Abstract
The control of organ size is a basic biological question. In the last several years, the Hippo signaling pathway has been delineated and shown to be critical in control of organ size in both Drosophila and mammals. Acting downstream of the Hippo pathway is the Yki/YAP/TAZ transcription co-activators. In mammalian cells, the Hippo pathway kinase cascade inhibits YAP and its paralog TAZ by phosphorylation and promotion of their cytoplasmic localization. The TEAD family transcription factors have recently been identified as evolutionarily conserved key mediators of YAP biological functions. yap is a candidate oncogene, and several other components of the Hippo pathway are tumor suppressors. Dysregulation of the Hippo pathway contributes to the loss of contact inhibition observed in cancer cells. Therefore, the Hippo-YAP pathway connects the regulation of organ size and tumorigenesis.
Introduction
People have long been interested in the precise regulation of body and organ size of multicellular organisms. However, silencing of basic developmental regulatory genes often leads to early lethality, which makes further characterization very difficult. This obstacle was overcome first in Drosophila by the development of technology generating genetic mosaics in developing tissue. The mosaic screen fueled discovery of many Drosophila tumor-suppressor genes including the Hippo pathway components, which form a kinase cascade in regulation of a transcription co-activator Yorkie (Yki) [1-6]. Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ, also called WWTR1), two Yki homologs in mammals, are phosphorylated and inhibited by the Hippo pathway through cytoplasmic retention [7-9]. The function of YAP in regulation of organ size is conserved from Drosophila Yki [10,11]. Furthermore, yap is a candidate oncogene amplified in human cancers [12,13]. In this review we discuss the regulation and downstream transcription factors of YAP and TAZ in mammalian cells emphasizing the connections between the Hippo pathway and cancer.
The Hippo pathway in Drosophila
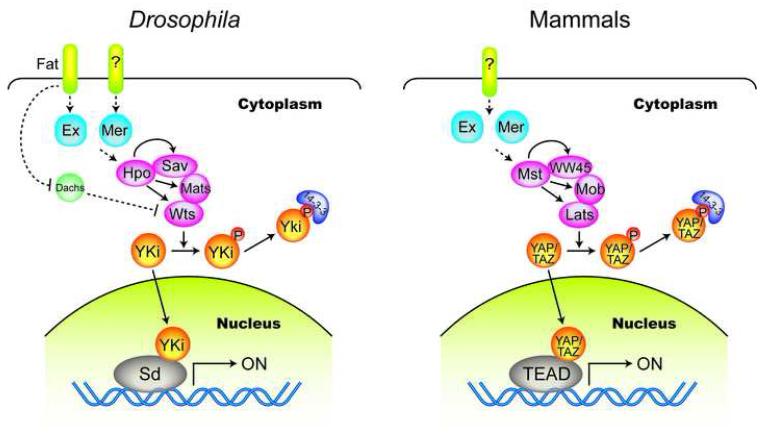
In 1995, The first Hippo pathway component, wts, was uncovered by genetic mosaic screens in Drosophila. wts encodes a nuclear Dbf-2-related (NDR) family protein kinase [14,15]. Mutation of wts leads to robust tissue overgrowth. Since 2002, similar screens have identified several other Hippo pathway components, including Salvador (Sav) [16,17], Hippo (Hpo) [18-22], and Mats[23]. Together they form the core of the Drosophila Hippo pathway in which Hpo kinase, in association with an adaptor protein Sav, phosphorylates and activates Wts kinase, which is associated with an activating subunit Mats (Fig.1). Upstream of that might be Merlin (Mer) and Expanded (Ex), two ERM (ezrin/radixin/moesin) family cytoskeleton-related proteins [24]. Fat, a protocadherin might be further upstream [25-29]. However, the biochemical mechanisms of Mer, Ex and Fat in regulation of the Hippo pathway core components are not clear.
Figure 1. The Hippo pathway in Drosophila and mammals.
Corresponding components in Drosophila and mammals are shown in the same color. The abbreviations used are as follows: Ex (Expanded), Mer (Merlin, also called NF2), Hpo (Hippo), Sav (Salvador), Mats (Mob as tumor suppressor), Wts (Warts), Yki (Yorkie), Sd (Scalloped), Mst (Mst1/2, also called STK4 and STK3, Hpo homolog), WW45 (Sav homolog), Mob (Mps One Binder kinase activator-like 1A/B, MOBKL1A/B, Mats homolog), Lats (Lats1/2, Wts homolog), YAP (Yes-associated protein, Yki homolog), TAZ (transcriptional co-activator with PDZ-binding motif, also called WWTR1, Yki homolog), and TEAD (TEA domain family member 1/2/3/4). Dashed arrows indicate unknown biochemical mechanism and question marks denote unknown components.
