Abstract
Constitutive defense mechanisms are critical to the understanding of defense mechanisms in conifers because they constitute the first barrier to attacks by insect pests. In interior spruce, trees that are putatively resistant and susceptible to attacks by white pine weevil (Pissodes strobi) typically exhibit constitutive differences in traits such as resin duct size and number, bark thickness, and terpene content. To improve our knowledge of their genetic basis, we compared globally the constitutive expression levels of 17,825 genes between 20 putatively resistant and 20 putatively susceptible interior spruce trees from the British Columbia tree improvement program. We identified 54 upregulated and 137 downregulated genes in resistant phenotypes, relative to susceptible phenotypes, with a maximum fold change of 2.24 and 3.91, respectively. We found a puzzling increase of resistance by downregulated genes, as one would think that “procuring armaments” is the best defense. Also, although terpenes and phenolic compounds play an important role in conifer defense, we found few of these genes to be differentially expressed. We found 15 putative small heat-shock proteins (sHSP) and several other stress-related proteins to be downregulated in resistant trees. Downregulated putative sHSP belong to several sHSP classes and represented 58% of all tested putative sHSP. These proteins are well known to be involved in plant response to various kinds of abiotic stress; however, their role in constitutive resistance is not yet understood. The lack of correspondence between transcriptome profile clusters and phenotype classifications suggests that weevil resistance in spruce is a complex trait.
Keywords: gene expression levels, microarray, constitutive resistance, sHSP
Introduction
The white pine weevil (Pissodes strobi) is a major pest of North American forests (Drouin and Langor 1991; Alfaro 1994; Hamid et al. 1995). The weevil primarily attacks Sitka spruce (Picea sitchensis), white spruce (Pi. glauca), and Engelmann spruce (Pi. engelmannii), but it can also attack several other pine and spruce species and even Douglas fir (Pseudotsuga menziesii). Adults lay eggs in the bark below the terminal bud cluster, and larva feed on the terminal leader. Such attacks can lead to leader death and consequential stem deformation, which is an economic cost to the forest industry (Alfaro 1994). Knowledge of the genetic mechanisms of weevil resistance in spruce would aid in developing marker-assisted breeding strategies for spruce and add to our knowledge about the diversity of resistance mechanisms in the plant kingdom.
In conifers as in other plants, resistance to insect pests involves both constitutive (pre-existing) and induced defenses. Constitutive defense mechanisms are both mechanical (resin ducts, parenchyma cells, and sclerenchyma) and chemical (oxalate crystals and accumulation of toxic or repellant molecules) (Hall et al. 2011). Induced defenses form a second line of defense, operating during or after pest attack. They are generally more specific in their action and include increases of resin flow and production of repellant or toxic chemicals or even de novo defenses (formation of traumatic resin ducts, callus formation, synthesis of new chemicals that are possibly specific to a given pest). Most workers regard induced defensive mechanisms to be the most important component of insect defense; however, constitutive resistance is less liable and easier to study and quantify in the context of quantitative genomics.
With regard to white spruce, several studies have identified constitutive features of resistance. Resistant trees possess a thinner bark, with a higher density of outer resin ducts and larger inner resin ducts (Tomlin and Borden 1994, 1997b; Alfaro et al. 2004). In interior spruce (Pi. glauca–engelmannii complex), resistance is positively correlated with tree growth (both height and trunk diameter), although weevils prefer to oviposit in longer leader shoots (Kiss and Yanchuk 1991; King et al. 1997). Gerson and Kelsey (2002) analyzed piperidine alkaloids contents of resistant and susceptible families of Sitka spruce, but they did not find any correlation with resistance to weevil ovipositing. With regard to terpenoids, Nault et al (1999) showed profiles to be good indicators of resistance in white spruce and Engelmann spruce. In Sitka spruce, resistant trees can show either a lower or a higher content of foliar terpenoids than susceptible trees, suggesting that they can use either repellency strategy (the tree try to repel the insects) or stealth strategy (the tree try to be less attractive to the insects; Tomlin et al. 1997). However, higher levels of a diterpene (dehydroabietic acid) and two monoterpenes ((+)-3-carene and terpinolene) are associated with resistance in Sitka spruce (Robert et al. 2010). Following this study, Hall et al. (2011) showed that the (+)-3-carene is produced by three different (+)-3-carene synthase genes. One was specific to resistant trees (PsTPS-3car2), one was specific to susceptible trees (PsTPS-3car3), and one is expressed in both phenotypes (PsTPS-3car1). They concluded that (+)-3-carene is explained by the variation in gene copy number, in gene sequence, in protein expression levels, and in enzyme activity levels.
The development of “omics” approaches and the development of several cDNA libraries within the Arborea I, II and Treenomix I, II spruce genome projects (http://www.arborea.ulaval.ca/; http://www.treenomix.ca; Pavy et al. 2005; Ralph, Yueh, et al. 2006; Ralph et al. 2008) opened insights into the nature of both constitutive and induced defense mechanisms in spruce. To date, most published studies have focused on induced defenses (Ralph, Yueh, et al. 2006; Lippert et al. 2007, 2009; Zulak et al. 2009; Robert et al. 2010; Hall et al. 2011). These studies compare the biological response with various types of induction (methyl jasmonate and chitosan elicitation, white pine weevil and western spruce budworm herbivory, mechanical wounding) at the transcriptome, proteome, and/or metabolome levels. However, induced and constitutive defenses are complementary and distinct defense mechanisms. Induced defenses take place when constitutive defenses have been defeated by an insect attack. Their primary function is to reinforce the constitutive defense mechanisms and add new barriers against the insect attack. Consequently, we might expect induced and constitutive defenses to have a different genetic basis. The purpose of this study was to investigate these differences.
The comparison of resistant and susceptible trees at the global transcriptome level has not yet been conducted, and such a comparison can provide fundamental and perhaps unexpected findings about the basis of insect resistance in conifers. Here, we present a comparative study of gene expression in interior spruce (Pi. glauca–engelmannii complex) aimed to identify candidate genes involved in constitutive defense against white pine weevil. We used a set of 180 trees previously ranked for resistance to this weevil by breeders in the British Columbia Ministry of Forests. Using a 17,825 member cDNA microarray, we compare gene expression levels between the 20 most resistant trees and the 20 most susceptible trees. Significantly upregulated and downregulated genes will identify a suite of genes involved in constitutive weevil resistance. Particular attention will be given to the putative small heat-shock proteins (sHSP) that evidently play an important role in constitutive defense.
Materials and Methods
Selection and Sampling
As part of the British Columbia (BC) interior spruce tree breeding program (Experimental Project EP 670), 180 trees were selected in wild stands across the Prince George region of central BC (fig. 1). The parent tree selection criteria were largely height superiority, stem form, branch size, and crown shape. Their ages varied from 100 to 200 years. Open-pollinated seeds were collected from each wild tree, and test seedlings for each parent tree were grown in nursery beds near Prince George. Progeny tests of all families were established in 1972 at Aleza Lake, near Prince George, and in 1973 at three other sites: the Prince George Tree Improvement Station (PGTIS), Quesnel, and Barbie Lake. In the mid-1980s, the PGTIS and Aleza Lake sites began to suffer severe and repeated attacks of white pine weevils. In 1988, presence or absence of weevil damage was recorded for all trees on both sites. Kiss and Yanchuk (1991) reported that family damage was consistent between the two sites (r = .71) and had a moderately strong genetic basis (hfamily2 = 0.77;hindividual2 = 0.18). King et al. (1997) reported similar results in other BC interior spruce populations. Based on these results, it appears that parental resistant scores can be readily estimated from weevil damage on their progenies. In 2003, all families on both sites were ranked according to the number of damaged trees, and the observed damage was used to estimate resistance levels of the 180 parent trees. In this study, the 20 least and 20 most damaged families were chosen as the resistant and susceptible families, respectively.
