Abstract
Schizosaccharomyces pombe, the fission yeast, cells alternate between P- and M-mating type, controlled by the alternate alleles of the mating-type locus (mat1). The mat1 switching occurs by replacing mat1 with a copy derived from a silenced “donor locus,” mat2P or mat3M. The mechanism of donor choice ensuring that switching occurs primarily and productively to the opposite type, called directionality, is largely unknown. Here we identified the mat1-Mc gene, a mammalian sex-determination gene (SRY) homolog, as the primary gene that dictates directionality in M cells. A previously unrecognized, shorter swi2 mRNA, a truncated form of the swi2, was identified, and its expression requires the mat1-Mc function. We also found that the abp1 gene (human CENPB homolog) controls directionality through swi2 regulation. In addition, we implicated a cis-acting DNA sequence in mat2 utilization. Overall, we showed that switching directionality is controlled by judicious expression of two swi2 transcripts through a cell-type-regulated dual promoter. In this respect, this regulation mechanism resembles that of the Drosophila sex-determination Slx gene.
FISSION yeast is primarily a haploid single-celled organism whose cells exist as one of two cell/mating types, called P (plus) or M (minus). Under nitrogen-starvation growth conditions, haploid cells of opposite type mate and the resulting diploid zygotic cell undergoes meiosis to produce two mat1P and two mat1M haploid spore segregants. This 2:2 Mendelian segregation pattern observed in meiosis shows that cell type is controlled by two alleles of a single mat1 locus. However, the culture starting from a single cell of either type contains a roughly equal proportion of cells of both mating types. This cell-type change is due to the efficient mating-type switching phenomenon, called homothallism, which operates during mitotic growth of the culture (Arcangioli and Thon 2003; Egel 2005; Klar 2007). The mating-type switching process replaces the existing mat1 locus through the gene conversion process with a copy derived from one of the two epigenetically silenced “donor loci,” mat2P and mat3M (Figure 1).
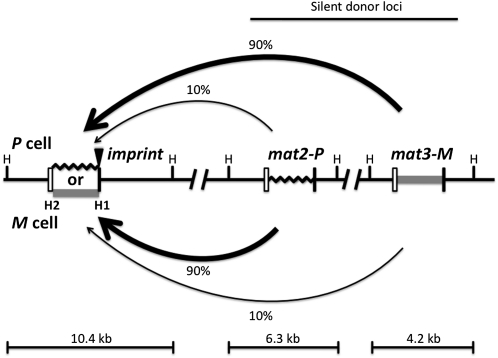
Figure 1 .
The directionality phenomenon of mating-type switching in fission yeast. The diagram shows mat1, mat2P, and mat3M genes located from left to right in chromosome 2. The mat1 locus is expressed and it confers cell type, while mat2P and mat3M, located in the silenced region, act only as donors for providing copies of genetic information for mat1 switching. The mat1 can be either P (black zigzag line, representing 1104-bp-long DNA sequence) or M (shaded bar, 1128 bp). Each mat cassette is flanked by the homology regions, boxes H2 (left open box, 135 bp) and H1 (right solid box, 59 bp), which are used for switching-promoted recombination. The imprint site (solid triangle) is located at the junction of the mat1 allele-specific and the H1 box sequences. The donor preference is determined by the mat1 cell type; mat1-P cells choose mat3 and mat1-M cells choose mat2, both with ∼90% preference (thick arrow) over the other, less-preferred (∼10%, thin arrow) donor locus. The HindIII restriction enzyme sites (H) flanking each cassette are shown. Digestion of yeast genomic DNA with the restriction enzyme generates three fragments of indicated sizes, each containing a specific mat locus. The figure is not drawn to scale.
The switching process is tied to the DNA replication cycle so that only one in four “grandchildren” of a nonswitchable cell switches in ∼90% of cell pedigrees (Miyata and Miyata 1981; Egel and Eie 1987; Klar 1990a). The switching process is initiated by a DNA strand-specific epigenetic entity, called an imprint (Klar 1987; Klar 1990b), found at the junction of the homology-box H1 and the mat1 allele-specific sequence (Figure 1). The imprint is either a strand- and sequence-specific nick and/or two ribonucleotides incorporated in DNA (Beach and Klar 1984; Egel et al. 1984; Nielsen and Egel 1989; Kaykov and Arcangioli 2004; Vengrova and Dalgaard 2004). Three genes (swi1, swi3, and swi7) are required for synthesis of the imprint (Egel et al. 1984; Singh and Klar 1993; Dalgaard and Klar 2000). The swi7, encoding DNA polymerase α, is an essential gene for cellular viability (Singh and Klar 1993). The swi1 and swi3 encode replication fork pause factors (Dalgaard and Klar 2000). DNA replication of the imprinted strand at mat1 is thought to create a transient double-strand break (DSB) in the resulting chromatid. The DSB is repaired by recombination by copying P or M information from one of the two donor loci through the synthesis-dependent strand-annealing (SDSA) mechanism (Arcangioli and De Lahondes 2000; Yamada-Inagawa et al. 2007). This strand-specific imprinting/segregation mechanism (Klar 1987; Klar 1990b) explains the generation of one-in-four-granddaughters switching pattern observed in cell pedigrees (Miyata and Miyata 1981; Egel and Eie 1987; Klar 1990a). Interestingly, a similar mechanism of asymmetric cell division, through epigenetic differentiation plus selective segregation of sister chromatids in mitosis, has been recently suggested for generating neuronal bilateral asymmetry in C. elegans (Nakano et al. 2011).
Interestingly, the donor locus selection is not random; mat1P prefers mat3M, and mat1M prefers the nearby mat2P in ∼90% of switches (Figure 1). Consequently, switches to the opposite type occur predominantly in standard mat2P and mat3M-containing strains, called h90 strains (for homothallic, ∼90% sporulation). This donor preference, called directionality of switching, is not because cells prefer the heterologous information-containing donor locus for switching, but it is because P cells prefer mat3 and M cells prefer mat2, regardless of the donors’ genetic content. The directionality control was demonstrated by swapping the donor loci genetic content to mat2M and mat3P (called h09 genotype). Notably, h09 cells switched preferentially by futile homologous information replacement (Thon and Klar 1993). Thus, the directionality control dictates mat1P to switch preferentially by choosing mat3, and mat1M by choosing mat2, regardless of the donors’ genetic content.
