Abstract
The hundreds of endemic species of cichlid fishes in the East African Great Lakes Tanganyika, Malawi, and Victoria are a prime model system in evolutionary biology. With five genomes currently being sequenced, eastern African cichlids also represent a forthcoming genomic model for evolutionary studies of genotype-to-phenotype processes in adaptive radiations. Here we report the functional annotation and comparative analyses of transcriptome data sets for two eastern African cichlid species, Astatotilapia burtoni and Ophthalmotilapia ventralis, representatives of the modern haplochromines and ectodines, respectively. Nearly 647,000 expressed sequence tags were assembled in more than 46,000 contigs for each species using the 454 sequencing technology, largely expanding the current sequence data set publicly available for these cichlids. Total predicted coverage of their proteome diversity is approximately 50% for both species. Comparative qualitative and quantitative analyses show very similar transcriptome data for the two species in terms of both functional annotation and relative abundance of gene ontology terms expressed. Average genetic distance between species is 1.75% when all transcript types are considered including nonannotated sequences, 1.33% for annotated sequences only including untranslated regions, and decreases to nearly half, 0.95%, for coding sequences only, suggesting a large contribution of noncoding regions to their genetic diversity. Comparative analyses across the two species, tilapia and the outgroup medaka based on an overlapping data set of 1,216 genes (∼526 kb) demonstrate cichlid-specific signature of disruptive selection and provide a set of candidate genes that are putatively under positive selection. Overall, these data sets offer the genetic platform for future comparative analyses in light of the upcoming genomes for this taxonomic group.
Keywords: Astatotilapia burtoni, Ophthalmotilapia ventralis, positive selection, EST, 454 sequencing, UTR
Introduction
Cichlid fishes from eastern African Great rift lakes and surrounding rivers represent a major model for rapid speciation in evolutionary biology (Kocher 2004; Seehausen 2006; Salzburger 2009). More than 1,500 endemic species have arisen in a few millions of years only, showing the most spectacular adaptive radiations known in vertebrates (Seehausen 2006). Explosive radiations in the cichlid species flocks of lakes Victoria, Malawi, and Tanganyika are mostly documented by paleo-geographical (i.e., the ages of the lakes) and molecular data. Lake Victoria, for example, is only between 200,000 and 500,000 years old and fell dry about 15,000 years ago (Johnson et al. 1996). Still, it harbors an endemic flock of several hundred species that are likely to have diversified in a maximum of about 100,000 years only (Verheyen et al. 2003). Accordingly, preliminary molecular data from partial genomes, nuclear and mitochondrial markers of East African cichlids have inferred a highly similar genetic background among species (Sturmbauer and Meyer 1993; Aparicio et al. 2002; Loh et al. 2008). This is in strong contrast with their tremendous diversity of morphotypes and ecological adaptations (Salzburger 2009) suggesting that, in cichlids, rapid phenotypic diversification is largely uncoupled from an equivalent molecular diversity in coding regions. Hence, cichlids represent an ideal system to dissect the genetic bases of several universal phenotypic traits (such as coloration, body morphology, color vision, etc.) and—more generally speaking—to explore the molecular evolutionary processes underlying diversification and ecological speciation.
An increasing number of studies in animals points to the diversity of transcriptomes and especially of the expression profiles (thus including regulation of gene expression) as the bridging link that translates highly similar genomes at protein-coding genes into the astonishing diversity of phenomes (i.e., set of phenotypes) (see, e.g., Cooper et al. 2003; Wray et al. 2003; Shapiro et al. 2004). In particular, regulatory changes involving a limited genetic diversity can affect the expression of alternatively spliced isoforms and may modulate timing, localization, and abundance of gene expression. These processes can be adaptive and, therefore, responsible for organismal diversification (reviewed by Fay and Wittkopp 2008).
To date, comparative transcriptome analyses of African cichlids have been limited in terms of species number and number of expressed sequence tags (ESTs) analyzed (Salzburger et al. 2008; Kobayashi et al. 2009; Lee et al. 2010). These studies, overall, revealed a high uniformity of the protein-coding sequences among closely related, yet phenotypically diverse species.
Here, we report more than a million new EST sequences, perform transcriptome analyses, and investigate the overall expression profiles of two African cichlid species, Astatotilapia burtoni (AB) and Ophthalmotilapia ventralis (OV). AB and OV are representatives of two main evolutionary cichlid lineages (tribes) from East Africa, the modern haplochromines and the more basal group of the ectodines, respectively (see, e.g., Salzburger et al. 2002, 2005). The two lineages are thought to have diverged several millions of years ago (Salzburger et al. 2005; Koblmuller et al. 2008). So far, comparative genetic studies between these two lineages were largely limited to a phylogenetic context (see, e.g., Salzburger et al. 2002, 2005; but see Salzburger et al. 2007), whereas genomic comparisons are lacking. The two species differ in body morphology, ecology, and behavior. AB is a mouth-brooding species found in rivers and estuaries around Lake Tanganyika and is characterized by the presence of “true” circular egg-spots on the anal fins of males. OV is also a mouth-brooding species endemic to lake Tanganyika but exhibits long ventral fins showing egg-dummies in form of yellow vessels at their tips. Functional egg-dummies are, hence, a feature that evolved several times during cichlid evolution in East Africa (Salzburger et al. 2007; Salzburger 2009).
For each of these two species, more than 647,000 ESTs were generated through 454 sequencing (Roche) and assembled in more than 46,000 contigs. These represent the first 454 data sets and the largest collection of EST available to date for African cichlids. This study also provides the first transcriptome data for a member of the ectodine lineage (OV). Functional annotation and comparative analyses were performed to explore major qualitative and quantitative differences of the two transcriptomes. Furthermore, comparative analyses were expanded to include additional species via the identification of more than a 1,200 orthologoues contigs across AB, OV, the Nile tilapia (Oreochromis niloticus) and medaka (Oryzia latipes) as outgroup. This allowed screening for differential substitution rates along lineages and for individual genes. Overall, our study provides an important molecular resource for comparative studies within cichlids and among fishes in general and will facilitate the assembling and annotation of the upcoming cichlid genomes (http://www.broadinstitute.org/models/tilapia).
Materials and Methods
Samples
Specimens from an inbreed laboratory strain of AB were kept at the University of Basel (Switzerland) under standard laboratory conditions. OV individuals were captured live in Mpulungu (Zambia), shipped to Basel, and kept at the same laboratory conditions for a week. For RNA isolation, individuals were euthanized with MS 222 using approved procedures (permit nr. 2317 issued by the cantonal veterinary office).
cDNA Library Construction and 454 Sequencing
From AB, we extracted total RNA from ten embryos, ten fish larvae, two juveniles, and two adults (one male and one female). From OV, we used four adults (three males and one female). For each species, specimens were pooled together, roughly chopped, and incubated for 2 h in 8 ml of trizol (Invitrogen). Samples were then ground to complete homogenization using a mortar and a pestel. RNA extraction was performed according to the manufacturer’s protocol. DNase treatment was carried out with the DNA-free Kit (Applied Biosystems). The quantity and quality of RNA were assessed by spectrophotometry and gel electrophoresis. One microgram of RNA of each sample was sent for commercial normalized library construction by Vertis Biotechnology AG (http://www.vertis-biotech.com/). From total RNA, first strand cDNA was synthetized using a reverse transcriptase, an N6 random primer and a small aliquot of an oligo(dT)-primer for enrichment of 3′ ends. 454 adapters A and B were ligated to the 5′ and 3′ ends of the cDNA. cDNAs were then amplified by polymerase chain reaction (PCR) (15 cycles) using a proofreading enzyme. Libraries were normalized by hydroxyl-apatite chromatography, and the single-stranded cDNA was amplified by PCR (nine cycles). cDNA was then selected with gel fractioning for fragments of sizes 500 to 700 bp.
