Abstract
Ubiquitination is a posttranslational modification that regulates a variety of cellular functions depending on timing, subcellular localization, and type of tagging, as well as modulators of ubiquitin binding leading to proteasomal or lysosomal degradation or nonproteolytic modifications. Ubiquitination plays an important role in the pathogenesis of acute lung injury (ALI) and other lung diseases with pathologies secondary to inflammation, mechanical ventilation, and decreased physical mobility. Particularly, ubiquitination has been shown to affect alveolar epithelial barrier function and alveolar edema clearance by targeting the Na,K-ATPase and epithelial Na+ channels upon lung injury. Notably, the proteasomal system also exhibits distinct functions in the extracellular space, which may contribute to the pathogenesis of ALI and other pulmonary diseases. Better understanding of these mechanisms may ultimately lead to novel therapeutic modalities by targeting elements of the ubiquitination pathway.
The Ubiquitin Proteasome System
The Discovery of the Ubiquitin Proteasome System
Protein turnover plays an important role in a broad spectrum of cellular processes. Notably, approximately 3% to 5% of our cellular proteins are degraded and resynthesized daily.1 Until the mid-1970s it was widely believed that all intracellular proteins were degraded at the same rate in lysosomes in a nonspecific manner, despite the fact that proteins have a wide range of half-lives.1 The existence of a novel, highly regulated, and specific degradation pathway was first proposed by Ciechanover and colleagues2 and Hershko and colleagues,3 which was in agreement with the discovery of an ATP-dependent proteolytic system in reticulocytes that lack lysosomes.4 As it was described 2 years later, this novel degradation pathway was mediated by a small protein of 76 amino acid residues, termed ATP-dependent proteolysis factor-1 or ubiquitin.5,6
The Ubiquitin Enzymatic Cascade
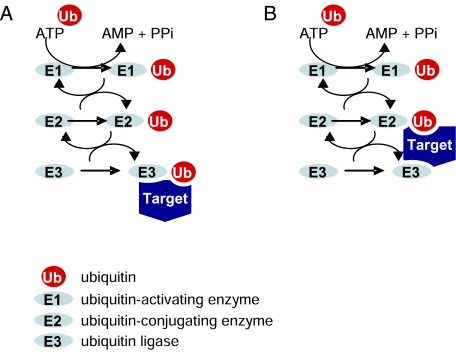
During ubiquitination, target proteins become “marked” as ubiquitin covalently conjugates via its C-terminal glycine to protein substrates at the ε-amino group of their lysine residues.7 Conjugation of ubiquitin to a target protein occurs via an enzymatic cascade.1,7 Ubiquitin is first activated by an ATP-dependent E1 ubiquitin-activating enzyme by the formation of a thiolester bond between the E1 enzyme and ubiquitin. Ubiquitin is then transferred via a transthiolation reaction to an E2 ubiquitin-conjugating enzyme, which in turn transfers ubiquitin onto the target protein. This conjugation step occurs in conjunction with E3 ubiquitin ligases, which provide the specificity of the cascade by recognizing the target proteins selected for ubiquitination. Three major E3 classes exist. The so-called zinc finger-containing really interesting new gene (RING) ligases facilitate transfer of ubiquitin from an E2 to substrates. In contrast, the homologous to E6-AP carboxyl terminus (HECT) ligases directly bind ubiquitin and transfer it onto the substrate.8,9 Recently, a novel E3 ligase family, the U box ubiquitin ligases, has been identified.10 A schematic representation of the ubiquitin enzymatic cascade is depicted in Figure 1.
Figure 1.
Schematic representation of ubiquitin enzymatic cascade. Ubiquitin is first activated by an ATP-dependent E1 ubiquitin-activating enzyme and then transferred to an E2 ubiquitin-conjugating enzyme, which in turn transfers ubiquitin onto the target protein. A, Homologous to E6-AP carboxyl terminus (HECT) HECT E3 ligases bind ubiquitin and subsequently transfer it onto the substrate. B, In contrast, really interesting new gene (RING) and U box ubiquitin ligases facilitate the transfer of ubiquitin from the E2 directly to the target. PPi = inorganic pyrophosphate.
The specificity and diversity of ubiquitination is a consequence of the hierarchical cascade of ubiquitin conjugation. Although in most eukaryotes there are only one or two E1s, the number of E2s is significantly larger (∼50-100 in mammals); it is estimated that >1,000 different E3 ligases exist, and the system comprises ∼5% of the genome.11 The diversity of the effects of ubiquitination is further enhanced by the various ways ubiquitin can tag a protein.12 In some cases, a single ubiquitin molecule attaches to one lysine (monoubiquitination) or to multiple lysine residues (multi-monoubiquitination) of the target protein; however, frequently polyubiquitin chains are generated that link several ubiquitin molecules via any of the seven lysine residues present in ubiquitin (K6, K11, K27, K29, K33, K48, and K63). Therefore, dependent on the internal linkages, the chains can be not only of varying lengths but also of varying types, further increasing the versatility of ubiquitination.13,14 Finally, the activity of approximately 100 deubiquitinating enzymes is responsible for the reversible nature of ubiquitination.15,16
Depending on subcellular localization and the number and topology of ubiquitin molecules conjugated to the target, the fate of the tagged molecule can be degradation or nonproteolytic modification.17 Degradation of damaged or abnormal proteins, as well as most proteins with short half-lives that control, for example, cell cycle, transcription or DNA repair is mediated by the 26S proteasome,18 a large 2.5 MDa protein complex consisting of two subcomplexes: the catalytic core 20S proteasome and the regulatory 19S proteasome.19 The proteolytic activity of the 20S proteasome is constituted by chymotrypsin-, trypsin-, and caspase-like activities.19 The proteasome cleaves tagged proteins into short oligopeptides while ubiquitin is detached and recycled.18 The proteasome also plays an important role in generating the major histocompatibility complex class 1-presented peptides by degrading proteins of intracellular pathogens.20
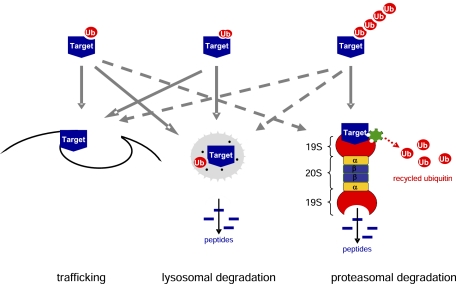
Usually, a polyubiquitin chain consisting of four or more ubiquitins confers recognition by the 26S proteasome; however, not all ubiquitinated targets are degraded in the proteasome (Fig 2).17 For example, a single ubiquitin tag often acts as an intracellular sorting signal affecting trafficking of the target molecule, such as internalization from the plasma membrane, sorting in endosomes, or Golgi-to-endosome trafficking.21 Moreover, the fate of the target protein is also determined by the type of conjugations between the ubiquitin molecule and the target and between the ubiquitin molecules. As described previously, conjugations between ubiquitin molecules may occur through any of the seven lysine residues inducing various effects; however, K48-linked polyubiquitination usually targets the protein to proteasomal degradation, whereas lysosomal sorting and trafficking are mainly regulated by K63-linked monoubiquitin or multi-monoubiquitin tags.12
Figure 2.