Several genes have been shown to effect directionality. Switching-recombination proteins Swi2 and Swi5 physically interact with each other and directly bind to the mat2/3 region to mediate switching (Akamatsu et al. 2003; Jia et al. 2004). The distribution pattern of the Swi2/Swi5 complex is cell-type regulated: in P cells, Swi2/Swi5 localizes only to the mat3 locus; in contrast, in M cells, it spreads to the mat2 locus (Jia et al. 2004). The precise mechanism of the cell-type control on the Swi2/Swi5 spread in the mat2/3 region is unknown, but the chromatin structure is thought to regulate the Swi2/Swi5 complex spreading because heterochromatin factors, such as Swi6, Clr4, Rik1, Sir2, Clr7, and Clr8, affect the mating-type switching efficiency (Ivanova et al. 1998; Shankaranarayana et al. 2003; Jia et al. 2004; Tuzon et al. 2004; Thon et al. 2005; Aguilar-Arnal et al. 2008). The fission yeast Abp1 (encoding autonomously replicating sequence-binding protein 1), one of the human centromere protein B (CENPB) homologs, has been reported to regulate the activation of DNA replication, to maintain genome integrity by forming heterochromatin at retrotransposons, and to protect replication forks during pausing (Murakami et al. 1996; Halverson et al. 1997; Okada et al. 2007; Cam et al. 2008; Zaratiegui et al. 2011). Interestingly, the Abp1 protein also controls switching directionality by regulating the Swi2/Swi5 spread on the mat2/3 region. The mechanism by which Abp1 controls Swi2/Swi5 spread at donor loci is also unknown. However, because Abp1 binds within the donor region, it has been postulated that such binding might regulate directionality (Aguilar-Arnal et al. 2008; Cam et al. 2008).
In this study, we discovered that mat1-Mc and abp1 genes are required for switching directionality. We showed that the swi2 gene produces two independent but overlapping transcripts by engaging two different promoters in M cells, but only the longer form is produced in P cells. The previously unrecognized short swi2 form (i.e., swi2S) is the one that controls directionality by promoting the mat2 donor locus selection in M cells. To some extent, such a two-promoter regulation mechanism resembles that of the sex-determination gene (Sxl) of Drosophila. Furthermore, we identified a mat2P cis-acting site (named Swi2-dependent recombinational enhancer 2, SRE2), which is critical for donor locus utilization for switching.
Materials and Methods
Strains, plasmids, and media
The S. pombe strains used in this study and their genotypes are listed in Table 1. The mat1 locus-specific primers (TAAGTGGGATGAGTGCTTGCTTTG and AGTGAGTATATTATGGTAGGGAGTGCGTAGCG) were used to amplify the mat1-mc− mutants. The mating-type donor region is silenced and prohibited from homologous recombination by an epigenetic mechanism (Egel 1984). To introduce mutations in donor loci by DNA-mediated transformation, we used a clr1-5 genotype that permits homologous recombination in the mat2/3 region (Thon and Klar 1992). All subsequent studies were conducted with strains from which the clr1-5 mutation was removed by genetic crosses. All mutations were generated by standard polymerase chain reaction (PCR) methods and confirmed by PCR or sequence analysis.
Table 1 . Fission yeast stains.
| Stain | Genotype | Reference |
|---|---|---|
| CY146 | h90, leu1-32, ura4-D18, ade6-210, Δste11::KAN | This study |
| CY148 | h09, leu1-32 ura4-Dl8, ade6-210 Δste11::KAN | This study |
| CY195 | h90, mat3Mi+mcop7, leu1-32, ura4-D18, ade6-216, his2 | This study |
| CY196 | h09, mat2Mi+mcop7, ade6-210 | This study |
| CY199 | h09, mat3pcop5Pi+, ade6-216 | This study |
| CY200 | h90, mat3mifs15Mc+, leu1-32, ura4-D18, ade-216, his2 | This study |
| CY202 | h09, mat2mifs15Mc+, ade6-210 | This study |
| CY204 | h90, mat2Pc+piop7, leu1-32, ura4-D18, his2, ade6-210 | This study |
| CY207 | h09, mat3Pc+piop7, leu1-32, ade6-216 | This study |
| CY212 | h90, ΔSRE2, leu1-32, ura4-D18, ade6-210, his2 | This study |
| CY294 | h90 ΔSRE1, leu1-32, ura4-D18, ade6-210, his2 | This study |
| CY301 | h90, mat2P::ura4::Δ121, leu1-32, ura4-D18, ade6-216, his2 | This study |
| CY303 | h90, mat2P::ura4::Δ109, leu1-32, ura4-D18, ade6-210, his2 | This study |
| CY308 | h90, mat2P::ura4, leu1-32, ura4-D18, ade6-216, his2 | This study |
| CY327 | h90, leu1-32, ura4-D18, ade6.210, Δabp1::KAN | This study |
| CY345 | h90, leu1-32, ura4-D18, ade6.210, swi2L::ura4 | This study |
| CY347 | mat1M, Δmat2/3::LEU2, leu1-32, ura4-D18, ade6-216, swi2HA | This study |
| CY348 | mat1P, Δmat2/3::LEU2, leu1-32, swi2HA, ura4-D18, ade6-216 | This study |
| PG19 | h09, leu1-32, ura4-D18, ade6-210 | Thon and Klar (1993) |
| SP713 | mat1P, Δmat2/3::LEU2, leul-32, ura4−, ade6-216 | Kelly et al. (1988) |
| SP714 | matlM, Δmat2/3::LEU2, leul-32, ura4−, ade6-216 | Kelly et al. (1988) |
| SP715 | mat1Mi+mcop7, Δmat2/3::LEU2, leul-32, ura4−, ade6-216 | Kelly et al. (1988) |
| SP716 | mat1mifsl5Mc+ Δmat2/3::LEU2, leul-32, ura4−, ade6-216 | Kelly et al. (1988) |
| SP717 | mat1Pc+piop7, Δmat2/3::LEU2, leul-32, ura4−, ade6-216 | Kelly et al. (1988) |
| SP718 | mat1pcop5Pi+ Δmat2/3::LEU2, leul-32, ura4−, ade6-216 | Kelly et al. (1988) |
| SP976 | h90, leu1-32, ura4-D18, ade6-210 | Thon and Klar (1993) |
| SPA160 | h90, leu1-32, ura4-D18, Δhis3, Δabp1::KAN | Aguilar-Arnal et al. (2008) |
| SP2534 | h90, mat1-Mc-fg2534, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2535 | h90, mat1-Mc-fg2535, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2536 | h90, mat1-Mc-fg2536, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2553 | h90, mat1-Mc-fg2553, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2830 | h90, mat1-Mc-fg2830, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2832 | h90, mat1-Mc-fg2832, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2833 | h90, mat1-Mc-fg2833, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| SP2834 | h90, mat1-Mc-fg2834, mat2P::ura4, ura4-D18, leu1-32, ade6-210, hsp1 | This study |
| YA664 | h90, leu1-32, ura4-D18, his3-D1, arg3-D1, swi2HA | Akamatsu et al. (2003) |
To construct plasmids pREP3-swi2L-HA, pREP3-swi2S-HA, and pREP3-swi2S, PCR fragments amplified from genomic DNA of YA664 (Akamatsu et al. 2003) or from a wild-type strain were inserted into the BalI–BamHI sites of the pREP3 vector (Maundrell 1993).
Strains were cultured in yeast extract with supplementary adenine-containing medium (YEA). For switching, sporulation, Northern blot, and Western blot experiments, Edinburgh minimal medium, supplemented with auxotrophic requirements of each strain (PMA), was used. Media and other standard conditions for growth and genetic analysis were employed as described previously (Moreno et al. 1991).
Genomic DNA preparation and Southern blot analysis
Genomic DNA preparation and Southern blot analyses with HindIII digested yeast genomic DNA were carried out as previously described (Moreno et al. 1991). The 10.6-kb mat1P-containing HindIII fragment was used as the probe.