Normalized cDNA libraries for the two species were sequenced with a Roche Genome Sequencer FLX system (Roche 454) in one Titanium FLX run (two lanes, one for each species) by Microsynth (http://www.microsynth.ch). Base calling was performed with Phred (Ewing et al. 1998). Reads were assembled with the GS De Novo Assembler version 2.0.0.22 using the default settings, a minimum overlap of 40 nucleotides and identity threshold of 90%.
ESTs Functional Annotation
Gene ontology (GO) annotation was conducted using Blast2GO version 2.4.4 (Conesa et al. 2005). Briefly, BlastX searches were performed against the nonredundant database (nr) using the QBlast for multiple queries, setting the e value to 1.0 × 10−6, the high scoring segment (HSP) length cut off greater than 33 and the number of hits to 5. GO annotation was done using the following settings: a pre-E-value-Hit-Filter of 1.0 × 10−6, a GO weight of 5, and the annotation cut off of 55. Contigs with no significant hits to the nr data set were BlastN searched against the nucleotide database (nt) for possible identification, setting the expected cut off value to 1.0 × 10−15.
Clustering of Orthologous Sequences
For the purpose of obtaining a data set suitable for comparative analyses, we generated three data sets, which included orthologous ESTs across AB and OV (data set #1), AB, OV, and O. niloticus (hereafter referred to as tilapia) (data set #2), and AB, OV, tilapia, and O. latipes (hereafter referred to as medaka) (data set #3). Data set #3 represented a subset of data set #2.
For the data set #1, identification of orthologous ESTs between the two species was performed using a bidirectional best hit (BBH) method (Overbeek et al. 1999). Reciprocal batch BlastN searches were carried out setting the expected value cut off to 1.0 × 10−50 to minimize significant matches to paralogous sequences. Outputs were analyzed using in-house R scripts. Hits with a bit score > 1,000 were retrieved for further analyses. Pairwise assemblies were performed using CodonCode Aligner version 3.7.1 (Codon Code Corporation) and aligned with MAFFT version 6.821b (Katoh et al. 2002) using a local pairwise method based on the Smith–Waterman algorithm.
For data set #2, a total of 117,222 tilapia ESTs were downloaded from GenBank in September 2010 (Lee et al. 2010). Among the total BBHs, we selected only annotated BBHs that had a length overlap > 400 bp and a bit score > 400. Contigs from both AB and OV belonging to this subset were batch BlastN searched against the tilapia data set, setting the expected value cut off value to 1.0 × 10−50. Corresponding best hits for the two species to the tilapia data set that had a length overlap > 150 bp were retrieved, assembled in CodonCode Aligner and aligned in MAFFT. Alignments were trimmed for full-length overlap.
Finally, for data set #3, all contigs belonging to the data set #2 (2,660) from AB were batch BlastX searched against complete protein data sets from Danio rerio and medaka (retrieved from the ENSEMBL database v59) using a cutoff of 1.0 × 10−50. Significant hits with concordant frames between D. rerio and medaka were chosen, and the corresponding cDNA sequences from medaka were retrieved from ENSEMBL. Clusters of orthologues cDNA sequences across medaka and the three cichlid species were generated and aligned using MAFFT. Danio rerio sequences were not included due to the high nucleotide divergence of this species with respect to the other species (Steinke et al. 2006). To obtain only open reading frames (ORFs), untranslated regions (UTRs) were trimmed from the alignments according to the corresponding medaka proteins. All frame-shifting indels introduced in Medaka sequences during the aligning process were trimmed to preserve medaka-reading frames. Alignments below 150 bp in length were discarded. Finally, all alignments were eye checked and refined manually.
The final data set #3 comprised 1,216 alignments of fully overlapping sequences starting with the correct reading frames. The pipeline was performed with in-house R and perl scripts.
Phylogeny, Genetic Distances, and Rates of Evolution
Maximum likelihood (ML) heuristic searches were performed on the concatenated alignment of 1,216 four-species clusters (526,113 bp) from data set #3 using RaxML version 7.0.4 (Stamatakis et al. 2005). We performed a rapid bootstrap analysis and search for the best ML tree employing the GTRGAMMA model. Indels were identified using the program SeqState (Muller 2005). All single and double indels present in cichlid sequences in the final alignments (36 and 5, respectively) were considered as sequencing errors and replaced with Ns. Two deletions longer than 100 bp identified in OV were attributed to a putative exon skipping (alternative spliced variants) and not to a genomic deletion and also replaced with Ns. Indels were then coded using the simple indel coding strategy (Simmons and Ochoterena 2000), implemented in SeqState, and mapped on the ML tree performing a maximum parsimony analysis in PAUP* v. 4.0b10 (Swofford 2000).
Uncorrected distance matrices were estimated for individual alignments using PAUP*. Pairwise synonymous and nonsynonymous substitution rates per site (Ks and Ka, dS and dN) were estimated under two methods; the Nei and Gojobori method (Nei and Gojobori 1986) implemented in the DNAStatistics package of Bioperl (http://www.bioperl.org/wiki/Main_Page) (Ks and Ka) and the Goldman and Yang method (Goldman and Yang 1994) using the program Codeml implemented in PAML version 4.4b (Yang 2007) (dS and dN).
Different rates of dN/dS for branches in the phylogenetic tree were investigated using the branch models from Codeml. dN/dS values were averaged across sites (NSsites = 0). Three models of molecular evolution were compared: 1) the one-ratio model (model = 0), allowing the same dN/dS value for all branches; 2) the two-ratio model, constraining the branches within the cichlid clade to one dN/dS ratio that was different from all the others (model = 2); and 3) the free-ratio model (model = 1), allowing one dN/dS ratio per each branch. Sites with ambiguous data were removed (cleandata = 1). The three models were compared (0 vs. 2 and 2 vs. 1) using a likelihood ratio test (LRT) with two and four degrees of freedom, respectively.
Positive selection acting on genes that showed average Ka/Ks values higher than one between species was further tested by estimating dN/dS for branches in individual gene phylogeny under the free-ratio model in Codeml.
Results
ESTs Sequence Annotation and Comparative Transcriptomics of AB and OV
The two EST libraries constructed for AB and OV yielded an equal number of reads (∼647,000), which were assembled in a similar number of contigs (>46,000, see table 1). The mean contig size was 585 bp for AB and 566 bp for OV, with 39% of the contigs having at least 500 bp.
Table 1.