The Hippo pathway limits organ size by inhibiting cell proliferation and promoting apoptosis [2]. Such regulation is achieved at least in part by transcriptional activation of target genes like cycE, diap1 [2] and bantam microRNA [30,31]. Logically, the Hippo pathway should target some transcription regulators. Indeed, Yki, ortholog of the mammalian YAP, a transcription co-activator, was identified as a Wts-interacting protein [32]. Yki regulates transcription of the Hippo pathway target genes, and its overexpression phenocopies the loss of Hippo pathway components. Further biochemical studies showed that Wts directly phosphorylates Yki, which leads to Yki cytoplasmic retention and inactivation [11,32].
The incorporation of Yki significantly advanced our understanding of the Hippo pathway. However, since Yki is a transcription co-activator, its promoter selectivity must be determined by its interacting transcription factors. It was recently reported that Scalloped (Sd), a critical regulator of proliferation and survival of wing imaginal disc cells [33,34], directly mediates Yki-induced gene expression and overgrowth phenotype [35-38]. However, Sd is expressed in a narrower spectrum of cells while Yki and the Hippo pathway functions more ubiquitously [39]; yki mutant clones have more severe growth defects than sd mutant clones [32,36]; and Sd-binding-defective Yki mutant elicits a reduced but still obvious overgrowth in Drosophila eyes and wings [37]. Therefore, other transcription factors mediating the function of Yki and the Hippo pathway likely exist.
The Hippo pathway in mammalian cells
Components of the Hippo pathway are highly conserved in mammals, including Mst1/2 (Hpo homolog), WW45 (also called Sav, Sav homolog), Lats1/2 (Wts homolog), Mob1 (Mats homolog), YAP and its paralog TAZ (both are Yki homologs), Mer (also called NF2, Mer homolog), and at a lesser degree FRMD6 (Ex homolog), and Fat4 (Fat homolog) (Fig.1). More strikingly, human YAP, Lats1, Mst2, and Mob1 can functionally rescue the corresponding Drosophila mutants in vivo, suggesting the functional conservation of these proteins in mammals [2]. As Hpo in Drosophila, Mst plays a key role in the mammalian Hippo pathway as it phosphorylates all three other core components. Lats1/2 is phosphorylated by Mst1/2 on the activation loop and hydrophobic motif, possibly with autophosphorylation involved [40]. WW45 interacts with Mst through the SARAH domains in each other, and is then phosphorylated by Mst [41]. Mob1 is also phosphorylated by Mst1/2, which enhances its interaction with Lats1 [42].
However, the mammalian Hippo pathway was not established until it was shown to inhibit YAP and TAZ. Mst, WW45, Lats, and Mob induce YAP phosphorylation, cytoplasmic translocation, and inhibition [8,9,43,44]. The mechanism of YAP regulation by the Hippo pathway is conserved in TAZ [7]. It was further shown that TEAD family transcription factors, homologs of the Drosophila Sd, are key mediators of YAP function in mammalian cells [37]. The function of the Hippo pathway in organ size control is also conserved in mammals because overexpression of YAP in mouse liver induces dramatic increase in liver size and eventually leads to tumor formation [10,11]. The regulation and function of YAP and TAZ will be discussed in detail below.
The functional conservation of the Hippo pathway upstream components Fat and Ex in mammalian cells is not clear. However, Mer has been shown to regulate YAP localization and inhibit its activity in cell culture [8]. In addition, RASSF, a subgroup of Ras effector proteins with inhibitory effect on the Hippo pathway in Drosophila [45], might be an activator of the Hippo pathway in mammals [46]. RASSF1A has been reported to activate Mst2, which may result in activation of YAP on p73 in the context of Fas-induced apoptosis [47]. However, the activation of YAP is difficult to interpret as the Hippo pathway activation was clearly shown to inhibit YAP activity. It will be important to clarify the role of RASSF in the Hippo pathway and YAP regulation.