Total RNA preparation and Northern blot analysis
Total RNA was extracted and purified using the TRIzol Plus RNA purification system (Invitrogen, no. 12183-555). Each lane was loaded with 20 µg total RNA of each sample for Northern blot analysis using the kit provided by Ambion (no. AM1940). The Swi2P probe consisted of the PCR fragment copied from genomic DNA with primers (ATGCCCATTGTGATGACCACCCA and AGGTAGAAAGACAGACATTAGAATAGTTTGG). The Swi2 5′P probe consisted of the PCR product copied from the yeast cDNA library with primers (TGTATTTCACAAAAGAGGGAGATTCAGT and cDNA 5′-end adaptor primer AAGCAGTGGTATCAACGCAGAGT).
RACE analysis
The mRNA was enriched from total yeast RNA with the Qiagen mRNA kit (Qiagen, no. 72022). A total of 200 ng mRNA was used for each RACE experiment. RACE analysis was performed according to instructions in the RACE cDNA amplification kit (Clonetaq, no. 634923). The Swi2-specific primers used for 5′ and 3′ RACE comprised TTACACTCCCCCTAAGTCTGCTACCTC and GGAGGTAGCAGACTTAGGGGGAGTGTAA, respectively.
Analysis of mat1 locus M/P ratio
To determine the genetic content at the mat1 locus, a quantitative multiplex PCR procedure was adapted from previous publications (Jia et al. 2004; Aguilar-Arnal et al. 2008). PCR-amplified products were run on a 1.5% agarose-TBE gel stained with 0.5 g/ml ethidium bromide and captured using a Typhoon scanning machine. Imagequant software was used for quantitative analysis.
Protein extraction and immunoblotting
Strain SP976 cells harboring pREP3-swi2L-HA, pREP3-swi2S-HA, or pREP3 plasmid was grown in PMA media lacking a leucine supplement for 1 day at 30°. The exponentially growing cells were collected by centrifugation, suspended in 5% trichloroacetic acid buffer, and disrupted by vortexing with glass beads. The extracted protein was subjected to immunoblotting. Mouse anti-HA antibody (Sigma, no. H9658) was used as the first antibody, and peroxidase-conjugated anti-mouse IgG antibody was used as the second antibody.
Results
Selection of mat1-mc− mutations
Ascospores synthesize starch during meiosis and sporulation, while nonsporulating cells do not. Therefore, efficiently switching colonies growing on sporulation medium (pombe minimal adenine, PMA, medium) stain black when exposed to iodine vapors because they contain ascospores, but colonies defective in switching do not stain (Leupold 1955). Using the iodine-staining procedure to screen for mating-type switching-defective mutants from a strain containing the hypersporulation 1 (hsp1) mutation (Michael et al. 1995), we found eight independent mutants with a reduced rate of switching. They were sterile for mating and also exhibited an unusual phenotype of repairing their mutations spontaneously. Both phenotypes might be explained should the inefficient mat1 switching process operate in them to repair/heal their mutations. We postulated that the mutations were located at the mat1 locus in regions that are replaced with wild-type donor information during the switching process. We also observed that in the hsp1 genetic background the switched and healed cells mate readily on PMA medium because of their hypersporulation property, and the resulting zygotic asci do not grow further. As a consequence, only the mat1 mutant cells multiply in PMA medium, making our mutation analyses possible despite the fact that mutations were subject to removal by the switching process (presented below). The DNA of the mat1 locus of the mutants was amplified with mat1-specific primers and sequenced. The eight mutations represented six different sites in the mat1-mc gene (Table 2) (Kelly et al. 1988). Because the Mc gene is required for both mating and sporulation (Kelly et al. 1988), both the sterility and mutations repair by switching are explained by their location within the mat1 cassette.
Table 2 . mat1-mc- mutations.
| Mutation name | Mutation in Mc RNA | Change in Mc protein |
|---|---|---|
| 2534, 2832 | T to C at 20 | ATG to ACG in initiation codon |
| 2535 | G to A at 21 | ATG to ATA in initiation codon |
| 2536, 2553 | G to A at 84 | Trp to TGA stop codon in codon 22 |
| 2833 | C insertion at 230 | Frameshift in codon 71 |
| 2834 | C deletion at 165 | Frameshift in codon 49 |
| 2830 | T to C at 443 | Trp to Arg in codon 141 |
The mat1 genes are not required for mat1 imprinting
Although our procedure favored selection of mutant cells during growth on PMA solid medium, the persistence of mutations during growth of the culture suggested that the cells were defective in some aspect of the switching pathway. Specifically, we wanted to understand why the switching process had not readily repaired the mat1-mc− mutations. Recombination required for mat1 switching is initiated with the synthesis of the mat1 imprint (Figure 1). The first possibility we considered was that the mat1-mc− mutations might reduce the imprint level. That is, the Mc transcription factor might be required for the imprinting process. It is not known whether mat1 genes influence the imprint level. The imprint forms a fragile DNA break site, resulting in a double-strand break (DSB) during the normal DNA extraction process, so the imprint level is usually evaluated by Southern blot analysis (Beach 1983; Beach and Klar 1984). Since our mutations were subject to repair during mitotic growth, for this analysis we employed an mc− mutant strain with deleted donor loci (Δmat2/mat3) (Klar and Miglio 1986). Because of their donor loci deletion, such strains do not switch at all, and we could thus directly assess the role of Mc in imprinting without the complication of mutations disappearing due to switching-promoted repair during our analysis. The imprint level observed in the mc− mutant was similar to that of the wild-type control (Figure 2A). The mat1P encodes mat1-Pc and mat1-Pi genes, and mat1-M encodes mat1-Mc and mat1-Mi genes (Kelly et al. 1988). The other three mating-type gene (pc−, pi−, mi−) mutants were also not affected in the imprinting process. These results suggest that none of the four mating-type genes is required for the imprinting step of the switching pathway (Figure 2A).
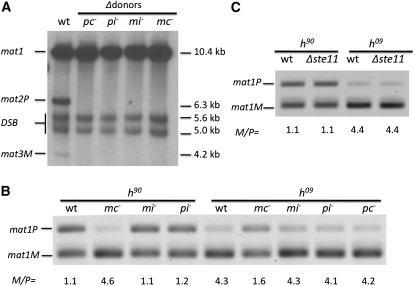
Figure 2 .