Summary of the ESTs Generated by 454 Sequencing in This Study
| AB | OV | |
| Summary run | ||
| Total number of reads | 647,219 | 647,816 |
| Average read length | 349.27 | 344.36 |
| Total number of bases | 226,048,424 | 223,072,738 |
| Summary assembly | ||
| Total number of contigs | 49,311 | 46,298 |
| Total number of large contigs (≥500 bases) | 19,408 | 17,207 |
| Average contig size | 585.84 | 566.33 |
| N50 contig sizea | 1,016 | 1,003 |
| Largest contig size | 8,335 | 7,430 |
Half of all bases reside in contigs of this size or longer.
Based on BlastX searches against the nr database, 19,121 AB (38.8% of the total) and 16,585 OV (35.8%) contigs had a significant hit above the cut off e value of 10−6 (table 2). These contigs corresponded to a total of 12,491 distinct accession numbers (AccNos) for AB and 11,269 AccNos for OV. Because the contigs are usually much shorter than the corresponding cDNA sequences, it is common that several contigs matched to the same gene, in spite of lacking adequate overlap to be assembled. For both species, the top-hit species for orthologue match was Tetraodon nigroviridis (approximately 35% of the contigs), followed by D. rerio (approximately 25%).
Table 2.
Summary of the ESTs Annotation Using Blast2GO
| AB | OV | |
| Number of ESTs returning BlastX hits | 19,121 (12,491 AccNos) | 16,582 (11,269 AccNos) |
| Number of ESTs with GO annotation | 11,956 (5,152 terms) | 10,250 (4,852 terms) |
| Biological process | 8,438 (2,974 terms) | 7,293 (2,732 terms) |
| Cellular component | 7,330 (616 terms) | 6,307 (623 terms) |
| Molecular function | 10,110 (1,562 terms) | 8,683 (1,497 terms) |
| Annotated protein-coding genes | 8,684 | 7,671 |
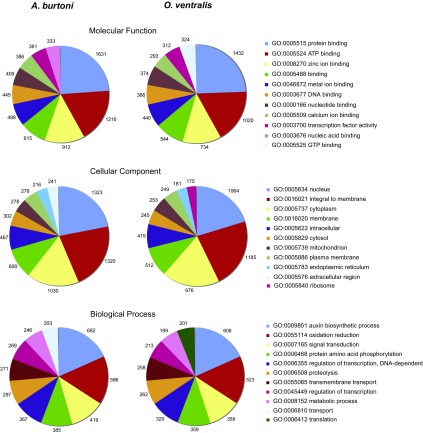
Of the contigs with significant BlastX hits, a total of 11,956 for AB and 10,250 for OV were annotated in 4,852 GO terms (24% of the total contigs) and 5,152 GO terms (22%), respectively. The GO terms were assigned to three biological categories that were equally represented in the two species (table 2). Relative and absolute abundance of the most represented GO terms per biological category were also comparable between AB and OV (fig. 1). The two species shared nine of ten terms in all three categories. The most represented terms for the molecular function category were associated to protein and nucleotide binding and transcription factor activity, whereas the predominant terms for the biological process category were involved in common enzymatic processes such as “auxin biosynthetic process,” “oxidation reduction,” and “signal transduction”. Finally, overrepresented GO terms for the cellular component category were mainly localized in the nucleus and membrane.
FIG. 1.—
Ten most represented GO terms per biological category and absolute number of ESTs assigned to each term. Overall representation of GO terms is nearly equal between AB and OV.
A large part of the contigs had no significant hit to the nr data set (above 60% for both species). These contigs were BlastN searched against the nt database for further identification. Only 9% of these contigs for both species (2,863 and 2,620 contigs for AB and OV, respectively) returned a significant hit to the nt database (1 × 10−15), with 609 unique AccNos shared between the two species (see supplementary table S1, Supplementary Material online). Of these AccNos, several (up to 100) mapped to noncoding regions, such as microsatellite sequences, pseudogenes, and transposons. We also retrieved genes predicted to play an important role in cichlid evolution, such as Bmp4, c-ski, pax genes, prolactin, Sox transcription factors, the vitellogenin receptor, among others. In terms of frequency of contigs per single hit, half of the total number of contigs mapped to the same two classes of genes in both species and with similar relative proportions (table 3): immune genes (MHC class, KLR, natural killer-like receptors), and patterning genes (Hox and ParaHox genes). This suggests that both a relatively high expression of these genes in the two species, as well as poor amino acid conservation outside the cichlid lineage that could explain why these contigs did not return any BlastX hit against the nr database. To some extent, this outcome might also be biased by the overrepresentation of these loci in GenBank.
Table 3.
Most Common Hits in the nt Database (cut off e value 1 × 10−15) for Contigs That Had No Hits in the nr Database
| Number of Contigs |
||||
| Hit Description | Species | AccNo | AB | OV |
| MHC class IA antigen UBA1, UBA2, UAA1 genes, UAA3 and UAA2 pseudogenes, UAA4, UAA5, and UAA6 pseudogene fragments | Oreochromis niloticus | AB270897.1 | 260 | 226 |
| Platelet-derived growth factor receptor beta b (pdgfrbb) and colony-stimulating factor 1 receptor b (csf1rb) genes | Astatotilapia burtoni | DQ386647.1 | 181 | 153 |
| Hoxba gene cluster | A. burtoni | EF594310.1 | 149 | 136 |
| KLR1 gene; KLR2 pseudogene, KLR3 and KLR4 genes; KLR5 gene, KLR6 and KLR7 pseudogenes | O. niloticus | AY495714.1 | 115 | 115 |
| Hoxdb gene cluster | A. burtoni | EF594316.1 | 84 | 59 |
| Platelet-derived growth factor receptor beta a (pdgfrba) and colony-stimulating factor 1 receptor a (csf1ra) genes | A. burtoni | DQ386648.1 | 60 | 43 |
| Gsh2 (gsh2), Pdgfra (pdgfra), and Kita (kita) genesKdrb (kdrb) gene; and Clock (clock) gene | A. burtoni | EF526075.2 | 57 | 64 |
| Hoxbb gene cluster | A. burtoni | EF594314.1 | 56 | 74 |
| Hoxab gene cluster, complete sequence | A. burtoni | EF594311.1 | 55 | 52 |
| KLR8 pseudogene; KLR9 gene, C-type lectin (CLECT2)-like protein pseudogene, and C-type lectin (CLECT2)-like protein gene; KLR10 pseudogene; C-type lectin natural killer cell receptor-like protein gene; and transposon TX1-like ORF2 pseudogene | O. niloticus | AY495715.1 | 45 | 47 |
| Hoxda gene cluster | A. burtoni | EF594315.1 | 31 | 32 |
| Hoxca gene cluster | A. burtoni | EF594312.1 | 22 | 30 |
| Hoxaa gene cluster | A. burtoni | EF594313.1 | 20 | 13 |
| Total number of contigs | 1,135 | 1,044 | ||
Comparative Transcriptomics within Cichlids
Using the BBH method, we identified 20,828 contigs that had best reciprocal hits between AB and OV. Of these, a total of 4,516 contigs that had a BlastN score bit ≥ 1,000 were selected to explore sequence diversity between the species (data set #1). These clusters of putatively orthologous sequences comprised a representation of all transcript types, such as annotated and nonannotated sequences, as well as coding and noncoding regions (including UTRs). The average alignment length was 1,463 bp with a mean pairwise nucleotide distance, excluding indels, of 0.0175 ± 0.0101, and a median of 0.0158 (table 4).