YAP is a transcription co-activator
YAP was first cloned as a protein bound to non-receptor tyrosine kinase c-Yes [48]. It has several distinct domains as the human YAP2 shown in Fig.2. YAP also exists as YAP1, another splicing variant missing the second WW domain. Regulation of the switch between the two YAP isoforms is not clear. In general, YAP mRNA is ubiquitously expressed in a wide range of tissues, except peripheral blood leukocytes [49]. YAP is also expressed in the full developmental stages from blastocyst to perinatal [50].
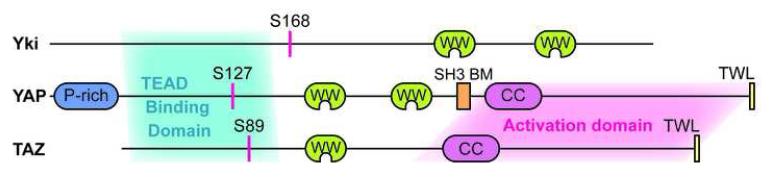
Figure 2. A schematic view of YAP, TAZ, and Yki.
YAP is a 65KDa protein with several distinct domains or motifs. It has a proline-rich (P-rich) region at the N-terminal, two tandem WW domains in the middle followed by a Src homology domain 3 binding motif (SH3 BM) PVKQPPPLAP, a coiled-coil domain (CC), and a C-terminal capped by TWL sequence, a PDZ domain ligand. The N-terminal (aa 47-154 in human YAP2, shaded in blue) of YAP was mapped to be the TEAD family transcription factors interaction domain [54], and the C-terminal of YAP (aa 292-488, shaded in pink) rich in serine, threonine, and acidic residues was shown to be a strong transcription activator [51]. The Lats phosphorylation and 14-3-3 binding critical S127 in human YAP2 and its equivalent in Yki and TAZ are also shown. The topology of Yki and TAZ are shown in similar fashion and the proteins are drawn in scale.
However, the function of YAP remained enigmatic until it was shown to be a transcription co-activator [51]. YAP interacts with the PPXY motif of transcription factor PEBP2α (RUNX1 and RUNX2) mainly through its first WW domain. More importantly, when fused to Gal4 DNA binding domain, YAP could activate luciferase reporter as strong as VP16, a potent transcription activator. The transcription activation domain of YAP was further mapped to the C-terminal region. Interestingly, this region was found to be truncated in possibly dominant-negative YAP isoforms specifically expressed in neurons [52]. However, YAP does not have any obvious DNA binding domain. Therefore, it is categorized as a transcription co-activator. YAP has also been reported to co-activate other PPXY-motif-containing transcription factors, including ErbB4 cytoplasmic domain [49] and p73 [53].
YAP also binds to TEAD family transcription factors [54], which have four highly homologous proteins sharing a conserved DNA-binding TEA domain in human and mouse. Most adult tissues express at least one TEAD gene. YAP was first identified as a TEAD-interacting protein by affinity purification [54]. Strikingly, about 75% of the purified TEAD2 are in complex with YAP. From a different direction, we screened for YAP targets in a Gal4-fusion transcription factor library, which covers about one third of potential transcription factors encoded by the human genome. This unbiased strategy identified TEAD2, TEAD3, and TEAD4 as the strongest positives [37]. TEAD1, which is not in the library, is activated by YAP in similar magnitude. Therefore, biochemical purification starts with TEAD and functional screen starts with YAP complement each other nicely in establishing a partnership between YAP and TEAD at least in cell culture.
More importantly, TEAD was shown to play a critical role in YAP function. In MCF10A human mammary epithelial cells, YAP and TEAD1 promoter occupancy is highly overlapped [37]. Knock-down of TEAD or introduction of a TEAD-binding-deficient mutation (serine 94 to alanine) in YAP aborts activation of a large fraction of YAP-inducible genes [37]. TEAD is further shown to be critical for YAP-induced overgrowth, epithelial-mesenchymal-transition (EMT), and oncogenic transformation in MCF10A cells [37]. Furthermore, the phenotype of TEAD1/TEAD2 double knockout mice resembles YAP knockout mice and evidence suggests that tead1/tead2 and yap genetically interact with each other in vivo [55]. In addition, TEAD1/TEAD2 double knockout embryos show decreased proliferation and increased apoptosis [55], a phenotype consistent with the Hippo pathway components mutants in Drosophila. Finally, the function of YAP and TEAD interaction in cell growth is implicated in human disease. Sveinsson’s chorioretinal atrophy is a human genetic disease caused by a heterozygous mutation of a highly conserved tyrosine in the YAP binding domain of TEAD1 [56]. Interestingly, mutation of this residue in TEADs abolished their interaction with and their activation by YAP [37,57], which may explain the atrophic phenotype. These observations support that TEAD is at the downstream of the Hippo pathway mediating YAP activity.
