Abstract
Recent results from Drosophila suggest that positive selection has a substantial impact on genomic patterns of polymorphism and divergence. However, species with smaller population sizes and/or stronger population structure may not be expected to exhibit Drosophila-like patterns of sequence variation. We test this prediction and identify determinants of levels of polymorphism and rates of protein evolution using genomic data from Arabidopsis thaliana and the recently sequenced Arabidopsis lyrata genome. We find that, in contrast to Drosophila, there is no negative relationship between nonsynonymous divergence and silent polymorphism at any spatial scale examined. Instead, synonymous divergence is a major predictor of silent polymorphism, which suggests variation in mutation rate as the main determinant of silent variation. Variation in rates of protein divergence is mainly correlated with gene expression level and breadth, consistent with results for a broad range of taxa, and map-based estimates of recombination rate are only weakly correlated with nonsynonymous divergence. Variation in mutation rates and the strength of purifying selection seem to be major drivers of patterns of polymorphism and divergence in Arabidopsis. Nevertheless, a model allowing for varying negative and positive selection by functional gene category explains the data better than a homogeneous model, implying the action of positive selection on a subset of genes. Genes involved in disease resistance and abiotic stress display high proportions of adaptive substitution. Our results are important for a general understanding of the determinants of rates of protein evolution and the impact of selection on patterns of polymorphism and divergence.
Keywords: dN/dS, neutral theory, purifying selection, translational selection, recurrent hitchhiking
Introduction
Achieving a better understanding of the factors that shape patterns of polymorphism and divergence across genomes is a central aim in evolutionary genetics. The importance of differences in functional constraint for rates of protein evolution has long been recognized (Kimura 1983), and purifying selection is clearly an important determinant of rates of protein evolution (Charlesworth B and Charlesworth D 2010). However, in contrast to predictions from the neutral theory, recent work in Drosophila suggests that adaptive evolution may also contribute substantially to protein evolution (Smith and Eyre-Walker 2002; Bierne and Eyre-Walker 2004; Andolfatto 2005; Welch 2006; Begun et al. 2007; Eyre-Walker and Keightley 2009; but see Sawyer et al. 2003) as well as to the evolution of noncoding regions (Andolfatto 2005; Begun et al. 2007; Haddrill et al. 2008; but see Kousathanas et al. 2011). Beyond patterns of divergence, recurrent selective sweeps may also impact levels of neutral polymorphism genome-wide (Andolfatto 2007; Macpherson et al. 2007; reviewed in Sella et al. 2009). Whether positive selection has a similarly broad impact on genomes of other groups of organisms is not yet clear, and estimates of the proportion of adaptive fixations for other taxa vary greatly, even between closely related species (e.g., Gossmann et al. 2010; Slotte et al. 2010; Strasburg et al. 2011). For example, so far, there is little evidence of widespread adaptive fixations in Arabidopsis protein-coding regions, whereas there seems to be a substantial proportion of adaptive nonsynonymous fixations in Capsella grandiflora, a close relative of Arabidopsis (Slotte et al. 2010). Genome-wide patterns of polymorphism and divergence in Arabidopsis might therefore be expected to differ from those in Drosophila (Wright and Andolfatto 2008).
While understanding the contribution of positive and purifying selection to protein evolution is an important aim, it is also of general interest to obtain a more detailed understanding of the extent of heterogeneity in the strength and direction of selection across the genome. Increasing data on genome-wide gene expression, protein dispensability, and protein–protein interactions have improved our prospects for this. A general conclusion is that gene expression explains a significant amount of variation in rates of protein evolution across a wide range of taxa (Rocha 2006; Drummond and Wilke 2008; Yang and Gaut 2011). In addition, several other factors are also frequently but weakly correlated with rates of protein evolution, such as, for example, gene essentiality, number of protein–protein interactions (Wang and Lercher 2011), gene length (Stoletzki and Eyre-Walker 2007), recombination, and GC content (Ratnakumar et al. 2010). Furthermore, depending on their biological function(s), genes are likely to vary in the strength of negative selection as well as rates of positive selection, making a global estimate of adaptive divergence somewhat misleading. This highlights the importance of characterizing how the properties of genes and genomic regions within a genome could influence selection (see, for instance, Obbard et al. 2009).
Here, we use published genomic polymorphism data from the model plant Arabidopsis thaliana as well as the recently sequenced genome of the closely related outcrosser Arabidopsis lyrata to characterize genomic patterns of polymorphism and divergence and test whether there is evidence for recurrent selective sweeps with effects on neutral polymorphism across the genome in Arabidopsis. In addition, we assess correlations between rates of protein evolution and genomic factors such as gene expression level and breadth, recombination rate, and GC content in order to understand what genomic factors underlie the variation in rates of protein evolution in Arabidopsis. Our results are important for a more general understanding of the genomic determinants of rates of protein evolution and the impact of selection on patterns of polymorphism and divergence.
Materials and Methods
Data
We used the Araly1 genome assembly of A. lyrata and the accompanying Filtered Models 6 annotation available at: http://genome.jgi-psf.org/Araly1/Araly1.download.ftp.html (Hu et al. 2011). For A. thaliana, we used the TAIR8 genome release (available at TAIR: http://www.arabidopsis.org). Alignments of A. lyrata and A. thaliana for broad-scale genomic analyses were kindly supplied by Yves van de Peer and Jeffrey Fawcett (University of Gent). Alignments were made using the A. thaliana genome as the template for aligning A. lyrata contigs, and further description of the alignments is available in Hu et al. (2011). Single nucleotide polymorphism (SNP) data were obtained from the Nordborg lab (the 250 k chip, from 8 November 2008) and consisted of the 363 accessions comprising a global sample. No explicit correction was made for ascertainment bias. An augmented version of the data (comprising 473 accessions) is further described in Li et al. (2010). The SNP data are available for download from http://borevitzlab.uchicago.edu/resources/genetic/hapmap/core473.
