Abstract
Cockroaches (Blattaria: Dictyoptera) harbor the endosymbiont Blattabacterium sp. in their abdominal fat body. This endosymbiont is involved in nitrogen recycling and amino acid provision to its host. In this study, the genome of Blattabacterium sp. of Cryptocercus punctulatus (BCpu) was sequenced and compared with those of the symbionts of Blattella germanica and Periplaneta americana, BBge and BPam, respectively. The BCpu genome consists of a chromosome of 605.7 kb and a plasmid of 3.8 kb and is therefore approximately 31 kb smaller than the other two aforementioned genomes. The size reduction is due to the loss of 55 genes, 23 of which belong to biosynthetic pathways for amino acids. The pathways for the production of tryptophan, leucine, isoleucine/threonine/valine, methionine, and cysteine have been completely lost. Additionally, the genes for the enzymes catalyzing the last steps of arginine and lysine biosynthesis, argH and lysA, were found to be missing and pseudogenized, respectively. These gene losses render BCpu auxotrophic for nine amino acids more than those corresponding to BBge and BPam. BCpu has also lost capacities for sulfate reduction, production of heme groups, as well as genes for several other unlinked metabolic processes, and genes present in BBge and BPam in duplicates. Amino acids and cofactors that are not synthesized by BCpu are either produced in abundance by hindgut microbiota or are provisioned via a copious diet of dampwood colonized by putrefying microbiota, supplying host and Blattabacterium symbiont with the necessary nutrients and thus permitting genome economization of BCpu.
Keywords: symbiosis, genome reduction, Blattabacterium, Bacteroidetes, metabolic pathway loss, wood-feeding
Introduction
Blattabacteria are obligatory endosymbionts of cockroaches that reside in the abdominal fat body. Cockroaches are generally considered as omnivorous, and the role of the endosymbiont has recently been revealed via genome sequencing as an involvement in the nitrogen metabolism of the host (López-Sánchez et al. 2009; Sabree et al. 2009). Blattabacteria contain a urea cycle as well as an active urease that converts urea to ammonia and therefore are likely responsible for the ammonotely of cockroaches, the excretion of ammonia as nitrogen waste product instead uric acid like most other insects (Cochran 1985; O'Donnell 2008). Furthermore, blattabacteria are also involved in the nitrogen recycling for the host throughout provision of essential amino acids and vitamins. The two blattabacteria genomes are about 640 kb in size with DNA G+C content of 27–28%, contain one plasmid, and are highly similar in gene content and order (Sabree et al. 2010).
In recent years, genome sequencing of the small genomes of insect endosymbionts has demonstrated an impressive versatility of mutualistic relationships (Moran et al. 2008; Moya et al. 2008). Symbionts studied so far belong to different bacterial phyla and their functions—although highly specific and diverse—are mostly related to synthesis and provision of nutrients deficient in the host's natural diet. The best described systems are those of hemiptera, namely the symbiosis between aphids and Buchnera aphidicola (Baumann 2005; Latorre et al. 2005; Brinza et al. 2009; Pérez-Brocal et al. 2011; Shigenobu and Wilson 2011), although in recent years, data have become available for other insect groups whose symbionts have been sequenced, for example, the carpenter ant Blochmannia sp. (Gil et al. 2003; Wernegreen and Wheeler 2009; Williams and Wernegreen 2010) and the tsetse fly-Wigglesworthia–Sodalis symbioses (Aksoy and Rio 2005; Belda et al. 2010). Among the Bacteroidetes to which the flavobacterial blattabacteria belong (Bandi et al. 1994), genomes have been sequenced of three other endosymbiont lineages: Candidatus Azobacteroides pseudotrichonymphae (Hongoh et al. 2008b), Candidatus Amoebophilus asiaticus (Schmitz-Esser et al. 2010), and Candidatus Sulcia muelleri (McCutcheon and Moran 2007). For Candidatus Sulcia muelleri, the closest relatives to blattabacteria so far, four genomes are known; however, they are much more reduced with genome sizes of only 244–277 kb (McCutcheon et al. 2009). Candidatus Sulcia muelleri has been found in phylogenetically distinct host groups, that is, sharpshooters, cicadas, and spittlebugs, moreover, it always coexists with another symbiont exhibiting intensive metabolic complementation (Lo, Engel, et al. 2007; McCutcheon and Moran 2010).