Regulation of YAP phosphorylation and localization
The Hippo pathway phosphorylates Yki to control organ size in Drosophila. Regulation on such a basic biological process would be expected to be conserved in higher organisms. Indeed, YAP is directly phosphorylated by Lats on serine residues in five conserved HXRXXS motifs [8,9], including S127 [11,44]. Phosphorylation by Lats on this residue generates a 14-3-3 binding site and induces YAP cytoplasmic translocation, and therefore, inactivation [8,9]. Such mechanism explains the Hippo pathway-dependent nuclear/cytoplasmic translocation of YAP based on cell density. Consistently, keratinocytes lacking Hippo pathway component WW45 lost the cytoplasmic translocation of YAP upon Ca2+ induced differentiation [58]. Removing the inhibitory phosphorylation sites disrupts the regulation on YAP localization and promotes YAP induced over-proliferation of NIH-3T3 cells (unpublished observation), oncogenic transformation of MCF10A cells [9], and overgrowth of Drosophila tissue in vivo [8]. In agreement with that, the transformation activity of YAP is inhibited by co-expression of Lats1 and Mst2 [9,43]. These studies support the evolutionarily conserved function of YAP in promotion of cell proliferation and oncogenic transformation under negative regulation by the Hippo pathway.
YAP S127 has also been suggested to be an Akt phosphorylation site [59]. However, the sequences around this site do not match the optimal Akt target site. YAP S127 phosphorylation is neither suppressed by PI3K inhibitors nor induced by EGF/ insulin stimulation or active Akt expression [8]. More importantly, YAP phosphorylation is not affected by knockout of PDK1, which is essential for Akt activity [8]. Consistent with that, the Drosophila Yki is not phosphorylated by Akt either [11]. All these results strongly indicate that YAP is not directly phosphorylated by Akt at least under most physiological conditions. However, it cannot be excluded that YAP is phosphorylated by Akt under some circumstances.
Besides the Hippo pathway mediated serine/threonine phosphorylation, YAP was recently shown to be regulated by tyrosine phosphorylation. A recent report from Dr. Shaul’s lab showed that c-Abl directly binds and phosphorylates YAP on Y357, which stabilizes YAP and confers selective binding of YAP to p73 and is required for cisplatin-induced apoptosis [60]. In contrast with previously suggested mechanism of YAP-p73 activation involving Akt or RASSF, the Y357 phosphorylation and stabilization of YAP was shown to be indeed induced by DNA damage. However, the biochemical mechanism of Y357 phosphorylation in YAP activity regulation is not yet clear, and it will be interesting to determine if there is any cross-talk between the Hippo pathway and c-Abl regulated YAP phosphorylation.
The Hippo pathway promotes YAP cytoplasmic retention. However, YAP does not have any obvious nuclear localization signal sequence. Therefore, it is not clear how YAP gets into the nucleus when the Hippo pathway is silenced. One possible mechanism is through interaction and co-transportation with target transcription factors, such as shown for Drosophila Yki, which is translocated from cytoplasm to nucleus by co-expression of Sd in S2 cells [35,38]. Such effect is overridden by the Hippo pathway as Hpo expression sequesters both Yki and Sd in the cytoplasm [35]. More importantly, Sd expression significantly potentiates the effect of wts mutation in inducing Yki nuclear localization in vivo [35]. Considering the functional conservation of Yki/Sd in the mammalian YAP/TEAD, such regulation likely exists for YAP, although it awaits confirmation. However, it has already been reported that upon cisplatin treatment, YAP translocates to the nucleus in a p73-dependent manner [61]. It will be important to examine the contribution of different transcription factors in regulation of YAP nuclear localization and determine the underlying mechanism.
