MRX, the favored repair pathway is HR. This model depicts SSA in which DSBR results in a deletion product and does not produce reciprocal product; however, MRX could also promote other types of HR repair. If repair occurs via Ku-dependent repair, the primary pathway would be classical NHEJ, in which the ends of the breaks are protected and religated together restoring the original sequence. Alternatively, degradation at the break site resulting in complete loss of the intervening sequence, subsequently followed by end binding and ligation, would result in a deletion product.
Competition of HR and NHEJ pathways for DSBs may also explain the significant decrease in detectable break products in the strains lacking the Ku complex. Binding of Ku to the ends of DNA breaks may block or delay the exonucleolytic processing of the induced DSB that is required to generate the 3′ single-strand DNA ends for initiation of HR, as has been observed for nuclear DSBs (Clerici et al. 2008; Shim et al. 2010). In the absence of Ku, repair would shift to HR, and more efficient processing of the 5′ ends would result in loss of a discrete break product in the DNA samples used for Southern blotting. We cannot rule out an alternative model in which the DNA is a poor cleavage substrate in the absence of Ku70/80; however, we favor the first hypothesis, since the presence of the KpnI endonuclease stimulates accumulation of the deletion product in these strains.
Our previous studies indicated that at least some of the DRMDs in wild-type yeast were generated by HR pathways that generate reciprocal products (Phadnis et al. 2005). Here, we see a reduction in reciprocal products of direct repeat recombination in the presence of DSBs targeted to the interrepeat sequence. This would be a logical outcome of driving break repair to the SSA pathway, which does not generate reciprocal products, as we see in the nucleus (Sugawara and Haber 1992). It may seem counterintuitive that the absence of the Ku complex would exacerbate the loss of reciprocal products. However, it is likely that reciprocal products are generated when recombination is initiated within the short, directly repeated sequences and not by breaks in the intervening sequence. Therefore, it may be that repair of the DSBs in the intervening sequence by NHEJ, without loss of the repeats, restores the reporter sequence as an available substrate for reciprocal recombination events.
Our studies are consistent with a direct role for the proteins of the MRX and Ku complexes in the repair of mitochondrial DSBs. The identification of Mre11p and Rad50p in the mitochondrial proteome of yeast further support this model (Sickmann et al. 2003). Targeting of the Ku complex proteins in yeast has not been demonstrated; however, an isoform of Ku80 is required for DNA end-binding activity in extracts of purified mouse mitochondria (Coffey and Campbell 2000). Other nonnuclear functions have been demonstrated for the Ku proteins in mammalian cells, and nuclear, cytoplasmic, and membrane localizations have all been reported (reviewed in Koike 2002). Further studies will examine the recruitment of repair proteins to mitochondrial DSBs and effects on DSB processing.
In our inducible system, KpnI endonuclease expression is leaky under repressed conditions. This leads to a low level of chronic breaks, even before we induce its expression, stimulating deletion events. Analysis of the DNA from these strains indicates no loss of mtDNA content relative to nuclear DNA, suggesting that the DNA is repaired and not degraded. Three hours after induction of DSBs, we continue to see maintenance of the mitochondrial genome at wild-type levels, in spite of the fact that visible break product accounts for, on average, 15% of the COX2 signal. In mice, induction of DSBs in the mitochondrial genome was shown to result in significant depletion of mtDNA (Srivastava and Moraes 2005; Bacman et al. 2007, 2009). This may reflect a difference in the efficiency of DSBR in yeast and mammals; however, significant differences in the experimental design may also explain our disparate observations. In the mouse model, multiple recognition sequences for the restriction endonuclease used are present in the mitochondrial genome (Srivastava and Moraes 2005; Bacman et al. 2007), while in our system a single site is available for cleavage, potentially affecting the efficiency of repair. It is possible that the location of the break in our system between two long direct repeats also improves repair of these lesions.
Although our work indicates that at least some of the same proteins are important for both nuclear and mitochondrial DSBR and DRMDs, the hierarchy of pathways may not be the same as in the nucleus, or some alternative nuclear pathways do not exist in mitochondria. At this point, the proteins required for mitochondrial reciprocal recombination events are not known, although other proteins involved in nuclear HR (Rad51, Rad51C, and Xrcc3) were recently identified in human mitochondria (Sage et al. 2010), suggesting that these recombination pathways may be conserved and should be targets of future study. Further characterization of the proteins involved in nuclear DSBR as well as mitochondrial-specific proteins may aid in unraveling the multiple pathways that can act in the formation of mtDNA deletions and the repair of mtDSBs in both yeast and mammals.
Acknowledgments
We thank V. Nagaraja for providing the pETRK and pACMK+ plasmids, S. A. Mookerjee for critical reading of the manuscript, and M. Fasullo for insightful suggestions. This work was supported by National Science Foundation grants MCB0543084 and MCB0841857 (E.A.S. and R.A.S.) and National Institutes of Health grant F31 GM078700 (L.K.).
