Abstract
Assembly of Human Immunodeficiency Virus Type-1 (HIV-1) particles is initiated in the cytoplasm by the formation of a ribonucleoprotein complex comprising the dimeric RNA genome and a small number of viral Gag polyproteins. Genomes are recognized by the nucleocapsid (NC) domains of Gag, which interact with packaging elements believed to be located primarily within the 5´-leader of the viral RNA. Recent studies revealed that the native 5´-leader exists as an equilibrium of two conformers, one in which dimer-promoting residues and NC binding sites are sequestered and packaging is attenuated, and one in which these sites are exposed and packaging is promoted. To identify the elements within the dimeric 5´-leader that are important for packaging, we generated HIV-1 5´-leader RNAs containing mutations and deletions designed to eliminate substructures without perturbing the overall structure of the leader and examined effects of the mutations on RNA dimerization, NC binding and packaging. Our findings identify a 159 residue RNA packaging signal that possesses dimerization and NC binding properties similar to those of the intact 5´-leader and contains elements required for efficient RNA packaging.
Keywords: retrovirus assembly, diploid genome packaging, RNA structure, ITC, nucleocapsid protein
Introduction
Like all retroviruses, the Human Immunodeficiency Virus (HIV) specifically packages two copies of its unspliced RNA genome, both of which are utilized for strand-transfer mediated recombination during reverse transcription.1,2 Although only one DNA allele is generated by this mechanism (and retroviruses are thus considered “pseudodiploid”),3 the production of recombinant proviruses from heterozygous virions appears to serve as a primary means for promoting genetic evolution under conditions of environmental and chemotherapeutic stresses.4–7 Genome selection is mediated by interactions between the nucleocapsid (NC) domains of a subset of the viral structural proteins (called Gag) and RNA elements located primarily within the 5´-leader region of the genome.8–18 Genetic studies indicate that genomes are selected for packaging as dimers,19,20 and there is now compelling evidence that dimerization and selection occur in the cytoplasm subsequent to nuclear export.21–23 HIV-1 genomes are trafficked to the plasma membrane by a small number of Gag proteins, where additional Gag proteins co-localize and virus assembly occurs.24,25 Subsequent to budding, the packaged RNA molecules exist as weakly formed, non-covalently linked dimers that form more stable dimers as the virus ages.26–31
Agents that disrupt the structure of the NC domains of Gag and interfere with genome packaging have potent antiviral properties,32–36 and a better understanding of the structural determinants of genome selection could facilitate the development of new therapeutic approaches for the treatment of AIDS. However, despite considerable effort, current understanding of the protein-RNA interactions37 and mechanisms that promote and/or regulate retroviral genome selection remains limited (for reviews see 8–17,38,39). For some retroviruses, including the Moloney Murine Leukemia Virus (MoMuLV) and Rous Sarcoma Virus (RSV), relatively small regions of the 5´-leader RNAs (fewer than 100 nucleotides) have been identified that are capable of directing the packaging of heterologous RNAs into virus-like particles (VLPs), and this has facilitated high resolution structural studies of the NC:RNA complexes responsible for RNA packaging.40–42 Although atomic-level structures have been determined for the HIV-1 NC protein bound to isolated hairpin and single-stranded RNAs,43–45 these studies did not explain how the virus discriminately packages its dimeric, unspliced genome. To date, no high-resolution structural information is available for a packaging competent HIV-1 RNA.
Efforts to identify a minimal HIV-1 packaging signal have been complicated by a number of factors. First, the 5´-leader contains elements that are important for functions that are unrelated to packaging but critical for replication (including transcriptional activation, primer binding during reverse transcription, splicing, and dimerization) (Fig. 1), and mutagenesis studies designed to test effects on packaging can adversely affect other functions. For example, mutagenesis studies originally suggested that the TAR hairpin is required for genome packaging,46–48 but more recent studies indicate that effects originally attributed to a packaging defect were actually due to dominant negative effects on viral gene expression.49 A second complication is that correct Rev-mediated RNA localization is a prerequisite to lentiviral RNA packaging, so that even RNAs that possess intact packaging signals are inefficiently packaged without Rev.50 A third issue is that HIV-1 VLPs can efficiently assemble in the absence of their native genomes and, in the process, package similar amounts of cellular RNAs.51–55 Another complication relates to the fact that some previous packaging studies were conducted using transfection experiments in which the cellular concentration of mutant vector RNAs was high. At high cellular concentrations, the mutant RNAs can be packaged at high levels, as long as they do not have to compete with RNAs containing the wild-type 5´-leader. In addition, efforts to determine the secondary structure of the HIV-1 5´-leader using chemical probing, mutagenesis and biochemical approaches, and phylogenetic analyses56–68 have led to multiple structural proposals and mechanistic predictions, thus hindering the design of mutations to properly test structure/function hypotheses.39
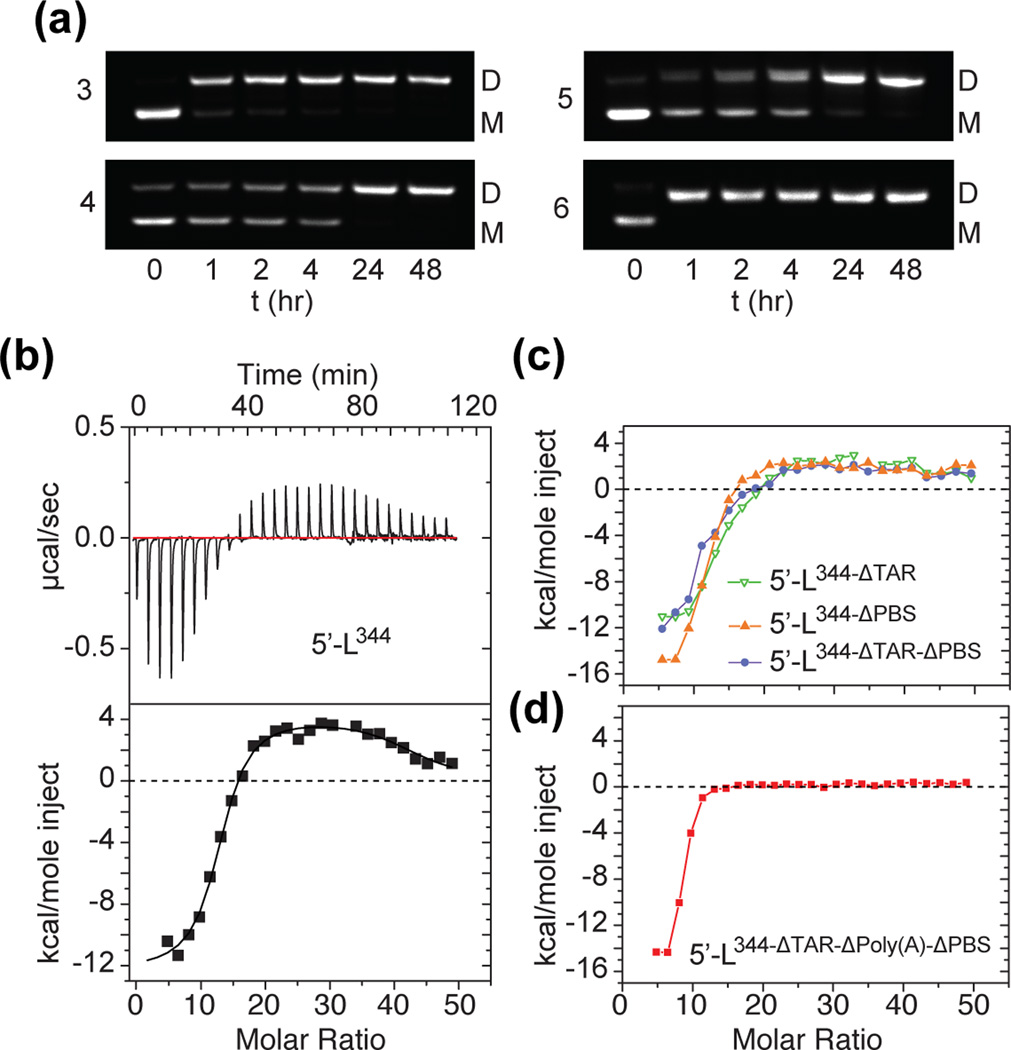
Fig. 1.