Genome-wide polymorphism counts were extracted from the data produced by Ossowski et al. (2008) and SNP density data from Clark et al. (2007). We used the AtGenExpress development array set (Schmid et al. 2005) as a source of data on gene expression. Recombination rate estimates and codon bias estimates (frequency of optimal codons; FOP) for A. thaliana were from Marais et al. (2004); for A. lyrata, we defined optimal codons as those corresponding to the most abundant transfer RNAs, as in Wright et al. (2004). Raw data on polymorphism and divergence were integrated into a genome browser, available as a java application, which was used to extract data for the analysis of broad-scale genomic patterns.
Analyses of Broad-Scale Patterns of Polymorphism and Divergence
Analysis of broad-scale patterns of genomic variation was carried out in the statistical programming language R using linear models on three genomic scales (20, 50, and 200 kb). At each scale, a data set comprising consecutive windows was extracted, yielding data sets consisting of n = 5,973 (20 kb), n = 2,390 (50 kb), and n = 598 (200 kb) windows. Measures of synonymous and nonsynonymous divergence (dN and dS) in exonic regions were computed on a per gene basis using the simple Nei and Gojobori (1986) method and were averaged in each window. Data were only collected from windows with more than 500 aligned bases, and subcentromeric windows (averaging dS > 0.22) were excluded from the analysis. Linear models were systematically checked for outliers and homogeneity (variance) of residuals. Some variables (see Results) were consequently log(1 + x) transformed.
We assembled counts of the number of sites exhibiting synonymous and nonsynonymous polymorphism in A. thaliana (genome-wide SNP data from Ossowski et al. (2008)) and A. thaliana–A. lyrata divergence. These counts—referred to as McDonald-Kreitman (MK) tables—were pooled according to gene ontology (GO) categories of “biological processes” as available from the TAIR database (annotations from CVS Version: 1.1328 downloaded from TAIR 27 August 2010) and chromosome, respectively. We used those counts and the maximum likelihood framework developed by Welch and implemented in the software MKTest (Welch 2006; see also Obbard et al. 2009) to infer from MK tables counts the proportion (1 − f) of amino acid (AA) changing mutation undergoing strong purifying selection and the proportion (α) of nonsynonymous divergence that was driven by positive selection. Given the sampling variance associated with these parameters (in particular α), when analyzing pooled MK tables by GO categories, we restricted our attention to the most abundant GO categories (i.e., n = 44 categories comprising at least 100 genes). We built a series of models, ranging from models assuming strict neutrality, where f and α are constrained to, respectively, 1 and 0, to models containing purifying or/and positive selection with varying intensities according to GO categories. To compare the fit of these alternative models, Mi, we computed the Akaike information criterion (AIC) of each model Mi as AIC(Mi) = −2 ln L(Mi) + 2 p(Mi), where ln L(Mi) is the (natural) log likelihood of Model Mi and p(Mi) the number of free parameters fitted. Previous work based on simulations suggests that AIC is a sensible choice for model selection in that context (Welch 2006).
Genomic Factors Correlated with Protein Evolution and Codon Bias
We extracted spliced coding sequences (CDS) for all genes with AGI locus tags from the A. lyrata genome assembly and aligned them with A. thaliana CDS sequences using transAlign (Bininda-Emonds 2005). For gene-level analyses, we estimated dN, dS, and dN/dS by the method of Goldman and Yang (1994) as implemented in Codeml (PAML v. 4.2b; Yang 2007). We assumed a transition/transversion bias, and codon frequencies were estimated from average nucleotide frequencies at the three codon positions (F3×4).
Gene expression data from AtGenExpress were separated into seven tissue clusters: root, stem, leaf, flower, pollen, seed, and apex. We used the maximum absolute gcRMA across all tissues as an estimate of expression level. Tissue expression bias was quantified using tau (Yanai et al. 2005), which ranges from 0 (equal expression across tissues) to 1 (tissue-specific expression).
To assess what genomic factors are correlated with evolutionary rates and codon bias, we estimated partial correlation coefficients and assessed their significance based on permutations as in Larracuente et al. (2008) for a data set containing a total of 6,768 genes. We assessed the degree of partial correlation between dN/dS and expression level, expression breadth (tau), gene length, and recombination rate. In correlation analyses with the degree of codon bias (FOP), we also included dN and dS as partial correlates. To test for effects of linked selection, we assessed partial correlations separately for genes in each of the four quartiles in terms of dN/dS.
GO List Enrichment Tests
We conducted list enrichment analyses in order to explore the biological functions of genes with extreme dN/dS values. For this purpose, we analyzed a set of 11,799 genes with matching AGI locus tags in A. thaliana and A. lyrata and more than 60 synonymous sites. We tested for enrichment of GO biological process terms among the 1,000 genes in this list with the highest and lowest dN/dS values using default options in DAVID (Huang et al. 2009).
Results
Broad-Scale Patterns of Divergence and Polymorphism
We examined the dependence of nonsynonymous divergence and intensity of purifying selection on various genomic factors using linear models. For these analyses, we used log-transformed dN as a measure of nonsynonymous divergence and dN/dS (restricted to windows with dN/dS < 1) as a measure of constraint. At all three scales examined (windows of 20, 50, and 200 kb), log-transformed dS, exonic GC content, and exon density were significant as predictors for both dN and dN/dS, whereas there was no consistent significant effect of recombination rate on any of these variables (table 1). Models with nonsynonymous divergence as the response variable had greater explanatory power than those with dN/dS as response variable, and explanatory power also increased with window size (table 1).