Footnotes
Communicating Editor: J. A. Nickoloff
Literature Cited
- Adams A., Gottschling D. E., Stearns T., Kaiser C. A., 1997. Methods in Yeast Genetics, 1997: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Bacman S. R., Williams S. L., Hernandez D., Moraes C. T., 2007. Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ’differential multiple cleavage-site’ model. Gene Ther. 14: 1309–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman S., Williams S. L., Moraes C. T., 2009. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 37: 4218–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley J., Diggle C. P., Harnden P., Knowles M. A., Kiltie A. E., 2004. DNA double strand break repair in human bladder cancer is error prone and involves microhomology associated end-joining. Nucleic Acids Res. 32: 5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi N. O., Bianchi M. S., Richard S. M., 2001. Mitochondrial genome instability in human cancers. Mutat. Res. 488: 9–23 [DOI] [PubMed] [Google Scholar]
- Boesch P., Weber-Lotfi F., Ibrahim N., Tarasenko V., Cosset A., et al. , 2011. DNA repair in organelles: pathways, organization, regulation, relevance in disease and aging. Biochim. Biophys. Acta 1813: 186–200 [DOI] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P., 1996. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15: 186–200 [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Guerini I., Lucchini G., Longhese M. P., 2008. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO J. 9: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clikeman J. A., Khalsa G. J., Barton S. L., Nickoloff J. A., 2001. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics 157: 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey G., Campbell C., 2000. An alternate form of Ku80 is required for DNA end-binding activity in mammalian mitochondria. Nucleic Acids Res. 28: 3793–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey G., Lakshmipathy U., Campbell C., 1999. Mammalian mitochondrial extracts possess DNA end-binding activity. Nucleic Acids Res. 27: 3348–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva N. I., Malide D., Burg M. B., 2011. Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA. Am. J. Physiol. Regul. Integr. Comp. Physiol.; Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B., 1981. Mitochondrial genetics and functions, pp. 505–635 Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Elson J. L., Andrews R. M., Chinnery P. F., Lightowlers R. N., Turnbull D. M., et al. , 2001. Analysis of European mtDNA for recombination. Am. J. Hum. Genet. 68: 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M. J., Sondheimer N., 2010. Mitochondrial genetic diseases. Curr. Opin. Pediatr. 22: 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D., Folley L. S., Mulero J. J., McMullin T. W., Thorsness P. E., et al. , 1991. Analysis and manipulation of yeast mitochondrial genes, pp. 149–165 Guide to Yeast Genetics and Molecular Biology, edited by Guthrie C., Fink G. R. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- Fukui H., Moraes C. T., 2009. Mechanisms of formation and accumulation of mitochondrial DNA deletions in aging neurons. Hum. Mol. Genet. 18: 1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Nordborg M., 2002. Recombination or mutational hot spots in human mtDNA? Mol. Biol. Evol. 19: 1122–1127 [DOI] [PubMed] [Google Scholar]
- Ivanov E. L., Sugawara N., Fishman-Lobell J., Haber J. E., 1996. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cervisiae. Genetics 142: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde L. B., Bamshad M., 2000. Questioning evidence for recombination in human mitochondrial DNA. Science 288: 1931a. [PubMed] [Google Scholar]
- Kajander O. A., Karhunen P. K., Holt I. J., Jacobs H. T., 2001. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2: 1007–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander O. A., Karhunen P. J., Jacobs H. T., 2002. The relationship between somatic mtDNA rearrangements, human heart disease and aging. Hum. Mol. Genet. 11: 317–324 [DOI] [PubMed] [Google Scholar]
- Kalifa L., Sia E. A., 2007. Analysis of Rev1p and Polζ in mitochondrial mutagenesis suggests an alternative pathway of damage tolerance. DNA Repair (Amst.) 6: 1732–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa L., Beutner G., Phadnis N., Sheu S. S., Sia E. A., 2009. Evidence for a role of FEN1 in maintaining mitochondrial DNA integrity. DNA Repair (Amst.) 8: 1242–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M., 2002. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. (Tokyo) 43: 223–236 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Schwartz M., Brown T. A., Ebralidse K., Kunz W. S., et al. , 2004. Recombination of human mitochondrial DNA. Science 304: 981. [DOI] [PubMed] [Google Scholar]
- Krishnan K. J., Reeve A. K., Samuels D. C., Chinnery P. F., Blackwood J. K., et al. , 2008. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 40: 275–279 [DOI] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S., 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38: 233–271 [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U., Campell C., 1999. Double-strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 27: 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea D. E., Coulson C. A., 1949. The distribution of the number of mutants in bacterial populations. J. Genet. 49: 264–284 [DOI] [PubMed] [Google Scholar]
- Lee H. C., Wei Y. H., 2007. Oxidative stress, mitochondrial DNA mutation, and apoptosis in aging. Exp. Biol. Med. (Maywood) 232: 592–606 [PubMed] [Google Scholar]