Table 4.
Average Pairwise Genetic Distance (Pi, Uncorrected) with Standard Deviation and Median Values Estimated from 4,516 BBHs between AB and OV (Data set #1) and from 2,660 Three-Species Alignments (AB, OV, and Tilapia; Data set #2)
| Pi | Median | Mean Length (Range), bp | ||
| Data set #1a | ||||
| AB | OV | 0.0175 ± 0.0101 | 0.0158 | 1,463 (516–6,837) |
| Data set #2 | ||||
| AB | OV | 0.0138 ± 0.0096 | 0.0117 | 541 (150–2,588) |
| Tilapia | AB | 0.0302 ± 0.0203 | 0.0261 | |
| Tilapia | OV | 0.0314 ± 0.0212 | 0.0268 |
Data set #1 includes both annotated and nonannotated ESTs, whereas data set #2 includes only annotated ESTs with UTRs.
Considering only annotated sequences, we generated 2,660 clusters of orthologous contigs among AB, OV and tilapia (data set #2) that could reliably be aligned. Average pairwise genetic distance was virtually the same between tilapia and both AB and OV (∼0.030) and more than twice as large as between OV and AB (0.0138) (table 4). Genetic distance between AB and OV was higher than the one calculated in the previous data set, likely because this second data set only included annotated sequences, thus excluding all novel, less conserved, and untranslated mRNA sequences (but yet including UTR regions).
We finally generated a third data set (#3) including orthologous sequences across the three cichlid species and the outgroup medaka. UTRs were trimmed using medaka proteins as reference. We obtained 1,409 clusters of fully overlapping orthologous sequences across AB, OV, tilapia, and medaka that included only ORFs. Inspection of the alignments revealed 191 clusters in which premature stop codons were present in one or more cichlid species but not in medaka. These stop codons could represent sequencing errors or real substitutions resulting in pseudogenization or truncation of proteins (with potential novel functions). At this stage, we could not tease apart the three scenarios and we therefore decided to exclude these clusters from the data set. The final data set #3 comprised 1,216 four-species alignments of ORFs, with a total length of 526,113 bp. Average length for individual alignments was 433 bp, varying between 153 and 741 bp. We used this data set for phylogenetic reconstructions and to investigate genetic diversity and levels of selection for each species pairwise comparison and along phylogenetic lineages.
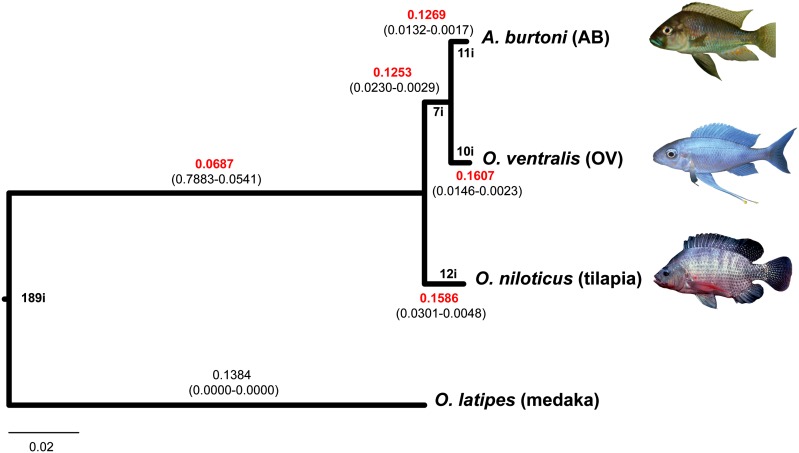
The ML phylogeny based on the concatenated data set is shown in figure 2. The tree is in accordance with previously reported phylogenetic relationships among the four species (Salzburger et al. 2005; Steinke et al. 2006): AB and OV grouped together and formed a well-supported monophyletic group with tilapia (bootstrap values = 100 for both nodes). The three cichlids showed similar genetic distance from the outgroup medaka.
FIG. 2.—
ML phylogeny based on four-species concatenated alignment of 1,216 genes (526,113 bp). The tree is rooted using medaka as outgroup. All nodes had a 100 bootstrap value support. For each branch, individual dN and dS values (in brackets, respectively) and the corresponding dN/dS ratios (in red) were calculated under the free-ratio model (codeml). Indel events per branch (specified by number followed by “i”) were mapped by maximum parsimony.
In accordance with the phylogenetic reconstruction, the shortest absolute genetic distance was found between AB and OV (0.0095), followed by tilapia versus these two species (0.0222 and 0.0230), with the longest distance occurring between medaka and the remaining three species (0.1605 and 0.1609) (table 5). Within cichlids, contribution of indels to the genetic diversity was low, with a total of 268 indel sites detected out of 524,047 nucleotides. These corresponded to a total of 38 distinct indel events equally distributed along the three cichlid lineages (AB, OV, and tilapia) (mapped in fig. 2). Six deletion events (3- to 6-bp deletions) were specific of the AB/OV clade and occurred in the following genes: the “low choriolytic enzyme precursor,” involved in the breakdown of the egg envelope, the “src kinase-associated phosphoprotein 2,” involved in the src signaling pathway, the “deoxyribonuclease tatdn3,” the “v-type atpase b subunit,” the “dna topoisomerase 2-beta,” and the “probable rna-binding protein eif1ad.” Further investigations are needed to clarify whether these amino acid deletions confer important biological changes to these proteins and are therefore involved in some cichlid-specific traits.
Table 5.