(a) Representation of HIV-1 viral genome showing relative positions of splice sites and coding versus non-coding regions. (b) Secondary structure of the 5´-L conformer that promotes dimerization, NC binding and genome packaging.69 Long range base pairing interactions between U5 (blue) and AUG (green) help expose the dimer-promoting GC-rich loop of the DIS hairpin (cyan). Residues in pairing patterns that give rise to resolved, upfield-shifted adenosine C2-H signals are highlighted (colored boxes). The 3´-most residues of AUG (light green) are deleted in the 5´-L344 construct. (c) Mutations in AUG that affect base pairing with U5 influence the monomer-dimer equilibrium. Lane 1: native 5´-L exists as a mixture of monomer and dimer when incubating within PI buffer. Lane 2: Substitution of 331-GAGAGAUGGGUGCGAGAGCGUCGGU-355 by 331-AACUAUACAAUCGGAGACGAUUGCA-355, which has been reported in our previous study known as 5´-LHP-AUG), favors the AUG hairpin structure and exists mainly as a monomer. This mutation has been shown to inhibit packaging.69 Lane 3: Substitution of 333-UGGGUGC-339 by 333-CUCUAGA-339, which favors the AUG hairpin structure, mainly exists as a monomer, and has been reported with packaging deficiency.120 Lane 4: Substitution of 337-UGGGUGCGAGAGCGUC-353 by 337-CGGGCACAAGAAAAAA-353 (5´-LU5:AUG) favors the formation of U5:AUG base pairs and promotes dimerization and packaging.69 Lane 5: 5´-L344, in which residue 345–356 was truncated from 5´-L. The AUG hairpin cannot form in 5´-L344, but the residues involved in U5:AUG base pairs are maintained. This truncation stabilizes the RNA as a dimer. (d) 5´-L344 readily forms a dimer upon incubation in PI buffer. (e) Portion of the 2D NOESY spectrum obtained for a dimeric 5´-L344 RNA sample containing protons on the non-exchangeable adenosine C-2 and ribose (A2,R), the guanosine ribose (GR), and the cytosine ribose (CR) carbons and deuterons at all other non-exchangeable aromatic and ribose carbons: A2,R,GR,CR-[5´-L344]2. NMR signal assignments were made based on comparisons with isolated oligo-RNAs corresponding to the TAR, lower-PBS, DIS, and Ψ-RNA hairpins (Lu, Heng and Summers, in preparation).
Recently, using an NMR approach for studying relatively large RNAs, we showed that the HIV-1 5´-leader exists as an equilibrium of two conformationally distinct species, one in which residues spanning the gag start codon (G328-A356; hereafter called “AUG”) form a hairpin and the GC-rich palindromic loop of the dimer promoting DIS hairpin is base paired with an upstream element (U5), and another in which AUG is base paired with U5 and the dimer promoting loop of DIS is exposed.69 U5:AUG formation, proposed originally on the basis of phylogenetic and biochemical analyses,62,70 promotes dimerization by displacing the DIS and simultaneously exposes high-affinity NC binding sites, thereby promoting the packaging of a dimeric genome.69 Having identified the conformer of the 5´-leader that directs packaging, we wished to explore the potential roles of specific elements within the leader on RNA packaging. Our present study identifies a 159 nucleotide segment of the 5´-leader that is sufficient for RNA dimerization and NC binding in vitro and is required for efficient and selective RNA packaging.
Results
Construct desig
A primary goal of these studies was to systematically remove elements from 5´-L (residues 1–356) without altering the folding of other structural elements, as assessed by the NC-binding, dimerization, and NMR spectroscopic properties of the RNA. Under conditions of physiological-like ionic strength (PI buffer = 140 mM KCl, 10 mM NaCl, 1 mM MgCl2, 10mM TRIS-HCl, pH 7.0) and at an RNA concentration of ~ 1.0 µM, 5´-L exists as an equilibrium mixture of two species: a monomeric conformation in which the GC-rich palindrome of the DIS hairpin is sequestered by base pairing with U5 and AUG forms a hairpin, and a dimeric conformation in which AUG base pairs with U5 and the DIS is exposed (Fig. 1c) (dimer dissociation constant Kd = 0.9 ± 0.1 µM).69 Mutations in AUG that favor U5:AUG base pairing promote dimerization and NC binding of 5´-L RNAs in vitro and the packaging of analogous vector RNAs into VLPs (Fig. 1c),69 whereas the complete truncation of the AUG hairpin inhibits dimerization by preventing U5:AUG dependent displacement of the DIS.69 The present studies focused on 5´-L constructs that lacked the 3’-most residues of AUG (A345-A356 deleted; 5´-L344) (Fig. 1b).69 This deletion allows U5:AUG formation while simultaneously precluding formation of the competitive AUG-hairpin structure, thereby shifting the equilibrium in favor of the dimer (Fig. 1c,d).
2D NOESY spectra obtained the fully protonated [5´-L344]2 RNA exhibited severe signal overlap and were unassignable using standard methodologies.69 However, a significant number of resolved adenosine C2-H signals were detectable in 2D NOESY spectra obtained for RNAs in which all non-exchangeable aromatic protons except the C2-H of the adenosines were substituted by deuterium. For example 1H NMR signals with chemical shifts and NOE cross peak patterns matching those observed for the isolated TAR, DIS and Ψ-site oligo-RNA hairpins were readily identified in 2D NOESY spectra obtained for a dimeric 5´-L344 sample that contained protons on the non-exchangeable adenosine C-2 and ribose (A2,R), the guanosine ribose (GR), and the cytosine ribose (CR) carbons and deuterons at all other non-exchangeable aromatic and ribose carbons (A2,R,GR,CR-[5´-L344]2, Fig. 1e) (8, 2, and R = protonated on the C-8, C-2, and ribose carbons, respectively). These outlier signals and their NOE interactions with H1´ protons were monitored to assess potential effects of mutations and deletions on the structure of the RNA.
Additional leader RNAs were prepared that contained deletions of the TAR hairpin (5´-L344-ΔTAR), the upper region of the PBS loop (5´-L344-ΔPBS), the TAR and upper PBS loop (5´-L344-ΔTAR-ΔPBS), and the TAR and Poly(A) hairpins and upper PBS loop (5´-L344-ΔTAR-ΔPoly(A)-ΔPBS) (Fig. 2a). RNA homogeneity was confirmed by denaturing polyacrylamide gel electrophoresis (PAGE, Fig. 2b). Base pairing patterns of the TAR and Poly(A) hairpins and the lower stem of the PBS loop have been supported by several studies,62,71–75 including compensatory mutagenesis-based packaging experiments.76 The PBS deletion construct was designed such that the entire upper region of the loop (residues A132–A216) was substituted by a GAGA tetraloop (a member of the well-characterized GNRA family77–84). Resolved A220 C2-H signals were observed for all constructs in partially deuterated samples (prepared as described above), including those containing both the native and truncated PBS loops (Fig. 2c), indicating that base pairing in the lower stem of the PBS loop is maintained in both the native and mutant 5´-L RNAs. 2D NOESY data obtained for the 5´-L344 mutants did not exhibit line broadening or additional NMR signals (other than those expected for the GAGA tetraloop) that would be indicative of alternative folding. In particular, the outlier adenosine C2-H signals of the most highly truncated construct, 5´-L344-ΔTAR-ΔPoly(A)-ΔPBS, exhibited 1H NMR chemical shifts and 2D NOESY cross peaks similar to those observed for the wild-type 5´-L344 RNA (Fig. 2d). Signals assigned to the TAR, Poly(A) and upper PBS loop structures were missing in spectra obtained for the corresponding deletion mutants, supporting the 1H NMR chemical shift assignments for these elements.
Figure 2.
(a) RNA constructs used in the present studies: (1) 5´-L, (2) 5´-L344, (3) 5´-L344-ΔTAR, (4) 5´-L344-ΔPBS, (5) 5´-L344-ΔTAR-ΔPBS, (6) 5´-L344-ΔTAR-ΔPolyA-ΔPBS. (b) Denaturing PAGE gels show sample purity and their relative electrophoretic migration. (c) Portions of 2D NOESY spectra showing the A220 C2-H peaks observe for (from left to right) A2,R,GR,CR- labeled [5´-L344]2, 5´-L344-ΔPBS, 5´-L344-ΔTAR-ΔPolyA-ΔPBS, and for a fully protonated oligoribonucleotide corresponding to the lower PBS hairpin (5´-ggCUCUGGUgagaGCCAGAGcc; lower case = non-native), showing that the lower PBS stem structure is maintained in these 5´-L constructs. (d) Chemical shifts of the C2-H signals for the PBS lower stem, Ψ, and DIS hairpins are unaffected by removal of the TAR, Poly(A) and PBS upper loop (gray and black spectra correspond to RNAs labeled 2 and 6 in panel (a), respectively.
Effects of TAR, Poly(A) and PBS deletions on dimerization and NC binding
The 5´-L344, 5´-L344-ΔTAR, 5´-L344-ΔPBS, 5´-L344-ΔTAR-ΔPBS and 5´-L344-ΔTAR-ΔPoly(A)-ΔPBS RNAs all exist predominantly as monomers at low ionic strength but form dimers upon incubation in PI buffer (Fig. 1d and Fig 3a), indicating that the TAR, Poly(A), and upper PBS hairpin structures do not play a significant role in dimerization. The potential roles of these structures in NC binding was assessed by isothermal titration calorimetry (ITC).69 As observed previously in ITC studies of native and mutant forms of 5´-L, the binding isotherms obtained for 5´-L344 upon NC titration in PI buffer included an initial exothermic component followed by an endothermic component (Fig.3b). The exothermic events are attributed to high affinity NC binding since similar ITC profiles have been observed for oligo-RNAs that contain a single high affinity NC binding site43–45. The endothermic events occur during the later injections at progressively higher [NC]:[RNA] ratios and are attributed to weak NC-RNA interactions and NC-induced RNA unfolding.69 5´-L344 exists as a dimer under conditions of the ITC experiments, and non-linear least squares fitting using a two-event binding model indicates that [5´-L344]2 binds 15 ± 1 NC molecules per RNA strand with high affinity (Kd = 17 ± 9 nM) (Fig. 3b and Table 1). Both the number of high affinity NC sites and the binding affinity are similar to those measured for 5´-L RNAs containing mutations in AUG that favor U5:AUG base pairing (5´-LU5:AUG; 16 ± 1 NC sites per strand; Table 1).69
Figure 3. Effects of TAR, PolyA and PBS deletions on NC binding.