Cockroaches (Blattaria) form a suborder of Dictyoptera, which comprises six families. To date, all examined species, but one (Lo, Beninati, et al. 2007) harbor blattabacteria as obligatory symbionts in their fat body. Within the Dictyoptera subsocial, wingless cockroaches of the family Cryptocercidae are considered a direct sister group of the Isoptera (termites), rendering the Blattaria paraphyletic (Cleveland et al. 1934; Nalepa and Bandi 1999; Inward et al. 2007; Lo, Engel, et al. 2007; Klass et al. 2008). Termites, except one species, the early divergent lineage Mastotermes darwiniensis, have lost their Blattabacterium endosymbiont later during evolution (Bandi et al. 1995; Clark et al. 2001; Maekawa et al. 2005). In contrast to other cockroaches that are true omnivores, the nine Cryptocercus species described are xylophageous, like termites, and harbor a complex microbiota of protists in their intestine that is responsible for digesting wood (Cleveland et al. 1934; Nalepa 1984). However, in contrast to the termites living in warm climates and feeding primarily on drywood, the five North American Cryptocercus species live in forest habitats in temperate climates and feed primarily on damp rotting wood (Nalepa and Bandi 1999; Kambhampati and Peterson 2007). Nevertheless, regarding their biology, wood roaches can be considered as a bridging state between the majority of cockroaches and termites (Nalepa et al. 2001; Bell et al. 2007).
The two sequenced genomes of the symbionts of Blattella germanica and Periplaneta americana (López-Sánchez et al. 2009; Sabree et al. 2009), hereon referred to as BBge and BPam, show complete colinearity, apart from an inverted 19 kb stretch (Sabree et al. 2010). Differential loss of only 18 genes in either one of the two lineages occurred after the split of the Blattellidae and Blattidae lineages, estimated to have taken place in the early Cretaceous (Lo et al. 2003; Maekawa et al. 2005).
Here, we report the sequence determination and genome analysis of the Blattabacterium symbiont of Cryptocercus punctulatus, one of four Cryptocercus species present in the Appalachians (Clark et al. 2001), as a representative of the Cryptocercidae lineage. Analysis reveals a reduced genome compared to the other two lineages but with fully conserved colinearity of gene order with regard to BBge.
Material and Methods
Genome Sequencing and Assembly
Endosymbionts were extracted from ten female and ten male adults of a 2N = 37 (male) population of C. punctulatus obtained from the Log Hollow site (Transylvania County, NC; Nalepa et al. 2002) and their genomic DNA isolated following protocols according to López-Sánchez et al. (2008). Freshly dead bodies were dissected, and fat body material was collected. First, bacterial cells were isolated following the procedure of Harrison et al. (1989) including a DNase digestion step (DNase I [Roche], 20 ng μl−1, 15 min 25 °C in 10 mM Tris pH 7.5, 8 mM MgCl2, and 2 mM CaCl2). Genomic DNA was isolated using a Cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson 1980). Genomic DNA was subjected to multiple displacement amplification (illustra GenomiPhi V2 DNA amplification kit, GE Healthcare, Little Chalfont, UK). Pyrosequencing with 454 technology in half-plate format was performed twice: once directly with genomic DNA using the standard assay (average read length: 224 bases) and once with an MDA-amplified sample applying the titanium assay (average read length: 344 bases). The two runs of pyrosequencing yielded a total of 709,186 reads.
Draft assemblies were constructed with MIRA (Version V3 release candidate 1; Chevreux et al. 1999) resulting in 884 initial contigs. Using the GAP program (version 4.10) of the Staden package a scaffold was obtained consisting of 43 contigs with 200,368 assembled reads (28.3%). The rest of sequence reads represented contaminating DNA, with no indication for the presence of another bacterium, such as a secondary symbiont in the fat body. The contigs showed high synteny with the genomes of BBge and BPam and represented the chromosome and a plasmid. Gaps between these contigs were closed via Sanger sequencing of bridging PCR products using primers designed for the ends of adjacent contigs. Sanger sequencing helped also to resolve homopolymer ambiguities. Direct sequencing of amplicons with sizes between 200 bp and 2.5 kb was performed with the ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit at an ABI PRISM 3730 Genetic Analyzer (Applied Biosystems). Final read coverage was 84 for the chromosome and 589 for the plasmid. The complete sequences of chromosome and plasmid of Blattabacterium sp. (C. punctulatus) BCpu were deposited at GenBank/DDBJ/EMBL and were assigned accession numbers CP003015 and CP003016, respectively.