Table 1.
Relative Importance of Genomic Factors for Nonsynonymous Divergence and dN/dS
| Response Variable | Adj. R2 | Scale (k) | log(1 + dS) | Rec (cM/Mb) | Exonic GC | Exon Density | Chromosome |
| log(1 + dN) | 0.24 | 20 | 0.93 | 0 | 0.05 | 0.01 | 0 |
| 0.32 | 50 | 0.93 | 0.01 | 0.05 | 0.01 | 0 | |
| 0.50 | 200 | 0.87 | 0.01 | 0.07 | 0.05 | 0.01 | |
| log(1 + dN/dS) | 0.03 | 20 | 0.35 | 0.03 | 0.51 | 0.08 | 0.05 |
| 0.05 | 50 | 0.57 | 0.06 | 0.28 | 0.04 | 0.05 | |
| 0.16 | 200 | 0.83 | 0.04 | 0.08 | 0.04 | 0.02 |
Note.—Results are presented for analyses at three spatial scales (windows of 20, 50, and 200 kb). The proportion of variance explained by each model is presented (Adj. R2) as well as the estimate of the relative portion of the variance explained by each predictor variable in the model (these may not sum exactly to 1 due to rounding of proportions). Entries in bold denote factors with associated significance level <0.001.
We tested whether there is evidence for a genome-wide negative correlation between nonsynonymous divergence and neutral polymorphism, as expected under recurrent hitchhiking, To do so, we used silent (nonexonic) heterozygosity in A. thaliana as dependent variable to qualify levels of neutral polymorphism. We did not find a negative relationship between either dN/dS or dN and silent polymorphism as expected under a recurrent hitchhiking scenario (table 2; fig. 1). Instead, the most important explanatory variable in these models (relative variance explained >80%) was synonymous divergence, and for all spatial scales examined, regions with higher synonymous divergence also had higher nonexonic heterozygosity. Other variables were also significant at some spatial scales but had little explanatory power (table 2). A similar pattern was seen for both synonymous and nonsynonymous heterozygosity (table 2). This suggests that variation in mutation rate is the main predictor of both functional and silent polymorphism in Arabidopsis at these broad spatial scales, although it should be noted that the overall explanatory power of these models is modest for patterns of divergence (table 1) and remarkably low for genome-wide level of polymorphism, where 90% of the variation remains unexplained (table 2).
Table 2.
Relative Importance of Genomic Factors for Non-exonic, Synonymous, and Nonsynonymous Nucleotide Diversity (π)
| Response Variable | Adj. R2 | Scale (k) | log(1 + dN/dS) | log(1 + dS) | Rec (cM/Mb) | Exonic GC | Exon Density | Chr |
| πNonexonic | 0.03 | 20 | 0.06 | 0.45 | 0.19 | 0.13 | 0.08 | 0.08 |
| 0.05 | 50 | 0.07 | 0.53 | 0.14 | 0.16 | 0.03 | 0.06 | |
| 0.07 | 200 | 0.02 | 0.51 | 0.16 | 0.07 | 0.14 | 0.09 | |
| πSyn | 0.01 | 20 | 0.49 | 0.12 | 0.26 | 0.08 | 0.05 | |
| 0.02 | 50 | 0.57 | 0.18 | 0.10 | 0.04 | 0.10 | ||
| 0.08 | 200 | 0.57 | 0.07 | 0.04 | 0.06 | 0.07 | ||
| πNonsyn | 0.01 | 20 | 0.63 | 0.27 | 0.06 | 0.01 | 0.04 | |
| 0.03 | 50 | 0.66 | 0.13 | 0.07 | 0.07 | 0.06 | ||
| 0.07 | 200 | 0.71 | 0.05 | 0.04 | 0.14 | 0.06 |
NOTE.—Results are presented for analyses at three spatial scales (windows of 20, 50, and 200 kb). The proportion of variance explained by each model is presented (Adj. R2) as well as the estimate of the relative portion of the variance explained by each predictor variable in the model (these may not sum exactly to 1 due to rounding of proportions). Entries in bold denote factors with associated significance level <0.001. Chr, chromosome.
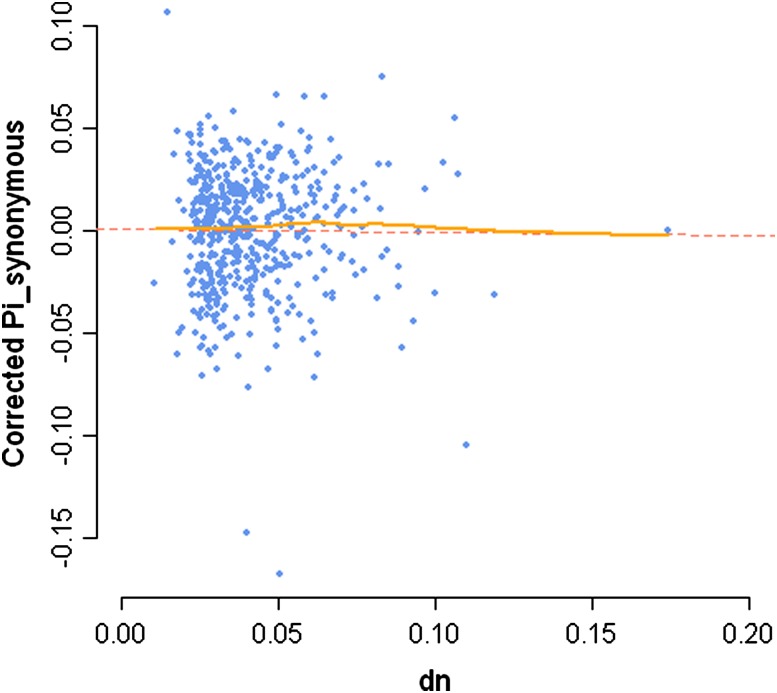
FIG. 1.—
The relationship between nonsynonymous divergence and synonymous polymorphism in Arabidopsis thaliana does not show a signature of recurrent hitchhiking. Levels of synonymous polymorphism—as measured through nucleotide diversity Pi—were corrected for the joint effect of dS and exon density per window (n = 515 windows of 200 kb are used here) using a linear model. The dotted line denotes the standard least square regression line through the data points, and the continuous line—almost superimposed—denotes a local robust “lowess” regression.