YAP as an oncoprotein
YAP is a potent growth promoter. Overexpression of YAP increases organ size in Drosophila and saturation cell density in NIH-3T3 cell culture [8]. However, yap was termed a candidate oncogene only after it was shown to be in human chromosome 11q22 amplicon, which is evident in several human cancers [12,13]. Consistently, yap was shown to be amplified in human primary intracranial ependymomas by clinical study [62]. Besides the genomic amplification, YAP expression and nuclear localization was also shown to be elevated in multiple types of human cancers [8,11,13,63]. Several experiments further confirmed that YAP has oncogenic function: YAP overexpression in MCF10A cells induces epithelial-mesenchymal transition (EMT), which is often associated with cancer metastasis [12]; YAP cooperates with myc oncogene to stimulate tumor growth in nude mice [13]; and more interestingly, transgenic mice with liver-specific YAP overexpression show a dramatic increase in liver size and eventually develop tumors [10,11]. The above evidence strongly indicates the function of yap as an oncogene. However, YAP was also reported to be a tumor suppressor as its gene locus is deleted in some breast cancers with a correlated loss of YAP expression [64]. Further experiments such as conditional knockout animal model will finally clarify the role of YAP in tumorigenesis.
The oncogenic function of YAP is further supported by the tumor suppressor function of its inhibitory upstream Hippo pathway components. Lats1 knockout leads to soft-tissue sarcoma and ovarian tumor development [65]. mob, an activating subunit of Lats, is mutated in both human and mouse cancer cells [23]. Loss-of-function mutation of WW45 has been observed in several human cancer cell lines [17]. Furthermore, a recent report showed that knockout of ww45 leads to hyperplasia and differentiation defects in mouse embryonic epithelial structures [58]. Mer, which is further upstream of the Hippo pathway, is a well-established human tumor suppressor [66]. Therefore, the Hippo pathway consists of many proven or candidate tumor suppressors that inhibit YAP oncoprotein.
Noteworthy, several studies showed a proapoptotic function of YAP, which was mainly explained by co-activation of p73 [44,47,60,61]. So far, the proapoptotic activity of YAP was only observed by overexpression of YAP or in response to strong apoptotic stimuli, such as Fas activation or DNA damage. However, the effect of YAP overexpression in vivo was shown to be an increase of organ size and finally tumor formation without accompanied increase of apoptosis. In fact, YAP overexpression protects liver tissue from Fas induced apoptosis [10,11]. On the other hand, the Drosophila genetic studies have clearly established that Yki inhibits aopotosis in vivo. It is still possible that under certain conditions like DNA damage, YAP was tyrosine phosphorylated by c-Abl, which selectively activates YAP transcriptional activity on p73 to induce apoptosis.
Contact inhibition of cell growth, often referred to as a hallmark of cancer cells, has long been a mystery. However, the Hippo pathway may have opened the window a little bit to understand this phenomenon. Several components of this pathway have been implicated in contact inhibition. Mer becomes dephosphorylated and activated in confluent cells [67,68], which has been reported to be both necessary and sufficient for contact inhibition. Lats2 and WW45 are also related to contact inhibition as their knockout MEF cells show loss of contact inhibition [58,69]. Finally, YAP is phosphorylated and translocated to the cytoplasm by the Hippo pathway at high cell density in a Mer-dependent manner [8]. More importantly, a dominant-negative form of YAP restores contact inhibition in ACHN [8], a cancer cell line with activation of YAP due to WW45 mutation. These observations suggest a critical role of YAP and the Hippo pathway in contact inhibition. Indentifying the upstream signal of this pathway might solve a long-standing mystery in cell biology.
Similarity and differences between TAZ and YAP
TAZ is a YAP paralog initially identified as a 14-3-3 binding protein [70]. In human and mouse, TAZ mRNA is expressed in all tissues except thymus and peripheral blood leukocytes, with the highest expression in kidney [70]. TAZ has approximately 50% sequence identity and very similar topology with YAP, although the differences are also apparent, including the lack of N-terminal proline-rich domain, the second WW domain, and the SH3 binding motif (Fig.2). This suggests both shared and distinct regulation/ function between TAZ and YAP.
TAZ is regulated by the Hippo pathway in a fashion similar to YAP. TAZ can be phosphorylated by Lats2 on serine residues in four HXRXXS motifs [7], including S89, the counterpart of YAP S127. Phosphorylation on TAZ S89 by Lats, creates a 14-3-3 binding site. Therefore, TAZ is sequestered in the cytoplasm and inactivated [7]. This model suggests that besides YAP inhibition, TAZ inactivation is also an important downstream output of the Hippo pathway.