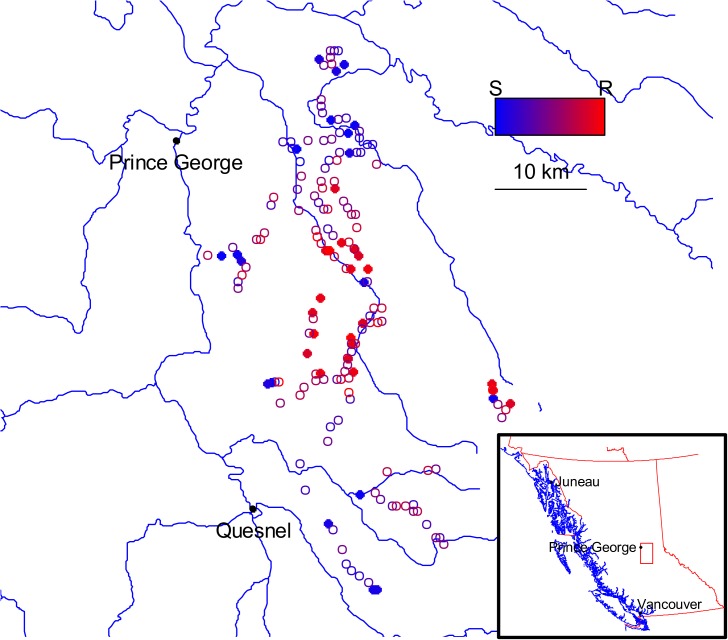
FIG. 1.—
Parent trees' origin within the Prince George area. The color scale (S–R) indicates the level of resistance of the trees, from highly susceptible to highly resistant, blue to red, respectively. Filled circles represent origin of the tree families used in the present microarray study. Open circles represent the origin of tree families not used in the microarray study but used for the resistance ranking (map layers from MapPlace Web site http://www.empr.gov.bc.ca/MINING/GEOSCIENCE/MAPPLACE/Pages/default.aspx).
In addition to collecting open-pollinated seed from the 180 parent trees in the wild, scionwood (i.e., shoot tips) was collected from each tree, and all trees were cloned by grafting and established in clone banks at Vernon, Barnes Creek (near Enderby, BC), and PGTIS. Samples used for genetic analysis in this study were collected from parent tree grafts at the Barnes Creek site. The use of cloned trees growing in the same location instead of wild trees located across a vast geographic area removes bias due to different environmental growth conditions.
RNA Extraction and Microarray Profiling
Bark samples were collected from lateral shoots of the trees from the Barnes Creek clone bank. Total RNA was extracted following Kolosova et al. (2004). RNA quantity and quality were assessed by measuring spectral absorbance between 200 and 350 nm and by visual assessment on a 1% agarose gel. cDNA synthesis was completed for each sample independently using Superscript II Reverse Transcriptase (Invitrogen) with an oligo dT12–18 primer. cDNA samples were hybridized using 3DNA Array 350 Expression Array Detection Kit (Genisphere) onto the Treenomix Spruce cDNA microarray (21.8K version) comprising 18,725 unique elements. A balanced design with dye swaps was used to make direct comparison of gene expression levels of resistant and susceptible trees. Each resistant tree was randomly contrasted with a susceptible tree.
Statistical Analysis
Slides were scanned, and spot intensity was quantified using ImaGene 6.0.1 software (BioDiscovery, Inc., El Segundo, CA). To correct for background intensity, the lowest 10% of median foreground intensities per subgrid was subtracted from the median foreground intensities. Data were then normalized slide by slide, by variance stabilizing normalization to compensate for nonlinearity of intensity distributions (Huber et al. 2002). A linear mixed effects model was fit to the data taking account of both resistance/susceptibility and dye effects. Fold change (FC) and P and Q values were computed for all genes. Genes were considered to have a significant differential expression level when their P value is below 0.05 and their fold change above 1.5.
Heat map and cluster analysis were performed on genes with P < 0.05 and FC > 1.5. Individuals and genes were clustered with Pearson correlation index and Spearman correlation index, respectively. Dendograms were drawn using the “hclust” function in R Script.
To identify major themes appearing among the differentially expressed genes, we used the software Blast2Go (Conesa et al. 2005; Götz et al. 2008) to test for statistical overrepresentation of gene ontology terms (GO terms) among genes up- and downregulated. A more detailed functional categorization was performed using BlastX and tBlastX search versus viridiplantae database on National Center for Biotechnology Information (NCBI). We considered only results with an E value lower than 10−10. Given the number of differently expressed putative sHSP, a particular emphasis has been given to this protein family. tBlastN searches using protein sequences of known sHSP of Arabidopsis thaliana and Oryza sativa (Scharf et al. 2001; Siddique et al. 2008; Sarkar et al. 2009) were performed over the whole microarray to identify putative members of the sHSP family. Sixty-one representative sequences of the 16 known sHSP classes from Ar. thaliana (Scharf et al. 2001; Siddique et al. 2008), Populus trichocarpa (Waters et al. 2008), and O. sativa (Sarkar et al. 2009) were added to this sequences data set. Sequences were first aligned using the online version of PROMALS3D (Pei et al. 2008) and then optimized manually. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965). Phylogenetic relationships were inferred based on amino acid sequences using the Neighbor-Joining method to determine the exact class of each sequence. Only the conserved C-terminal sequences have been considered (see supplementary tables 1 and 2, Supplementary Material online). The reliability of the inferred tree was tested by bootstrap analysis with 1,000 replicates (Felsenstein 1985).
Raw data and normalized data are uploaded to the Gene Expression Omnibus with accession number GSE27476 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE27476). Sequences for array clones are found in NCBI using the clone IDs given in Table 2 and Table 3 and supplementary table S1 (Supplementary Material online).
Table 2.
Summary of t-Test Comparisons between Resistant and Susceptible Trees of Clusters #1 and #2 (=references)
| 18,725 Analyzed Genes | Resistant (20) vs. Group S1 (11) |
Resistant (20) vs. Group S2 (9) |
||
| Upregulated | Downregulated | Upregulated | Downregulated | |
| Genes with P value < 0.05 | 1,778 | 1,709 | 305 | 337 |
| (FDR = 18.9%) | (FDR = 18.9%) | (FDR = 100%) | (FDR = 100%) | |
| Genes with FC > 1.5 | 326 | 482 | 79 | 56 |
| Maximum FC | 3.39 | 10.04 | 2.84 | 3.22 |
| Significant genes(P < 0.05 and FC > 1.5) | 274 | 430 | 15 | 15 |
NOTE.—FDR, false discovery rate.
Table 3.