Effect of mat1 mutations on mat1 imprint and on directionality of switching. (A) Imprint level determined by Southern blot analysis. The wild-type (wt, SP976; see Table 2 for complete genotype) was the homothallic h90 strain, which contains all three cassettes diagrammed in Figure 1. The stocks with mating-type gene mutations in pc− (SP718), pi− (SP717), mi− (SP716), mc− (SP715) had been deleted for both donor loci; thus bands reflecting mat2P and mat3M genes were lacking in them. Such strains lacking donor loci cannot switch, and therefore stably maintain their mat1 mutations. Genomic DNA was digested with HindIII and the blot was probed with mat1P gene-containing 10.6 kb long probe (Figure 1). Locations of the HindIII sites flanking each cassette are shown in Figure 1. The 5.6-kb and 5.0-kb bands result from the double-strand break (DSB) generated by shearing imprinted DNA during its preparation. The DSB reflects the level of mat1 imprint (Beach and Klar 1984). (B) Effect of mat1 genes mutations on directionality of h90 (wt, SP976; mc−, CY195; mi−, CY200; pi−, CY204) and h09 (wt, PG19; mc−, CY196; mi−, CY202; pi−, CY207; pc−, CY199) strains. Quantitative multiplex PCR analysis of cultures showed M/P ratio of the mat1 locus of strains in which mutations introduced in donor loci have been transposed to mat1 through switching. The PCR reaction included one primer from outside the mat1 allele-specific sequences and another one from the mat1-P or mat1-M allele-specific sequences (see Materials and Methods ). The mat1 allele-specific PCR product bands are indicated. (C) Effect of Δste11 (h90, CY146; h09, CY148) on directionality. The mat1 M/P ratio of cultures was determined as described above.
mat1-Mc gene controls the directionality of switching
We next entertained an alternative model for explaining the persistence of our mutations in mat1-mc mutant stocks; the mat1-Mc gene might dictate mat2P donor preference for switching the mat1-M allele. In this hypothesis, our mat1-mc− mutants might switch by a default mode, preferring the mat3M as a donor instead of mat2P; accordingly, homologous information would be transferred to the mat1 locus. We previously showed that only a portion of the cassette information can be replaced by switching during the homologous cassette replacement, rather than always replacing the entire cassette (Yamada-Inagawa et al. 2007). Accordingly, each switch event might not have repaired mat1-mc− mutations. To determine whether the Mc gene dictates mat2 utilization, we engineered a mc– mutation in the mat3M donor locus. Cells of this culture should alternate between mat1-P and mat1-mc− alleles. The wild-type culture contains an equal proportion of mat1P and mat1M cells due to the efficient switching of both mat1 alleles, either one to the opposite mat1 allele. In contrast, according to our hypothesis, the mutant strain should predominate with mat1-mc− cells. To test this possibility, we analyzed the mat1 locus M/P ratio by PCR as described previously (Jia et al. 2004; Aguilar-Arnal et al. 2008). For wild-type h90 cells, the mat1P amount was observed to be nearly equal to the amount of mat1M, as expected (Figure 2B). By comparison, the mc− mutant exhibited much more mat1M content. This result suggested that mat1-Mc controls directionality by promoting mat2 utilization in M cells.
We previously showed that donor preference depends on specific chromosomal location and not on the genetic content of donor loci (Thon and Klar 1993). To explore the directionality issue further, we checked the effect of a mc− mutation on the mat1 content of an h09 strain (mat2-mc− and mat3P) whose genetic contents have been swapped with respect to those of standard h90 cells. Here, the mat1P allele was greatly enriched in the mat3-mc− mutant in comparison to the wild-type cells (Figure 2B). This result confirmed that the mat2 donor locus is used inefficiently in mat1-mc− h09 cells, a result similar to that presented above with h90 cells.
Similarly, we also determined whether the other three mating-type genes are involved in switching directionality. Our results showed that none of the three genes we tested (pc, pi, and mi) was involved in the directionality of h90 and h09 cells (Figure 2B). We did not check the mat2-pc− mutant in an h90 cell background because a previous report indicated that the mat2-pc− mutant does not affect the mat1 locus M/P ratio (Ruusala 1991). Thus, the mat1-Mc gene is the only mat1 gene that controls specific donor-locus choice in both h90 and h09 genetic backgrounds.
Mc is a known transcriptional factor regulating many M cell-type-specific genes in combination with another DNA-binding protein partner named Ste11 (Kjaerulff et al. 1997). Is Ste11 also required for switching directionality? We found that the Δste11 mutation did not affect the mat1 M/P ratio in both h90 and h09 backgrounds (Figure 2C). This result is consistent with the notion that Mc uses a Ste11-independent pathway to control directionality. However, because the efficiency of switching cannot be determined directly in ste11 mutants owing to their sterile phenotype, in this case only knowing their mat1 M/P ratio is not sufficient to assess whether Ste11 controls directionality (see below).
A previously unknown, shorter swi2 (swi2S) transcript requires Mc and Abp1 factors for expression
Next, we addressed how Mc might control the switching direction. As described in the Introduction, assembly of the Swi2/Swi5 complex spreads to both donor loci in M cells, while it is localized at the mat3 locus in P cells. The precise mechanism of this cell-type-specific spread of the Swi2/Swi5 complex is not known. Because mc− mutants are defective in mat2 donor choice, we considered the possibility that such mutants may be defective in the spreading of the Swi2/Swi5 complex at the mat2/3 region. We therefore examined the swi2 and swi5 expression using Northern blot analysis of mRNA isolated from P and M cells, which lacked both donor loci and thus stably maintained their cell type (Klar and Miglio 1986). Analysis of swi5 did not produce any clear signal, possibly due to its weak expression. swi2 showed one transcript that was 2.5 kb long (swi2L, L for longer) in P cultures, plus another, shorter one that was 2.0 kb long (swi2S, S for shorter) in M cultures (Figure 3, A and B). Furthermore, the amount of swi2S transcript was about twice the amount of swi2L in M cultures. Most interestingly, the swi2S species was absent in the mc− mutant. This result showed that expression of the swi2S transcript was Mc dependent. Furthermore, these modes of expression were unaffected whether or not the cells contained donor loci (Figure 3B). In addition, since Mc is partially repressed in nitrogen-rich medium (Kelly et al. 1988), the swi2S transcript level was reduced in rich medium in comparison to the level found in nitrogen-starved conditions (Figure 3B). We concluded that Mc controls the swi2S mRNA expression.
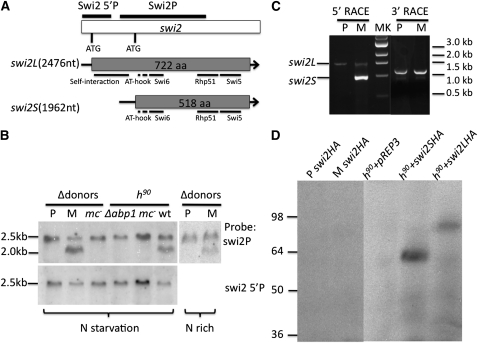
Figure 3 .
Characterization of the swi2L and swi2S products. (A) The swi2 gene, mRNAs, and predicted proteins. Upper drawing shows the swi2 gene’s DNA structure indicated along with its two translation initiation ATG codons. The Northern blot probes, Swi2 5′P, Swi2P, we used are also indicated. The two lower drawings indicate mRNA and ORF length of swi2L and swi2S forms of the swi2 gene. Both swi2L and swi2S predicted proteins share the same open reading frame (ORF) in regions where the genes overlap. The Swi2 protein domains, such as for other proteins interaction, self-interaction, and two AT-hook motifs, are marked (Akamatsu et al. 2003). The Swi2S lacks the self-interaction domain. The GenBank accession nos. for swi2L and swi2S are JQ308182 and JQ308183. (B) Northern blot of swi2 transcripts. The cultures with indicated genotype were grown in PMA media (nitrogen-starvation condition) and YEA media (nitrogen-rich condition). Strains: P, SP713; M, SP714; wt, SP976; mc− in h90, CY195; mc− in Δdonors, SP715; Δabp1, spA160. (C) Rapid amplification of swi2 cDNA ends (RACE). Results show agarose gel electrophoresis of RACE products obtained by amplifying 5′- and 3′-end products of swi2 cDNA. (D) Western blot analysis of the Swi2L and Swi2S proteins. Analysis of the swi2HA tagged gene from the endogenous locus in donors-deleted P (CY347) and M (CY348) cells is presented in the left two lanes. The remaining lanes reflect analysis of swi2L-HA and swi2S-HA tagged genes that were expressed from a plasmid vector in h90 cells (SP976). The anti-HA antibody was used for Swi2 detection. The h90 wild-type culture harboring empty pREP3 vector was used as a negative signal detection control.