Types of ubiquitin tagging and fate of the target protein. A, A single ubiquitin molecule may attach to one lysine (monoubiquitination) of the target protein; or B, A single ubiquitin molecule may attach to multiple lysine residues (multi-monoubiquitination) of the target protein. C, Alternatively, polyubiquitin chains are generated that link several ubiquitin molecules via lysine residues present in ubiquitin. Usually, monoubiquitination and multi-monoubiquitination promote trafficking or lysosomal degradation of the target protein, whereas polyubiquitin chains consisting of four or more ubiquitins confer recognition by the 26S proteasome; however, trafficking or lysosomal degradation of proteins after polyubiquitination and proteasomal degradation after monoubiquitination have also been reported. See Figure 1 legend for expansion of abbreviation.
Role of Ubiquitination in Respiratory Diseases
Role of Ubiquitination in Chronic Inflammation, Fibrosis, and Muscle Dysfunction
Although ubiquitination has been demonstrated to play a role in a myriad of various cellular functions, its function in respiratory disease and particularly in acute lung injury (ALI) remains poorly understood. Only in the last few years have we started to gain insights into some of these mechanisms. We now know that regulation of inflammation by ubiquitination plays an important role in the pathogenesis of asthma and COPD. For example, the nuclear factor κ-light-chain-enhancer of activated B cells pathway, which has been implicated with a key role in the pathogenesis of both asthma and COPD by playing a pivotal role in adaptation to noxious stimuli and inflammation, is critically regulated by ubiquitination.22 Another mechanism that may contribute to chronic inflammation in these patient populations is the dysregulation of histone deacetylation. Histone deacetylation exerts antiinflammatory effects by repressing inflammatory genes. However, in patients with COPD and smoking patients with asthma, oxidative and nitrative stress results in the formation of peroxynitrite, which nitrates tyrosine residues on histone deacetylase-2, thereby resulting in its ubiquitination and degradation and, thus, chronic inflammation.23 Moreover, regulation of the human platelet-activating factor receptor by ubiquitination has also been implied in the pathogenesis of asthma.24 Furthermore, increasing evidence suggest that regulation of the transforming growth factor-β signaling cascade by ubiquitination plays an important role in the pathogenesis of lung fibrosis.25
Several studies focused on the role of ubiquitination in muscle dysfunction in respiratory diseases. Muscle wasting and impaired muscle function have been well documented in several chronic pulmonary diseases, such as COPD, interstitial lung disease, and cystic fibrosis, and also represent typical consequences of delayed recovery in critically ill patients.26,27 It is now increasingly evident that muscle weakness during respiratory disease is not a passive event but is rather mediated by ubiquitination and subsequent degradation of muscle fibers.28 For example, in an experimental rat ICU model, in which animals were mechanically ventilated, sedated, and pharmacologically paralyzed for up to 14 days, severe muscle wasting and paralysis similar to that observed among ICU patients was reported.28 Interestingly, the marked decrease in muscle fiber force generation capacity was associated with a sequential change in the localization of the muscle-specific RING finger proteins 1/2 leading to degradation and transcriptional downregulation of the molecular motor protein myosin, suggesting that ubiquitination played a central role in both muscle proteolysis and synthesis.28
Furthermore, it is well known that diaphragmatic function is critically important for successful weaning of patients from mechanical ventilation. However, mechanical ventilation itself has been shown to result in a rapid loss of diaphragmatic strength. Notably, several recent studies suggested an involvement of ubiquitination in diaphragmatic weakness in mechanically ventilated patients.29-31 In a recent study, rapidly progressive diaphragmatic weakness and injury in ventilated patients was reported, which was associated with high expression of ubiquitin and other proteolysis-related factors.29 Another study also identified a marked increase in the ubiquitin-proteasome activity as well as atrophic protein kinase B-forkhead Box O signaling, leading to degradation of myosin and thus resulting in myofiber atrophy and reduced diaphragm force-generating capacity in patients on mechanical ventilation.30 Moreover, a recent manuscript reported the potential involvement of the autophagy-lysosomal pathway secondary to activation of the E3 ligase, muscle-specific RING finger protein 1, in diaphragmatic protein degradation in ventilated patients.31
Ubiquitination in ALI
Role of Ubiquitination in Acute Lung Inflammation and Surfactant Dysregulation
Ubiquitination may also regulate the inflammatory responses initiating ALI.32 The E3 ubiquitin ligase Cblb, which plays an important role in inflammation and autoimmunity, has been shown to regulate the Toll-like receptor 4-mediated acute lung inflammation induced by polymicrobial sepsis, thereby impairing lung injury and survival, suggesting that ubiquitination may play a role in the initiation of ALI secondary to sepsis and lung inflammation.32
Ubiquitination has also been proposed to play a role in the pathomechanism of ALI by altering surfactant function. In a recent study it has been demonstrated that lipopolysaccharide causes polyubiquitination and subsequent degradation of acyl-CoA:lysophosphatidylcholine acyltransferase, an enzyme critically involved in the synthesis of the bioactive surfactant phospholipid, dipalmitoylphosphatidylcholine, using the Skp1-Cullin-F-box ubiquitin E3 ligase component, β-transducin repeat-containing protein.33 Furthermore, in a murine model of Pseudomonas aeruginosa-induced surfactant deficiency, which was characterized by the loss of CTP:phosphocholine cytidylyltransferase α, a nuclear enzyme that catalyzes the rate-limiting step in phosphatidylcholine synthesis, lung injury could be prevented by inhibition of the Skp1-Cullin-F-box E3 ligase, which was shown to monoubiquitinate CTP:phosphocholine cytidylyltransferase α leading to its degradation. Thus, ubiquitination may be critically important in surfactant homeostasis and ALI.34
Ubiquitination in Alveolar Epithelial Barrier Dysfunction