Average Pairwise Genetic Distances (Pi, Uncorrected), Rates of Synonymous and Nonsynonymous Substitutions Per Site and Relative Ratio Estimated for Both Individual and Concatenated 1,216 Four-Species Alignments (526,113 bp, Data set #3)
| Individual Alignments |
Concatenated Alignments |
||||||||||
|
Nei and Gojobori
(1986) |
Nei and Gojobori
(1986) |
Goldman and Yang
(1994) |
|||||||||
| Pi | Ks | Ka | Ka/Ks | Ks | Ka | Ka/Ks | dS | dN | dN/dS | ||
| AB | OV | 0.0095 ± 0.0072 | 0.0289 ± 0.0001 | 0.0048 ± 4.7 × 10−06 | 0.1856 ± 0.2688 | 0.0288 | 0.0057 | 0.1979 | 0.0288 | 0.0039 | 0.1358 |
| Tilapia | AB | 0.0222 ± 0.0207 | 0.0732 ± 0.0006 | 0.0096 ± 1 × 10−05 | 0.1753 ± 0.2124 | 0.0685 | 0.0103 | 0.1504 | 0.0686 | 0.0091 | 0.1323 |
| Tilapia | OV | 0.0230 ± 0.0210 | 0.0746 ± 0.0005 | 0.0102 ± 0.0000 | 0.1827 ± 0.2349 | 0.0699 | 0.0117 | 0.1674 | 0.0700 | 0.0097 | 0.1387 |
| Medaka | Tilapia | 0.1609 ± 0.0496 | 0.8657 ± 0.0197 | 0.065 ± 0.0002 | 0.0810 ± 0.0977 | 0.8128 | 0.0672 | 0.0827 | 0.8160 | 0.0607 | 0.0744 |
| Medaka | AB | 0.1605 ± 0.0497 | 0.8695 ± 0.0125 | 0.0644 ± 0.0002 | 0.0806 ± 0.1171 | 0.8167 | 0.0665 | 0.0814 | 0.8201 | 0.06 | 0.0731 |
| Meakda | OV | 0.1605 ± 0.0497 | 0.8681 ± 0.016 | 0.0647 ± 0.0002 | 0.0810 ± 0.1062 | 0.8143 | 0.0676 | 0.0830 | 0.8182 | 0.0603 | 0.0737 |
UTRs Contribution to Cichlid Genetic Divergence
To check for the specific contribution of UTRs to the genetic diversity between cichlid species, pairwise genetic distances were calculated on the same gene data set as data set #3 (thus excluding nonannotated sequences) before and after trimming for fully coding sequences (table 6). The average length of the 1,216 alignments among the three cichlid species including partial or full UTRs was 536 bp, ranging between 156 and 1,746 bp, for a total of 652,849 bp. Inclusion of UTRs was responsible for a total increase of approximately 0.002 of the genetic divergence compared with the data set including ORFs only (table 6). This corresponds to a relative increase of about 17%, 12.6%, and 8.7% in the genetic divergence between AB and OV and tilapia versus AB and OV, respectively. For the same gene data set, we also retrieved full-length contigs from AB and OV in order to extend our analysis of UTRs to longer sequences (thus excluding tilapia which reduced the length overlap across AB and OV in the previous data set). Average length of the 1,216 AB-OV pairwise alignments was 925 bp, nearly double the length for the two species considering ORFs only, and ranged between 402 and 3,669 bp, for a total of 1,122,962 bp. Average pairwise divergence between the two species was 0.0133, corresponding to an increase of 40% of their genetic divergence with respect to alignments including ORFs only (table 6). This last value is likely an underestimate of UTR contribution to the genetic divergence between AB and OV; indeed, in some cases, these longer alignments also include additional coding regions that were trimmed in data set#3 because they did not fully overlap with tilapia sequences (which are, on average, shorter than our AB and OV contigs).
Table 6.
Average Pairwise Genetic Distances (Pi, Uncorrected) Estimated for 1,216 Individual Four-Species Alignments (Gene Data set #3) before and after Trimming UTRs
| Pi |
||||
| ORFs only | ORFs + UTRsa | ORFs + UTRsb | ||
| AB | OV | 0.0095 ± 0.0072 | 0.0112 ± 0.0077 | 0.0133 ± 0.0080 |
| Tilapia | AB | 0.0222 ± 0.0207 | 0.0250 ± 0.0171 | na |
| Tilapia | OV | 0.0230 ± 0.0210 | 0.0250 ± 0.0171 | na |
Total length: 652,849 bp.
Total length: 1,122,962 bp.
Rates of Evolution and Signature of Disruptive Selection in the Cichlid Lineage
We used ORFs from data set#3 to estimate rates of evolution within cichlids (table 5). Based on the mean pairwise estimates on single alignments, the smallest average Ks value was found for the AB/OV comparison (0.0289), followed by similarly low values between tilapia and both OV and AB (0.0732 and 0.0746) and between medaka and all other species comparisons (0.8657–0.8695). The average Ks values calculated from the concatenated alignment were comparable (table 5).
Within cichlids, the average pairwise Ka/Ks ratios across the three species were also similar (0.175–0.186) but at least two times higher than for all pairwise comparisons between medaka and the three cichlids (0.081) (Whitney–Mann test, P < 0.001), suggesting disruptive selection in the cichlid lineage. Estimates of Ka/Ks based on average individual and concatenated alignments using the Nei and Gojobori method were similar and comparable to the estimates obtained using the more sophisticated model of substitutions from Goldman and Yang (1994) implemented in Codeml (Yang 2007).
We further tested the hypothesis for differential selective forces among lineages by comparing several branch models implemented in Codeml (PAML). Among the models tested, the free-ratio model, allowing one dN/dS for each branch, was significantly better than both the one-ratio model, which assigned the same dN/dS value to all branches, and the two-ratio model, which assigned to the medaka lineage a dN/dS value that differed from all other branches (LRT, P < 0.001 in both comparisons). According to the free-ratio model, the branches within the cichlid clade evolved with at least as twice as large dN/dS (0.1269–0.1607) compared with the branch at the base of the clade (0.0687) (fig. 2). The branch leading to medaka also showed a dN/dS value similar to that of cichlids; however, individual values of dN and dS were extremely low (<10−4), impeding a reliable estimate of the ratio.
Positively Selected Genes
We screened all individual 1,216 alignments for pairwise Ka/Ks values higher than one and obtained a set of 33 genes that are putatively under positive selection in at least one pairwise comparison (table 7). Individual inspection of these gene alignments ruled out possible misalignments or chimeric structures. All 33 genes showed Ka/Ks > 1 exclusively within cichlids comparisons: 14 genes between AB and OV, 13 between tilapia and either AB or OV, five in two pairwises and one gene for all three-cichlids pairwise comparisons. No genes showed values of Ka/Ks > 1 between medaka and any of the three cichlid species. This is compatible with the lower dN/dS value assigned to the branch leading to the cichlid clade (reported above).
Table 7.