Gene Identification and Annotation
Four gene prediction algorithms were applied: Glimmer 3.02 (Delcher et al. 1999), GeneMark.hmm for Prokaryotes (Version 2.8; Lukashin and Borodovsky 1998), the heuristic model of GeneMark.hmm (Besemer and Borodovsky 1999) and Prodigal (Hyatt et al. 2010). Predicted open reading frames (ORFs) were manually examined and compared with the genomes of BBge and BPam using genomic Blast (NCBI). Start and stop sites were curated manually. tRNA genes were searched using tRNAscan-SE (Schattner et al. 2005) and Aragorn (Laslett and Canback 2004) and additionally analyzed using the TFAM Webserver 1.3 (Tåquist et al. 2007). CAT anticodon tRNAs were classified according to Silva et al. (2006). Based on automatic annotations using Blast2GO (Conesa et al. 2005) and BASys (Van Domselaar et al. 2005), protein and RNA genes were annotated with references to the following databases: COG, KEGG, MBGD, Pfam, BioCyc/MetaCyc, and Interpro. Pseudogenes were annotated according to the narrow approach used for the annotation of both the BBge and the BPam genomes considering only almost intact ORFs that contained single or few inline stops or frame shift mutations. Multiple remnants of lost genes, that is, detectable by BlastX, found in zones with already advanced gene erosion were not annotated as pseudogenes.
Structural and Functional Analysis
Gene contents of blattabacteria genomes were compared for orthologous genes using BlastP. Genes were identified as orthologous with the genes of BBge and BPam applying an e value cutoff of 10−6 and a minimum overlap of 50%. Functional analysis was done on the model presented for BBge (López-Sánchez et al. 2009) using the following databases: CDD, KEGG, MBGD, Uniprot, Expasy, Pfam, EcoCyc/MetaCyc, Interpro, BRENDA, Bacteriome.org, PortEco, EcoliWiki, and iHOP. Codon usage analysis and distribution of amino acids in CDS were calculated with Artemis (Rutherford et al. 2000).
Results
Genome Structure
The genome of BCpu consists of a chromosome of 605,745 bp with 23.8mol% GC content of DNA and a plasmid of 3,816 bp (30.5mol% GC) (Table 1). The chromosome of BCpu contains 585 genes in total, 545 protein-coding, 38 noncoding RNAs (32 tRNAs, 3 rRNAs, tmRNA, the signal recognition particle RNA, and the RNA component of the RNase P) and two pseudogenes, which are ygfA and lpxP (see supplementary table 1, Supplementary Material online). The plasmid contains three genes, all orthologous to genes found in the plasmids of BPam and BBge: dut, nrdF, and a hypothetical protein. A fourth plasmid-coded hypothetical protein gene is pseudogenized in BCpu as BLBCPU_p002ps.
Table 1.
Comparison of Genomes of BCpu, BBge, and BPam
| Blattabacterium sp. host, symbiont abbreviation |
Cryptocercus punctulatus |
Blattella germanica |
Periplaneta americana |
| BCpu | BBge | BPam | |
| GenBank and RefSeq accession number | CP003015 | NC_013454 | NC_013418 |
| CP003016 | NC_015679 | NC_013419 | |
| Genome size (bp) | 609,561 | 640,935 | 640,442 |
| Chromosome size (bp) | 605,745 | 636,850 | 636,994 |
| Plasmid size (bp) | 3,816 | 4,085 | 3,448 |
| GC content (%)a | 23.8/30.5 | 27.1/29.8 | 28.2/28.5 |
| Total number of genes | 589 | 635 | 636 |
| CDSsa | 545 + 3 | 590b + 4 | 587c + 4 |
| rRNAs | 3 | 3 | 3 |
| tRNAs | 32 | 34 | 33 |
| Other RNAs: tmRNA, ffs, rnpB | 3 | 3 | 3d |
| Pseudogenesa | 2 + 1 | 1 | 6 |
The first and the second values refer to chromosome and plasmid, respectively.