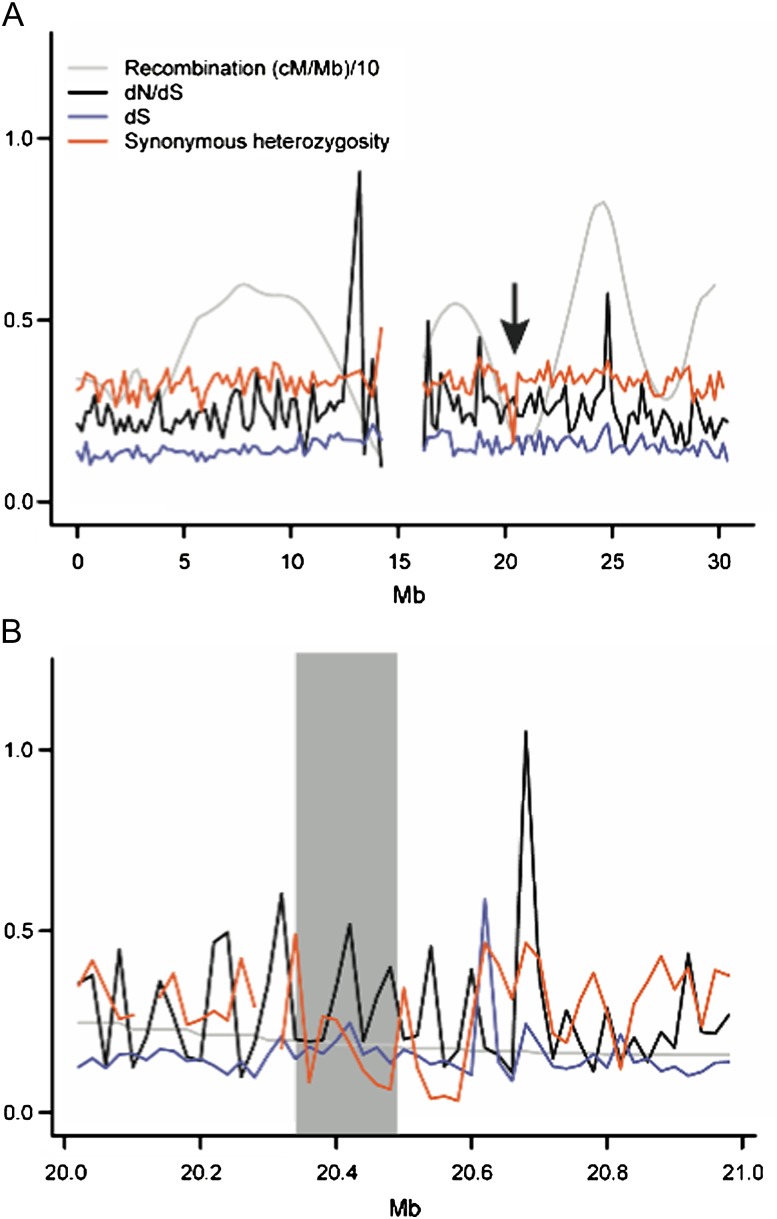
A previous study identified several recent selective sweeps in A. thaliana (Clark et al. 2007). In particular, a region between positions 20.34 and 20.49 Mb on chromosome 1 exhibited nearly complete homozygosity over almost all accessions and was hypothesized to represent a recently swept region. To examine whether a local reduction in mutation rate can explain the reduced polymorphism in this region, we assessed levels of synonymous divergence. We did not find evidence for reduced synonymous divergence in the swept region, and the mean dS in this region (0.18) was actually slightly higher than that of the flanking regions (0.15). Synonymous divergence in the putatively swept region was also not extreme when considering the distribution of dS in windows of the same size across chromosome 1 (90% of noncentromeric chromosome 1 windows show a dS ≤ 0.18) (fig. 2). The lack of a local reduction in synonymous divergence thus supports the interpretation that this region has recently undergone a selective sweep in A. thaliana.
FIG. 2.—
(A) Levels of synonymous divergence, dN/dS, recombination rate, and synonymous heterozygosity plotted in 200-kb windows across Arabidopsis thaliana chromosome 1. There is no reduction in synonymous divergence at a previously identified swept region in A. thaliana (indicated by arrow at ∼20 Mb). (B) A close-up of the putatively swept region shows that there is no evidence for a reduction in synonymous divergence (20-kb windows).
Next, we used genome-wide patterns of polymorphism and divergence at synonymous versus nonsynonymous sites as a way of estimating what amount of nonsynonymous divergence between A. thaliana and A. lyrata can be attributed to positive selection. We first obtained estimates of the amount of positive selection for genome-wide levels. Consistent with the window-based analysis above, we find little evidence for adaptive evolution when averaging over the genome. Analysis of MK table counts of polymorphism and divergence at the chromosome level (table 3) suggests genome-wide purifying selection eliminating approximately 3/4 of new AA changing mutation, but little or no positive selection (all chromosome-wide α estimates are very close to zero) in line with previous reports suggesting very little adaptive evolution in A. thaliana (Bustamante et al. 2002).