Ubiquitination has been proposed to play an important role in alveolar epithelial dysfunction during ALI. Accumulating data suggest that ubiquitination may regulate structural components of the alveolar epithelial monolayer. Structural integrity of epithelial cells and intercellular junctions play an important role in the maintenance of alveolar epithelial barrier integrity. For example, keratin intermediate filaments, a major cytoskeletal component of epithelial cells, are critically involved in alveolar barrier function. Interestingly, in response to injury, keratin intermediate filaments may undergo rapid structural reorganization, thereby transmitting signals from the cell surface to intracellular regions of cell.35 This structural reorganization of keratin intermediate filaments, mediated by the UbcH5 family members and Ubc3 E2 ubiquitin-conjugating enzymes, has been recently shown to lead to disassembly and degradation of the keratin intermediate filaments, thereby contributing to barrier dysfunction.36 Similarly, it has been recently proposed that both assembly and degradation of tight junction and adherens junction proteins are tightly regulated by ubiquitination.37-39 Because of the potential therapeutic value, additional research addressing these events in the context of ALI is of high priority.
A key function of the alveolar epithelium is to maintain optimal alveolar fluid balance.40 Under physiologic conditions, the alveolar epithelial barrier is highly impermeable, and its apical surface is covered by a thin liquid film, the alveolar epithelial lining fluid (ELF), which facilitates maintenance of surface tension, gas exchange and host defense.41 During ALI and its more severe form, ARDS, which are characterized by an increase in alveolo-capillary barrier permeability resulting in flooding of the alveolar space, a life-threatening impairment of gas exchange occurs.42 Therefore, effective control of the ELF volume is of critical importance. Optimal ELF volume is achieved by alveolar fluid reabsorption, which is driven by the active transport of Na+ from the airspaces into the lung interstitium and the pulmonary circulation.40 This vectorial transport is contributed by the apically located epithelial Na+ channels (ENaC) and basolateral Na,K-ATPase, which generate an osmotic gradient leading to the movement of water from the alveolar space into the interstitium.40,43 Importantly, in the majority of patients with ALI/ARDS, the Na+ transport and thus the capacity to clear pulmonary edema is markedly impaired, which is associated with worse outcomes.43-45
Regulation of Na,K-ATPase and ENaC Function by Ubiquitination
Recently, ubiquitination has been implicated with a key role in regulating Na,K-ATPase46 and ENaC,47 thereby effecting alveolar epithelial barrier function and integrity. The Na,K-ATPase (also known as the Na+ pump) is a heterodimeric holoenzyme composed of a catalytic α and a regulatory β subunit.43 The Na,K-ATPase represents the only transporter by which alveolar epithelial cells actively remove sodium; furthermore, its expression and function are also intimately coupled to that of junctional complexes.43,48 Therefore, the Na,K-ATPase plays a central role in both prevention of alveolar edema formation and clearance of lung edema fluid. Ubiquitination of the catalytic α1 and α2 subunits of the Na,K-ATPase was first reported almost 15 years ago in transformed kidney fibroblast cells, and although its physiologic role was not experimentally demonstrated, the authors hypothesized that ubiquitination may drive degradation of misassembled Na,K-ATPase at the endoplasmic reticulum and/or endocytosis and degradation of Na,K-ATPase located at the plasma membrane.49 Another study established that oxidative stress-induced Na,K-ATPase degradation could be prevented by inhibiting either lysosomal or proteasomal degradation in kidney proximal tubule cells.50
The regulation of Na,K-ATPase function by ubiquitination in the alveolar epithelium has been extensively studied in the context of hypoxia, a hallmark of ALI/ARDS.43 The first of these studies showed that during hypoxia the α1 subunit of the Na,K-ATPase underwent polyubiquitination at the plasma membrane leading to Na,K-ATPase degradation that could be prevented by using a mutant of the ubiquitin-activating enzyme E1.51 Interestingly, Na,K-ATPase degradation could be prevented by both proteasomal and lysosomal inhibitors, suggesting that perhaps both proteasomal and lysosomal degradation pathways are involved in the hypoxia-induced Na,K-ATPase down-regulation.51,52 Another study further characterized the nature of ubiquitination and degradation of the Na,K-ATPase and demonstrated the critical involvement of four lysine molecules on the N-terminus of the α1 subunit of the Na,K-ATPase in its ubiquitination.53 Interestingly, these lysines were in the immediate proximity of the S18 residue, the phosphorylation site for protein kinase C-ζ, a key regulator of Na,K-ATPase cell surface abundance.54-57 Furthermore, mutation of the S18 residue to alanine, which prevented phosphorylation of the Na,K-ATPase by protein kinase C-ζ, also prevented the hypoxia-induced ubiquitination, endocytosis, and subsequent degradation of the Na+ pump, suggesting a cross-talk between phosphorylation and ubiquitination.53
Although it is evident that the α subunit of the Na,K-ATPase undergoes ubiquitination upon exposing the alveolar epithelium to hypoxia, the E3 ligase promoting ubiquitination of the Na,K-ATPase remains unidentified. It has been previously reported that Ubc5, an E2 ubiquitin-conjugating enzyme, is necessary for the hypoxia-induced trafficking and degradation of the Na+ pump. Notably, Ubc5 is the upstream regulator of the E3 ligase, von Hippel Lindau protein (pVHL), a key regulator of hypoxia that is involved in the ubiquitination and degradation of the hypoxia-inducible factor-1, and thus it was hypothesized that pVHL may also serve as the E3 ligase for the Na,K-ATPase.58 This hypothesis was further supported by the observation that pVHL was required for the degradation of the Na+ pump upon exposure to hypoxia.58 However, this study also demonstrated that pVHL did not directly ubiquitinate the Na,K-ATPase, suggesting that pVHL was probably involved in the ubiquitination of an adaptor protein or a protein upstream of the Na,K-ATPase.58 Thus, the E3 ligase for the Na,K-ATPase remains to be identified. Since the Na,K-ATPase plays a central role in alveolar epithelial barrier integrity, and its dysfunction is associated with worse outcomes in ALI/ARDS, identification of the E3 ligase that mediates Na,K-ATPase degradation may be of high clinical relevance.