Genes Under Putative Positive Selection Based on Pairwise Ka/Ks Values > 1
| Pairwise | Gene | Length, bp | Pi | Ks | Ka | Ka/Ks |
| Single | ||||||
| AB versus OV | Aquaporin fa-chip | 396 | 0.0202 | 0.0103 | 0.0273 | 2.650 |
| Succinate dehydrogenase | 450 | 0.0178 | 0.0085 | 0.0214 | 2.518 | |
| 20-beta-hydroxysteroid dehydrogenase | 501 | 0.0140 | 0.0084 | 0.0159 | 1.893 | |
| 26s proteasome nonatpase regulatory subunit 9 | 636 | 0.0173 | 0.0140 | 0.0227 | 1.621 | |
| Muscle-type creatine kinase ckm1 | 438 | 0.0092 | 0.0098 | 0.0151 | 1.541 | |
| Darmin protein | 363 | 0.0083 | 0.0061 | 0.0090 | 1.475 | |
| Serine hydrolase-like protein | 489 | 0.0226 | 0.0180 | 0.0247 | 1.372 | |
| Tetratricopeptide repeat protein 35 | 600 | 0.0034 | 0.0078 | 0.0107 | 1.372 | |
| Transmembrane protein 16f | 357 | 0.0114 | 0.0120 | 0.0148 | 1.233 | |
| Dead (asp-glu-ala-asp) box polypeptide 56 | 537 | 0.0075 | 0.0080 | 0.0098 | 1.225 | |
| Novel protein (zgc:100919) | 384 | 0.0131 | 0.0116 | 0.0139 | 1.198 | |
| loc733309 protein | 363 | 0.0138 | 0.0128 | 0.0142 | 1.109 | |
| Alpha-sialyltransferase st3gal v | 345 | 0.0116 | 0.0111 | 0.0119 | 1.072 | |
| Trypsinogen 2 | 540 | 0.0315 | 0.0311 | 0.0325 | 1.045 | |
| Tilapia versus OV | Beta-galactoside-binding lectin | 378 | 0.0212 | 0.0119 | 0.0243 | 2.042 |
| Decaprenyl-diphosphate synthase subunit 2 | 348 | 0.0201 | 0.0120 | 0.0231 | 1.925 | |
| Elastase 2-like protein | 540 | 0.0225 | 0.0152 | 0.0253 | 1.664 | |
| cdc42-interacting protein 4 homolog | 306 | 0.0132 | 0.0157 | 0.0167 | 1.064 | |
| Cytochrome c oxidase subunit 4 isoform mitochondrial precursor | 516 | 0.0177 | 0.0178 | 0.0182 | 1.022 | |
| Regulator of g-protein signaling 18 | 417 | 0.0240 | 0.0218 | 0.0283 | 1.298 | |
| Serum paraoxonase arylesterase 2 | 435 | 0.0300 | 0.0305 | 0.0336 | 1.102 | |
| hbaa_serqu ame: full = hemoglobin subunit alpha-a ame: full = hemoglobin alpha-a chain ame: full = alpha-a-globin | 426 | 0.0423 | 0.0403 | 0.0445 | 1.104 | |
| Suppression of tumorigenicity 14 (colon epithin) | 477 | 0.0359 | 0.0266 | 0.0400 | 1.504 | |
| Tilapia versus AB | Signal sequence alpha | 528 | 0.0076 | 0.0078 | 0.0126 | 1.615 |
| Nadh dehydrogenase 1 alpha subcomplex subunit mitochondrial precursor | 330 | 0.0182 | 0.0135 | 0.0199 | 1.474 | |
| mgc85594 protein | 402 | 0.0150 | 0.0123 | 0.0159 | 1.293 | |
| ca++ cardiac fast twitch 1 like | 447 | 0.0201 | 0.0180 | 0.0212 | 1.178 | |
| Two | ||||||
| Tilapia versus OV | Annexin a4 | 534 | 0.0356 | 0.0361 | 0.0366 | 1.014 |
| Tilapia versus AB | 0.0300 | 0.0279 | 0.0314 | 1.125 | ||
| Tilapia versus OV | Lipid phosphate phosphohydrolase 2 | 258 | 0.0233 | 0.0149 | 0.0268 | 1.799 |
| Tilapia versus AB | 0.0233 | 0.0149 | 0.0268 | 1.799 | ||
| AB versus OV | 39s ribosomal protein mitochondrial precursor | 318 | 0.0126 | 0.0138 | 0.0165 | 1.196 |
| Tilapia versus AB | 0.0189 | 0.0138 | 0.0207 | 1.500 | ||
| AB versus OV | Ubiquinol-cytochrome c rieske iron-sulfur polypeptide 1 | 441 | 0.0136 | 0.0096 | 0.0150 | 1.563 |
| Tilapia versus OV | 0.0159 | 0.0096 | 0.0181 | 1.885 | ||
| AB versus OV | Epithelial cadherin precursor | 651 | 0.0691 | 0.0671 | 0.0742 | 1.106 |
| Tilapia versus OV | 0.0799 | 0.0807 | 0.0857 | 1.062 | ||
| Three | ||||||
| AB versus OV | Cell cycle control protein 50a | 372 | 0.0162 | 0.0109 | 0.0218 | 2.000 |
| Tilapia versus OV | 0.0431 | 0.0218 | 0.0558 | 2.560 | ||
| Tilapia versus AB | 0.0457 | 0.0439 | 0.0483 | 1.100 |
Note.—Of the 33 genes, 27 were found with Ka/Ks > 1 only in single cichlid pairwises, five in two pairwises, and one in all three pairwise comparisons.
To further confirm these findings, all 33 individual genes were tested for positive selection in the framework of a phylogeny using the branch free-ratio model in Codeml. dN/dS was larger than one in one or more lineages in all the 33 genes, supporting the above results.
Discussion
Coverage and Functional Annotation of the Two Transcriptomes
Our transcriptome-wide study provides the first high-throughput 454 sequencing data available for eastern African cichlids and the largest current EST data set for cichlids. With nearly 647,000 reads assembled in more than 46,000 contigs, this data set offers the very first extensive genetic resource for a member of the Ectodini tribe, O. ventralis (OV), for which current molecular data were limited to few mitochondrial and nuclear genes only (see, e.g., Clabaut et al. 2005; Salzburger et al. 2007; Koblmuller et al. 2008). It also largely integrates current EST data available for A. burtoni (AB) (Salzburger et al. 2008). Comparative analysis of the new EST data set generated for this species (49,311 contigs) with the one already available in GenBank (10,312 contigs) via BlastN searches (1 × 10−50) indicates an overlap of 6,935 contigs between data sets. More than 70% of the hits showed a sequence identity between 98% and 100%, confirming the quality of our EST sequences and providing a further coverage for a subset of them. Overall, combining the two data sets, the ESTs generated in this study contributed to more than 70% of unique new sequences, greatly enlarging the current coverage of the transcriptome for AB.
Based on comparison of the number of proteins predicted for closely related fishes with those identified in our two EST libraries, the transcriptomes generated for both AB and OV cover at least half of their total proteomes. Specifically, the number of protein-coding genes ranges from a minimum of 18,523 in the highly compact genome of Takifugu (Aparicio et al. 2002) to up to 24,147 in D. rerio (http://www.sanger.ac.uk/Projects/D_rerio/). Taking these two values as a reference range for the expected number of protein-coding genes, ESTs from AB cover between 52% and 67% of the total protein-coding genes diversity (with 8,684 predicted proteins, see table 2), whereas ESTs from OV cover between 47% and 61% (7,671 proteins). It is, however, important to consider that ESTs represent, in most cases, partial transcripts, with a typical 3′-UTR bias introduced during the sequencing process, and thus, the actual coverage obtained for a full proteome (total length of the cDNA sequences transcribed) of both species is likely lower.
Comparative Transcriptomics between AB and OV
Comparative analyses of the functional annotation of more than 10,000 EST contigs for both AB and OV showed highly similar transcriptomes between the two species, in terms of both types and relative frequencies of GO categories expressed. The ten most represented GO terms per category were typically the same for both species, with very similar relative and absolute frequencies (fig. 1). An analogous comparative transcriptome analysis was recently performed for two closely related Central America cichlids (Elmer et al. 2010) and also showed a comparable functional annotation of their transcriptomes, with similar coverage of expressed GO categories (both as types and frequencies) between species. These categories are however differently represented compared with our data set, suggesting quite divergent transcriptome features between Central American and eastern African cichlids, although further analyses are needed to explore these differences.