(a) Native gel electrophoresis data (2% high-gelling-temperature agarose gel containing TRIS-borate buffer and ethidium bromide for RNA visualization) showing that 5´-L344-ΔTAR, 5´-L344-ΔPBS, 5´-L344-ΔTAR-ΔPBS and 5´-L344-ΔTAR-ΔPolyA-ΔPBS (labeled 3–6, respectively, as in Fig. 2) form dimers after incubation in PI buffer (140 mM KCl, 10 mM NaCl, 1 mM MgCl2, pH 7.0), indicating that deletions of TAR, PolyA and PBS do not affect RNA dimerization (b) ITC data obtained upon titration of NC into 5´-L344. The raw data are shown in the top panel, and the integrated heat changes are shown in the bottom panel. First two calorimetric data points were deleted before non-linear least squares isotherm fitting using “Two-Set of Sites” mode (MicroCal Origin 5.0). The first set of binding parameters were utilized to characterize the high-affinity interactions between 5´-L344 and NC (Table 1). (c) ITC NC titration data obtained for 5´-L344-ΔTAR, 5´-L344-ΔPBS, 5´-L344-ΔTAR-ΔPBS, showing that these deletions have modest effects on high affinity NC binding. (d) ITC NC titration data obtained for 5´-L344-ΔTAR-ΔPolyA-ΔPBS. The post-high affinity endothermic contributions to NC binding observed for 5´-L344 (and at diminished levels in the other 5´-L344 mutants) are nearly undetectable.
Table 1.
Thermodynamic parameters from ITC measurementsa
| N1 | Kd1 (nM) | ΔH1 (Kcal/mol) |
−TΔS1 (Kcal/mol) |
N2 |
Kd2 (mM) |
ΔH2 (Kcal/mol) |
−TΔS2 (Kcal/mol) |
|
|---|---|---|---|---|---|---|---|---|
| 5´-L344 | 15 ± 1 | 17 ± 9 | −12.5 ± 0.2 | 1.7 ± 0.5 | 31 ± 4 | 1.0 ± 0.5 | −2.3 ± 9.0 | −8.3 ± 8.6 |
| 5´-L344-ΔTar | 13 ± 1 | 17 ± 15 | −14.0 ± 2.0 | 2.8 ± 2.9 | 30 ± 3 | 0.6 ± 0.4 | 1.5 ± 1.9 | −12.6 ± 0.6 |
| 5´-L344-ΔPBS | 11 ± 1 | 16 ± 15 | −16.7 ± 1.8 | 5.7 ± 1.3 | 43 ± 9 | 4.2 ± 6.0 | 3.4 ± 0.3 | −11.4 ± 1.0 |
| 5´-L344-ΔTar-ΔPBS | 9 ± 1 | 21 ± 15 | −15.5 ± 1.9 | 4.7 ± 1.9 | 31 ± 3 | 0.8 ± 0.6 | 3.1 ± 0.2 | −11.7 ± 0.7 |
| 5´-L344-ΔTar-ΔPolyA-ΔPBS | 8 ± 1 | 26 ± 39 | −11.7 ± 6 | 4.4 ± 1.4 | a | a | a | a |
Two-sites binding mode was utilized to fit the ITC isotherms from NC titrations.
The magnitude of the endothermic component was too small for reliable calculation.
The ITC isotherms obtained upon titration of 5´-L344-ΔTAR, 5´-L344-ΔPBS, 5´-L344-ΔTAR-ΔPBS and 5´-L344-ΔTAR-ΔPoly(A)-ΔPBS with NC were similar in appearance to those obtained for 5´-L and 5´-L344 (Fig. 3b,c), except that these truncated RNAs bound fewer NC molecules with high affinity. Deletion of the TAR and PBS loops resulted in losses of 2 ± 1 and 4 ± 1 NC binding sites, respectively, and the simultaneous deletion of both the TAR and PBS loops eliminated 6 ± 1 NC sites (Table 1), indicating that NC binds independently and non-cooperatively to these hairpins. The isolated oligo-TAR RNA was previously shown to interact with NC,85 and an oligo-RNA corresponding to the isolated TAR hairpin was found by ITC to bind a single NC molecule with high affinity (Kd = 110 ± 2 nM) (Heng & Summers, unpublished results). Removal of the Poly(A) hairpin from 5´-L344-ΔTAR-ΔPBS resulted in the loss of a single additional high affinity NC site (Table 1). Thus, TAR, PBS and Poly(A) hairpins appear to contribute approximately 2, 4, and 1 high affinity NC binding sites, respectively. Interestingly, we also observed small but systematic reductions in the magnitude of the endothermic component of the ITC isotherms upon removal of TAR, Poly(A) and PBS elements (Fig. 3b,c), with isotherms for 5´-L344-ΔTAR-ΔPoly(A)-ΔPBS exhibiting only a slight endothermic feature at the higher NC/RNA ratios (Fig. 3c). Thus, the NC-induced RNA unwinding that occurs after saturation of the high affinity NC binding sites appears to diminish with decreasing size of the RNA.
Effects of TAR, Poly(A) and PBS deletions on RNA packaging
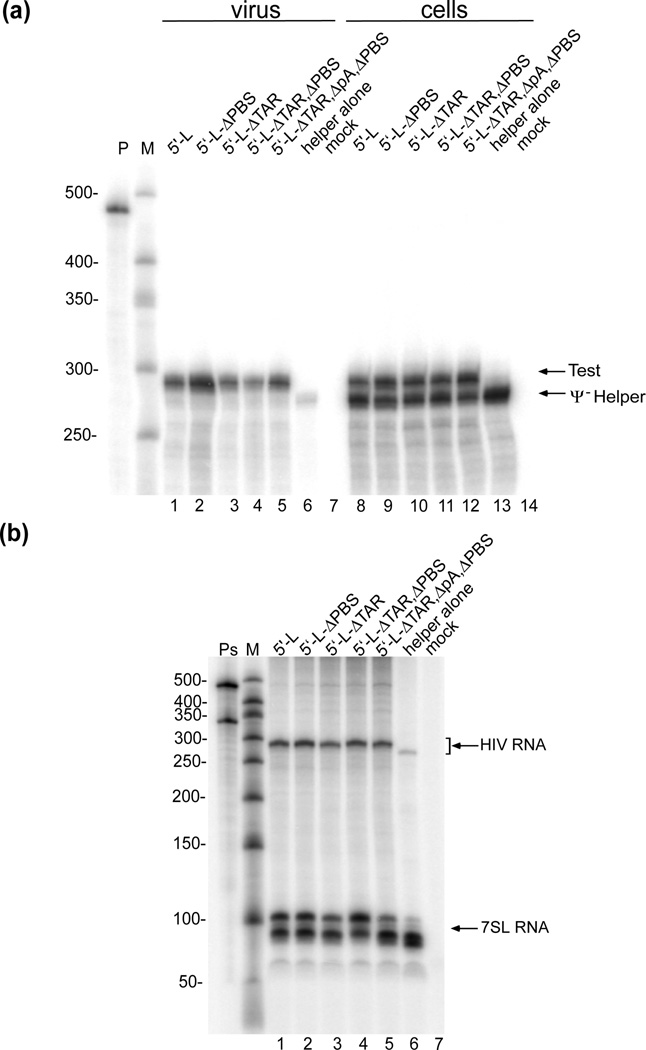
When HIV-1 particles bud from cells in the absence of genomic RNAs with packaging signals, these particles encapsidate “random” host mRNAs roughly in proportion to their intracellular concentrations.86 In contrast, most such adventitiously packaged mRNAs are displaced when RNAs with authentic packaging signals are present, due to the specificity of HIV-1 for its own genome. Here, a packaging assay based on these observations was used to address whether or not the Tar, Poly(A), and PBS loop deletion RNAs studied above possessed information sufficient to mediate authentic HIV-1 RNA packaging. In these experiments, the RNA transcribed from the HIV-1-based helper function plasmid pCMVΔR8.2 (herein called Ψ(−) helper),87 which expresses all virion proteins except env but lacks all HIV-1 5´ leader sequences upstream of the gag start codon, served as an internal control for the extent of packaging of an RNA that lacks all HIV-1 packaging elements. In the present studies, we adjusted transfection levels such that the test and Ψ(−) helper RNAs were transcribed in cells at similar levels, thereby enabling a more quantitative assessment of relative packaging efficiencies, Fig. 4. RNA packaging levels were measured by using RT activities (Fig. 4a) or viral 7SL RNA levels (Fig. 4b) as controls. 7SL RNA is a cellular transcript that is incorporated independent of genomic RNA and can be used as an internal control for quantitating packaging efficiencies.88 Relative efficiencies were determined by comparing packaging levels of test RNAs that contained 5´-leader mutations with those that contained the intact, wild-type leader and gag sequences. All test vectors contained Rev responsive elements (RREs) and were expressed in the presence of Rev, and thus Rev-dependent RNA localization, which is required for efficient packaging,50 was not a variable in these experiments.