Functional Categorization of Differentially Expressed Genes (P value < 0.05 and FC > 1.5)
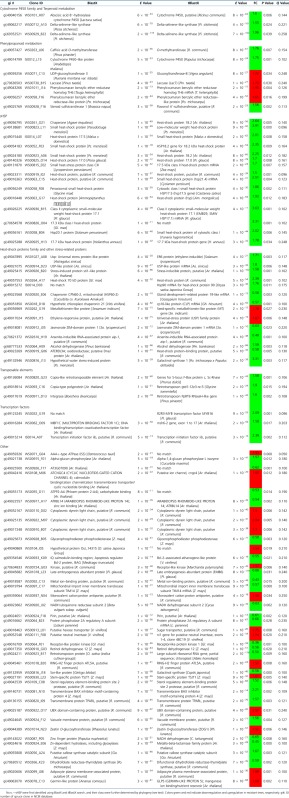 |
Results
Resistance Levels
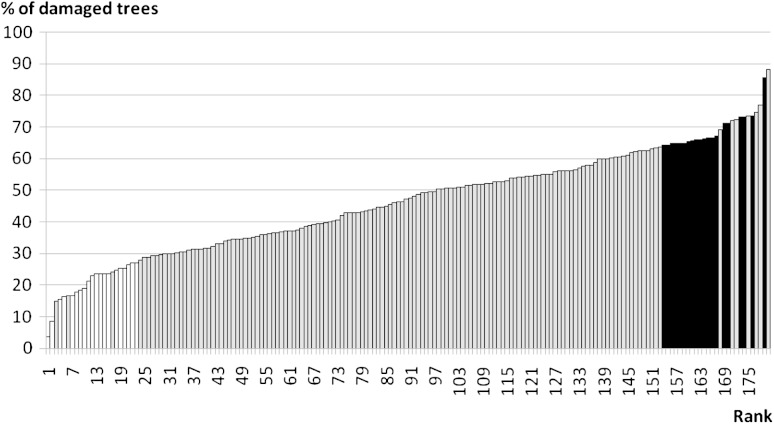
The percentage of trees damaged by weevils was significantly higher among susceptible trees (68%) than among resistant trees (21%; P < 2.2 × 10−16; fig. 2). No difference was found between susceptible and resistant trees neither in size nor in survival. Supplementary table S1 (Supplementary Material online) summarizes the observed damages. These results show that we have a valid comparison of phenotypic differences between two classes of trees that differ in resistance to the weevil.
FIG. 2.—
Percentage of damaged trees among progenies. Progenies are ordered from the least damaged to the most damaged. Resistant and susceptible families are located on the left and on the right, respectively. White bars and black bars show selected families for the present study.
Gene Expression Profiles
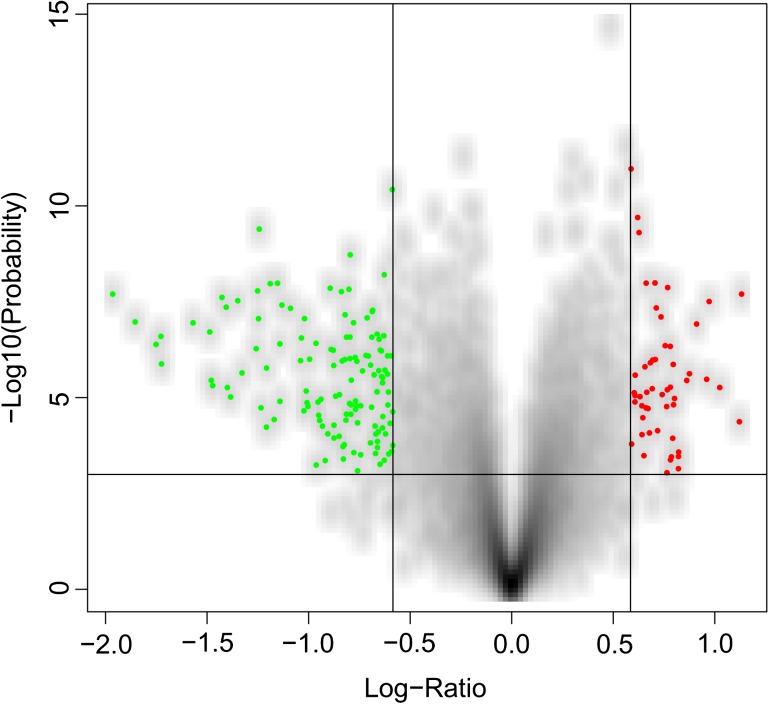
Among the 18,725 genes on our microarray chip, 2,499 showed a P value less than 0.05 for significant differences of gene expression between the two classes of trees that differ in resistance (table 1). The highest Q value observed among these genes was 0.282 but only one gene showed a Q value less than 0.05. FC were low with the maximums FC of 2.24 and 3.91 in upregulated and downregulated genes, respectively (table 1 and fig. 3). Consequently, we considered gene expression to be significantly different if the P value was less than 0.05 and FC was greater than 1.5. With such a rigorous criteria, we identified 54 genes as upregulated and 137 genes as downregulated, in resistant trees compared with susceptible trees, for a total of 191 significant genes.
Table 1.
Summary of t-Test Comparisons between Resistant and Susceptible Trees (=reference)
| 18,725 Analyzed Genes | Upregulated | Down-regulated |
| Genes with P value < 0.05 | 1,225 (FDR = 28.2%) | 1,274 (FDR = 28.2%) |
| Genes with FC > 1.5 | 60 | 151 |
| Maximum FC | 2.24 | 3.91 |
| Significant genes (P < 0.05 and FC > 1.5) | 54 | 137 |
NOTE.—FDR, false discovery rate.
FIG. 3.—
Smoothed densities color representation of volcano plot, showing the differential expression levels of 18,725 genes between resistant and susceptible trees. Significant downregulated and upregulated genes are shown in blue and red, respectively.
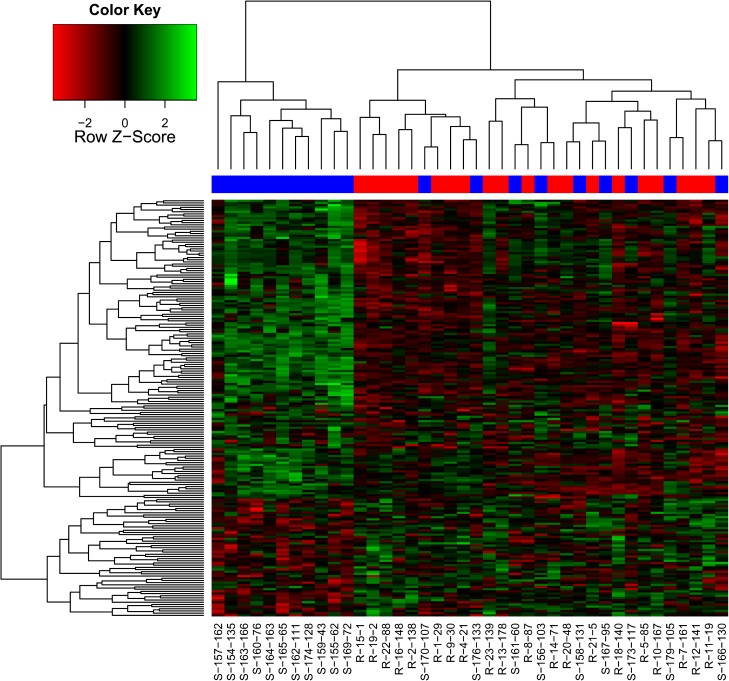
As a further verification of differential gene expression, we performed cluster analysis and heat map based on the 191 significant genes (fig. 4). The cluster analysis indicates two groups, however, they do not match the resistant/susceptible classification; cluster #1 contained 11 susceptible trees, whereas cluster #2 contained 9 susceptible trees and 20 resistant trees. There is no evidence of a link between the resistance levels and the classification of susceptible trees in two distinct groups. The heat map (fig. 4) confirms the differences in gene expression profiles between the two clusters and suggests no difference between susceptible and resistant trees in cluster #2. Genes cluster in two main groups: 1) downregulated genes and 2) upregulated genes.