It was previously known that Abp1 regulates switching directionality, but the molecular mechanism of its action is unknown (Aguilar-Arnal et al. 2008). We found that the swi2S transcript was absent in the Δabp1 mutant. Collectively, these results led us to entertain a model in which both Mc and Abp1 factors affect directionality by regulating the swi2S gene expression.
Next we determined the ends of both transcripts. We conducted rapid amplification experiments of cDNA ends (RACE) and cloned the PCR products (Scotto-Lavino et al. 2006) (Figure 3C). Sequencing results showed that the common swi2L forms found in both P and M cells were identical. swi2L is a 2476-nt-long transcript capable of encoding a 722-amino-acid-long open reading frame (ORF) (Figure 3A). The swi2S is 1962-nt-long transcript capable of encoding a predicted 518-amino-acid-long ORF. The 3′ ends of both forms were identical. Overall, the swi2S form is a 5′-end truncated species of the swi2L form, and these species share the same ORF in overlapping regions of the gene. Northern blot analysis with the 5′-end-specific probe from the swi2L form (Swi2 5′P) did not detect the swi2S signal (Figure 3B), confirming our RACE results.
Swi2S and Swi2L proteins detected when expressed from a strong promoter
We tested whether the Swi2S form is expressed as an ORF predicted protein in yeast cells. We could not detect hemagglutinin (HA)-tagged Swi2 protein when it was expressed at the genomic locus from its indigenous promoter (Figure 3D), a result in accord with a previous report suggesting that the level of expression is too low to be detected by Western blot analysis (Akamatsu et al. 2003). Then we attempted to express both swi2L and swi2S genes with a stronger nmt1 promoter of a pREP3 plasmid vector (Maundrell 1993). Two bands of about 80 kDa and 60 kDa, corresponding to the 722-aa and 518-aa predicted proteins, respectively, were detected (Figure 3D). These results suggested that the swi2S encodes an N-terminal truncated protein form of the Swi2L protein when these genes were expressed from the nmt1 promoter.
swi2S gene plays a key role in regulating directionality
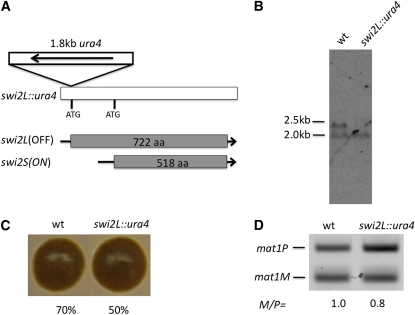
Our molecular work predicted that swi2S is essential for directionality of switching. We genetically tested this possibility. We inserted the 1.8-kb DNA fragment, containing the ura4 gene in a transcription orientation reversed with that of the swi2 gene, into the swi2L translation initiation site in the genome (Figure 4A). The resulting strain was named swi2L::ura4. Northern blot analysis showed that the DNA insert did not alter the swi2S expression, but the swi2L form was missing (Figure 4B). This result demonstrated that the swi2S mRNA is independently transcribed from the swi2L form and that the swi2S promoter is probably located in the 5′-coding region of the swi2L gene. The iodine vapor dark-staining phenotype of the swi2L::ura4 strain was indistinguishable from that of the wild-type genotype, but the sporulation frequency of strains was somewhat reduced (50% vs. 70%) (Figure 4C). Also, the mat1P content was increased relative to the mat1M content in the swi2L::ura4, in comparison to that of the wild-type cultures (Figure 4D). A colony of the swi2L::ura4 genotype stains dark (Figure 4C), and in comparison, the Δswi2 or Δabp1 colonies were reported previously to stain lightly (Akamatsu et al. 2003; Aguilar-Arnal et al. 2008). These results indicated that the swi2S form indeed provides a mat1 switching function and that it facilitates mat2 donor utilization over mat3.
Figure 4 .
Effect of Δswi2L on mating-type switching. (A) Structure of the swi2L::ura4 allele is diagrammed. A 1.8-kb ura4 HindIII fragment was inserted in the translation initiation site (ATG) of the swi2L gene. (B) Northern blot analysis of wild-type (SP976) and the swi2L::ura4 (CY345) cultures. RNAs prepared from cultures grown in PMA medium were analyzed with the Swi2P probe (Figure 3A). (C) Iodine-staining colony phenotype. Numbers below each colony reflect their efficiency of sporulation. (D) The mat1 M/P ratio of cultures was determined with the method described in Figure 2B.
Mc and abp1 genes regulate directionality specifically by inducing swi2S expression
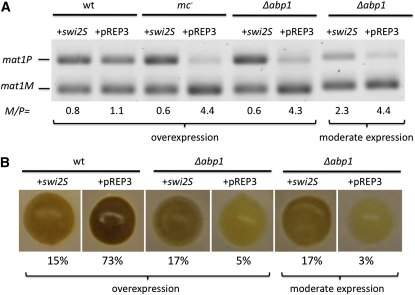
Do Mc and Abp1 control directionality specifically by promoting the expression of swi2S? We conducted complementation experiments to address this question. We expressed swi2S through the nmt1 promoter from an ectopic pREP3 vector (Maundrell 1993). Ectopic expression of swi2S led to increased mat1P content over mat1M (Figure 5A), and consequently, otherwise wild-type colonies stained lighter and a reduced level of sporulation was observed in them (Figure 5B). swi2S expression in mc− or Δabp1 mutants led to a dramatic mat1 M/P ratio reversal from >4.0 to 0.6. These results indicated that increased swi2S expression greatly suppressed the mc− and Δabp1 mutants’ switching defect. Specifically, the mat2P donor utilization defect of mc− and Δabp1 mutants was suppressed through swi2S expression (Figure 5B). Expression from the nmt1 promoter can be inhibited by adding 0.05 μM thiamine to the medium (Javerzat et al. 1996). Moderating swi2S expression with the thiamine addition partially complemented the Δabp1-staining phenotype, and consequently, the mat1 M/P ratio changed from 4.4 to 2.3 (Figure 5A). These results strongly suggest that Mc and abp1 genes regulate directionality specifically, or predominantly, by inducing swi2S expression.
Figure 5 .