Figure 4. TAR, Poly(A) and PBS deletions do not affect RNA packaging.
RNA packaging efficiency of 5´-L variants were monitored by RNase protection assay. (a) RNA samples obtained from the co-expression of a series of packaging test vectors with CMVΔR8.2, an HIV-1 Ψ- helper construct. Lanes 1–5: products of RNA samples harvested from media of cells transfected with the indicated constructs plus pCMVΔR8-2 helper; lane 6: from cells transfected with pCMVΔR8-2 alone; lane 7: from mock-transfected cells. Lanes 8–14 are RNase protection assay products of cellular samples from the same transfections used in lanes 1–7. P, undigested riboprobe; ΔpA = Δpoly(A). The mobilities of size standards are indicated to the left; the mobilities of riboprobe fragments protected by transcripts from each of the test vectors and from the Ψ- helper are indicated at right. (b) RNase protection assay of viral samples using two different riboprobes specific to 7SL RNA or HIV-1 vectors RNA (Ps = the two undigested riboprobes). Lanes 1–5: products of RNA samples harvested from media of cells transfected with the indicated constructs plus CMVΔR8-2 helper; lane 6: from cells transfected with CMVΔR8-2 alone; lane 7: from mock-transfected cells. The mobilities of riboprobe fragments protected by transcripts from each of the test vectors and the Ψ- helper are indicated at right as HIV RNA; the mobilities of riboprobe products protected by 7SL RNA indicated as 7SL RNA
As shown in the RNase protection assay in Fig. 4a, when Ψ(−) helper was transfected into 293T cells in the absence of RNAs possessing HIV-1 packaging signals, the transcript was highly expressed in cells (lane 13) and was encapsidated into virions that were released from these cells with modest efficiency (lane 6). In contrast, whereas both transcripts were detected at similar levels within cells (lane 8), when CMVΔR8.2 was co-transfected with an HIV-1 test vector possessing the native NL4-3 5´-leader (Native), RNAs with native NL4-3 packaging elements were significantly enriched among virion RNAs, demonstrating that the leaderless Ψ(−) helper transcripts could not compete for packaging (lane 1). TAR, Poly(A), and PBS loop deletion test RNAs also were co-expressed with Ψ(−) helper. Although these vectors lacked TAR regulatory elements, they were well-expressed under the transfection conditions used, which were selected to ensure that the test vector and Ψ(−) helper RNA levels were similar to one another within cells (lanes 9–12). However, similar to results obtained for the native NL4-3 leader, the Ψ(−) helper RNAs were unable to compete efficiently with these test RNAs (lanes 2–5).
Similar results were obtained in RNase protection assays that quantified vector RNA packaging by normalizing to the co-packaged host RNA 7SL, which serves as a surrogate for virion copy number89 (Fig. 4b). Similar amounts of 7SL RNA-protected products were present in all viral samples from cells transfected with CMVΔR8.2, whether alone or in the presence of test plasmids (lanes 1–6). As expected, HIV-1 specific transcripts were robustly packaged in the sample from cells co-transfected with CMVΔR8.2 plus the parental 5´-L HIV-1 test vector (lane 1), and a significant drop in packaging was observed for the Ψ- transcript produced when CMVΔR8.2 was expressed alone (lane 6). In contrast to this Ψ- transcript, 5´-L RNAs from which either the PBS loop or TAR were deleted (lanes 2–3), those that lacked both TAR and the PBS loop (lane 4), and the TAR, Poly(A), and PBS loop triple-deletion test RNA (lane 5), were all encapsidated at levels similar to those of native 5´-L RNA (lane 1). As quantified in Table 2, only small decreases in overall packaging were observed, with all of these deleted 5´-L RNAs displaying packaging ≥ 80% that observed for the native 5´-leader.
Table 2.
Packaging efficiencies of vector RNAs containing native and mutant HIV-1 5´-leader sequencesa
| Test RNA | %Packaging/RTb | %Packaging/Ψ−helperc | %Packaging/7SLd |
|---|---|---|---|
| 5´-L (Native) | 100 | 100 | 100 |
| 5´-L-ΔTAR | 41 ± 32 | 98 ± 4 | 86 ± 4 |
| 5´-L-ΔPBS | 133 ± 77 | 100 ± 3 | 94 ± 4 |
| 5´-L-ΔTAR-ΔPBS | 37 ± 20 | 90 ± 6 | 82 ± 1 |
| 5´-L-ΔTAR-ΔPoly(A)-ΔPBS | 86 ± 59 | 100 ± 7 | 89 ± 11 |
| 5´-L350 | 101 ± 5 | 100 ± 1 | e |
Data reported as mean ± standard deviation obtained from 3 independent experiments except data for 5´-L350, which is from 2 independent experiments.
%Test RNA/virion based on RT activity to quantify virus production. Data normalized relative to Test RNA containing the native 5´-leader.
%Test RNA relative to Ψ− helper RNA in particles normalized relative to Test RNA containing the native 5´-leader.
%Test RNA/virion using 7SL packaging to quantify virus production. Data normalized relative to Test RNA containing the native 5´-leader.
Not measured.
Effects of gag deletion on packaging
To determine if elements in gag downstream of the AUG are important for packaging, we generated packaging vector variants that retained the 5´ leader sequences studied by NMR above, but that lacked HIV-1 gag-derived sequences downstream of position +350 that were present in the native 5´-L vectors examined above. As shown in RNase protection assay results in Figure 5, co-expression of either the original 5´-L or a truncated 5´-L350 vector lacking gag sequences with the CMVΔR8.2 helper construct yielded indistinguishable RNA levels in cells (lanes 5 and 6) and identical strong packaging levels in virions (lanes 1 and 2; Table 2). These findings demonstrate that sequences in gag are not required for efficient packaging of HIV-1 5´-Leader containing RNAs into VLPs.
Figure 5. Effect of gag deletion on RNA packaging.
RNA samples obtained from the co-expression of a series of native and 3´ truncated packaging test vectors with CMVΔR8.2. Lanes 1–2: RNase protection assay products of RNA samples harvested from media of cells transfected with the indicated constructs plus pCMVΔR8-2 helper; lane 3: from cells transfected with pCMVΔR8-2 alone; lane 4: from mock-transfected cells. Lanes 5–8 are RNase protection assay products of cellular samples from the same transfections used in lanes 1–4.
Effect of TAR deletion on 5´-leader structure
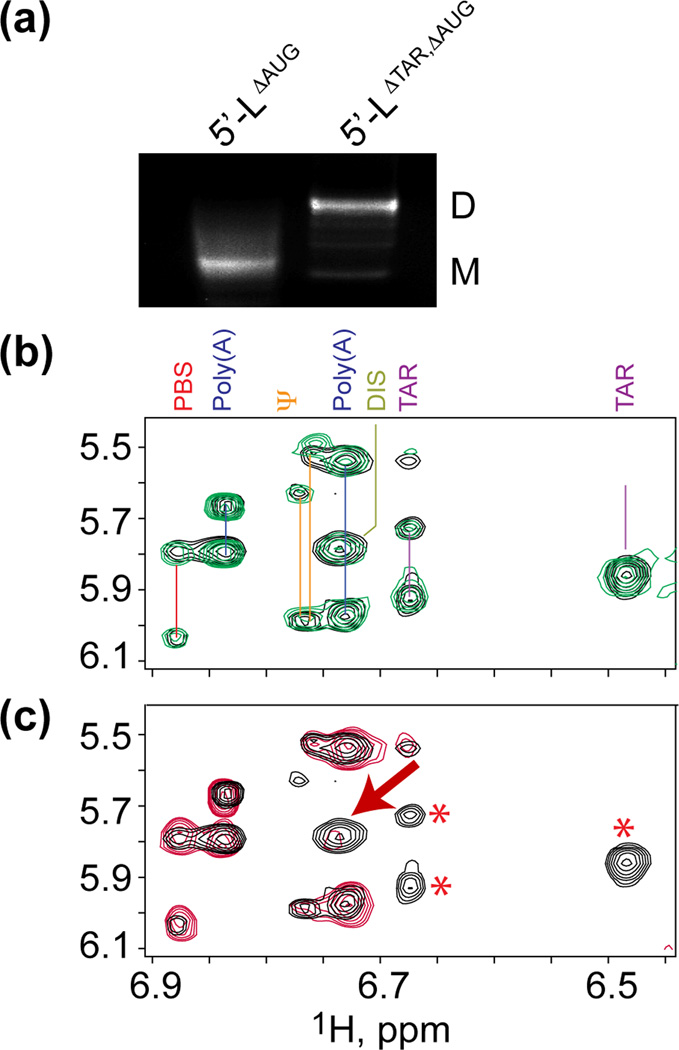
The observation that deletion of TAR results in a moderate but reproducible reduction in RNA packaging, whereas deletion of the PBS loop does not affect packaging, is difficult to reconcile with our finding that the PBS upper loop contains a greater number of high affinity NC binding sites. 2D NOESY spectra obtained for [5´-L344-ΔTAR]2 appeared similar to spectra obtained for the parent [5´-L344]2 RNA (except for the missing TAR signals; data not shown), suggesting that the TAR deletion does not affect the overall fold of the dimeric RNA. To determine if the TAR deletion affects the folding of the monomeric form of the leader, we examined the dimerization properties and 2D NOESY spectra of leader RNAs in which AUG was fully truncated (5´-LΔAUG). As indicated above, this RNA lacks the 5´-residues of AUG necessary to displace and expose the DIS hairpin, and like full-length 5´-L RNAs containing mutations that stabilize the AUG hairpin, 5´-LΔAUG exists predominantly as a monomer in PI buffer (Fig. 6a). Unexpectedly, deletion of the TAR hairpin from 5´-LΔAUG (5´-LΔAUG -ΔTAR) strongly promoted dimerization in PI buffer (Fig. 6b). In addition, whereas the outlying adenosine C2-H signals in 2D NOESY spectra obtained for 5´-LΔAUG were similar in appearance to spectra obtained for [5´-L344]2 (Fig. 6b), the spectrum obtained for 5´-LΔAUG-ΔTAR lacked several expected signals, including the A268 C2-H signal associated with the properly folded form of the DIS hairpin (Fig. 6c). These findings indicate that removal of TAR leads to non-native like folding and dimerization of the 5´-leader.