FIG. 4.—
Heat map of the 191 significantly differently expressed genes between susceptible and resistant trees to the white pine weevil. Blue and red squares at the top of the heat map indicate susceptible and resistant trees, respectively. Tree labels are indicated at the bottom as follow: the tree phenotype (R = resistant, S = susceptible), the family rank in progeny tests for resistance (1 = the most resistant; 179 = the most susceptible), and then the family number.
To find differences that might exist between resistant trees and susceptible trees of clusters #1 and #2 (fig. 4), we performed a complementary analysis. We fitted the data as previously described to a mixed linear model but considered three groups of trees: group S1 = cluster #1 (S-157-162, S-154-135, S-163-166, S-160-176, S-164-163, S-165-65, S-162-111, S-174-128, S-159-43, S-155-62, S-169-72), group S2 = susceptible individuals of cluster #2 (S-170-107, S-176-133, S-161-60, S-156-103, S-158-131, S-167-95, S-173-117, S-179-105, S-166-130, see fig. 4), and group R = resistant individuals (of cluster #2). This approach is not compatible with our experimental design as this analysis consists of three groups, and the experimental design was made to compare two groups. Hence, individuals are not properly balanced over dyes and groups. Moreover, this statistical approach is not adequate as we predefine groups according to their gene expression profiles prior to the statistical comparisons based on the gene expression profiles. So results should be taken with caution. Only 30 genes are significantly differently expressed (FC up to 3.52) between group R and group S2 according to the criteria P < 0.05 and FC > 1.5 but with a Q value of 1 (table 2). This tends to confirm the low levels of difference between these groups. By contrast, the observed differences between group S1 and group R are high with 274 upregulated (FC up to 10.05) and 430 downregulated genes (FC up to 3.40) in group S1.
Functional Characterization
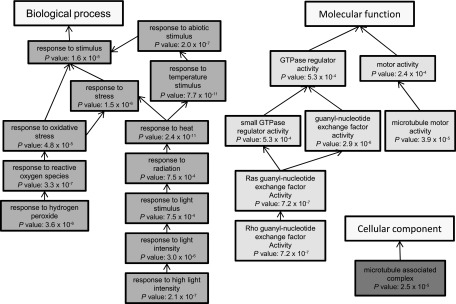
Using Blast2Go, we tested the occurrence of overrepresented GO terms among the set of significant genes arising from the comparison of resistant and susceptible trees compared with the entire microarray. Among the biological processes, only a few categories were overrepresented (fig. 5): “response to hydrogen peroxide,” “response to heat,” and “response to high light intensity” and several higher categories. All belong to the wider category “response to stimulus.” Among cellular components, the only overrepresented category is “microtubule-associated complex.” Among molecular functions, the two lowest overrepresented categories are “Rho guanyl-nucleotide exchange factor” and “microtubule motor.” Although the trees were not stimulated, the overrepresented GO terms suggest that differentially expressed genes are involved in stress or stimulus responses, but their molecular functions remain obscure.
FIG. 5.—
Significantly overrepresented GO terms of genes among significant upregulated or downregulated genes between susceptible and resistant trees, respectively. Fisher’s exact tests with multiple testing corrections were performed using Blast2GO software. Only GO categories with false detection rate lower than 0.05 are shown.
To complete analysis of the GO terms, BlastX and tBlastX searches were preformed against Viridiplantae on NCBI to deduce the functions of these putative genes, using E values less than 10−10. One hundred and six clones gave no results or matched sequences with unknown functions. We did find 85 matches with annotations using either BlastX or tBlastX. Genes with significant blast results are presented in Table 3. Differentially expressed genes belong to various gene families with few apparent links, except for putatively stress-related genes (including the putative sHSP). Three genes were annotated as putative transcription factors and three genes are annotated as part of putative transposable elements, but their possible function here is unknown.
Of the 191 genes either up- or downregulated between resistant and susceptible trees, we found very few differentially expressed genes to be putatively involved in phenylpropanoid and terpenoid metabolisms. Only four genes were putatively assigned to the terpenoid metabolism: one putative cytochrome P450, two putative delta-selinene–like synthases that were downregulated, and one putative zeatin O-glucosyltransferase that was upregulated. Eight to nine genes were putatively directly related to phenylpropanoid metabolism: a putative UDP-glycosyltransferase, a putative laccase, two putative phenylcoumaran benzylic esther reductase, a putative zeatin O-glucosyltransferase, a putative caffeic acid O-methyltransferase, a putative Flavonol 4′-sulfotransferase and a putative cytochrome P450, and eventually the putative transcription factor (MYB16) that might be linked to phenylpropanoid or terpenoid metabolism (Bedon et al. 2007).
Differential Expression of sHSP and Stress-Related Proteins
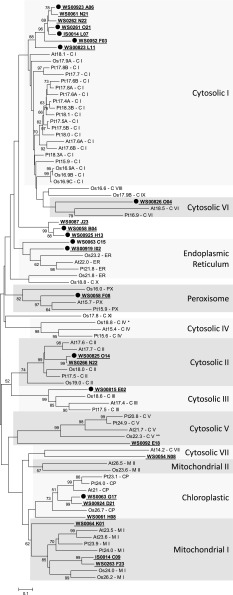
Of the 26 putative sHSP printed on our microarray chips, 15 were downregulated in resistant trees. We compared their sequences with Ar. thaliana, Po. trichocarpa, and Zea mays sHSP sequences allowing class determination of the majority of these genes (fig. 6). The phylogeny is congruent with previous classifications of sHSP (Scharf et al. 2001; Siddique et al. 2008; Waters et al. 2008; Sarkar et al. 2009) with the exception of Os21.8 ER, which was previously characterized as a member of the endoplasmic reticulum group of sHSP but clustered here with Os 18.8 of the cytoplasmic class X. Most of these spruce sHSP sequences cluster within the classes of sHSP previously identified in Ar. thaliana, Po. trichocarpa, and Z. mays. Those that failed to cluster might belong to new sHSP classes.
FIG. 6.—
Phylogenetic analysis of spruce sHSP. The tree was derived by Neighbor-Joining method with bootstrap analysis (1,000 replicates) from alignment of amino acid sequences of sHSP of rice, Arabidopsis, and poplar. Bootstrap values higher than 50% are shown next to the branches. Phylogenetic analyses were conducted in MEGA4. EST clones ID of Picea are indicated in bold and underlined. Downregulated sHSP are indicated by a closed black circle.