Ectopically expressed swi2S suppresses mc− (CY195) and Δabp1 (CY327) mutants’ switching defect. (A) The mat1 M/P ratio of cultures was determined with the method described in Figure 2B. Analysis of an empty pREP3 vector transformant was included for comparison, as a negative control (wt, SP976). (B) Iodine-staining colony phenotype. Moderate swi2S gene expression was achieved by 0.05 μM thiamine addition to the medium and thiamine was not added to achieve overexpression.
As shown above, Δste11 did not influence the P/M cell ratio in both h90 and h09 cells (Figure 2C). As we have implicated swi2S in directionality, we tested whether Ste11 is required for swi2S expression. The Northern blot result showed that the Δste11 mutation reduced the swi2S mRNA level as compared to that of the swi2L form in both h90 and h09 background (Supporting Information, Figure S1). The reduction of swi2S is more likely due to reduction of the Mc gene expression, which is regulated by Ste11 (Kjaerulff et al. 1997; Xue-Franzen et al. 2006). Although swi2S expression is partially reduced, the P/M cell ratio is not affected by the ste11 mutation (Figure 2C). We think that there are two possible explanations for it: one is that the reduction of swi2S amount is not enough to affect switch directionality; the other is that Ste11 may control other target genes that can compensate for the swi2S reduction.
A mat2 cis-acting SRE2 element implicated in donor preference
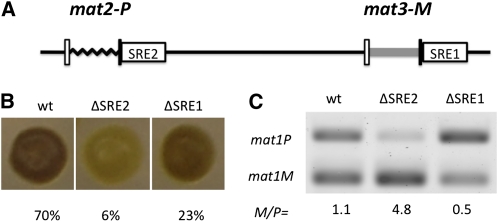
A previous report identified an SRE (Swi2-dependent recombinational enhancer) element, located centromere-distal to the mat3 locus as the Swi2/Swi5 complex loading site (Jia et al. 2004). Here we refer to it as the SRE1 element. Similarly, we discovered a sequence located centromere-distal to the mat2 locus that promotes its utilization as a donor (Figure 6A). A 400-bp deletion mutation of mat2:H1 distal region greatly reduced both colony staining and sporulation (Figure 6B). We named the region defined by the deletion as the SRE2 element. Analysis of the mat1 locus composition showed that mat1P predominates in ΔSRE1 and mat1M predominates in ΔSRE2 strains (Figure 6B). This work demonstrated that both mat2 and mat3 loci use flanking elements for their utilization as donors for switching, and they probably help recruit the recombination Swi2/Swi5 complex in a cell-type-specific fashion. This idea is supported by studies reporting the recruitment of Swi2/Swi5 at these two sites (Jia et al. 2004; Aguilar-Arnal et al. 2008).
Figure 6 .
Effect of ΔSRE (switching recombinational enhancer) deletion on mating-type switching. (A) The diagram shows location of SRE1 and SRE2 elements with respect to donor loci. ΔSRE2 (CY212) comprises a 400-bp deletion located distal to the H1 region of mat2P. The SRE1 is defined by a 454-bp deletion of sequences (ΔSRE1, CY294) located distal to H1 region of mat3M (Jia et al. 2004). The iodine staining colony phenotype (B) and the mat1 M/P ratio (C) of indicated strains are presented.
Discussion
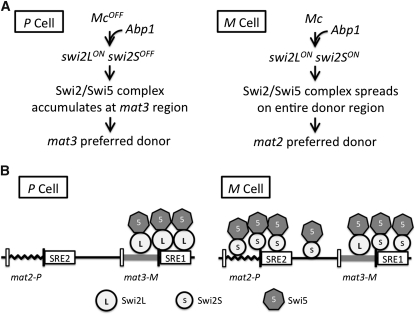
In this study, we found evidence that the mat1-Mc gene dictates directionality in M cells such that mat2 is preferred over mat3 as the donor for switching. Moreover, we discovered the mechanism of this preference. Specifically, Mc induces the transcription of a shorter form of swi2 mRNA by activating a previously uncharacterized promoter, which lies within the 5′-end side of the ORF of the longer form of the swi2 gene. We also found that Abp1 similarly dictates directionality by inducing swi2S mRNA synthesis. Furthermore, a new cis-acting element, SRE2, located next to mat2 was identified, and that also facilitates mat2 utilization. A diagram of the genetic pathway summarizing our results is presented in Figure 7A.
Figure 7 .
The mechanism directionality of switching. (A) Summary of genetic determinants controlling directionality of mat1 switching. In P cells, because Mc factor is absent, only swi2L form is expressed ON. In M cells, the Mc factor, in combination with the Abp1 factor, activates the swi2S gene. (B) Proposed role of Swi2L and Swi2S in directionality of switching. In P cells, the Swi2L protein promotes localization of the Swi2/Swi5 complex to the mat3 region to make it as the preferred donor. In M cells, perhaps Swi2S and Swi2L cooperate to promote assembly of the Swi2/Swi5 complex across the entire silenced region to make mat2 as the preferred donor. The two SRE elements might facilitate enrichment of Swi2/Swi5 complexes at donor loci in cell-type specific fashion.
A model for Swi2L and Swi2S factors controlling directionality
A model for directionality controlled by two forms of the swi2 gene in P and M cells is proposed in Figure 7B. In mat1-M cells, the directionality is primarily governed by the mat1-Mc gene, which induces the M-cell-type-specific swi2S expression at the transcriptional level. It is not known whether the Mc factor regulates swi2S expression directly or indirectly through regulating other gene(s). Similarly, the abp1 gene is required for swi2S expression, also by acting either directly or indirectly. We postulate that, owing to their protein sequence differences, the Swi2L and Swi2S proteins possess different Swi2/Swi5 complex-spreading abilities at the donor-loci region. The Swi2L form contains a strong self-interaction domain at its N-terminal end, and it was postulated that this form determines the cell-type specific pattern of Swi2/Swi5 complex distribution (Akamatsu et al. 2003; Haruta et al. 2008). The Swi2L protein prefers to accumulate at the mat3 region, and it does not spread to other regions. The Swi2S protein lacks the strong self-interaction domain, and it might facilitate the spread to the entire donor loci heterochromatic region. It is also possible that the Swi2S helps bind the complex, preferably to the mat2 locus, by binding to the SRE2 element (Figure 7B). The proposed model is in accord with our observations and those of others, including: (1) in Δabp1 background, the Swi2L is the only form expressed in P as well as in M cells, and the Swi2/Swi5 complex binds to the mat3 locus, as shown previously by chromatin immunoprecipitation experiments (Jia et al. 2004; Aguilar-Arnal et al. 2008); (2) the Swi2S is the predominant form in M cells, and the Swi2/Swi5 complex is assembled on the entire region to make mat2 as the preferred donor; (3) in the Δswi2L (swi2L::ura4) strain, where only the Swi2S form is expressed, the mat2 is a preferred donor when compared with wild-type cells; (4) in the Δswi2 strain, where both Swi2L and Swi2S forms are lacking, the mat3 donor is not preferred because Swi2L is absent. In this case, however, it is not possible to comment on the donor locus preference because of the very low rate of switching (Jia et al. 2004; Aguilar-Arnal et al. 2008). In Δswi2 cells, the mat1-P allele predominates, perhaps because the nearby mat2 is at an advantage over the more distant mat3 locus for recombination with mat1.