Table 3.
Strength of Purifying Selection (1 − f) and Levels of Adaptive Evolution (α) in the Arabidopsis thaliana versus Arabidopsis lyrata Divergence. Chromosome-Wide Estimates of Constraint and Adaptive Evolution
| Chromosome | α | 1 − f |
| 1 | 0.05 ± 0.04 | 0.76 ± 0.008 |
| 2 | −0.01± 0.05 | 0.75 ± 0.010 |
| 3 | −0.08± 0.05 | 0.73 ± 0.013 |
| 4 | 0.07 ± 0.04 | 0.76 ± 0.010 |
| 5 | 0.10 ± 0.04 | 0.77 ± 0.008 |
NOTE.—Estimates are given ± 2 standard errors (SEs). Approximate SEs were estimated via 50 stratified bootstrap samples. 1 − f quantifies the intensity of purifying selection through the fraction of new AA changing mutation subjected to strong purifying selection. α is the fraction of divergence attributable to adaptive evolution (driven by positive selection on new AA changing mutations).
We examined if, by focusing on specific gene categories and examining patterns of polymorphism and divergence, we could pinpoint other clear instances of adaptive evolution. When focusing on the most abundant GO categories (comprising at least 100 genes), we find evidence that both the strength of purifying selection (1 − f) and the proportion of adaptive evolution, α, vary significantly with the class of biological processes considered (table 4). This implies that there is significant evidence for positive selection on some classes of protein (table 5). These GO categories comprise a number of biological processes that have previously been reported as undergoing positive selection (such as genes involved in plant immune response or abiotic stress response). Thus, despite the genome-wide picture of fairly weak purifying selection and near absence of adaptive evolution, there is evidence for positive selection on some protein functions in the genome of A. thaliana, although these instances remain proportionally rare.
Table 4.
Strength of Purifying Selection (1 − f) and Levels of Adaptive Evolution (α) in the Arabidopsis thaliana versus Arabidopsis lyrata Divergence. Fit of Alternative Models for the 44 Most Abundant GO Categories
| Model Name and Description | f | α | LogL | AIC |
| M0: Strict selective neutrality | 1 | 0 | −419,123 | 838,336 |
| M1: Homogenous purifying selection + No adaptive evolution | 0.27 | 0 | −57637.4 | 115,367 |
| M2: Purifying selection with variable intensity + No adaptive evolution | f estimates per GO category | 0 | −6132.64 | 12,443 |
| M3 Homogeneous purifying selection + homogenous levels of adaptive evolution | 0.22 | 0.2 | −56682.4 | 113,459 |
| M4 Purifying selection with variable intensity + homogenous levels of adaptive evolution | f estimates per GO category | 0.1 | −5945.6 | 12,071 |
| M5: Purifying selection with variable intensity + varying levels of adaptive evolution | f estimates per GO category | α estimate per GO category | −4431.5 | 9,129 |
NOTE.—α is the fraction of divergence attributable to adaptive evolution (driven by positive selection on new AA changing mutations). The model with the best AIC, here M5, is highlighted in bold.
Table 5.
Estimates of Constraint (1 − f) and Proportion of Adaptive Evolution, α, in the Abundant GO Categories Undergoing Most Adaptation
| GOs | GO Term | Ngenes | α | 1 − f |
| GO:0006869 | Lipid transport | 137 | 0.60 ± 0.36 | 0.71 ± 0.10 |
| GO:0009414 | Response to water deprivation | 221 | 0.57 ± 0.34 | 0.86 ± 0.02 |
| GO:0045087 | Innate immune response | 135 | 0.49 ± 0.10 | 0.76 ± 0.02 |
| GO:0009737 | Response to abscisic acid stimulus | 312 | 0.41 ± 0.10 | 0.86 ± 0.02 |
| GO:0006508 | Proteolysis | 460 | 0.35 ± 0.18 | 0.79 ± 0.04 |
| GO:0006281 | DNA repair | 140 | 0.34 ± 0.36 | 0.69 ± 0.07 |
| GO:0006457 | Protein folding | 269 | 0.30 ± 0.10 | 0.78 ± 0.04 |
| GO:0005975 | Carbohydrate metabolic process | 437 | 0.24 ± 0.23 | 0.81 ± 0.02 |
| GO:0006950 | Response to stress | 133 | 0.23 ± 0.36 | 0.83 ± 0.07 |
| GO:0008152 | Metabolic process | 939 | 0.13 ± 0.15 | 0.80 ± 0.02 |
| GO:0009753 | Response to jasmonic acid stimulus | 171 | 0.08 ± 0.16 | 0.81 ± 0.03 |
| GO:0009409 | Response to cold | 298 | 0.19 ± 0.14 | 0.87 ± 0.04 |
NOTE.—Estimates ± 2 SEs. Approximate SEs on estimates obtained through 100 bootstrap samples (re-sampling stratified by GO category).