As in other species, the most diverse class of putative sHSP in spruce is the nucleocytoplasmic class I, represented by seven putative clones (WS0052_F03, WS00923_A06, WS0061_N21, WS0262_N22, IS0014_L07, WS0261_O21, and WS00823_L11; fig. 6). Nucleocytoplasmic classes II and III are represented by two putative clones (WS0266_N22 and WS00825_O14) and one putative clone (WS00815_E02), respectively. WS0058_F08 putatively belongs to the peroxisomal class and WS0063_C15 and WS00919_I02 putatively belong to the endoplasmic reticulum class. WS0087_J23, WS0058_B04, and WS00925_H13 do not cluster within any classes of either reference species. They may belong to a new class, specific to conifers. Six clones are found within a clade consisting of mitochondrial (group I) and chloroplastic sHSP. IS0014_C09 and WS0263_F23 unambiguously cluster within the mitochondrial group I of sHSP. Similarly, WS0063_G17 and WS00924_D21 unambiguously cluster within chloroplastic sHSP. Because WS0064_K01 and WS0061_H08 are branched between mitochondrial group I and chloroplastic sHSP within the large clade consisting of both mitochondrial and chloroplastic sHSP, they cannot be assigned with high confidence to either class. WS0092_E18, WS00826_O04, and WS0054_N08 do not match any known class of sHSP. Nevertheless, they are putatively related to the cytosolic classes V, VI, and VII, respectively, and are tentatively assigned to these groups of sHSP. WS00930_B15 cannot be assigned to any sHSP class because the clone sequence is too short, even though tBlastN and tBlastX searches place it as a putative sHSP.
The downregulated putative sHSP belong to several classes working in different cellular compartments: nucleocytoplasmic (nine putative sHSP of classes I–VI), endoplasmic reticulum (two putative sHSP), peroxisome (one putative sHSP), and chloroplast (one putative sHSP). Two of the downregulated putative sHSP could not be assigned to a particular class and operate in an unknown cellular compartment and seem to belong to the new sHSP class. In addition to these putative sHSP, 14 putative stress-related proteins of various gene families are differentially expressed (12 downregulated and 2 upregulated in resistant trees), including 3 putative heat-shock proteins and at least 2 putative universal stress proteins.
Discussion
Differences between Resistant and Susceptible Trees
Our comparison gene expression for 18,725 genes between 20 susceptible and 20 resistant trees found 54 upregulated genes and 137 downregulated genes in resistant trees as compared with the susceptible trees. As presented in the introduction, several studies have shown that differences exist between resistant and susceptible phenotypes at the morphological, chemical, and genetic levels. Moreover, previous studies have shown several hundred genes are involved in induced defenses in both Sitka spruce and Norway spruce (Ralph, Yueh, et al. 2006; Lippert et al. 2009). Therefore, the number of differentially expressed genes (i.e., with FC higher than 1.5) was expected to be greater than 211 that found in this study (191 statistically significant).
Such a low number of differentially expressed genes suggests that differences between resistant and susceptible phenotypes are linked more to variation in gene sequences, translation, and/or variation of catalytic efficiencies than to regulatory differences. Hall et al. (2011) showed that differences in (+)-3-carene levels can be explained by variation in 1) the number of gene copies, 2) protein expression levels, 3) gene sequences, and 4) catalytic efficiencies. Such differences can also be expected in other gene families, and the observed differences of gene expression levels may not explain all of the observed phenotypic differences.
Another possible explanation for the low number of differentially expressed genes is that in conifers, several gene families are composed of a large number of closely related genes: terpenoid synthases (Martin et al. 2004; Keeling et al. 2008), cytochrome P450 monooxygenases (Hamberger and Bohlmann 2006), dirigent proteins (Ralph, Park, et al. 2006; Ralph et al. 2007), and MYB transcription factors (Bedon et al. 2007, 2010). Therefore, we can expect that some spots of the microarray hybridize with transcripts of two, or even several, similar genes. In these cases, the observed gene expression levels are the average of the respective gene expression levels (i.e., upregulated genes cancel the effect of the downregulated genes). The low number of differentially expressed genes can also be linked to the existence of disparate strategies of resistance (e.g., stealth or repellent) (see Phenotype Prediction and Efficiency of the Approach).
Previous comparisons between resistant and susceptible trees have shown that resistant phenotypes in spruce are better “armed” to defend against weevils; however, these results are inconsistent. Tomlin and Borden (1994, 1997b) and Alfaro et al. (2004) found that resistant trees possessed more and larger resin ducts, whereas Tomlin et al. (1997) and Nault et al. (1999) reported no clear link between terpene profiles and resistance. Only one study suggested the existence of a stealth strategy (Tomlin et al. 1997). In the case that procuring “armaments” is the most common defense strategy, we might expect a majority of upregulated genes in resistant phenotypes. However, most of the differentially expressed genes in this study were downregulated (72%). This suggests that resistance could be linked more to a stealth strategy than to a repellent strategy. The silencing of certain genes may reduce the probability of detection and attack by weevils. Moreover, because resistance is useful only when weevils are present, the cost of a constant expression of genes involved in resistance might be higher than the associated benefit.
The comparison of resistant trees and the 11 susceptible trees of cluster #1 lead to a higher number of differentially expressed genes than the comparison of the 20 resistant and 20 susceptible trees. It suggests that more genes might show differences in constitutive expression levels. However, we cannot link the classification of the trees in three groups to a classification of phenotypes. Because this statistical approach is not adequate, we will not talk more about these results and we just mention them as further analyses.
Terpenoid and Phenylpropanoid Pathways: Few Genes Were Constitutively Differently Expressed in Resistant Spruces
Only three differentially expressed genes have been found across the terpenoid metabolic pathways. Only two putative delta-selinene–like synthases are downregulated in resistant trees. In grand fir, delta-selinene synthase use farnesyl pyrophosphate as substrate to produce 34 different sesquiterpene olefins (Steele et al. 1998). The downregulated gene annotated as putative abscisic acid 8′-hydroxylase belongs to the wide super family of cytochrome P450. This enzyme degrades abscisic acid into 8′-hydroxyabscic acid (Nambara and Marion-Poll 2005). Abscisic acid is an important terpenoid phytohormone involved in many plant developmental processes and plant responses to environmental stress and pathogens (Seo and Koshiba 2002). In particular, abscisic acid regulates the opening of stomates and thus the loss of water in cells. Pei et al. (2000) showed abscisic acid also triggers an increase in cytosolic calcium in guard cells. In Pistia stratiotes, the Ca2+ channels play an important role in calcium oxalate crystals formation (Volk et al. 2004). We might hypothesize that the reduced catabolism of abscisic acid is linked to an increase in the production of the toxic calcium oxalate crystals. However, more research is needed to confirm this hypothesis.
There are seven differently expressed genes that can be putatively assigned to phenylpropanoid metabolism. First, a putative caffeic O-methyltransferase (COMT) is downregulated in resistant trees. This enzyme is known to be involved in methylation of precursors of both syringyl- and guaiacyl-lignin subunits in angiosperms (Baucher et al. 2003; Do et al. 2007; Vanholme et al. 2008; Tu et al. 2010). Several studies showed that downregulation of COMT leads to syringyl/guaiacyl-lignin ratio change or event suppression of syringyl-lignin. COMT downregulation also leads to the incorporation of 5′-hydroxy-guaiacyl units in lignin. However, syringyl-lignin does not exist in conifers, and we found no studies that show an effect of COMT downregulation on 5′-hydroxy-guaiacyl production in conifers. Because guaiacyl-lignin is the dominant lignin type in conifers, a decrease of COMT expression level could be associated with a decrease of lignin synthesis.