Mc and Abp1 may physically bind to a putative swi2S promoter to activate expression
We learned here that the swi2 gene expresses two swi2S and swi2L forms. The swi2S is more crucial than swi2L for efficient mating-type switching. The swi2S expression requires Mc and Abp1. Mc is a transcriptional factor and regulates M-cell-specific genes in conjunction with Ste11 (Kjaerulff et al. 1997). Our study showed that Δste11 reduced swi2S expression, perhaps indirectly through controlling mat1-Mc expression, rather than by directly controlling swi2S expression. We do not think the Ste11 directly regulate swi2S expression because Δste11 still show reduced levels of swi2S expression. We investigated the sequence upstream of the swi2S transcription start site to look for a putative promoter element that might exist there. A short 12-bp motif (5′-ACAATGCCCATTGT) was found 45 bp upstream of the swi2S transcript start site, and this motif contains two standard, inverted Mc-binding sites (ACAATG) (Kjaerulff et al. 1997). This finding reminds us of the mating-type switching-directionality regulation of the distantly related budding yeast, Saccharomyces cerevisiae. Budding yeast mating-type MATα2 factor binds to two closely located recognition motifs at RE (recombinational enhancer) region as a diamer to repress the RE function for donor choice during mating-type switching (Houston et al. 2004; Coic et al. 2006). The Mc binding to this putative promoter might activate swi2S transcription. A recent chromatin immunoprecipitation analysis showed that Mc physically interacts with the swi2S promoter region, as predicted here (Matsuda et al. 2011). Whether Mc binding depends on the two inverted presumptive recognition sites needs further investigation.
The abp1 gene was originally identified as the ARS-binding protein (Murakami et al. 1996) and it is probably involved in DNA replication initiation. Abp1 and the other two CENPB homologs (Cbh1 and Cbh2) bind to the centromere, helping centromeric heterochromatin assembly to promote centromere function (Nakagawa et al. 2002). More recently, genome-wide Abp1 distribution mapping showed that Abp1 also binds to retrotransposons to help maintain genome stability by silencing transposons and by controlling replication fork pause release (Cam et al. 2008; Zaratiegui et al. 2011). The Abp1 was previously implicated in the directionality of switching by promoting Swi2/Swi5 distribution at donor loci (Aguilar-Arnal et al. 2008). Since there is a strong Abp1 binding signal near the CenH sequence in the mat2/3-region, it was postulated that Abp1 might control Swi2/Swi5 binding by directly binding to this site (Aguilar-Arnal et al. 2008). We deleted the region containing the putative Abp1-binding site, but no obvious colony-staining defect was observed (Figure S2). This result indicated that the putative Abp1 binding site is not required for switching directionality. Instead, we found that Abp1 regulates directionality of switching primarily by regulating swi2S expression. According to the high-resolution mapping data (S. pombe epigenome website: http://pombe.nci.nih.gov/index.html?org=S.+pombe&db=pombe&hgsid=59268), there is about a 2.5-fold Abp1 enrichment signal found at the putative swi2S promoter region. Thus, it is possible that Abp1 induces the swi2S transcript by directly binding to our predicted swi2S gene promoter region. We speculate that Abp1 helps load Mc to the swi2S promoter to activate swi2S expression (Figure 7A).
Regulation of the swi2 gene by two separate cell-type-activated promoters
Different-sized transcripts and the encoded proteins are usually produced by alternative mRNA splicing. In the database, the swi2 gene (GenBank Accession no. NM_001019690) is listed as encoding a 722-amino-acid ORF, lacking introns reflecting the swi2L species. We found two swi2 species resulting from two promoters with overlapping transcripts that differ from each other only at the 5′ end. A precedent for one gene producing two overlapping transcripts, initiated from two different promoters, is found with the Drosophila sex-lethal (Sxl) gene. Although alternatively spliced products of Sxl eventually regulate sexual identity during development, the initial decision for sexual identity occurs through a choice between the maintenance promoter (Pm) and establishment promoter (Pe) (Keyes et al. 1992). The Sxl-Pm is transcribed in both sexes, but the Sxl-Pe, located in the region encoding the first intron of the Sxl-Pm transcript, shows female-specific expression. The protein product from Sxl-Pe regulates the Sxl-Pm splicing to produce the SXL protein in female embryos. The Sxl-Pm transcript found in males includes an extra exon, which renders the transcript nonfunctional (Salz and Erickson 2010). Similarly, we found that swi2L expresses in both yeast cell types, but the swi2S expresses only in M cells and at a level higher than that of swi2L. In this respect, swi2 regulation is analogous to that of the Sxl gene of Drosophila. However, both of the swi2L and swi2S species are functional and bind to the mating-type donor region with alternative distribution patterns to regulate directionality of switching. In sum, the two swi2 forms cooperate to regulate donor choice such that most switches to the opposite mating type occur in cells of either mating type.
We note that previously the swi2S form has escaped detection by microarray analysis or genome-wide sequence analysis possibly because it was regarded as a partial degradation product of the swi2L form. As a consequence, previous studies (Akamatsu et al. 2003; Jia et al. 2004) performed to define the function of swi2 in switching considered only the swi2L form and here we found that the swi2S form is a major determinant of directionality of switching.
While we were preparing this manuscript for publication, it was reported (Matsuda et al. 2011) that the Mc and Abp1 genes control the switching recombination directionality, that Mc protein directly binds to the Swi2 gene regulatory region, and that binding requires Abp1. Results of that report are complementary to some parts of our study and support our conclusions. However, the discovery of the Swi2S form and its role in directionality is presented here only in our study.
Supplementary Material
Acknowledgments
We thank Sharon Moore, Ken Ishikawa, Stephan Sauer, and Jeff Strathern’s laboratory members for discussions and advice. We also thank Drs. Fernando Azorin and Hiroshi Iwasaki for providing yeast strains. The Intramural Research Program of the National Cancer Institute of the National Institutes of Health supported this work. C.Y. and A.K. designed and performed the experiments, analyzed the data, and wrote the manuscript; M.B. provided technical support for this work. The authors declare that they have no conflict of interest.