- Liefshitz B., Parket A., Maya R., Kupiec M., 1995. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics 140: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Demple B., 2010. DNA repair in mammalian mitochondria: Much more than we thought? Environ. Mol. Mutagen. 51: 417–426 [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbruck M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour W. Y., Schumacher S., Rosskopf R., Rhein T., Schmidt-Petersen F., et al. , 2008. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 36: 4088–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A., Brisson N., 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Morel F., Renoux M., Lachaume P., Alziari S., 2008. Bleomycin-induced double-strand breaks in mitochondrial DNA of Drosophila cells are repaired. Mutat. Res. 637: 111–117 [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., 2001. Regulation of intracellular localization of human MTH1, OGG1, and MYH proteins for repair of oxidative damage. Prog. Nucleic Acid Res. Mol. Biol. 68: 75–94 [DOI] [PubMed] [Google Scholar]
- Oliveira M. T., Garesse R., Kaguni L. S., 2010. Animal models of mitochondrial DNA transactions in disease and ageing. Exp. Gerontol. 45: 489–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F., Haber J. E., 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Sia E. A., 2004. Role of the putative structural protein Sed1p in mitochondrial genome maintenance. J. Mol. Biol. 342: 1115–1129 [DOI] [PubMed] [Google Scholar]
- Phadnis N., Sia R. A., Sia E. A., 2005. Analysis of repeat-mediated deletions in the mitochondrial genome of Saccharomyces cerevisiae. Genetics 171: 1549–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau G., Eyre-Walker A., 2004. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity 92: 282–288 [DOI] [PubMed] [Google Scholar]
- Poulton J., Deadman M. E., Bindoff L., Morten K. L., Brown G., 1993. Families of mtDNA rearrangements can be detected in patients with mtDNA deletions: duplications may be a transient intermediate form. Hum. Mol. Genet. 2: 23–30 [DOI] [PubMed] [Google Scholar]
- Prado F., Cortes-Ledesma F., Huertas P., Aguilera A., 2003. Mitotic recombination in Saccharomyces cerevisiae. Curr. Genet. 42: 185–198 [DOI] [PubMed] [Google Scholar]
- Sage J. M., Gildemeister O. S., Knight K. L., 2010. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J. Biol. Chem. 285: 18984–18990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan M., Bujnicki J. M., Cymerman I. A., Rao D. N., Nagaraja V., 2004. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 32: 6129–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C., Gibson J. L., Darrow M. C., Gonzalez C., Rosenberg S. M., 2011. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 108: 13659–13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H. V., 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112: 441–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim E. Y., Chung W.-H., Nicolette M. L., Zhang Y., Davis M., et al. , 2010. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29: 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Kokoska R. J., Dominska M., Greenwell P., Petes T. D., 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Butler C. A., Dominska M., Greenwell P., Fox T. D., et al. , 2000. Analysis of microsatellite mutations in the mitochondrial DNA of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A., Reinders J., Wagner Y., Joppich C., Zahedi R., et al. , 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Moraes C. T., 2005. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Genet. 14: 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele D. F., Butler C. A., Fox T. D., 1996. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. USA 93: 5253–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Shafer B. K., McGill C. B., 1995. DNA synthesis errors associated with double-strand break repair. Genetics 140: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E., 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12: 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Ira G., Haber J. E., 2000. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20: 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B., Padua R. A., Campbell C., 1996. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 271: 27536–27543 [DOI] [PubMed] [Google Scholar]
- Wallace D. C., 2010. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 51: 440–450 [DOI] [PubMed] [Google Scholar]
- White D. J., Gemmell N. J., 2009. Can indirect tests detect a known recombination event in human mtDNA. Mol. Biol. Evol. 26: 1435–1439 [DOI] [PubMed] [Google Scholar]
- Wiuf C., 2001. Recombination in human mitochondrial DNA? Genetics 159: 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., DeLuca S. Z., O’Farrell P. H., 2008. Manipulating the metazoan mitochondrial genome with targeted restriction enzymes. Science 321: 575–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. L., Weissman L., Bohr V. A., Mattson M. P., 2008a Mitochondrial DNA damage and repair in neurodegenerative disorders. DNA Repair (Amst.) 7: 1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sterling J., Storici F., Resnick M. A., Gordenin D. A., 2008b Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 4: e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Chatterjee A., Singh K. K., 2006. Saccharomyces cerevisiae polymerase ζ functions in mitochondria. Genetics 172: 2683–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]