A large portion of the transcriptomes of AB and OV (64 and 75% of the contigs, respectively) could not be annotated or had no BlastX matches to the protein nr database, suggesting that these sequences might represent novel proteins, unique to cichlids, fast evolving genes or UTRs. Recent studies in humans indicate that large parts of transcriptomes are indeed noncoding, although this remains unclear in fishes (Cheng et al. 2005). Further identification of these contigs via BlastN searches in the nt database provided a significant match only for 9% of these contigs in both species, suggesting that the large majority of these sequences (either translated or untranslated) might indeed be cichlid-specific, as result, for instance, of accelerated sequence evolution. Among those contigs that returned a significant match in the nt database, roughly half of them matched to 13 unique hits (i.e., AccNos), represented solely by two gene categories, immune and patterning genes, both in AB and OV. The two species also paired in terms of relative frequencies of these most represented contigs, indicating similar high expression levels of these transcripts in AB and OV. Among other hits that were less represented in terms of number of contig per hit, several matched to genes that are known to play a crucial role in rapid species evolution, such as bmP4, pax6, and color genes. Overall, this suggests that genes implied in key features of (cichlid) species, such as body morphology, coloration, development, and immunity represent a variable portion of the cichlid transcriptome (i.e., genes under accelerated evolution) with respect to other species, as predicted based on their function in processes typically under strong natural selection. Nevertheless, these findings should be taken with caution as we cannot exclude a bias in the type of sequences available in the nt database for closely related species to AB and OV, which would also bias the results of the BlastN searches.
Genetic Diversity between AB and OV
The two new transcriptomes presented here show up to 0.0175 uncorrected genetic divergence based on >4,000 pairwise alignments of putatively orthologues ESTs identified through a best reciprocal hit approach. It should be noted that all the alignments included both annotated and nonannotated sequences. When only annotated sequences are considered (using data set #2), the genetic diversity drops to 0.0138 between OV and AB. Furthermore, when only ORFs are considered (data set #3), the genetic diversity drops to nearly half (0.0095), suggesting that noncoding regions and nonannotated coding genes, such as putative novel or fast evolving genes, contributed to at least half of the total transcriptome divergence. In particular, UTR regions appear to carry a great proportion of variable sites between the two species. Comparative analysis of the same gene data set before and after trimming UTRs indicates a 40% increase of genetic divergence between AB and OV when UTRs are included. Similarly, an increase in genetic divergence, although smaller (likely due to shorter sequences), is seen when UTRs are included in pariwise comparisons between tilapia and both AB and OV.
It has been proposed that large part of the phenotypic variation found among closely related species is associated to changes at the regulatory regions affecting the expression profiles (e.g., cis-regulatory elements; Fay and Wittkopp 2008). In cichlids, this scenario is mainly supported by the indirect finding of very limited or no genetic diversity at the protein-coding regions among phenotypically diverse species (see Kobayashi et al. [2009] for lake Victoria species and Elmer et al. [2010] for Central American cichlids). Direct evidence of adaptive variation at noncoding regions comes from recent data showing that cichlid 3′-UTRs contain target sites for fast evolving microRNA. These sites present elevated SNP densities in response to the rapid diversification of these miRNA, clearly pointing to a prominent role of UTRs in cichlid evolution (Loh et al. 2011).
In our data set, part of the observed UTR diversity might simply result from weaker functional constraints and therefore be nonadaptive. Future investigations targeting, for example, the functional role of divergent UTRs found in association with highly conserved protein-coding sequences will shed light on the contribution of UTRs in cichlids evolution.
Evolutionary Divergence and Mutational Rates among Cichlids
In order to address more specific questions on genetic diversity, substitution rates and selection within the cichlid clade, we expanded our comparative transcriptome analyses to include EST data publicly available for another cichlid, tilapia (Lee et al. 2010), which is a representative of a distinct and more ancestral cichlid lineage, as well as cDNA from medaka, which is presently the closest fully sequenced outgroup to cichlids (Steinke et al. 2006).
We were able to generate a total of 1,216 clusters of aligned sequences (up to 526 Kb) containing exclusively ORFs that fully overlapped across the four species (data set #3). The stringent criteria used for clustering, including cut off e values for Blast searches set to 1.0 x 10−50, best reciprocal Blast hits and removal of sequence clusters with stop codons in cichlids (putatively pseudogenes or novel truncated genes) likely prevented inclusion of paralogous sequences, providing a reliable data set for molecular evolution analyses. Nevertheless, inference of orthology should be taken with caution as transcriptomes are partial and might not represent all sequences belonging to a gene family, causing reciprocal best BlastN hits between paralogous sequences. Although we can largely exclude clustering of cichlid paralogous sequences that are members of old gene families (formed before the cichlid radiation), we cannot rule out clustering of sequences derived from more recent lineage-specific duplications for which only one copy was present in individual species data sets.
Within cichlids, the nucleotide diversity Pi, Ka, and Ks between tilapia and both OV and AB was approximately the same but more than 2-fold higher than between OV and AB (table 5). This is also confirmed by the ML phylogeny reconstructed based on the concatenated data set, which shows equal branch length between tilapia and both AB and OV. Nucleotide diversity estimates based on nuclear data are available for other cichlids, too, albeit based on much smaller samples of orthologous genes. Specifically, genetic distances are reported for three members of the Lake Victoria region superflock, which range between 0.00339 and 0.00346 based on 68 genes (Kobayashi et al. 2009). An average genetic distance of 0.0026 was detected among five Malawi species, based on partial genomic data with low coverage (Loh et al. 2008).
Assuming a divergence time between tilapia and the remaining cichlids of 10.51 to 29.43 Ma (average of 19.44 Ma; Matschiner et al. 2011) and using the neutral Ks divergence estimated on the concatenated alignment by Codeml (accounting for transition/transversion rates and base-frequency dependency), we calculated a mutation rate ranging from 1.2 to 3.3 × 10−9 substitutions per silent site per year (average of 1.8 × 10−9 substitutions per silent site per year) for both comparisons of tilapia to OV and AB. This mutational rate is in accordance to the average mammalian genome mutation rate of 2.2 × 10−9 per base pair per year (Kumar and Subramanian 2002), but it could represent an underestimate because we did not correct for multiple hits. Using the linear equations of time versus Ks given by tilapia comparisons to AB and OV and considering a Ks value between AB and OV of 0.0288 (table 5), we estimated a divergence time for the AB-OV split of between 4.4 and 12 Ma (average of 8 Ma). This dating roughly coincides with the onset of truly lacustrine conditions in Lake Tanganyika (ca. 6 Ma), which is when the primary lacustrine radiation of cichlids is thought to have started and the main cichlid lineages, including the haplochromines and ectodines, emerged (see, e.g., Salzburger et al. 2002; Koblmuller et al. 2008).
Signature of Positive Selection in the Cichlid Lineage
Ka/Ks values estimated for all cichlid pairwise comparisons were at least two times greater (0.175–0.186) than those calculated between medaka and the three cichlids, which were nearly the same (0.081). This argues for homogeneous substitution rates within cichlids, independent of genetic divergence.
Looking at substitution rates in the framework of a phylogeny, dN/dS values per branch estimated under the best branch model (i.e., free-ratio model) confirmed a higher dN/dS for branches within the cichlid clade with respect to the outgroup medaka. This is largely concordant with previous findings for closely related Malawi cichlids, where cichlids showed a much higher Ka/Ks (up to five times) than the one estimated between more distant outgroups (such as between Fugu and Tetraodon or among Danio strains) (Loh et al. 2008). Taken as a whole, these studies provide good evidence for a relatively higher rate of fixation of nonsynonymous substitutions in cichlids, likely driven by disruptive selection. Alternatively, such elevated Ka/Ks might result, in part, from a relaxed purifying selection, due for instance to smaller effective population size of the cichlid ancestor.