Footnotes
Communicating Editor: F. Winston
Literature Cited
- Aguilar-Arnal L., Marsellach F. X., Azorin F., 2008. The fission yeast homologue of CENP-B, Abp1, regulates directionality of mating-type switching. EMBO J. 27: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu Y., Dziadkowiec D., Ikeguchi M., Shinagawa H., Iwasaki H., 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100: 15770–15775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B., de Lahondes R., 2000. Fission yeast switches mating type by a replication-recombination coupled process. EMBO J. 19: 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B., Thon G., 2003. Mating-types cassettes: structure, switching and silencing, pp. 129–147 Molecular Biology of Schizosaccharomyces pombe, edited by Egel R. Springer Verlag, Berlin [Google Scholar]
- Beach D. H., 1983. Cell type switching by DNA transposition in fission yeast. Nature 305: 682–688 [Google Scholar]
- Beach D. H., Klar A. J., 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 3: 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam H. P., Noma K., Ebina H., Levin H. L., Grewal S. I., 2008. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature 451: 431–436 [DOI] [PubMed] [Google Scholar]
- Coic E., Sun K., Wu C., Haber J. E., 2006. Cell cycle-dependent regulation of Saccharomyces cerevisiae donor preference during mating-type switching by SBF (Swi4/Swi6) and Fkh1. Mol. Cell. Biol. 26: 5470–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard J. Z., Klar A. J., 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751 [DOI] [PubMed] [Google Scholar]
- Egel R., 1984. Two tightly linked silent cassettes in the mating-type region of Schizosaccharomyces pombe. Curr. Genet. 8: 199–203 [DOI] [PubMed] [Google Scholar]
- Egel R., 2005. Fission yeast mating-type switching: programmed damage and repair. DNA Repair 4: 525–536 [DOI] [PubMed] [Google Scholar]
- Egel R., Eie B., 1987. Cell lineage asymmetry in Schizosaccharomyces pombe: unilateral transmission of a high-frequency state for mating-type switching in diploid pedigrees. Curr. Genet. 12: 429–433 [Google Scholar]
- Egel R., Beach D. H., Klar A. J., 1984. Genes required for initiation and resolution steps of mating-type switching in fission yeast. Proc. Natl. Acad. Sci. USA 81: 3481–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson D., Baum M., Stryker J., Carbon J., Clarke L., 1997. A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J. Cell Biol. 136: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta N., Akamatsu Y., Tsutsui Y., Kurokawa Y., Murayama Y., et al. , 2008. Fission yeast Swi5 protein, a novel DNA recombination mediator. DNA Repair 7: 1–9 [DOI] [PubMed] [Google Scholar]
- Houston P., Simon P. J., Broach J. R., 2004. The Saccharomyces cerevisiae recombination enhancer biases recombination during interchromosomal mating-type switching but not in interchromosomal homologous recombination. Genetics 166: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A. V., Bonaduce M. J., Ivanov S. V., Klar A. J., 1998. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 19: 192–195 [DOI] [PubMed] [Google Scholar]
- Javerzat J. P., Cranston G., Allshire R. C., 1996. Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 24: 4676–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S., Yamada T., Grewal S. I., 2004. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 119: 469–480 [DOI] [PubMed] [Google Scholar]
- Kaykov A., Arcangioli B., 2004. A programmed strand-specific and modified nick in S. pombe constitutes a novel type of chromosomal imprint. Curr. Biol. 14: 1924–1928 [DOI] [PubMed] [Google Scholar]
- Kelly M., Burke J., Smith M., Klar A., Beach D., 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J. 7: 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes L. N., Cline T. W., Schedl P., 1992. The primary sex determination signal of Drosophila acts at the level of transcription. Cell 68: 933–943 [DOI] [PubMed] [Google Scholar]
- Kjaerulff S., Dooijes D., Clevers H., Nielsen O., 1997. Cell differentiation by interaction of two HMG-box proteins: Mat1-Mc activates M cell-specific genes in S. pombe by recruiting the ubiquitous transcription factor Ste11 to weak binding sites. EMBO J. 16: 4021–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., 1987. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature 326: 466–470 [DOI] [PubMed] [Google Scholar]
- Klar A. J., 1990a. The developmental fate of fission yeast cells is determined by the pattern of inheritance of parental and grandparental DNA strands. EMBO J. 9: 1407–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J., 1990b. Regulation of fission yeast mating-type interconversion by chromosome imprinting. Dev. Suppl. 1990: 3–8 [PubMed] [Google Scholar]
- Klar A. J., 2007. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu. Rev. Genet. 41: 213–236 [DOI] [PubMed] [Google Scholar]
- Klar A. J., Miglio L. M., 1986. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell 46: 725–731 [DOI] [PubMed] [Google Scholar]
- Leupold U., 1955. Methodisches zur Genetik von Schizosaccharomyces pombe. Schweiz. Z. Allg. Pathol. Bakteriol. 18: 1141–1146 [PubMed] [Google Scholar]
- Matsuda E., Sugioka-Sugiyama R., Mizuguchi T., Mehta S., Cui B., et al. , 2011. A homolog of male sex-determining factor SRY cooperates with a transposon-derived CENP-B protein to control sex-specific directed recombination. Proc. Natl. Acad. Sci. USA 108: 18754–18759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130 [DOI] [PubMed] [Google Scholar]
- Michael H., Fecke H. C., Fleck O., Gutz H., 1995. Hyperspeckled mutants of Schizosaccharomyces pombe: frequent mating-type switching without detectable double-strand breaks. Mol. Gen. Genet. 249: 297–300 [DOI] [PubMed] [Google Scholar]
- Miyata H., Miyata M., 1981. Mode of conjugation in homothallic cells of Schizosaccharomyces pombe. J. Gen. Appl. Microbiol. 27: 365–371 [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Murakami Y., Huberman J. A., Hurwitz J., 1996. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 93: 502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Lee J. K., Hurwitz J., Allshire R. C., Nakayama J., et al. , 2002. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 16: 1766–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S., Stillman B., Horvitz H. R., 2011. Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in C. elegans. Cell 147: 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O., Egel R., 1989. Mapping the double-strand breaks at the mating-type locus in fission yeast by genomic sequencing. EMBO J. 8: 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Ohzeki J., Nakano M., Yoda K., Brinkley W. R., et al. , 2007. CENP-B controls centromere formation depending on the chromatin context. Cell 131: 1287–1300 [DOI] [PubMed] [Google Scholar]
- Ruusala T., 1991. The mating type in fission yeast is switched independently of its expression. Curr. Genet. 20: 379–383 [DOI] [PubMed] [Google Scholar]
- Salz H. K., Erickson J. W., 2010. Sex determination in Drosophila: the view from the top. Fly 4: 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto-Lavino E., Du G., Frohman M. A., 2006. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 1: 2555–2562 [DOI] [PubMed] [Google Scholar]
- Shankaranarayana, G. D., M. R. Motamedi, D. Moazed, and S. I. Grewal, 2003 Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr. Biol. 13: 1240–1246. [DOI] [PubMed]
- Singh J., Klar A. J., 1993. DNA polymerase-alpha is essential for mating-type switching in fission yeast. Nature 361: 271–273 [DOI] [PubMed] [Google Scholar]
- Thon G., Klar A. J., 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131: 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G., Klar A. J., 1993. Directionality of fission yeast mating-type interconversion is controlled by the location of the donor loci. Genetics 134: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G., Hansen K. R., Altes S. P., Sidhu D., Singh G., et al. , 2005. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 171: 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzon C. T., Borgstrom B., Weilguny D., Egel R., Cooper J. P., et al. , 2004. The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J. Cell Biol. 165: 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova S., Dalgaard J. Z., 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue-Franzen Y., Kjaerulff S., Holmberg C., Wright A., Nielsen O., 2006. Genomewide identification of pheromone-targeted transcription in fission yeast. BMC Genomics 7: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada-Inagawa T., Klar A. J., Dalgaard J. Z., 2007. Schizosaccharomyces pombe switches mating type by the synthesis-dependent strand-annealing mechanism. Genetics 177: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M., Vaughn M. W., Irvine D. V., Goto D., Watt S., et al. , 2011. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature 469: 112–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.