Figure 6.
(a) Non-denaturing gel electrophoresis (2% agarose) results showing that 5´-LΔAUG exists predominantly as a monomer, as expected, and that truncation of TAR (5´-LΔAUG-ΔTAR) strongly promotes dimerization. (b) [5´-L344]2 (green) versus 5´-LΔAUG (black). (c) 5´-LΔAUG (black) versus 5´-LΔAUG-ΔTAR (red); The star and arrow symbols highlight the expected loss of the TAR signals (stars) and unexpected loss of the DIS A268-C2-H signal (arrow) that were observed upon deletion of TAR. These data indicate that removal of TAR leads to misfolding of DIS hairpin and induces non-native dimerization of the monomeric form of the 5´-leader.
DISCUSSION
The TAR, Poly(A) and PBS loops have each been implicated in at least some studies to play roles in genome dimerization, NC binding, and packaging. Isolated TAR oligo-RNA hairpins can form dimers in the presence of NC, and electron microscopy (EM) images of NC incubated HIV-1 5´-RNA fragments (nucleotides 1–744) exhibit distinct circular structures that have been interpreted in terms of a TAR-mediated dimer interface.90 Some studies suggested that deletion of TAR promotes the packaging of monomeric RNAs, to an extent similar to that observed upon deletion of the DIS hairpin.91 On the other hand, truncation of the TAR hairpin from a 290 nucleotide 5´-fragment of the HIV-1 leader promoted, rather than inhibited, dimerization of this RNA.92,93 The present studies show that 5´-L344, which spans the entire HIV-1 5´-UTR, forms a stable dimer even in the absence of the TAR hairpin. 2D NOESY spectra are relatively unperturbed (except for the loss of the TAR signals) upon removal of TAR, indicating that dimerization does not result from artificial misfolding of the dimeric leader. The fact that the A46 C2-H signals of TAR are relatively intense in both the monomeric and dimeric forms of 5´-L RNAs (Fig. 1e) suggests that the hairpin is relatively mobile and probably does not participate in long-range interactions of the dimeric 5´-leader (although TAR could participate in intermolecular RNA-RNA interactions in the more stable dimer that is formed in virions during viral maturation91,94).
Deletion of TAR was shown in one study to reduce packaging efficiency by 74% compared to the packaging of a parent RNA derived from the intact viral genome that encoded a stop codon within the capsid domain of gag.47 Although TAR plays a critical role in transcriptional activation via its interactions with the viral Tat protein95,96 these studies concluded that reductions in packaging were too great to be attributed to the relatively modest reductions in cellular RNA levels.47 Mutations that destabilized the stem of TAR reduced packaging to 70% of wild-type levels (after controlling for reductions in cellular RNA levels)48 and altered the ratio of spliced/unspliced viral RNAs that were packaged.97 Interestingly, using a proviral expression system that is largely independent of Tat for transcriptional activation, Rekosh and co-workers found that missense mutations in the upper stem of TAR had little to no effect on packaging, but mutations at the base of the TAR stem could lead to significant reductions in packaging.46 Compensatory mutations in the TAR stem restored packaging to near wild-type levels, indicating that it is the structure of TAR, and not the TAR sequence, that is important for packaging.46 A more recent study showed that the complete removal of TAR had little impact on replication as long as other changes were made to overcome the requirement for TAR-mediated transcriptional activation (packaging efficiencies were not reported).49 These studies also showed that there is evolutionary pressure to restore base pairing at the 5´-terminal end of the viral RNA,49 and supported proposals that packaging defects induced by mutations in TAR result from misfolding of the 5´-leader. Our findings are consistent with this latter hypothesis. Thus, deletion of TAR did not significantly affect the NC binding properties of the 5´-leader RNA and also did not significantly impair vector RNA packaging. The modest packaging defects that we observed upon deletion of TAR are most likely due to misfolding of the monomeric form of the 5´-leader, which we believe leads to a reduction in the cellular levels of the packageable, dimeric form of the RNA.
The Poly(A) hairpin has also been proposed to play roles in both genome dimerization and packaging. Although early computer-modeling studies and EM imaging of the HIV-1 genome suggested that the palindromic 5´-AAGCUU-3’ loop of Poly(A) might form a second dimer linkage structure,98 subsequent mutagenesis studies indicated that the Poly(A) palindrome does not influence RNA dimerization.99 Here we showed that deletion of the TAR and Poly(A) hairpins does not affect the dimerization properties of 5´-L344, in support of the latter conclusion. Poly(A) was originally thought to be important for RNA packaging because removal of the hairpin reduced packaging to ~10% of wild type levels.47,100 Mutations that disrupt the Poly(A) hairpin structure were also shown to inhibit genome packaging and viral replication, whereas compensatory mutations restored packaging to near wild-type levels indicating that it is the structure, rather than specific sequence of the Poly(A) hairpin that is required for packaging.97 However, more recent studies indicated that disruption of the Poly(A) hairpin structure can lead to reductions in cellular levels of the viral RNA due to reductions in RNA stability, and it was suggested that the Poly(A) hairpin may not actually play a significant role in genome packaging.101 As shown in the present studies, removal of Poly(A) from 5´-L344-ΔTAR does not significantly affect its structure, dimerization or NC binding properties. Furthermore, vector RNAs in which the TAR, Poly(A) and PBS loops are deleted were packaged as well as RNAs that contained only TAR and PBS deletions, indicating that Poly(A) does not play a significant role in RNA packaging.
The PBS element contains a sequence that binds human tRNALys, a requirement for initiation of reverse transcription.102–106 Numerous secondary structural models of PBS have been proposed based on mutational studies, enzymatic probing, UV-cross linking, phylogenetic studies and chemical modification assays (reviewed in 13,39,62,70,72,73,75,107). Deletions within the upper loop of the PBS generally do not significantly impair genome packaging, whereas the lower stem loop is required for RNA dimerization and efficient packaging76,108,109. Our PBS deletion mutant was designed to maintain native base pairing in the lower stem of the PBS loop structure. Significantly, the NMR data presented here show that A220 C2-H signal pattern was observed in [5´-L344]2 and all the PBS upper loop truncated constructs, indicating that the lower stem of the PBS element adopts the predicted secondary structure. Removal of the upper PBS loop structure eliminated ~ 4 high affinity NC binding sites but did not impair RNA packaging, suggesting that these NC sites may play roles in tRNA binding and/or reverse transcription but are not critical for packaging.
In vitro biochemical studies and phylogenetic predictions led to the proposal that loop residues of PBS base pair with residues G443-C449 in the matrix (MA) encoding region of gag.110 These putative long range interactions have been supported by chemical probing and modeling studies,62,74,75 but their role in packaging has not been established. Early packaging studies suggested that efficient HIV-1 genome packaging requires residues within the first several hundred nucleotides of gag,111 and could therefore be dependent on Poly(A)-gag interactions. However, the present studies show that RNAs containing the native 5´-leader (nucleotides 1–355, which include residues required for U5:AUG base pairing) but missing nearly the entire gag open reading frame are packaged with the same efficiency as RNAs containing the intact gag gene. Thus, if the Poly(A)-gag interactions exist in vivo, they likely play roles in functions unrelated to RNA packaging.