In contrast, the E3 ligase mediating ubiquitination of ENaC was identified more than 15 years ago.59 Two excellent recent reviews by Rotin and Staub47,60 detailed this discovery together with our current understanding of ENaC regulation by ubiquitination; therefore, we will only mention some key aspects of this important research in the current review. ENaC is comprised of an α, a β, and a γ subunit with a short proline-rich sequence, called the PY motif, present at the C terminus of each subunit, which serves as the binding site for the E3 ligase Nedd4-2.47,59 It has been demonstrated that Nedd4-2 can ubiquitinate ENaC at the apical plasma membrane of epithelial cells, which leads to internalization of the channel.60 The nature of ENaC ubiquitination remains unclear. Although most studies suggest that ENaC undergoes either monoubiquitination or multi-monoubiquitination at the cell surface and subsequent endocytosis, some reports also demonstrated that ENaC may be polyubiquitinated and degraded in the proteasome.47 Of note, proteolytic cleavage of the ectodomain of ENaC by serine proteases leads to activation of the channel by increasing its open probability. Interestingly, ubiquitination seems to regulate the conformation of the extracellular domain and thus the accessibility of protease sites for cleavage. However, since the protease cleavage releases the inhibitory tracts of ENaC, the cleaved subunits are constitutively active, and this activity can only be inhibited if the channel was removed from the cell surface, which is also exhibited by ubiquitination.60 Therefore, ubiquitination plays a role in both activation of ENaC by promoting its maturation and deactivation/inhibition of the channel by initiating its endocytosis.
The Extracellular Proteasome
Biologic Roles and Potential Therapeutic Value of the Extracellular Proteasome System
Although the classic description of the proteasome degradation pathway is limited to the intracellular environment, recent evidence points to the presence and possible activity of extracellular proteasome.25,61,62 This has been surmised to represent a marker of certain diseases63-65; however, studies have suggested a potential immunomodulatory role for extracellular proteasome in certain groups of patients.66 In order to demonstrate that the proteasome might have a specific extracellular biologic function, as opposed to being a marker of cell death, multiple studies have shown that proteasome particles are bound to membranes. One found that when lipopolysaccharide was added to a macrophage membrane preparation, it was specifically bound to 20S proteasome attached membranes.61,67 Other studies have demonstrated that proteasomes bind both endoplasmic reticulum and mitochondrial membranes.61,68-70 Nakagawa and colleagues70 have established that FK506-binding protein 38, a member of the immunophilin family of endoplasmic reticulum and mitochondrial membrane proteins, binds multiple subunits of the 26S proteasome and acts to anchor the proteasome at the organelle membrane. In addition, proteasomes have been found on the cell membrane surface, detectable by proteasome subunit-specific antibodies and flow cytometry on the surface of nonpermeabilized human leukemic cell line U937 and human T and B lymphocytes.61,71,72
Despite research showing that proteasomes interact with phospholipid membranes, there is no evidence that explains how proteasomes may be secreted or released from cells, except as a result of cell death. Zoeger and colleagues73 compared the function and subtype of circulating proteasomes in patients with rheumatoid arthritis, patients with systemic lupus erythematosus, and healthy volunteers. Circulating proteasomes from all groups were found to be intact and enzymatically active. However, when subtypes were analyzed with high-resolution anion exchange chromatography, the subtypes from patients with rheumatoid arthritis and systemic lupus erythematosus were found to be distinct from those from proteasome recovered from healthy volunteer hematologic cells.73 Related to the finding of increased circulating 20S proteasome, increased serum or plasma levels of ubiquitin have been detected in multiple conditions, including parasitic infection, renal failure and hemodialysis, cirrhosis, type 2 diabetes mellitus, hairy cell leukemia, and sepsis.61,74,75 In addition, it has been proposed that extracellular ubiquitin and proteasome may have therapeutic potential. One study of swine infused with endotoxin found that exogenous ubiquitin infusion reduced mortality, respiratory failure, fluid requirements, erythema, and edema over placebo.76 Intriguingly, there is also evidence that extracellular ubiquitin may have antimicrobial properties as well.77 As there is documented ubiquitin protease activity in several viruses, bacteria, and protozoa,78 the potential for uncovering a pathogen-host ubiquitin and proteasome pathway interaction is promising.
Extracellular Proteasome Function in Sepsis and ALI
One inflammatory state in which the presence of extracellular proteasome has been evaluated is critical illness. A small case-control study of critically ill patients found significantly higher serum levels of 20S proteasome in 15 patients admitted to the ICU with a diagnosis of sepsis and 13 with a diagnosis of trauma compared with 15 patients admitted for elective abdominal surgery and 15 healthy volunteers.79 A mouse model of sepsis found that infusion of the proteasome inhibitor MG-132 significantly dampened tumor necrosis factor-α, IL-1, and IL-10 levels and prolonged survival compared with placebo.80 This finding may suggest that circulating proteasome interacts with the inflammatory response in sepsis, implying that the proteasome may have important immunomodulatory functions in patients with critical illness such as sepsis.