Major Correlates of Protein Evolution and Codon Bias
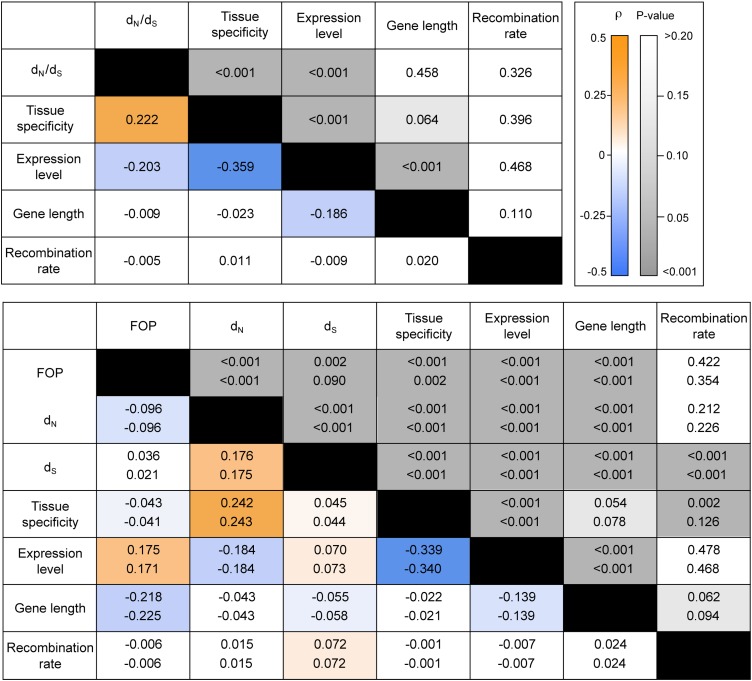
To obtain a more detailed understanding of the relative role of different genomic factors for variation in protein evolution and codon bias, we conducted partial correlation analyses for a data set of 6,768 genes. We assessed the degree of partial correlation between dN/dS and expression level, breadth of expression, gene length, and recombination rate. Similar analyses were conducted for codon bias (FOP) in A. lyrata and A. thaliana.
In agreement with previous studies (see, for instance, Ingvarsson 2007), we find a strong and significant negative partial correlation between expression level and dN/dS, that is, highly expressed genes are more constrained. Expression breadth is also significantly correlated with dN/dS, with broadly expressed genes being more constrained on average (fig. 3). Expression level and gene length are the strongest predictors of codon bias, and this is evident in both A. thaliana and A. lyrata (fig. 3). We also see a very weak but significant negative correlation between dN and codon bias, but there is no significant effect of recombination rate on codon bias in either species. This argues against high selfing rates eroding the expected correlation between recombination and codon bias in A. thaliana, as had been previously suggested (Marais et al. 2004).
FIG. 3.—
The upper panel shows partial correlation coefficients for an analysis aimed at understanding the genomic factors correlated with variation in dN/dS, and the lower panel shows results from an analysis of genomic factors correlated with codon bias. Partial correlation coefficients (below diagonal) are color coded according to sign and degree of correlation, and P values (above diagonal) are color coded by significance level. In the lower panel, the upper value in each cell comes from analysis of codon bias in Arabidopsis thaliana, whereas the lower value corresponds to results for Arabidopsis lyrata. FOP is the frequency of optimal codons.
Analyzing all genes together, we do not find a significant correlation between dN/dS and either recombination rate or gene length. To examine whether this could be due to opposing effects for genes under positive and purifying selection, we conducted separate partial correlation analyses on genes in the upper and lower 25% in terms of dN/dS. There was no significant partial correlation between recombination rate and dN/dS in the low dN/dS set, and although there was a significant partial correlation in the high dN/dS set, it was very weak (partial correlation coefficient: 0.0008, P = 0.04). However, in both of these analyses, the major correlate was gene length, and the sign of the correlation was reversed for the two sets (low dN/dS set; partial correlation coefficient: 0.19, P < 0.001, high dN/dS set: partial correlation coefficient: −0.11, P < 0.001) (supplementary fig. S1, Supplementary Material online). There were no significant partial correlations with either gene length or recombination rate for genes in the two middle quartiles in terms of dN/dS. If gene length can be taken as a proxy for the density of selected sites, these observations are therefore consistent with expectations under Hill–Robertson interference; for genes that are mostly under purifying selection, a reduction in the efficacy of selection leads to an increase in dN/dS, whereas for genes undergoing frequent positive selection, a reduction leads to a decrease in dN/dS.
Functional Classification of Genes with High versus Low dN/dS
To explore the biological characteristics of genes in the tails of the genome-wide distribution of dN/dS values, we conducted list enrichment tests. For genes with high dN/dS values, GO terms corresponding to functions in regulation of transcription and RNA metabolism were overrepresented (table 6; see Materials and Methods for details). In contrast, GO biological process terms related to functions in translation, protein localization, and chromatin structure were overrepresented for the most highly constrained genes (table 5).
Table 6.