The present findings also provide insights into the number and general locations of the high-affinity NC bindings sites. Early studies using UV-cross linking indicated that NC is the only major protein that binds to the genomic RNA in HIV-1 virions, and it was estimated that approximately 15 NC molecules are bound per RNA strand.112 In contrast, in vitro cross-linking experiments involving a fragment of the 5´-leader spanning from DIS through residue 170 of gag were interpreted to indicate that only ~2–3 NC or Gag proteins bind to each RNA strand,113 and RNA footprinting studies performed with a 5´-leader construct that included the intact 5´-UTR and 66 nucleotides of gag were similarly interpreted in terms of only two high affinity NC or Gag binding sites.72 In addition, quantitative filter-binding and ITC assays indicated that NC can bind with high affinity (Kd ≈ 55–400 nM) to fragments of the HIV-1 5´-leader between the DIS and AUG hairpins,43–45,64,76 Our ITC data of 5´-L and the truncated mutants titration with NC show that they all have high affinity NC binding sites with Kd in the same range (Table 1). Our current and recent studies69 are most consistent with the earlier work of Darlix,112 and indicate that the 5´-leader contains approximately 16 high affinity NC binding sites per RNA strand. The locations of the high-affinity NC binding sites were probed recently by monitoring the effects of zinc ejection on SHAPE activity.75 These studies suggested that the 5´-leader contains 7 NC binding sites, none of which were proposed to reside within the TAR, PolyA or upper PBS loops.75 Although these results are incompatible with our present findings, we cannot rule out the possibility that structural changes associated with viral maturation, or constraints within the context of the packaged RNA, could lead to altered NC binding.
Sakuragi and coworkers have used a novel approach to identify the minimal determinants of HIV-1 genome dimerization and packaging that involves duplication of the 5´-leader in downstream (ectopic) regions of the RNA, which can lead to the unusual packaging of monomeric RNAs (in addition to dimeric RNAs).114 In this system, RNAs with ectopic sequences lacking the Poly(A) and SD hairpins (to prevent unwanted splicing and polyadenylation) were efficiently packaged as monomers.114 This group subsequently showed that RNAs lacking the native 5´-leader, but containing two downstream copies of a sequence that includes TAR through the first half of gag, but that lacks the Poly(A) and PBS hairpins (a 19-nucleotide deletion in the upper loop to eliminate ectopic initiation of reverse transcription), exclusively packages monomeric RNAs, although virion production and packaging efficiency were reduced to ~20% and ~63% of wild-type levels, respectively.115 Using a similar packaging system that included a much smaller portion of gag, these investigators showed that deletions in the upper loop of the PBS do not significantly affect dimerization or packaging, but deletions in U5 and/or the lower PBS stem lead to significant reductions in packaging.109 Surprisingly, severe modifications and deletions within the DIS hairpin led to relatively modest reductions in packaging levels (to 18–50 % of WT levels).109 A systematic deletion mutagenesis study revealed that relatively efficient packaging of monomeric RNAs can be achieved by RNAs that contain a fully native 5´-leader and an ectopic element that lacks the TAR, Poly(A), upper PBS loop, and SD sequences.107 Since the RNAs employed contained a native 5´-leader sequence, these studies do not rule out the possibility that packaging could depend on the presence of at least one fully native leader RNA per dimer. However, the findings are nevertheless fully compatible with our observation that, under the experimental conditions used here, relatively efficient packaging can be achieved by RNAs that lack the TAR, Poly(A) and upper PBS loops.
In summary, our findings identify 5´-L344-ΔTAR-ΔPoly(A)-ΔPBS as an essential HIV-1 core encapsidation element. This fragment of the 5´-leader exhibits 1H NMR spectral features similar to those of the intact 5´-leader, possesses native-like in vitro dimerization and NC binding properties, and is required for promoting RNA packaging into VLPs. Packaging deficiencies observed here and by others46–48,97 upon deletion of TAR are likely due to misfolding of the monomeric form of the 5´-leader. The clustering of high affinity NC binding sites, coupled with the known self-association propensities of the CA domain of Gag, could function synergistically to promote cooperative assembly of the Gag:RNA complex that is trafficked to the plasma membrane and nucleates virus assembly. Since the minimal packaging signal comprises only 159 nucleotides, high-resolution structural studies by NMR or combined NMR/cyro-electron tomographic approaches116 should now be feasible.
MATERIALS AND METHODS
Plasmid construction
The 5´-L, 5´-LU5:AUG and 5´-LHP-AUG mutants were cloned into PUC19 as described.69 5´-L344 and 5´-L344-ΔTAR were subcloned from the 5´-L plasmid using 5´-CGG TCG AAT TCT AAT ACG ACT CAC TAT AGG TCT CTC TGG TTA GAC CAC ATC TGA GCC TGG GAG CTC TCT GGC-3’ and 5´-CGG TCG AAT TCT AAT ACG ACT CAC TAT AGG CAC TGC TTA AGC CTC AAT AAA GCT TGC CTT GAG ATC-3’ as forward primer, respectively, and 5´-CCC TAG GAT CCC CCG GGC GCA CCC ATC TCT CTC CTT CTA-3’ as their mutual reverse primer. 5´-L344-ΔPBS was made by MegaPrimer mutagenesis method using 5´-L344 as template. The four primers used are 5´-CGG TCG AAT TCT AAT ACG ACT CAC TAT AGG TCT CTC TGG TTA GAC CAG ATC-3’, 5´-CCT GCG TCG AGA GAT CTC CTC TGG TCT CCC AGA GTC ACA CAA CAG ACG GGC ACA CAC TAC TTT GAG CA-3’, 5´-GAG ACC AGA GGA GAT CTC TCG ACG CAG GAC TCG GCT TGC TGG AGA CGG CAA GAG GCG AGG GGC GGC GAC-3’, and 5´-CCC TAG CTA GCT TCT GCC TCC GCT AGT CAA AAT TTT TGG CGT ACT CAC CA-3’. 5´-L344-ΔTAR,-ΔPBS and 5´-L344-ΔTAR-ΔPoly(A)-ΔPBS were then cloned from 5´-L344-ΔPBS plasmid using the forward primer 5´-CGG TCG AAT TCT AAT ACG ACT CAC TAT AGG CAC TGC TTA AGC CTC AAT AAA GCT TGC CTT GAG ATC-3’ and 5'-CGG TCG AAT TCT AAT ACG ACT CAC TAT AGG TGT GCC CGT CGT TGT GTG ACT CTG GTG AGA GCC AGA GGA GA-3', respectively, and the mutual reverse primer 5'-CCC TAG GAT CCC CCG GGC GCA CCC ATC TCT CTC CTT CTA-3'. All of the mutations were sent to the DNA sequencing facility (University of Maryland, Baltimore County) to ensure the desired mutations were obtained.
T-LNL4-3 - - leader region sequences in place of the corresponding sequences in HIV-lac.117 Variants with 5´-LΔTAR, 5´-LΔTAR-ΔPBS or 5´-LΔTAR-ΔPoly(A)-ΔPBS mutations were constructed by introducing mutations into the - -leader region using overlap extension PCR, and using these to replace the corresponding regions of 5´-LNL4-3. Correct structures of all plasmids were confirmed by sequencing. For the initial series of test vectors, a stop codon was introduced after the 85th codon in the gag open reading frame. For gag deletion mutants, PCR mutagenesis was used to delete sequences downstream of the 15th nucleotide in gag and fuse the 15th residue to the Not I restriction site of HIV-lac.117 Riboprobe RNAs to detect helper and test vector RNAs were templated by pEG676-1, a derivative of pBluescript KS (Stratagene), which includes sequences complementary to 286 bases of CMV promoter shared by helper and test vectors plus additional 14 bases sequences unique to the test vectors, preceded by a T7 promoter.
RNA packaging experiments
Human 293T cells were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Gemini). Cells were grown at 37°C under 5% CO2. To generate virions for RNA packaging analysis, 293T cells were co-transfected with a total of 20 µg of plasmid DNA that included one test vector plasmid and pCMVΔR8.2, a Ψ-negative HIV-1 helper vector driven by the cytomegalovirus (CMV) promoter87, at an approximate 1:3 molar ratio on 100-mm plates. Modest empirically determined variation in co-transfection ratios was introduced for non-native vectors, to achieve similar test/helper expression ratios for all samples. For transfection by PEI, plasmid DNA was mixed with 4 µg of polyethyleneimine (Polysciences) per µg of DNA in 1 ml of 150 mM NaCl by vortexing at medium speed for 10 s. After room temperature incubation for 15 min, the mixture was added dropwise to medium on cells. The transfection mixture was replaced with fresh DMEM medium 24 h after transfection.
Tissue culture medium was harvested at 48 h post-transfection, pooled, and filtered through 0.2-µm-pore-size filters. Virus-containing medium was concentrated by centrifugation through a 2-ml 20% sucrose cushion in phosphate-buffered saline (PBS) for 2 h at 4°C and 25,000 rpm using a Sorvall Surespin 630 rotor in a Sorvall Discovery 90 ultracentrifuge. Viral pellets were suspended in 0.5 ml of Trizol reagent (Invitrogen), and RNA was isolated according to the manufacturer's instructions. Cellular RNA was obtained by harvesting cells 48 h after transfection by scraping cells into 2 ml of Trizol reagent per 100-mm plate, and RNA was isolated according to the manufacturer's instructions.