Similar to YAP, TAZ also functions as a transcriptional co-activator [70]. TAZ interacts with TEAD [71], and based on the screen of a human transcription factor library and tandem affinity purification of TAZ-interaction proteins, we actually observed TEADs as the major transcription factor targets of TAZ (unpublished data). TAZ has also been reported to interact with several other transcription factors such as RUNX2 [70]. At this point, it is apparent that YAP and TAZ share many transcription factor targets such as TEAD and RUNX. However, their contribution to the functions of those shared transcription factors is not clear. Nor is the activation of unique targets in defining the distinct physiological functions of YAP and TAZ.
YAP increases organ size and functions as an oncogene [8,11,12]. Similarly, TAZ also promotes cell proliferation, induces EMT, increases cell migration and invasion [7,72], and is shown to be overexpressed in approximately 20% of breast cancer samples [72]. Experiments are needed to show if the TAZ gene locus is also amplified in cancer and if TAZ overexpression also leads to increase in organ size and tumor development.
In spite of these similarities, existing evidence suggests that YAP and TAZ do not compensate each other. First, YAP and TAZ knockout mice show different phenotypes: YAP knockout animals are embryonic lethal and show shortened body axis and defects in yolk sac vasculogenesis [50]. In contrast, TAZ knockout mice are viable and are characterized by renal cysts which lead to end stage kidney disease [73,74]. Second, in many reports, the phenotype of YAP or TAZ knockdown were not masked by the presence of the other [37,72,75,76]. Such differences can be explained by differential spatial/temporal regulation of YAP and TAZ activity or different downstream targets, which require further study.
Function of TAZ in stem cells
The balance between cell proliferation and differentiation is implicated not only in normal tissue development but also in tumorigenesis. Mesenchymal stem cells (MSCs) are pluripotent precursor cells with ability to differentiate into several distinct lineages. A recent study showed that TAZ functions as a transcriptional modulator of MSC differentiation by promoting osteoblast differentiation while repressing adipocyte differentiation [75]. More interestingly, TAZ has recently been shown to maintain human embryonic stem cell (hESC) pluripotency [76]. TAZ binds heteromeric Smad2/3-4 upon TGFβ stimulation, and plays an essential role in Smad nuclear accumulation. In hESCs, TAZ is required to maintain self-renewal markers and loss of TAZ leads to inhibition of TGFβ signaling and differentiation of hESCs into a neuroectoderm lineage. Coincidently, YAP has also been implicated in stem cell maintenance. In mouse intestine, expression of endogenous YAP is restricted to the progenitor/stem cell compartment, and YAP overexpression expands multipotent undifferentiated progenitor cells, which differentiate upon cessation of YAP expression [10]. Consistent with the role of YAP and TAZ in maintaining stemness, mice lacking WW45 showed immature differentiation and hyperplasia likely due to defective cell-cycle exit in epithelial progenitor cells [58]. Therefore, the Hippo pathway in control of YAP and TAZ may regulate stem cell renewal and differentiation, although the underlying mechanism is not yet clear.
Key questions to be addressed
Genetic, cell biology, and biochemical studies have established the novel Hippo tumor suppressor pathway. Inhibition of YAP and TAZ transcription co-activators is the major target of the Hippo pathway to regulate cell proliferation, apoptosis, and organ size in mammals [77]. In spite of rapid progresses in the field, many key questions remain to be answered. Perhaps the most interesting question in the Hippo pathway is the upstream signals that activate the core components. The sensing of organ size in vivo and cell confluence in vitro are long-standing mysteries. It is reasonable to speculate that such a signal may act upstream of the Hippo pathway.
Equally important is what are the other transcription factors mediating the biological function of YAP and TAZ. The PPXY-motif-containing transcription factors may interact with YAP WW domains, and are therefore possible candidates. A related question is how YAP and TAZ activate transcription. Although largely unknown, current evidence suggests mechanisms such as recruitment of histone modification factors or Mediator complex. Answering these questions is important in understanding the mechanism of YAP and TAZ in control of cell growth and organ size.