Research has also focused on extracellular proteasome activity in patients with the ALI. A small study illustrates the presence and activity of proteasome in the alveolar space.81 All three proteasome activities were identified in a cell-free supernatant of BAL fluid and were inhibited by epoxomicin. Cell lysis was also ruled out as a potential source of alveolar proteasome. Finally, BAL supernatant successfully cleaved albumin in an ATP- and ubiquitin-independent manner.81 These data point to the presence and biologic activity of proteasome in the alveolar space; furthermore, they imply a unique, ubiquitin-independent role for extracellular proteasome activity. The previous investigation could have profound implications on human pulmonary diseases, particularly those that involve an alveolar filling process that alters intra-alveolar protein makeup or oncotic pressure. This was explored in a remarkable study by the same authors in patients with ALI showing a significantly increased alveolar proteasome concentration but 17-fold decreased proteasomal activity compared with healthy control subjects. In addition, alveolar proteasome activity in BAL from healthy patients was found to be inhibited by the addition of BAL from patients with ALI.82 Thus, an inhibitor of proteasome activity may exist in the alveolar space, which could play a role in the increased intraalveolar protein concentration and oncotic pressure in ALI.
Potential Therapeutic Approaches Targeting Elements of the Ubiquitin System
Since ubiquitination plays a central role in the pathogenesis of several disease states, it is logical that pharmacologic inhibitors of the elements of the ubiquitination cascade may serve as novel therapeutic modalities. The first pharmacologic agents developed targeted the proteasome. These typically short peptide molecules mimic the substrate and inhibit the threonine residue in the 20S active site of the proteasome. Several synthetic proteasome inhibitors are available, which can be distinguished by their chemical properties. For example, MG-132, a cell-permeable peptide aldehyde, which is an effective and reversible inhibitor of the proteasome, is widely used for in vitro studies; however, its nonspecificity (inhibition of serine and cysteine proteases) precludes its clinical application.25 More importantly, Bortezomib, a boronic acid peptide that specifically and reversibly inhibits the chymotrypsin-like activity of the 26S proteasome in mammalian cells, has recently been approved as the first proteasomal inhibitor by the US Food and Drug Administration for the treatment of multiple myeloma and mantel cell lymphoma.83 Two novel and irreversible nonpeptide protease inhibitors, carfilzomib (also known as PR-171) and salinosporamide A (also known as NPI-0052), which were derived from lactacystin, a naturally occurring proteasome inhibitor produced by Streptomyces lactacystinaeus, are currently in clinical trials.84
Notably, not all proteins ubiquitinated undergo proteasomal degradation. Thus, to specifically interfere with ubiquitination of the target protein it appears logical that the interaction between the specific E3 ligase and the substrate must be inhibited. However, targeting E3 ligase-substrate associations remains challenging because of the limited information on the various partnering motifs and the weak nature of these interactions, which often occur along large surfaces.83
Most recent studies targeted regulatory elements of the ubiquitin cascade. One such study targeted the neural precursor cell expressed, developmentally downregulated 8 (NEDD8)-activating enzyme, an essential component of the NEDD8 conjugation pathway that controls the activity of the cullin-RING subtype of E3 ligases, the substrates of which have important roles in cancer cell growth. Interestingly, MLN4924, a potent and selective novel inhibitor of the NEDD8-activating enzyme, disrupted the cullin-RING ligase-mediated protein turnover, thereby suppressing tumor growth in mice.85 Thus, targeting elements of the ubiquitin proteasome system may hold promise for the treatment of various diseases.
Conclusions
Ubiquitination is a posttranslational modification that regulates a wide variety of cellular functions. The versatility of ubiquitination effects relies on timing, subcellular localization, and type of tagging, as well as modulators of ubiquitin binding. Upon ubiquitination, tagged proteins may undergo proteasomal or lysosomal degradation or other nonproteolytic modifications. Ubiquitination appears to play an important role in the pathogenesis of ALI and alveolar epithelial barrier dysfunction upon injury. Particularly, the regulation of alveolar edema clearance by targeting ENaC and the Na,K-ATPase has been a focus of intense research in the last few years. Interestingly, the proteasomal system also exhibits distinct functions in the extracellular space, thereby contributing to ALI and other pulmonary diseases, which may have therapeutic implications. This is a relatively new field in the research of pulmonary and critical care diseases and provides a platform for better understanding of mechanisms, which may lead to novel therapeutic modalities by targeting elements of the highly specific ubiquitination pathway.
Acknowledgments
Financial/nonfinancial disclosures: The authors have reported to CHEST the following conflicts of interest: Dr Sznajder receives a stipend from the American Thoracic Society as the editor of the American Journal of Respiratory and Critical Care Medicine. Drs Vadász and Weiss have reported that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: The sponsors had no role in the design of the study, the collection and analysis of the data, or in the preparation of the manuscript.
Abbreviations
- ALI
acute lung injury
- ARDS
acute respiratory distress syndrome
- ELF
epithelial lining fluid
- ENaC
epithelial Na+ channels
- NEDD8
neural precursor cell expressed, developmentally downregulated 8
- pVHL
von Hippel Lindau protein
- RING
really interesting new gene
Footnotes
Funding/Support: Research in the authors’ laboratories addressing ubiquitination was supported by the Deutsche Forschungsgemeinschaft [Grant DFG/IRTG1062] to Drs Vadász and Sznajder, the University Medical Center Giessen and Marburg [Grant 62589064] to Dr Vadász, and the National Institutes of Health [Grants HL-71643, R37 48129, HL-85534, and HL-T32 76139] to Drs Weiss and Sznajder. Dr Vadász is supported by the Else Kröner Memorial Award, and Dr Weiss is the recipient of Parker B. Francis Fellowship.
Reproduction of this article is prohibited without written permission from the American College of Chest Physicians (http://www.chestpubs.org/site/misc/reprints.xhtml).