RNase protection assays were performed and quantified by phosphorimager as previously described.89 Riboprobes templated by pEG676-1 (see above) were used in Figures 4 and 5 experiments. This riboprobe was designed to contain 14 bases unique to the helper as well as 268 shared bases of complementarity, to allow simultaneous detection of both vector and helper RNAs with a single probe. The experiment shown in Figure 4b included a second riboprobe templated by pBru7SL,88 which detects 7SL RNA. The signals in the cluster of bands detected with the 7SL probe were summed: the appearance of multiple bands is typical and results from allelic heterogenity in this multi-copy gene and some probe nicking.88 For each experiment, the amount of riboprobe loaded in the undigested probe lane was 1% of that used in the hybridization reaction for each sample lane. Protein was removed from the aqueous phase by ATrizol (confirmed by spectrophotometric analysis of the RNA).
RNA in vitro transcription
The PUC19 vectors containing different RNA clones were transformed into DH5α cells. Large scale of DNA template preparation was performed using QIAGEN Plasmid Mega Kit (Qiagen). In vitro transcription by T7 polymerase118 is performed in solution with a mixture of transcription buffer, MgCl2, NTPs, DNA template. The amount of MgCl2 and NTPs were optimized for each RNA sample in order to guarantee best RNA yield. The RNA was then purified using 6% denaturing PAGE gels and recovered via Elutrap Electroelution System (Whatman).
NC purification
The 55-residue NC protein (from HIV-1 strain NL4–3) was expressed from the bacteria vector pRD2, which was transformed into BL21 (DE3) pLysE. The protein was purified under non-denaturing conditions as described 119.
Dimerization assay
RNA was prepared in 10 mM Tris-HCl first, heat denatured for 3 minutes, and cooled on ice. 10× ITC salt was then added to RNA sample to reach a final salt concentration of 140 mM KCl, 10 mM NaCl and 1 mM MgCl2. After incubating at 37 °C for 1, 2, 4, 24 and 48 hours, the RNA samples were loaded to 2% agarose gel prestained with ethidium bromide, and run at 110 volts in 1× TB buffer (44.5 mM Tris-boric, pH 7.5) on ice to prevent thermal denaturation during electrophoresis.
Isothermal titration calorimetry
ITC experiments were carried out using a VP-Isothermal Titration Calorimeter MicorCalorimeter (VP-ITC) (MicroCal Corp. Northampton, MA; 1989). NC (180 µM) in ITC buffer (10 mM Tris-HCl, pH 7.0, 140 mM NaCl, 10 mM NaCl, 1 mM MgCl2, and 100 µM β-mercaptoethanol) was loaded into the injection syringe. The calorimetry cell was loaded with RNA (0.8 µM). The RNA sample (in 10 mM Tris-HCl) was heat denatured, then cooled on ice. 10× ITC salt was then added to reach a final salt concentration of 140 mM KCl, 10 mM NaCl and 1 mM MgCl2. The RNA sample was then incubated overnight at 37 °C before loading into the calorimetry cell. After thermal equilibration at 30 °C and the initial 60 seconds delay, 28 serial injections of NC protein (10 µl each) were made into calorimetry cell. The thermodynamic parameters of the NC-RNA interaction were determined by a nonlinear-squares fit of the data using two-sites binding mode.
Highlights.
A 159 nucleotide region of the HIV-1 5´-leader is identified that exhibits NMR spectral features and dimerization and NC binding properties similar to those of the intact leader.
The 159 nucleotide “core encapsidation signal” is required for efficient and selective packaging of heterologous RNAs into virus particles.
Packaging defects observed here and elsewhere upon deletion of the TAR hairpin are due to unexpected misfolding of the monomeric form of the 5´-leader.
Acknowledgments
This work was supported by a grant from the National Institute of General Medical Sciences (NIGMS; GM 42561). K.E. was supported by a grant from the NIGMS for enhancing minority access to research careers (MARC U*STAR 2T34 GM008663). We thank HHMI staff at UMBC for technical assistance.
Abbreviations used
- HIV-1
human immunodeficiency virus 1
- NC
nucleocapsid
- 5´-L
5´-leader
- ITC
isothermal titration calorimetry
- RPA
RNase protection assay
- RT
reverse transcriptase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hu W-S, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proceedings of the National Academy of Sciences U.S.A. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 3.Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc Natl Acad Sci U S A. 2000;97:14566–14571. doi: 10.1073/pnas.97.26.14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, Meese E, Wain-Hobson S, Meyerhans A. Recombination: Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 6.Quinones-Mateu ME, Arts EJ. Recombination in HIV-1: Update and Implications. AIDS Rev. 1999;1:89–100. [Google Scholar]
- 7.McCutchan FE. Global epidemiology of HIV. J Med Virol. 2006;78(Suppl 1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 8.Rein A. Retroviral RNA packaging: A review. Arch. Virology. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immun. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 10.Greatorex J, Lever A. Retroviral RNA dimer linkage. J. Gen. Virol. 1998;79:2877–2882. doi: 10.1099/0022-1317-79-12-2877. [DOI] [PubMed] [Google Scholar]
- 11.Paillart J-C, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 12.Jewell NA, Mansky LM. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 2000;81:1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 13.Paillart J-C, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nature Reviews Microbiology. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 14.Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes,no, probably? Retrovirology. 2004;1 doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greatorex J. The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology. 2004;1 doi: 10.1186/1742-4690-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Souza V, Summers MF. How retroviruses select their genomes. Nature Reviews Microbiology. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 17.Jouvenet N, Laine S, Pessel-Vivares L, Mougel M. Cell biology of retroviral RNA packaging. RNA Biology. 2011;1 doi: 10.4161/rna.8.4.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011 doi: 10.1016/j.jmb.2011.03.037. in press. [DOI] [PubMed] [Google Scholar]
- 19.Chin MPS, Rhodes TD, Chen J, Fu W, Hu W-S. Identification of a major restriction in HIV-1 intersubtype recombination. Proc. Natl. Acad. Sci. USA. 2005;102:9002–9007. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu W-S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhodes T, Wargo H, Hu W-S. High rates of human immunodeficiency virus type 1 recombination: Near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 2003;77:11193–11200. doi: 10.1128/JVI.77.20.11193-11200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onafuwa A, An W, Robson ND, Telesnitsky A. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 2003;77:4577–4587. doi: 10.1128/JVI.77.8.4577-4587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000627. e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bender W, Chien Y-H, Chattopadhyay S, Vogt PK, Gardner MB, Davidson N. High-molecular-weight RNAs of AKR, NZB and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J. Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kung H-J, Hu S, Bender W, Baily JM, Davidson N, Nicolson MO, McAllister RM. RD-114, baboon and wolly monkey viral RNAs compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 28.Maisel J, Bender W, Hu S, Duesberg PH, Davidson N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J. Virol. 1978;25:384–394. doi: 10.1128/jvi.25.1.384-394.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murti KG, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. Journal of Virology. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. Journal of Virology. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oritz-Conde BA, Hughes SH. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J. Virol. 1999;73:7165–7174. doi: 10.1128/jvi.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice WG, Schaeffer CA, Harten B, Villinger F, South TL, Summers MF, Henderson LE, Bess JW, Jr, Arthur LO, McDougal JS, Orloff SL, Mendeleyev J, Kun E. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 33.Rice WG, Turpin JA, Schaeffer CA, Graham L, Clanton D, Buckheit RW, Zaharevitz D, Summers MF, Wallqvist A, Covell DG. Evaluation of selected chemotypes in coupled cellular and molecular target-based screens identifies novel HIV-1 zinc finger inhibitors. J. Med. Chem. 1996;39:3606–3616. doi: 10.1021/jm960375o. [DOI] [PubMed] [Google Scholar]
- 34.Grigorov B, Bocquin A, Gabus C, Avilov S, Mely Y, Agopian A, Divita G, Gottikh M, Witvrouw M, Darlix JL. Identification of a methylated oligoribonucleotide as a potent inhibitor of HIV-1 reverse transcription complex. Nucleic Acids Res. 2011;39:5586–5596. doi: 10.1093/nar/gkr117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avilov SV, Boudier C, Gottikh M, Darlix JL, Mely Y. Characterization of the inhibition mechanism of the HIV-1 nucleocapsid protein chaperone activities by methylated oligoribonucleotides. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.05614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldschmidt V, Miller LM, Dde Rocquigny H, Darlix J-L, Mely Y. The nucleocapsid protein of HIV-1 as a promising therapeutic target for antiviral drugs. HIV Therapy. 2010;4:179–198. [Google Scholar]
- 37.De Guzman RN, Turner RB, Summers MF. Protein-RNA recognition. Biopolymers. 1999;48:181–195. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 38.Lever AM. HIV-1 RNA packaging. Advances in pharmacology. 2007;55:1–32. doi: 10.1016/S1054-3589(07)55001-5. [DOI] [PubMed] [Google Scholar]