In Drosophila, Yki activates expression of many genes, including cycE, diap1 and bantam microRNA. However, in mammalian cells, cycE is not induced by YAP, and the bantam microRNA is not conserved, while induction of birc5, an IAP family member, is insufficient to explain the increased proliferation and organ size. CTGF is recently shown to be a direct YAP target gene important for YAP function in mammalian cells [37]. However, there is no evidence that CTGF homolog is an Yki target gene in Drosophila. It would be very interesting if common genes in Drosophila and mammals mediate the Hippo pathway functions, especially, if there is a functional equivalent of the bantam microRNA in mammals.
In the next few years, one can expect exciting discoveries in the Hippo pathway. Advances in this field may not only solve the puzzle of size control and contact inhibition, but also provide new targets for treatment of human diseases such as atrophy and cancer.
Acknowledgements
We apologize for the many important contributions to the Hippo pathway field that could not be cited due to space constraints. We thank Karen Tumaneng and Kristen Slanina for critical reading of the manuscript. This work is supported by grants from NIH (KLG), National High Technology Research and Development Program of China (Grant No. 2004BA711A18, 2006AA02A308) and National Natural Science Foundation of China (Grant No. 30600112) (QYL), and University of Michigan Rackham Predoctoral Fellowship (BZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
” of special interest
””of outstanding interest
- 1.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 2.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 4.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 5.Hariharan IK. Growth regulation: a beginning for the hippo pathway. Curr Biol. 2006;16:R1037–1039. doi: 10.1016/j.cub.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Buttitta LA, Edgar BA. How size is controlled: from Hippos to Yorkies. Nat Cell Biol. 2007;9:1225–1227. doi: 10.1038/ncb1107-1225. [DOI] [PubMed] [Google Scholar]
- 7.Lei Q, Zhang H, Zhao B, Zha ZY, Bai F, Pei XH, Zhao S, Xiong Y, Guan KL. TAZ promotes cell proliferation and EMT and is inhibited by the Hippo pathway. Mol Cell Biol. 2008 doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ”8.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. This study and references 9 and 43 elucidated a biochemical mechanism of YAP inhibition by Lats. This paper demonstrated that YAP localization is regulated by cell density and the Hippo pathway plays a key role in cell contact inhibition.
- ”9.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- ”10.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. This report showed that in intestine, expression of endogenous YAP is restricted to the progenitor/ stem cell compartment, and transgenic expression of YAP expands multipotent undifferentiated progenitor cells, which differentiate upon cessation of YAP expression.
- ”11.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. This report showed the size control function of the Hippo pathway is conserved in mammals by a liver-specific YAP transgenic mouse model.
- ””12.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. This study and reference 13 identified amplification of a genomic region containing yap in both mouse cancer model and human cancers with concomitant elevation of YAP protein level. These were the first reports of yap as a candidate oncogene.
- ””13.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 16.Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Sharpei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 17.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 18.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 20.Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 21.Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 22.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 24.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 25.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 26.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 27.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;vol 16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- ””32.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. The authors identified Yki as a Wts-interacting protein and established the role of Yki at the downstream of the Hippo pathway by Drosophila genetics studies. This study established a link between the Hippo pathway and transcriptional regulation.
- 33.Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A, Carroll S. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 1998;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD, Bell JB. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ”35.Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF Family of Transcription Factor Scalloped Mediates Hippo Signaling in Organ Size Control. Dev Cell. 2008 doi: 10.1016/j.devcel.2008.01.006. This paper and references 36 and 38 established the role of Scalloped in mediating the Hippo pathway function in size control. This study further demonstrated the regulation of Yki subcellular localization by Scalloped.
- ”36.Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF Family Protein Scalloped Mediates Transcriptional Output of the Hippo Growth-Regulatory Pathway. Dev Cell. 2008 doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- ”37.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. This study established the role TEAD family transcription factors in mediating the function of YAP and the Hippo pathway in mammalian cells.