References
- 1.Ciechanover A. The ubiquitin proteolytic system: from an idea to the patient bed. Proc Am Thorac Soc. 2006;3(1):21–31. doi: 10.1513/pats.200510-106JH. [DOI] [PubMed] [Google Scholar]
- 2.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81(4):1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A, Rose IA. Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc Natl Acad Sci U S A. 1979;76(7):3107–3110. doi: 10.1073/pnas.76.7.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etlinger JD, Goldberg AL. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc Natl Acad Sci U S A. 1977;74(1):54–58. doi: 10.1073/pnas.74.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciechanover A, Elias S, Heller H, Ferber S, Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem. 1980;255(16):7525–7528. [PubMed] [Google Scholar]
- 6.Wilkinson KD, Urban MK, Haas AL. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980;255(16):7529–7532. [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102(5):549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 9.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92(11):5249. doi: 10.1073/pnas.92.11.5249-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302(4):635–645. doi: 10.1016/s0006-291x(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 11.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 12.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2(3):169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 13.Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010;38(5):627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 14.Xu P, Duong DM, Seyfried NT, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papa FR, Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366(6453):313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 16.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao M, Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol Cell. 2005;19(5):581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Nickell S, Beck F, Scheres SH, et al. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci U S A. 2009;106(29):11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie Y. Structure, assembly and homeostatic regulation of the 26S proteasome. J Mol Cell Biol. 2010;2(6):308–317. doi: 10.1093/jmcb/mjq030. [DOI] [PubMed] [Google Scholar]
- 20.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 21.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 22.Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2(3):a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes PJ. Role of HDAC2 in the pathophysiology of COPD. Annu Rev Physiol. 2009;71:451–464. doi: 10.1146/annurev.physiol.010908.163257. [DOI] [PubMed] [Google Scholar]
- 24.Dupré DJ, Chen Z, Le Gouill C, et al. Trafficking, ubiquitination, and down-regulation of the human platelet-activating factor receptor. J Biol Chem. 2003;278(48):48228–48235. doi: 10.1074/jbc.M304082200. [DOI] [PubMed] [Google Scholar]
- 25.Weiss CH, Budinger GR, Mutlu GM, Jain M. Proteasomal regulation of pulmonary fibrosis. Proc Am Thorac Soc. 2010;7(1):77–83. doi: 10.1513/pats.200906-055JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rüegg MA, Glass DJ. Molecular mechanisms and treatment options for muscle wasting diseases. Annu Rev Pharmacol Toxicol. 2011;51:373–395. doi: 10.1146/annurev-pharmtox-010510-100537. [DOI] [PubMed] [Google Scholar]
- 27.Herridge MS, Tansey CM, Matté A, et al. Canadian Critical Care Trials Group. Functional disability 5 years after acute respiratory distress syndrome. N Engl J Med. 2011;364(14):1293–1304. doi: 10.1056/NEJMoa1011802. [DOI] [PubMed] [Google Scholar]
- 28.Ochala J, Gustafson AM, Diez ML, et al. Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. J Physiol. 2011;589(Pt 8):2007–2026. doi: 10.1113/jphysiol.2010.202044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaber S, Petrof BJ, Jung B, et al. Rapidly progressive diaphragmatic weakness and injury during mechanical ventilation in humans. Am J Respir Crit Care Med. 2011;183(3):364–371. doi: 10.1164/rccm.201004-0670OC. [DOI] [PubMed] [Google Scholar]
- 30.Levine S, Biswas C, Dierov J, et al. Increased proteolysis, myosin depletion, and atrophic AKT-FOXO signaling in human diaphragm disuse. Am J Respir Crit Care Med. 2011;183(4):483–490. doi: 10.1164/rccm.200910-1487OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain SN, Mofarrahi M, Sigala I, et al. Mechanical ventilation-induced diaphragm disuse in humans triggers autophagy. Am J Respir Crit Care Med. 2010;182(11):1377–1386. doi: 10.1164/rccm.201002-0234OC. [DOI] [PubMed] [Google Scholar]
- 32.Bachmaier K, Toya S, Gao X, et al. E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat Med. 2007;13(8):920–926. doi: 10.1038/nm1607. [DOI] [PubMed] [Google Scholar]
- 33.Zou C, Ellis BM, Smith RM, et al. Acyl-CoA:lysophosphatidylcholine acyltransferase I (Lpcat1) catalyzes histone protein O-palmitoylation to regulate mRNA synthesis. J Biol Chem. 2011;286(32):28019–28025. doi: 10.1074/jbc.M111.253385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen BB, Coon TA, Glasser JR, Mallampalli RK. Calmodulin antagonizes a calcium-activated SCF ubiquitin E3 ligase subunit, FBXL2, to regulate surfactant homeostasis. Mol Cell Biol. 2011;31(9):1905–1920. doi: 10.1128/MCB.00723-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogel MR, Jaitovich A, Ridge KM. The role of the ubiquitin proteasome pathway in keratin intermediate filament protein degradation. Proc Am Thorac Soc. 2010;7(1):71–76. doi: 10.1513/pats.200908-089JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaitovich A, Mehta S, Na N, Ciechanover A, Goldman RD, Ridge KM. Ubiquitin-proteasome-mediated degradation of keratin intermediate filaments in mechanically stimulated A549 cells. J Biol Chem. 2008;283(37):25348–25355. doi: 10.1074/jbc.M801635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traweger A, Fang D, Liu YC, et al. The tight junction-specific protein occludin is a functional target of the E3 ubiquitin-protein ligase itch. J Biol Chem. 2002;277(12):10201–10208. doi: 10.1074/jbc.M111384200. [DOI] [PubMed] [Google Scholar]