The upregulated putative laccase enzyme belongs to the wide super family of the multicopper oxidase (Nakamura and Go 2005). In plants, some laccase enzymes are involved in lignin biosynthesis, although they have a large spectrum of substrates and form a large family of genes. In loblolly pine, eight laccase genes have been described and two of them have been functionally characterized (Sato et al. 2001; Sato and Whetten 2006). Both enzymes were able to oxidize coniferyl alcohol and produce dimers of coniferyl alcohol and as a consequence are involved in lignin biosynthesis.
Two other upregulated genes in our constitutive samples are annotated as putative phenylcoumaran benzylic ether reductase. Phenylcoumaran benzylic ether reductases are involved in phenolic secondary metabolism and convert 8'5′-linked lignin dihydroconiferyl alcohol into isodihydrodehydrodiconiferyl alcohol by the reduction of benzylic ether functionality (Gang et al. 1999). A previous study showed that a phenylcoumaran benzylic ether reductase is involved in induced conifer defense following either mechanical wounding or weevil attack (Lippert et al. 2007).
The upregulated gene annotated as putative UDP-glucosyltransferase plays an important role in lignin biosynthesis. After their biosynthesis, the monomers of lignin (i.e., p-coumaryl, coniferyl, and sinapyl alcohols according to plant species) have to be translocated to the cell wall for the next oxidation step of lignin biosynthesis. The 4-O-β-D-glucosides of cinnamyl alcohols have been considered as the transport forms of coniferyl and sinapyl alcohols. A UDPG:coniferyl alcohol glucosyltransferase from Pinus strobus has been able to convert cinnamyl aldehydes as well as coniferyl and dihydroconiferyl alcohols into their corresponding O-β-D-glucosides in vitro (Steeves et al. 2001). However, because coniferyl and sinapyl alcohols might be able to freely diffuse through the plasma membrane, it has been suggested that these glucosides play no role in monolignol export for developmental lignin (Boija and Johansson 2006; Vanholme et al. 2008). Another noteworthy gene is annotated as putative MYB16, a member of the family of transcription factors. MYB16 belongs to the R2R3-MYB family and was shown to accumulate transiently in response to wounding in white spruce (Bedon et al. 2010)
At least two genes are annotated within the flavonoid metabolism. First, an upregulated gene was annotated as a putative flavonoid 3′-monooxygenase that belongs to the cytochrome P450 superfamily. This gene is involved in central flavonoid metabolism, the leading precursors of flavones, anthocyanins, and proanthocyanidins pathways (Winkel-Shirley 2001). Anthocyanins can play various roles, including the resistance mechanisms toward insect pests (Steyn et al. 2002). The second gene within the flavonoid metabolism is downregulated and annotated as a putative flavonol 4′-sulfotransferase. Ralph et al. (2006) found that several genes of flavonoid metabolism, including a Flavonoid 3′-monooxygenase (=hydroxylase), are upregulated after white pine weevil herbivory, mechanical wounding, or western spruce budworm (Choristoneura occidentalis, Lepidoptera) feeding.
Many Stress-Related Proteins Exist for Weevil Resistance
Our study shows that 15 of the 26 putative sHSP and several other stress-related genes are downregulated in resistant trees. sHSP belong to a large family of proteins. They are highly variable, but they share a conserved α-crystallin domain of approximately 100 residues (Caspers et al. 1995; de Jong et al. 1998; Fu et al. 2006). sHSP are classified into at least 11 subfamilies localized in different cell compartments: cytosol, mitochondria, chloroplasts, endoplasmic reticulum, and peroxisome (Vierling 1991b; Helm et al. 1993; Waters et al. 1996; Scharf et al. 2001; Siddique et al. 2003; Ma et al. 2006; Waters et al. 2008). The 15 downregulated putative sHSP belong to class I, class II, class III, chloroplastic endoplasmic reticulum, or cannot be assigned with confidence to a known class. The role of sHSP has been widely studied in plants. They are involved in plant response to various kinds of stress such as heat, cold, drought, heavy metals, salinity, oxidative, and osmotic stress (Vierling 1991a; Waters et al. 1996; Wang et al. 2004; Haslbeck et al. 2005; Sun and MacRae 2005; Nakamoto and Vigh 2007). sHSP are also involved in normal development of plants, during embryo development, seed germination, somatic embryogenesis, pollen development, and fruit maturation (Sun et al. 2002 and references therein). sHSP usually play a protection role (Haslbeck et al. 2005; Nakamoto and Vigh 2007). They can form stable complexes with denatured proteins to prevent its aggregation. sHSP also form soluble aggregates with substrate proteins, creating a transient reservoir of substrates. Release and refolding of both complexes and aggregates need the cooperation of ATP-dependent chaperone systems. sHSP also play a role in membrane quality control and are potential membrane stabilizing factors.
Several sHSP were previously shown to be involved in conifer defense. Lippert et al. (2007) showed that weevil feeding induces the overexpression of seven sHSP at the protein level (up to 6-fold induction) in Sitka spruce. They also showed that transcript and protein expression levels are not correlated as six of the seven sHSP corresponding transcripts are not upregulated following weevil feeding. The 2-fold upregulation of the seventh sHSP transcript (class I) is comparable to the upregulation of the associated protein. Nevertheless, they observed that all the seven sHSP transcripts are constitutively expressed to high levels in bark tissue. Such constitutive expression of sHSP has also been observed in Ar. thaliana (Siddique et al. 2008), but the constitutive role of sHSP remains unknown. The results of Lippert et al. (2007) suggested that sHSP transcripts accumulate in transient stocks and that sHSP expression is post-transcriptionally controlled. Recent studies have shown that RNA-binding proteins can regulate the stability, translation, or localization of mRNA (Glisovic et al. 2008; Hogan et al. 2008; Babitzke et al. 2009). sHSP activity is also regulated at the protein level by phosphorylation or oligomer reorganization. As a consequence, the expression levels of sHSP transcripts do not necessarily correlate with the sHSP expression at the protein level. sHSP may not play a role in constitutive defense and, in fact, may be involved in induced defense, among other biological processes. However, the test of this hypothesis needs a time-series comparison of both the transcriptome and the proteome after induction (e.g., weevil feeding), based on both susceptible and resistant strains of spruce. Together with 15 putative sHSP, 12 putative stress-related proteins are constitutively upregulated in susceptible trees. Their potential role is yet to be discovered.