- ”38.Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED Interacts with YORKIE, the Nuclear Effector of the Hippo Tumor-Suppressor Pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 39.Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M, Chovnick A. The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 1992;6:367–379. doi: 10.1101/gad.6.3.367. [DOI] [PubMed] [Google Scholar]
- 40.Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 41.Callus BA, Verhagen AM, Vaux DL. Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 2006;273:4264–4276. doi: 10.1111/j.1742-4658.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- 42.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ”43.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 44.Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of YAP. J Biol Chem. 2008 doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 45.Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Curr Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matallanas D, Romano D, Hamilton G, Kolch W, O’Neill E. A Hippo in the ointment: MST signalling beyond the fly. Cell Cycle. 2008;7:879–884. doi: 10.4161/cc.7.7.5630. [DOI] [PubMed] [Google Scholar]
- 47.Matallanas D, Romano D, Yee K, Meissl K, Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W, O’Neill E. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell. 2007;27:962–975. doi: 10.1016/j.molcel.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 49.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 50.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O’Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. Embo J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoshino M, Qi ML, Yoshimura N, Miyashita T, Tagawa K, Wada Y, Enokido Y, Marubuchi S, Harjes P, Arai N, et al. Transcriptional repression induces a slowly progressive atypical neuronal death associated with changes of YAP isoforms and p73. J Cell Biol. 2006;172:589–604. doi: 10.1083/jcb.200509132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 54.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ”55.Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. The authors generated tead1/tead2 double knockout mice. The embryo shows a similar phenotype as yap knockout embryo with reduced cell proliferation and increased apoptosis.
- 56.Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, Gulcher JR, Stefansson K. A novel TEAD1 mutation is the causative allele in Sveinsson’s chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 57.Kitagawa M. A Sveinsson’s chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem Biophys Res Commun. 2007;361:1022–1026. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- 58.Lee JH, Kim TS, Yang TH, Koo BK, Oh SP, Lee KP, Oh HJ, Lee SH, Kong YY, Kim JM, et al. A crucial role of WW45 in developing epithelial tissues in the mouse. EMBO J. 2008;27:1231–1242. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- ”60.Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. The authors showed that c-Abl directly phosphorylates YAP on Y357, which stabilizes YAP and confers selective binding of YAP to p73 and is required for cisplatin-induced apoptosis. This is the first report of YAP regulation by tyrosine phosphorylation.
- 61.Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, Mantovani F, Damalas A, Citro G, Sacchi A, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 62.Modena P, Lualdi E, Facchinetti F, Veltman J, Reid JF, Minardi S, Janssen I, Giangaspero F, Forni M, Finocchiaro G, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24:5223–5233. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 63.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008 doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan M, Tomlinson V, Lara R, Holliday D, Chelala C, Harada T, Gangeswaran R, Manson-Bishop C, Smith P, Danovi SA, et al. Yes-associated protein (YAP) functions as a tumor suppressor in breast. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.108. [DOI] [PubMed] [Google Scholar]
- 65.St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, McGrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 66.Evans DG, Sainio M, Baser ME. Neurofibromatosis type 2. J Med Genet. 2000;37:897–904. doi: 10.1136/jmg.37.12.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shaw RJ, McClatchey AI, Jacks T. Regulation of the neurofibromatosis type 2 tumor suppressor protein, merlin, by adhesion and growth arrest stimuli. J Biol Chem. 1998;273:7757–7764. doi: 10.1074/jbc.273.13.7757. [DOI] [PubMed] [Google Scholar]
- 68.Morrison H, Sherman LS, Legg J, Banine F, Isacke C, Haipek CA, Gutmann DH, Ponta H, Herrlich P. The NF2 tumor suppressor gene product, merlin, mediates contact inhibition of growth through interactions with CD44. Genes Dev. 2001;15:968–980. doi: 10.1101/gad.189601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McPherson JP, Tamblyn L, Elia A, Migon E, Shehabeldin A, Matysiak-Zablocki E, Lemmers B, Salmena L, Hakem A, Fish J, et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. Embo J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. Embo J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahoney WM, Jr., Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–225. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W, Zeng Q, Hong W. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi: 10.1158/0008-5472.CAN-07-2696. [DOI] [PubMed] [Google Scholar]
- 73.Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, Mitani A, Nagase T, Yatomi Y, Aburatani H, et al. Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol. 2008;294:F542–553. doi: 10.1152/ajprenal.00201.2007. [DOI] [PubMed] [Google Scholar]
- 74.Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, Qi Z, Ponniah S, Hong W, Hunziker W. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, Mueller E, Benjamin T, Spiegelman BM, Sharp PA, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- ”76.Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. The authors showed that TAZ binds heteromeric Smad2/3-4 upon TGFβ stimulation, and plays an essential role in Smad nuclear accumulation and human embryonic stem-cell self-renewal. This study established a direct link between TAZ and stem cell function.
- 77.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]