- 38.Théard D, Labarrade F, Partisani M, et al. USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 2010;29(9):1499–1509. doi: 10.1038/emboj.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y, Hirsch DS, Sasiela CA, Wu WJ. Cdc42 regulates E-cadherin ubiquitination and degradation through an epidermal growth factor receptor to Src-mediated pathway. J Biol Chem. 2008;283(8):5127–5137. doi: 10.1074/jbc.M703300200. [DOI] [PubMed] [Google Scholar]
- 40.Matthay MA, Folkesson HG, Clerici C. Lung epithelial fluid transport and the resolution of pulmonary edema. Physiol Rev. 2002;82(3):569–600. doi: 10.1152/physrev.00003.2002. [DOI] [PubMed] [Google Scholar]
- 41.Weibel ER. Morphological basis of alveolar-capillary gas exchange. Physiol Rev. 1973;53(2):419–495. doi: 10.1152/physrev.1973.53.2.419. [DOI] [PubMed] [Google Scholar]
- 42.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med. 2000;342(18):1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 43.Vadász I, Raviv S, Sznajder JI. Alveolar epithelium and Na,K-ATPase in acute lung injury. Intensive Care Med. 2007;33(7):1243–1251. doi: 10.1007/s00134-007-0661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ware LB, Matthay MA. Alveolar fluid clearance is impaired in the majority of patients with acute lung injury and the acute respiratory distress syndrome. Am J Respir Crit Care Med. 2001;163(6):1376–1383. doi: 10.1164/ajrccm.163.6.2004035. [DOI] [PubMed] [Google Scholar]
- 45.Sznajder JI. Alveolar edema must be cleared for the acute respiratory distress syndrome patient to survive. Am J Respir Crit Care Med. 2001;163(6):1293–1294. doi: 10.1164/ajrccm.163.6.ed1801d. [DOI] [PubMed] [Google Scholar]
- 46.Helenius IT, Dada LA, Sznajder JI. Role of ubiquitination in Na,K-ATPase regulation during lung injury. Proc Am Thorac Soc. 2010;7(1):65–70. doi: 10.1513/pats.200907-082JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86(2):669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- 48.Vadász I, Schermuly RT, Ghofrani HA, et al. The lectin-like domain of tumor necrosis factor-alpha improves alveolar fluid balance in injured isolated rabbit lungs. Crit Care Med. 2008;36(5):1543–1550. doi: 10.1097/CCM.0b013e31816f485e. [DOI] [PubMed] [Google Scholar]
- 49.Coppi MV, Guidotti G. Ubiquitination of Na,K-ATPase alpha1 and alpha2 subunits. FEBS Lett. 1997;405(3):281–284. doi: 10.1016/s0014-5793(97)00182-8. [DOI] [PubMed] [Google Scholar]
- 50.Thévenod F, Friedmann JM. Cadmium-mediated oxidative stress in kidney proximal tubule cells induces degradation of Na+/K(+)-ATPase through proteasomal and endo-/lysosomal proteolytic pathways. FASEB J. 1999;13(13):1751–1761. doi: 10.1096/fasebj.13.13.1751. [DOI] [PubMed] [Google Scholar]
- 51.Comellas AP, Dada LA, Lecuona E, et al. Hypoxia-mediated degradation of Na,K-ATPase via mitochondrial reactive oxygen species and the ubiquitin-conjugating system. Circ Res. 2006;98(10):1314–1322. doi: 10.1161/01.RES.0000222418.99976.1d. [DOI] [PubMed] [Google Scholar]
- 52.Lecuona E, Sun H, Vohwinkel C, Ciechanover A, Sznajder JI. Ubiquitination participates in the lysosomal degradation of Na,K-ATPase in steady-state conditions. Am J Respir Cell Mol Biol. 2009;41(6):671–679. doi: 10.1165/rcmb.2008-0365OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dada LA, Welch LC, Zhou G, Ben-Saadon R, Ciechanover A, Sznajder JI. Phosphorylation and ubiquitination are necessary for Na,K-ATPase endocytosis during hypoxia. Cell Signal. 2007;19(9):1893–1898. doi: 10.1016/j.cellsig.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dada LA, Chandel NS, Ridge KM, Pedemonte C, Bertorello AM, Sznajder JI. Hypoxia-induced endocytosis of Na,K-ATPase in alveolar epithelial cells is mediated by mitochondrial reactive oxygen species and PKC-zeta. J Clin Invest. 2003;111(7):1057–1064. doi: 10.1172/JCI16826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Briva A, Vadász I, Lecuona E, et al. High CO2 levels impair alveolar epithelial function independently of pH. PLoS ONE. 2007;2(11):e1238. doi: 10.1371/journal.pone.0001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vadász I, Dada LA, Briva A, et al. AMP-activated protein kinase regulates CO2-induced alveolar epithelial dysfunction in rats and human cells by promoting Na,K-ATPase endocytosis. J Clin Invest. 2008;118(2):752–762. doi: 10.1172/JCI29723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vadász I, Morty RE, Olschewski A, et al. Thrombin impairs alveolar fluid clearance by promoting endocytosis of Na+,K+-ATPase. Am J Respir Cell Mol Biol. 2005;33(4):343–354. doi: 10.1165/rcmb.2004-0407OC. [DOI] [PubMed] [Google Scholar]
- 58.Zhou G, Dada LA, Chandel NS, et al. Hypoxia-mediated Na-K-ATPase degradation requires von Hippel Lindau protein. FASEB J. 2008;22(5):1335–1342. doi: 10.1096/fj.07-8369com. [DOI] [PubMed] [Google Scholar]
- 59.Staub O, Dho S, Henry P, et al. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15(10):2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 60.Rotin D, Staub O. Role of the ubiquitin system in regulating ion transport. Pflugers Arch. 2011;461(1):1–21. doi: 10.1007/s00424-010-0893-2. [DOI] [PubMed] [Google Scholar]