Within our data set, we also specifically identified a set of 33 genes putatively under positive selection that represent potential candidates for a more thoroughly experimental and computational investigation. We note that the data set used for our estimates derived from randomly pooled ESTs that contained an ORF and showed a good level of amino acid conservation to return a significant BlastX hits to the medaka proteome, thus we do not expect any particular bias in the gene pooling. Nevertheless, these estimates should be taken with caution, as other types of biases should be considered. First, this data set comprises relatively short alignments of partial ORFs (mean was 433 bp), mostly due to a 3′-UTR bias introduced during the EST sequencing process. This decreases the power for testing positive selection in individual genes. Moreover, the cichlid radiation occurred in a very short evolutionary time frame and deleterious nonsynonymous mutations might not yet have been removed, which could affect proper estimates of Ka/Ks (Rocha et al. 2006; Wolf et al. 2009). Finally, 454 sequencing is known to have a high single read accuracy of > 99.5%, whereas the consensus read accuracy within an assembly with a coverage >20x is >99.99%. Nonetheless, even considering slightly higher error rates, we do not expect any systematic bias in the substitutions pattern that could have specifically affected the nonsynonymous rates.
Conclusions
Using the new 454 pyro-sequencing technology, we have provided the so far largest collection of new ESTs for cichlid species. Our functional annotation and expanded comparative transcriptome analysis, including a third cichlid lineage (tilapia) and the outgroup medaka, have shown a signature of disruptive selection in the cichlid lineage and pointed to a prominent contribution of UTRs in cichlid genetic diversity, potentially involved in regulatory changes of the expression profiles underlying their large phenotypic diversity. The new transcriptomes provide an important reference to now target more specific transcriptome-to-phenome comparative analyses aimed to investigate, for instance, the molecular bases of single and multiple traits diversity in more closely related species or shared traits among more distantly related species. Genome sequencing projects are currently ongoing for tilapia and four other cichlid species, including AB, Metraclinia (Maylandia) zebra, Pundamilia nyererei, and Neolamprologus brichardi (http://cichlid.umd.edu/CGCindex.html). Together with these, the partial genomic data and the EST resource already existing for cichlids and close outgroups, the transcriptome data sets reported here will provide the scientific community with a valuable resource for comparative analyses of both genetic and expression profiles within cichlids and among closely related species that will address crucial questions on the molecular bases of adaptive radiation and explosive speciation.
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We would like to thank Christof Wunderlin (Microsynth) for technical support on the 454 sequencing data and Michael Matschiner and Hugo Gante for helpful comments on the manuscript. This work was supported by a doctoral research fellowship from the Fundação para a Ciência e a Tecnologia [SFRH/BD/43421/2008 to E.S.], the European Research Council [Starting Grant ‘INTERGENADAPT’ to W.S.], and the Swiss National Science Foundation [grant 3100A0_122458 to W.S.].
References
- Aparicio S, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Clabaut C, Salzburger W, Meyer A. Comparative phylogenetic analyses of the adaptive radiation of Lake Tanganyika cichlid fish: nuclear sequences are less homoplasious but also less informative than mitochondrial DNA. J Mol Evol. 2005;61:666–681. doi: 10.1007/s00239-004-0217-2. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, et al. Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Mol Ecol. 2010;19(Suppl 1):197–211. doi: 10.1111/j.1365-294X.2009.04488.x. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wittkopp PJ. Evaluating the role of natural selection in the evolution of gene regulation. Heredity. 2008;100:191–199. doi: 10.1038/sj.hdy.6801000. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Johnson TC, et al. Late Pleistocene desiccation of Lake Victoria and rapid evolution of cichlid fishes. Science. 1996;273:1091–1093. doi: 10.1126/science.273.5278.1091. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Watanabe M, Horiike T, Kohara Y, Okada N. Extensive analysis of EST sequences reveals that all cichlid species in Lake Victoria share almost identical transcript sets. Gene. 2009;441:187–191. doi: 10.1016/j.gene.2008.11.023. [DOI] [PubMed] [Google Scholar]
- Koblmuller S, et al. Age and spread of the haplochromine cichlid fishes in Africa. Mol Phylogenet Evol. 2008;49:153–169. doi: 10.1016/j.ympev.2008.05.045. [DOI] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5:288–298. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BY, et al. An EST resource for tilapia based on 17 normalized libraries and assembly of 116,899 sequence tags. BMC Genomics. 2010;11:278. doi: 10.1186/1471-2164-11-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, et al. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Yi SV, Streelman T. Evolution of microRNAs and the diversification of species. Genome Biol Evol. 2011;3:55–65. doi: 10.1093/gbe/evq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M, Hanel R, Salzburger W. On the origin and trigger of the notothenioid adaptive radiation. PLoS One. 2011;6:e18911. doi: 10.1371/journal.pone.0018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K. SeqState: primer design and sequence statistics for phylogenetic DNA datasets. Appl Bioinformatics. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol Ecol. 2009;18:169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- Salzburger W, Braasch I, Meyer A. Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 2007;5:51. doi: 10.1186/1741-7007-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Mack T, Verheyen E, Meyer A. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol Biol. 2005;5:17. doi: 10.1186/1471-2148-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Meyer A, Baric S, Verheyen E, Sturmbauer C. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Syst Biol. 2002;51:113–135. doi: 10.1080/106351502753475907. [DOI] [PubMed] [Google Scholar]
- Salzburger W, et al. Annotation of expressed sequence tags for the East African cichlid fish Astatotilapia burtoni and evolutionary analyses of cichlid ORFs. BMC Genomics. 2008;9:96. doi: 10.1186/1471-2164-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. African cichlid fish: a model system in adaptive radiation research. Proc Biol Sci. 2006;273:1987–1998. doi: 10.1098/rspb.2006.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst Biol. 2000;49:369–381. [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Steinke D, Salzburger W, Meyer A. Novel relationships among ten fish model species revealed based on a phylogenomic analysis using ESTs. J Mol Evol. 2006;62:772–784. doi: 10.1007/s00239-005-0170-8. [DOI] [PubMed] [Google Scholar]
- Sturmbauer C, Meyer A. Mitochondrial phylogeny of the endemic mouthbrooding lineages of cichlid fishes from Lake Tanganyika in eastern Africa. Mol Biol Evol. 1993;10:751–768. doi: 10.1093/oxfordjournals.molbev.a040042. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland (MA): Sinauer Associates; 2000. [Google Scholar]
- Verheyen E, Salzburger W, Snoeks J, Meyer A. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science. 2003;300:325–329. doi: 10.1126/science.1080699. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Kunstner A, Nam K, Jakobsson M, Ellegren H. Nonlinear dynamics of nonsynonymous (dN) and synonymous (dS) substitution rates affects inference of selection. Genome Biol Evol. 2009;1:308–319. doi: 10.1093/gbe/evp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]