GO Biological Process Categories That Were Overrepresented (≤5% false discovery rate) among Sets of Genes in the Tails of the dN/dS Distribution
| Gene Set | GO Term | Description | Ngenes | Fold Enrichment | P Value | FDR |
| High dN/dS | GO:0006355 | Regulation of transcription, DNA dependent | 57 | 2.1 | 9.80 × 10−08 | 1.49 × 10−04 |
| GO:0051252 | Regulation of RNA metabolic process | 57 | 2.1 | 1.16 × 10−07 | 1.76 × 10−04 | |
| GO:0045449 | Regulation of transcription | 86 | 1.7 | 2.97 × 10−07 | 4.51 × 10−04 | |
| GO:0006350 | Transcription | 58 | 1.8 | 1.51 × 10−05 | 2.29 × 10−02 | |
| Low dN/dS | GO:0007264 | Small GTPase mediated signal transduction | 33 | 5.4 | 1.43 × 10−17 | 2.35 × 10−14 |
| GO:0006412 | Translation | 84 | 2.3 | 2.50 × 10−14 | 4.09 × 10−11 | |
| GO:0045184 | Establishment of protein localization | 66 | 2.3 | 1.17 × 10−10 | 1.92 × 10−07 | |
| GO:0015031 | Protein transport | 66 | 2.3 | 1.17 × 10−10 | 1.92 × 10−07 | |
| GO:0006091 | Generation of precursor metabolites and energy | 59 | 2.4 | 1.88 × 10−10 | 3.08 × 10−07 | |
| GO:0008104 | Protein localization | 66 | 2.2 | 6.94 × 10−10 | 1.14 × 10−06 | |
| GO:0010038 | Response to metal ion | 57 | 2.3 | 2.88 × 10−09 | 4.72 × 10−06 | |
| GO:0046686 | Response to cadmium ion | 52 | 2.3 | 7.86 × 10−09 | 1.29 × 10−05 | |
| GO:0010035 | Response to inorganic substance | 66 | 2.0 | 4.62 × 10−08 | 7.57 × 10−05 | |
| GO:0031497 | Chromatin assembly | 19 | 4.0 | 1.65 × 10−07 | 2.71 × 10−04 | |
| GO:0051258 | Protein polymerization | 12 | 6.1 | 2.90 × 10−07 | 4.75 × 10−04 | |
| GO:0006323 | DNA packaging | 19 | 3.7 | 6.01 × 10−07 | 9.85 × 10−04 | |
| GO:0034728 | Nucleosome organization | 18 | 3.9 | 6.47 × 10−07 | 1.06 × 10−03 | |
| GO:0006334 | Nucleosome assembly | 18 | 3.9 | 6.47 × 10−07 | 1.06 × 10−03 | |
| GO:0065004 | Protein–DNA complex assembly | 18 | 3.8 | 9.87 × 10−07 | 1.62 × 10−03 | |
| GO:0034622 | Cellular macromolecular complex assembly | 32 | 2.5 | 1.04 × 10−06 | 1.70 × 10−03 | |
| GO:0034621 | Cellular macromolecular complex subunit organization | 34 | 2.5 | 1.08 × 10−06 | 1.77 × 10−03 | |
| GO:0006333 | Chromatin assembly or disassembly | 20 | 3.4 | 1.62 × 10−06 | 2.65 × 10−03 | |
| GO:0009628 | Response to abiotic stimulus | 122 | 1.5 | 2.04 × 10−06 | 3.34 × 10−03 | |
| GO:0043933 | Macromolecular complex subunit organization | 37 | 2.1 | 9.54 × 10−06 | 1.56 × 10−02 | |
| GO:0065003 | Macromolecular complex assembly | 35 | 2.2 | 1.08 × 10−05 | 1.76 × 10−02 | |
| GO:0006119 | Oxidative phosphorylation | 16 | 3.5 | 1.30 × 10−05 | 2.13 × 10−02 | |
| GO:0030244 | Cellulose biosynthetic process | 11 | 4.8 | 2.47 × 10−05 | 4.05 × 10−02 |
Discussion
We have analyzed genomic data from A. thaliana and A. lyrata in order to understand the genomic and selective determinants of patterns of sequence variation and evolution at a genome-wide scale in a pair of plant species. Our results suggest that mutation rate variation across the genome has a profound influence on patterns of sequence evolution in Arabidopsis, as both silent and nonsynonymous polymorphism as well as nonsynonymous divergence mainly correlates with synonymous divergence.
Silent polymorphism is not negatively correlated with AA divergence, unlike in Drosophila, and thus, we do not detect a clear footprint of recurrent hitchhiking across the genome. Our results are in agreement with previous studies that found a prevalence of weak purifying selection on nonsynonymous sites (Weinreich and Rand 2000; Bustamante et al. 2002; Foxe et al. 2008) and a low proportion of adaptive nonsynonymous fixations (Foxe et al. 2008; Slotte et al. 2010) and are consistent with the hypothesis that neutral and nearly neutral processes dominate patterns of sequence evolution in Arabidopsis (Wright and Andolfatto 2008), at least in the noncentromeric regions that we have analyzed.
Our results do not imply that positive selection is unimportant in Arabidopsis. Indeed, we find support for an extensive selective sweep on chromosome 1 reducing heterozygosity over 500 kb identified by Clark et al. (2007), as there is no evidence of a locally reduced mutation rate in this region. Such recent sweeps can clearly contribute to sequence polymorphism in Arabidopsis without necessarily producing a signal of recurrent hitchhiking. We also find that certain GO categories exhibit higher rates of protein evolution, including those involved in biotic defense, and undergo substantial adaptive evolution as measured by the parameter α.
Recurrent hitchhiking may have a limited impact on the genome of A. thaliana for several reasons, including extensive linkage disequilibrium (and thus interference between selected sites), a low effective population size, and extensive population structure (Wright and Andolfatto 2008). It is currently unclear which of these factors is most important. However, recent results for the forest tree Populus tremula (Ingvarsson 2010), the sunflower genus Helianthus (Strasburg et al. 2011), and the crucifer C. grandiflora (Slotte et al. 2010; Slotte T, Platts A, Hazzouri KM, Cai S, Lu A, Wright SI, unpublished data) suggest that patterns similar to those in Drosophila can be found in plants with life-history features that would be expected to render species-wide natural selection more efficient.
Our partial correlation analyses also provided some evidence for Hill–Robertson interference reducing the efficacy of selection on AA mutations, if gene length can be taken as a proxy for the density of selected sites. However, similar to an analysis of human data (Bullaughey et al. 2008), we could not see an effect of recombination rate, which is unexpected because interference should be dependent on genetic distance as well as the physical density of selected sites. To fully examine the relationship between recombination rates and efficacy of selection in Arabidopsis, higher resolution maps and/or population recombination rate estimates are needed.