- 61.Sixt SU, Dahlmann B. Extracellular, circulating proteasomes and ubiquitin–incidence and relevance. Biochim Biophys Acta. 2008;1782(12):817–823. doi: 10.1016/j.bbadis.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Sixt SU, Peters J. Extracellular alveolar proteasome: possible role in lung injury and repair. Proc Am Thorac Soc. 2010;7(1):91–96. doi: 10.1513/pats.200906-035JS. [DOI] [PubMed] [Google Scholar]
- 63.Ostrowska H, Hempel D, Holub M, Sokolowski J, Kloczko J. Assessment of circulating proteasome chymotrypsin-like activity in plasma of patients with acute and chronic leukemias. Clin Biochem. 2008;41(16-17):1377–1383. doi: 10.1016/j.clinbiochem.2008.08.063. [DOI] [PubMed] [Google Scholar]
- 64.Henry L, Lavabre-Bertrand T, Vercambre L, et al. Plasma proteasome level is a reliable early marker of malignant transformation of liver cirrhosis. Gut. 2009;58(6):833–838. doi: 10.1136/gut.2008.157016. [DOI] [PubMed] [Google Scholar]
- 65.Lavabre-Bertrand T, Henry L, Carillo S, et al. Plasma proteasome level is a potential marker in patients with solid tumors and hemopoietic malignancies. Cancer. 2001;92(10):2493–2500. doi: 10.1002/1097-0142(20011115)92:10<2493::aid-cncr1599>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 66.Egerer K, Kuckelkorn U, Rudolph PE, et al. Circulating proteasomes are markers of cell damage and immunologic activity in autoimmune diseases. J Rheumatol. 2002;29(10):2045–2052. [PubMed] [Google Scholar]
- 67.Qureshi N, Perera PY, Shen J, et al. The proteasome as a lipopolysaccharide-binding protein in macrophages: differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J Immunol. 2003;171(3):1515–1525. doi: 10.4049/jimmunol.171.3.1515. [DOI] [PubMed] [Google Scholar]
- 68.Hori H, Nembai T, Miyata Y, Hayashi T, Ueno K, Koide T. Isolation and characterization of two 20S proteasomes from the endoplasmic reticulum of rat liver microsomes. J Biochem. 1999;126(4):722–730. doi: 10.1093/oxfordjournals.jbchem.a022509. [DOI] [PubMed] [Google Scholar]
- 69.Khan MT, Joseph SK. Characterization of membrane-associated proteasomes in WB rat liver epithelial cells. Arch Biochem Biophys. 2001;385(1):99–107. doi: 10.1006/abbi.2000.2116. [DOI] [PubMed] [Google Scholar]
- 70.Nakagawa T, Shirane M, Iemura S, Natsume T, Nakayama KI. Anchoring of the 26S proteasome to the organellar membrane by FKBP38. Genes Cells. 2007;12(6):709–719. doi: 10.1111/j.1365-2443.2007.01086.x. [DOI] [PubMed] [Google Scholar]
- 71.Bureau JP, Olink-Coux M, Brouard N, et al. Characterization of prosomes in human lymphocyte subpopulations and their presence as surface antigens. Exp Cell Res. 1997;231(1):50–60. doi: 10.1006/excr.1996.3453. [DOI] [PubMed] [Google Scholar]
- 72.Henry L, Baz A, Château MT, Scherrer K, Bureau JP. Changes in the amount and distribution of prosomal subunits during the differentiation of U937 myeloid cells: high expression of p23K. Cell Prolif. 1996;29(11):589–607. doi: 10.1111/j.1365-2184.1996.tb00974.x. [DOI] [PubMed] [Google Scholar]
- 73.Zoeger A, Blau M, Egerer K, Feist E, Dahlmann B. Circulating proteasomes are functional and have a subtype pattern distinct from 20S proteasomes in major blood cells. Clin Chem. 2006;52(11):2079–2086. doi: 10.1373/clinchem.2006.072496. [DOI] [PubMed] [Google Scholar]
- 74.Akarsu E, Pirim I, Capoğlu I, Deniz O, Akçay G, Unüvar N. Relationship between electroneurographic changes and serum ubiquitin levels in patients with type 2 diabetes. Diabetes Care. 2001;24(1):100–103. doi: 10.2337/diacare.24.1.100. [DOI] [PubMed] [Google Scholar]
- 75.Akarsu E, Pirim I, Selçuk NY, Tombul HZ, Cetinkaya R. Relation between serum ubiquitin levels and KT/V in chronic hemodialysis patients. Nephron. 2001;88(3):280–282. doi: 10.1159/000046005. [DOI] [PubMed] [Google Scholar]
- 76.Majetschak M, Cohn SM, Nelson JA, Burton EH, Obertacke U, Proctor KG. Effects of exogenous ubiquitin in lethal endotoxemia. Surgery. 2004;135(5):536–543. doi: 10.1016/j.surg.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 77.Kieffer AE, Goumon Y, Ruh O, et al. The N- and C-terminal fragments of ubiquitin are important for the antimicrobial activities. FASEB J. 2003;17(6):776–778. doi: 10.1096/fj.02-0699fje. [DOI] [PubMed] [Google Scholar]
- 78.Edelmann MJ, Kessler BM. Ubiquitin and ubiquitin-like specific proteases targeted by infectious pathogens: Emerging patterns and molecular principles. Biochim Biophys Acta. 2008;1782(12):809–816. doi: 10.1016/j.bbadis.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roth GA, Moser B, Krenn C, et al. Heightened levels of circulating 20S proteasome in critically ill patients. Eur J Clin Invest. 2005;35(6):399–403. doi: 10.1111/j.1365-2362.2005.01508.x. [DOI] [PubMed] [Google Scholar]
- 80.Safránek R, Ishibashi N, Oka Y, Ozasa H, Shirouzu K, Holecek M. Modulation of inflammatory response in sepsis by proteasome inhibition. Int J Exp Pathol. 2006;87(5):369–372. doi: 10.1111/j.1365-2613.2006.00490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sixt SU, Beiderlinden M, Jennissen HP, Peters J. Extracellular proteasome in the human alveolar space: a new housekeeping enzyme? Am J Physiol Lung Cell Mol Physiol. 2007;292(5):L1280–L1288. doi: 10.1152/ajplung.00140.2006. [DOI] [PubMed] [Google Scholar]
- 82.Sixt SU, Adamzik M, Spyrka D, et al. Alveolar extracellular 20S proteasome in patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2009;179(12):1098–1106. doi: 10.1164/rccm.200802-199OC. [DOI] [PubMed] [Google Scholar]
- 83.Navon A, Ciechanover A. The 26 S proteasome: from basic mechanisms to drug targeting. J Biol Chem. 2009;284(49):33713–33718. doi: 10.1074/jbc.R109.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103(13):1007–1017. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- 85.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]