- 39.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011;410:609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101 nucleotide core encapsidation signal of the Moloney Murine Leukemia Virus. J. Mol. Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 41.D'Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Bean RL, Vogt VM, Summers MF. Solution structure of the Rous sarcoma virus nucleocapsid protein:uY RNA packaging signal complex. J. Mol. Biol. 2006;365:453–467. doi: 10.1016/j.jmb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 44.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor K, Wu Z-R, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 45.Spriggs S, Garyu L, Connor R, Summers MF. Potential intra- and intermolecular interactions involving the Unique-5´ Region of the HIV-1 5´-UTR. Biochemistry. 2008;46:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helga-Maria C, Hammarskjold M-L, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type-1 genomic RNA. J. Virol. 1999;73:4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das AT, Klaver B, Berkhout B. The 5´ and 3´ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus Type 1 can be deleted when not required for Tat-mediated activation of transcription. J. Virol. 2007;81:7742–7748. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandt S, Blissenbach M, Grewe B, Konietzny R, Grunwald T, Uberla K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007;3:e54. doi: 10.1371/journal.ppat.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poon DTK, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type-1 nucleocapsid P7 protein involved in RNA pacakaging and infectivity. J. Virol. 1996;70:6607–6617. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. of Virology. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S-W, Aldovini A. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 2002;76:11853–11865. doi: 10.1128/JVI.76.23.11853-11865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muriaux D, Mirro J, Nagashima K, Harvin D, Rein A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J. Virol. 2002;76:11405–11413. doi: 10.1128/JVI.76.22.11405-11413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrison GP, Lever AML. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. Journal of Virology. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 58.Baudin F, Marquet R, Isel C, Darlix J-L, Ehresmann B, Ehresmann C. Functional sites in the 5´ region of human immunodeficiency virus type 1 RNA form defined structural domains. Journal of Molecular Biology. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi T, Ueno Y, Okamoto T. Elucidation of a conserved RNA stem-loop structure in the packaging signal of human immunodeficiency virus type 1. FEBS. 1993;327:213–218. doi: 10.1016/0014-5793(93)80172-q. [DOI] [PubMed] [Google Scholar]
- 60.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McBride MS, Panganiban AT. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5´ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Harrison GP, Miele G, Hunter E, Lever AML. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998;72:5886–5896. doi: 10.1128/jvi.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5´ packaging signal of Human Immunodeficiency Virus Type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clever JL, Parslow TG. Mutant Human Immunodeficiency Virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berkhout B, Klaver B, Das AT. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–281. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 68.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodefieiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu K, Heng X, Garyu L, Monti S, Garcia E, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Sachin Divakaruni S, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5´-leader RNA that regulate genome packaging. Science. 2011;344:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in Human Immunodeficiency Virus Type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 71.Berkhout B. Prog. Nucl. Acid Res. and Mol. Biol. Vol. 54. Academic Press, Inc.; 1996. Structure and function of the human immunodeficiency virus leader RNA; pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 72.Damgaard CK, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and HIV-2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paillart JC, Dettenhofer M, Yu X-F, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 74.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biology. 2008;6:883–899. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clever JL, Miranda JD, Parslow TG. RNA structure and packaging signals in the 5´ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woese CR, Winder S, Gutell RR. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc. Natl. Acad. Sci. USA. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. A Network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 79.Szewczak AA, Moore PB. The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 80.Szewczak AA, Moore PB, Chan YL, Wool IG. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orita M, Nishikawa F, Kohno T, Senda T, Mitsui Y, Endo Y, Taira K, Nishikawa S. Highresolution NMR study of GdAGA tetranucleotide loop that is an improved substrate for ricin, a cytotoxic plant protein. Nucleic Acids Res. 1996;24:611–618. doi: 10.1093/nar/24.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orita M, Nishikawa F, Shimayama T, Taira K, Endo Y, Nishikawa S. High-resolution NMR study of a synthetic oligonucleotide with a tetranucleotide GAGA loop that is a substrate for the cytotoxic protein, ricin. Nucleic Acids Res. 1993;21:5678–5678. doi: 10.1093/nar/21.24.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Correll CC, Munishkin A, Chan Y, Ren Z, Wool IG, Steitz TA. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RA, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1 Ψ-RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J. Mol. Biol. 2001;314:961–969. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- 85.Kanevsky I, Chaminade F, Ficheux D, Moumen A, Gorelick R, Negroni M, Darlix JL, Fosse P. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. Journal of molecular biology. 2005;348:1059–1077. doi: 10.1016/j.jmb.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 86.Rulli J, S J, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 88.Onafuwa-Nuga AA, Telesnitsky A, King SR. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA. 2006;12:542–546. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Onafuwa-Nuga AA, King SR, Telesnitsky A. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anderson ES, Contera SA, Knudsen B, Damgaard C, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 2004;279:22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 91.Song R, Kafaie J, Laughrea M. Role of the 5´ TAR stem-loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008;47:3283–3293. doi: 10.1021/bi7023173. [DOI] [PubMed] [Google Scholar]
- 92.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huthoff H, Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic acids research. 2001;29:2594–2600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007;371:1084–1096. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 95.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 96.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Clever JL, Eckstein DA, Parslow TG. Genetic dissociation of the encapsidation and reverse transcription functions in the 5´ R region of human immunodeficiency virus type 1. J. Virol. 1999;73:101–109. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Höglund S, Öhagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 99.Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the U5 RNA sequences. Virology. 2002;303:152–163. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 100.Das AT, Klaver B, Klasens BI, van Wamel JL, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Das AT, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Isel C, Lanchy JM, Le Grice SF, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–924. [PMC free article] [PubMed] [Google Scholar]
- 103.Huang Y, Khorchid A, Wang J, Parniak MA, Darlix JL, Wainberg MA, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160(gag-pol) and tRNA(Lys) incorporation into human immunodeficiency virus type 1. J. Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Z, Yu Q, Kang SM, Buescher J, Morrow CD. Preferential completion of human immunodeficiency virus type 1 proviruses initiated with tRNA3Lys rather than tRNA1,2Lys. J Virol. 1998;72:5464–5471. doi: 10.1128/jvi.72.7.5464-5471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beerens N, Berkhout B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goldschmidt V, Rigourd M, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J Biol Chem. 2002;277:43233–43242. doi: 10.1074/jbc.M205295200. [DOI] [PubMed] [Google Scholar]
- 107.Sakuragi J, Sakuragi S, Shioda T. Minimal region sufficient for genome dimerization in the human immunodeficiency virus type 1 virion and its potential roles in the early stages of viral replication. J. Virol. 2007;81:7985–7992. doi: 10.1128/JVI.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vicenzi E, Dimitrov DS, Engelman A, Migone TS, Purcell DF, Leonard J, Englund G, Martin MA. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J. Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sakuragi J-I, Ueda S, Iwamoto A, Shioda T. Possible role of dimerization in human immunodeficiency virus Type-1 genome RNA packaging. J. Virol. 2003;77:4060–4069. doi: 10.1128/JVI.77.7.4060-4069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5´-untranslated and matrix coding regions of the HIV-1 genomic RNA. J. Biol. Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 111.Berkowitz RD, Hammarskjold M-L, Helga-Maria C, Rekosh D, Goff SP. 5´ Regions of HIV-1 RNAs are not sufficient for encapsidation: Implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 112.Darlix J-L, Gabus C, Nugeyre M-T, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 113.Roldan A, Russell RS, Marchand B, Gotte M, Liang C, Wainberg MA. In vitro identification and characterization of an early complex linking HIV-1 genomic RNA recognition and Pr55Gag multimerization. J. Biol. Chem. 2004;279:39886–39894. doi: 10.1074/jbc.M405632200. [DOI] [PubMed] [Google Scholar]
- 114.Sakuragi J-I, Shioda T, Panganiban AT. Duplication of the primary encapsidation and dimer linkage region of Human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 2001;75:2557–2565. doi: 10.1128/JVI.75.6.2557-2565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sakuragi J-I, Iwamoto A, Shioda T. Dissociation of genome dimerization from packaging functions and virion maturation of Human Immunodeficiency Virus Type 1. J. Virol. 2002;76:959–967. doi: 10.1128/JVI.76.3.959-967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Manty A, Iyalla K, Loeliger K, D'Souza V, Khant H, Schmid MF, Garcia E, Telesnitsky A, Chiu W, Summers MF. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J. Mol. Biol. 2010;404:751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.An W, Telesnitsky A. Frequency of direct repeat deletion in a human immunodeficiency virus type 1 vector during reverse transcription in human cells. Virology. 2001;286:475–482. doi: 10.1006/viro.2001.1025. [DOI] [PubMed] [Google Scholar]
- 118.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Meth. Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 119.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 120.Nikolaitchik O, Rhodes TD, Ott D, Hu W-S. Effects of mutations in the Human Immunodeficiency Virus Type 1 gag gene on RNA packaging and recombination. J. Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]