Phenotype Prediction and Efficiency of the Approach
As in previous studies based on morphological features or terpene contents (Tomlin et al. 1997; Tomlin and Borden 1997a; Alfaro et al. 2004), our goal was to determine if the transcriptome profiling is able to predict resistance levels in interior spruce. To determine whether the observed gene expression profiles corresponded to the observed phenotype (i.e., resistant/susceptible), we performed a hierarchical clustering (fig. 4). Although the individuals clustered into two groupings, they did not match with the phenotype classification. One cluster contained 11 susceptible trees and a second cluster contained the remaining trees, that is, both susceptible and resistant trees. The heat map clearly shows that 11 susceptible trees have a distinct profile of gene expressions compared with the other 29 trees. Therefore, it might be possible to identify certain susceptible phenotypes by analyzing the transcriptome profiles, but it will not be possible to identify resistant trees with a high degree of certainty using this approach. Four hypotheses could explain this pattern but at least three of them can be rejected.
First, the resistance levels might be inaccurately assessed for some progenies. The family size of all the examined trees varied between 14 and 175 trees (see supplementary table S1, Supplementary Material online). Among the families used in the transcriptome comparison, six families (five susceptible and on resistant) contained fewer than 80 individuals: S-165-65, S-161-60, S-166-130, S-170-107, S-179-105, and R-11-19 (respectively, 42, 41, 30, 63, 14, and 42 trees). Four of them are considered susceptible and clustered with resistant trees in the cluster #2. Consequently, the assessment of the resistance levels of these progenies might be questionable. However, this does not explain why susceptible progenies (with more than 80 individuals) S-176-133, S-156-103, S-158-131, S-167-95, and S-173-117 cluster with resistant trees. However, the original assessment of damage was based on natural levels of weevil attack. Attack patterns are rarely uniform in the wild, and all trees do not have the same probability of attack (He and Alfaro 1997). Therefore, some of the undamaged trees could have been “escapes” and never subject to attack, leading to some bias in the resistance levels assessment, particularly in the small progenies.
Second, the differences in the observed damages caused by weevils can be explained by environmental factors such as growth conditions. This hypothesis seems improbable because all the parent trees were collected within the same region (Prince Georges area) and the progenies were randomly mixed across several stands. All of them were grown in the same standard conditions. Moreover, as the trees used for gene expression profiling were grafted on the same rootstock, we do not expect high difference due to misadaptation to local soil conditions.
Third, as the collected seeds were open pollinated in the wild, we know only the mother and have no information about the fathers of the progenies used for resistance scoring. This may induce a bias if parents have very different levels of resistance. However, a previous study has shown a high family heritability (h2 = 0.70) in a similar experiment design (King et al. 1997), and crosses between susceptible and resistant trees would lead to intermediate levels of resistance (Alfaro et al. 2004). As a consequence, a bias induced by the uncertainty of fatherhood of the progenies seems improbable.
Finally, the resistance or susceptibility may be based on several different strategies, involving different sets of genes. In this case, our experimental design does not allow us to identify genes involved only in rare strategies. If resistance can be associated with, for example, ten different profiles of gene expression, we can expect only a few trees for each strategy to be present in our sampling. In such a case, the differences in gene expression profiles will be confused with individual variations because we did not classify the trees according to their strategy but according to their phenotype.
Supplementary Material
Supplementary table S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Leyla Tabanfar for technical help and Nima Farzaneh for help in bioinformatics analysis. This work was supported by Genome British Columbia and Genome Canada as part of the Treenomix II project (www.treenomix.ca).
References
- Alfaro RI. The white pine weevil in British Columbia: biology and damage. 1994 In: Proceedings of a Symposium on the White Pine Weevil: Biology, Damage and Management. Richmond, British Columbia; 1994 January 19–21. FRDA Report Vol. 226. Victoria (BC): Pacific Forestry Centre. p. 7–22. [Google Scholar]
- Alfaro RI, Vanakker L, Jaquish B, King J. Weevil resistance of progeny derived from putatively resistant and susceptible interior spruce parents. For Ecol Manag. 2004;202:369–377. [Google Scholar]
- Babitzke P, Baker CS, Romeo T. Regulation of translation initiation by RNA binding proteins. Annu Rev Microbiol. 2009;63:27–44. doi: 10.1146/annurev.micro.091208.073514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Halpin C, Petit-Conil M, Boerjan W. Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol. 2003;38:305–350. doi: 10.1080/10409230391036757. [DOI] [PubMed] [Google Scholar]
- Bedon F, et al. Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses. J Exp Bot. 2010;61:3847–3864. doi: 10.1093/jxb/erq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedon F, Grima-Pettenati J, Mackay J. Conifer R2R3-MYB transcription factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca) BMC Plant Biol. 2007;7:17. doi: 10.1186/1471-2229-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija E, Johansson G. Interactions between model membranes and lignin-related compounds studied by immobilized liposome chromatography. Biochim Biophys Acta. 2006;1758:620–626. doi: 10.1016/j.bbamem.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Caspers G-J, Leunissen JAM, Jong WW. The expanding small heat-shock protein family, and structure predictions of the conserved “α-crystallin domain”. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- de Jong WW, Caspers GJ, Leunissen JAM. Genealogy of the alpha-crystallin—small heat-shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- Do C-T, et al. Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226:1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- Drouin JA, Langor DW. White pine weevil. 1991. Forestry leaflet 8. Edmonton (AB): Forestry Canada, Northwest Region, Northern Forestry Centre. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class A. J Mol Evol. 2006;62:257–266. doi: 10.1007/s00239-005-0076-5. [DOI] [PubMed] [Google Scholar]
- Gang DR, et al. Evolution of plant defense mechanisms. J Biol Chem. 1999;274:7516–7527. doi: 10.1074/jbc.274.11.7516. [DOI] [PubMed] [Google Scholar]
- Gerson EA, Kelsey RG. Piperidine alkaloids in Sitka spruce with varying levels of resistance to white pine weevil (Coleoptera: Curculionidae) J Econ Entomol. 2002;95:608–613. doi: 10.1603/0022-0493-95.3.608. [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, et al. High-throughput functional annotation and data mining with the Blast 2 GO suite. Nucleic Acids Res. 2008;36:3420. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DE, et al. An integrated genomic, proteomic, and biochemical analysis of (+)-3-carene biosynthesis in Sitka spruce (Picea sitchensis) genotypes which are resistant or susceptible to white pine weevil. Plant J. 2011;65:936–948. doi: 10.1111/j.1365-313X.2010.04478.x. [DOI] [PubMed] [Google Scholar]
- Hamberger B, Bohlmann J. Cytochrome P450 mono-oxygenases in conifer genomes: discovery of members of the terpenoid oxygenase superfamily in spruce and pine. Biochem Soc Trans. 2006;34:1209–1214. doi: 10.1042/BST0341209. [DOI] [PubMed] [Google Scholar]
- Hamid A, O'Dell TM, Katovich S. White pine weevil. Leaflet 21. Broomall (PA): U.S. Department of Agriculture, Forest Service,; 1995. Northern Area State & Private Forestry. [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- He F, Alfaro RI. White pine weevil (Coleoptera: Curculionidae) attack on white spruce: spatial and temporal patterns. Environ Entomol. 1997;26:888–895. [Google Scholar]