The clearest pattern emerging from our partial correlation analyses was a strong and significant correlation between dN/dS and expression level and tissue specificity of expression, with genes that are highly and/or broadly expressed being more constrained overall. These results are consistent with those for Drosophila (Larracuente et al. 2008) and previous results for Arabidopsis (Wright et al. 2004; Foxe et al. 2008). They are also in line with a recent genome-wide analysis of A. thaliana and A. lyrata (Yang and Gaut 2011), although in our case, considerably most of the variance in rates of protein evolution was explained, and our results are more comparable to reports from Drosophila (Larracuente et al. 2008). Although it is not clear why the conclusions differ, we take a partial correlation approach that is more comparable with the Drosophila analyses than the principal components regression analysis used by Yang and Gaut (2011). Codon bias was also correlated with expression level but only weakly with tissue specificity of expression in Arabidopsis. Thus, our results are in good general agreement with the finding that gene expression is a key correlate of rates of protein evolution across a broad range of organisms, possibly as a result of selection for translational robustness (Drummond and Wilke 2008).
Conclusions
Our analyses of broad-scale patterns of polymorphism and divergence in Arabidopsis suggest that regional mutation rate variation has a major effect on levels of nonsynonymous divergence and silent polymorphism in noncentromeric regions of the Arabidopsis genome. Expression level and specificity are major correlates of rates of protein evolution and codon bias, in agreement with results for other taxa that suggest a key role for translational selection in determining rates of protein evolution.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Ya-Long Guo for helpful comments on this manuscript. T.B. and M.H.S. acknowledge support from the Danish Council for Independent Research (Natural Sciences).
References
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 2007;17:1755–1762. doi: 10.1101/gr.6691007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Eyre-Walker A. The genomic rate of adaptive amino acid substitution in Drosophila. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds ORP. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics. 2005;6:156. doi: 10.1186/1471-2105-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullaughey K, Przeworski M, Coop G. No effect of recombination on the efficacy of natural selection in primates. Genome Res. 2008;18:544–554. doi: 10.1101/gr.071548.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, et al. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Elements of evolutionary genetics. Greenwood Village (CO): Roberts & Company Publishers; 2010. [Google Scholar]
- Clark RM, Schweikert G, Toomajian C, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol Biol Evol. 2009;26:2097–2108. doi: 10.1093/molbev/msp119. [DOI] [PubMed] [Google Scholar]
- Foxe JP, et al. Selection on amino acid substitutions in Arabidopsis. Mol Biol Evol. 2008;25:1375–1383. doi: 10.1093/molbev/msn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang ZH. Codon-based model of nucleotide substitution for protein-coding DNA-sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gossmann TI, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27:1822–1832. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Bachtrog D, Andolfatto P. Positive and negative selection on noncoding DNA in Drosophila simulans. Mol Biol Evol. 2008;25:1825–1834. doi: 10.1093/molbev/msn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DV, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Gene expression and protein length influence codon usage and rates of sequence evolution in Populus tremula. Mol. Biol. Evol. 2007;24:836–844. doi: 10.1093/molbev/msl212. [DOI] [PubMed] [Google Scholar]
- Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shape patterns of polymorphism in Populus tremula. Mol Biol Evol. 2010;27:650–660. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kousathanas A, Oliver F, Halligan DL, Keightley PD. Positive and negative selection on non-coding DNA close to protein-coding genes in wild house mice. Mol Biol Evol. 2011 doi: 10.1093/molbev/msq299. 28(3):1183–1191. [DOI] [PubMed] [Google Scholar]
- Larracuente AM, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:21199–21204. doi: 10.1073/pnas.1007431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson JM, Sella G, Davis JC, Petrov DA. Genomewide spatial correspondence between nonsynonymous divergence and neutral polymorphism reveals extensive adaptation in Drosophila. Genetics. 2007;177:2083–2099. doi: 10.1534/genetics.107.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G, Charlesworth B, Wright SI. Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 2004;5:R45. doi: 10.1186/gb-2004-5-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim KW, Jiggins FM. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5:e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar A, et al. Detecting positive selection within genomes: the problem of biased gene conversion. Philos Trans R Soc Lond B Biol Sci. 2010;365:2571–2580. doi: 10.1098/rstb.2010.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC. The quest for the universals of protein evolution. Trends Genet. 2006;22:412–416. doi: 10.1016/j.tig.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Sawyer SA, Kulathinal RJ, Bustamante CD, Hartl DL. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J Mol Evol. 2003;57(Suppl 1):S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Foxe JP, Hazzouri KM, Wright SI. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27:1813–1821. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, et al. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol Biol Evol. 2011;28:1569–1580. doi: 10.1093/molbev/msq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GZ, Lercher MJ. The effects of network neighbours on protein evolution. PLoS One. 2011;6:e18288. doi: 10.1371/journal.pone.0018288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich DM, Rand DM. Contrasting patterns of nonneutral evolution in proteins encoded in nuclear and mitochondrial genomes. Genetics. 2000;156:385–399. doi: 10.1093/genetics/156.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ. Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics. 2006;173:821–837. doi: 10.1534/genetics.106.056911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Andolfatto P. The impact of natural selection on the genome: emerging patterns in Drosophila and Arabidopsis. Annu Rev Ecol Evol Syst. 2008;39:193–213. [Google Scholar]
- Wright SI, Yau CB, Looseley M, Meyers BC. Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol Biol Evol. 2004;21:1719–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang L, Gaut BS. Factors that contribute to variation in evolutionary rate among Arabidopsis genes. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr058. 28(8):2359–2369. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]