- Helm KW, LaFayette PR, Nagao RT, Key JL, Vierling E. Localization of small heat shock proteins to the higher plant endomembrane system. Mol Cell Biol. 1993;13:238–247. doi: 10.1128/mcb.13.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18:S96. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Weisshaar S, Lin RPC, Bohlmann J. Functional plasticity of paralogous diterpene synthases involved in conifer defense. Proc Natl Acad Sci U S A. 2008;105:1085. doi: 10.1073/pnas.0709466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JN, Yanchuk AD, Kiss GK, Alfaro RI. Genetic and phenotypic relationships between weevil (Pissodes strobi) resistance and height growth in spruce populations of British Columbia. Can J For Res. 1997;27:732–739. [Google Scholar]
- Kiss GK, Yanchuk AD. Preliminary evaluation of genetic variation of weevil resistance in interior spruce in British Columbia. Can J For Res. 1991;21:230–234. [Google Scholar]
- Kolosova N, et al. Isolation of high-quality RNA from gymnosperm and angiosperm trees. Nat Rev Genet. 2004;2:353–359. doi: 10.2144/04365ST06. [DOI] [PubMed] [Google Scholar]
- Lippert D, et al. Conifer defense against insects: proteome analysis of Sitka spruce (Picea sitchensis) bark induced by mechanical wounding or feeding by white pine weevils (Pissodes strobi) Proteomics. 2007;7:248–270. doi: 10.1002/pmic.200600525. [DOI] [PubMed] [Google Scholar]
- Lippert DN, et al. Quantitative iTRAQ proteome and comparative transcriptome analysis of elicitor-induced Norway spruce (Picea abies) cells reveals elements of calcium signaling in the early conifer defense response. Proteomics. 2009;9:350–367. doi: 10.1002/pmic.200800252. [DOI] [PubMed] [Google Scholar]
- Ma C, Haslbeck M, Babujee L, Jahn O, Reumann S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006;141:47–60. doi: 10.1104/pp.105.073841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Faldt J, Bohlmann J. Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol. 2004;135:1908–1927. doi: 10.1104/pp.104.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto H, Vigh L. The small heat shock proteins and their clients. Cell Mol Life Sci. 2007;64:294–306. doi: 10.1007/s00018-006-6321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell Mol Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- Nault JR, Manville JF, Sahota TS. Spruce terpenes: expression and weevil resistance. Can J For Res. 1999;29:761–767. [Google Scholar]
- Pavy N, et al. Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics. 2005;6:144. doi: 10.1186/1471-2164-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, et al. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Ralph S, Park JY, Bohlmann J, Mansfield SD. Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound-and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.) Plant Mol Biol. 2006;60:21–40. doi: 10.1007/s11103-005-2226-y. [DOI] [PubMed] [Google Scholar]
- Ralph S, et al. A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis) BMC Genomics. 2008;9:484. doi: 10.1186/1471-2164-9-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SG. Conifer defence against insects: microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristo- neura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant Cell Environ. 2006;29:1545–1570. doi: 10.1111/j.1365-3040.2006.01532.x. [DOI] [PubMed] [Google Scholar]
- Ralph SG, Jancsik S, Bohlmann J. Dirigent proteins in conifer defense II: extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (Picea spp.) Phytochemistry. 2007;68:1975–1991. doi: 10.1016/j.phytochem.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Robert JA, et al. Terpenoid metabolite profiling in Sitka spruce identifies association of dehydroabietic acid, (+)-3-carene, and terpinolene with resistance against white pine weevil. Botany. 2010;88:810–820. [Google Scholar]
- Sarkar NK, Kim YK, Grover A. Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics. 2009;10:393. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Whetten RW. Characterization of two laccases of loblolly pine (Pinus taeda) expressed in tobacco BY-2 cells. J Plant Res. 2006;119:581–588. doi: 10.1007/s10265-006-0020-9. [DOI] [PubMed] [Google Scholar]
- Sato Y, Wuli B, Sederoff R, Whetten R. Molecular cloning and expression of eight laccase cDNAs in loblolly pine (Pinus taeda) J Plant Res. 2001;114:147–155. [Google Scholar]
- Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins) Cell Stress Chaperon. 2001;6:225–237. doi: 10.1379/1466-1268(2001)006<0225:tefoat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–48. doi: 10.1016/s1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- Siddique M, Gernhard S, Koskull-Döring P, Vierling E, Scharf K-D. The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperon. 2008;13:183–197. doi: 10.1007/s12192-008-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique M, et al. Tomato heat stress protein Hsp16.1-CIII represents a member of a new class of nucleocytoplasmic small heat stress proteins in plants. Cell Stress Chaperon. 2003;8:381–394. doi: 10.1379/1466-1268(2003)008<0381:thsphr>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R. Sesquiterpene synthases from grand fir (Abies grandis) J Biol Chem. 1998;273:2078–2089. doi: 10.1074/jbc.273.4.2078. [DOI] [PubMed] [Google Scholar]
- Steeves V, Förster H, Pommer U, Savidge R. Coniferyl alcohol metabolism in conifers—I. Glucosidic turnover of cinnamyl aldehydes by UDPG: coniferyl alcohol glucosyltransferase from pine cambium. Phytochemistry. 2001;57:1085–1093. doi: 10.1016/s0031-9422(01)00107-8. [DOI] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. Anthocyanins in vegetative tissues: a proposed unified function in photoprotection. New Phytol. 2002;155:349–361. doi: 10.1046/j.1469-8137.2002.00482.x. [DOI] [PubMed] [Google Scholar]
- Sun W, Van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants. BBA – Gene Struct Expr. 2002;1577:1–9. doi: 10.1016/s0167-4781(02)00417-7. [DOI] [PubMed] [Google Scholar]
- Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin ES, Borden JH. Relationship between leader morphology and resistance or susceptibility of Sitka spruce to the white pine weevil. Can J For Res. 1994;24:810–816. [Google Scholar]
- Tomlin ES, Borden JH. Multicomponent index for evaluating resistance by Sitka spruce to the white pine weevil (Coleoptera: Curculionidae) J Econ Entomol. 1997a;90:704–714. [Google Scholar]
- Tomlin ES, Borden JH. Thin bark and high density of outer resin ducts: interrelated resistance traits in Sitka spruce against the white pine weevil (Coleoptera: Curculionidae) J Econ Entomol. 1997b;90:235–239. [Google Scholar]
- Tomlin ES, Borden JH, Pierce HD. Relationship between volatile foliar terpenes and resistance of Sitka spruce to the white pine weevil. For Sci. 1997;43:501–508. [Google Scholar]
- Tu Y, et al. Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne) Plant Cell. 2010;22:3357–3373. doi: 10.1105/tpc.109.072827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Curr Opin Plant Biol. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1991a;42:579–620. [Google Scholar]
- Vierling E. The roles of heat shock proteins in plants. Annu Rev Plant Biol. 1991b;42:579–620. [Google Scholar]
- Volk GM, Goss LJ, Franceschi VR. Calcium channels are involved in calcium oxalate crystal formation in specialized cells of Pistia stratiotes L. Ann Bot. 2004;93:741–753. doi: 10.1093/aob/mch092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes identifies new subfamilies and reveals diverse evolutionary patterns. Cell Stress Chaperon. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER, Lee GJ, Vierling E. Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot. 1996;47:325. [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- Zulak KG, et al. Targeted proteomics using selected reaction monitoring reveals the induction of specific terpene synthases in a multi-level study of methyl jasmonate-treated Norway spruce (Picea abies) Plant J. 2009;60:1015–1030. doi: 10.1111/j.1365-313X.2009.04020.x. [DOI] [PubMed] [Google Scholar]