Includes 4 additional protein genes that are not contained in the annotation NC_013454.
Includes 9 additional protein genes that are not contained in the annotation NC_013418.
Includes genes for ffs and rnpB that are not contained in the annotation NC_013418.
The gene order in BCpu is conserved compared with BBge and BPam, although the genome is approximately 31 kb smaller due to the loss of 54 and 48 functional genes in the BCpu chromosome compared with those of BBge and BPam, respectively. Regarding the inversion of a 19-kb fragment in the origin region, BCpu has the same order as that found in BBge.
Similarly to other AT-rich endosymbiont genomes, homopolymers were observed, causing frame shifts in pseudogenes and were especially frequent in regions of lost genes.
Gene Prediction
The four applied prediction programs all resulted in an overestimation of ORFs with 571, 557, 566, and 561 predicted protein genes compared with the 548 annotated protein-coding genes plus three pseudogenes. In comparison to the manually curated annotation, the four algorithms predicted 88.7%, 89.5%, 94.0%, and 94.0% of the ORFs with positions of start and stop codons correctly. Overestimation resulted from false-positive detection of short ORFs, usually pseudogenic remnants of lost genes. Some cases of short ORFs had no homologous ORFs in BBge or BPam, nor was there sufficient support in database comparisons (BlastP, TBlastX, Pfam, CDD) for them to be annotated as CDS.
Functional Analysis
The principal difference of the BCpu genome in comparison with the other two blattabacterial genomes is the loss of several biosynthetic pathways (fig. 1 and supplementary fig. 1, Supplementary Material online), especially the complete loss of all genes for the synthesis of the amino acids tryptophan (trpABCDEF), leucine (leuABCD), threonine/isoleucine/valine (ilvABCDGI and thrBC), methionine (metBCE), and cysteine (cysEK). All these genes are present in both BBge and BPam. Two other pathways, those for arginine and lysine, are affected by gene losses. In both cases, only the genes argH and lysA coding for the enzymes catalyzing the last steps have been lost.
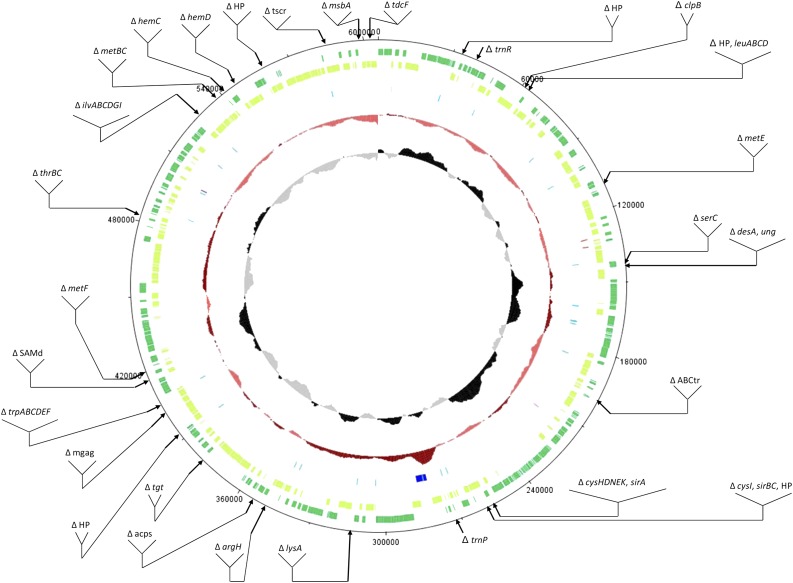
FIG. 1.—
Genome presentation of Blattabacterium sp. BCpu and indication of localization of lost genes present in BBge and BPam. From outward to inward are represented: CDS genes on the plus and minus strands, pseudogenes, RNA genes (rRNAs in dark blue, tRNAs in light blue, tmRNA, SRP RNA, and RNase P RNA in light, medium, and dark pink, respectively), DNA GC content (dark red above, light red below average), and GC skew (black skew above, light gray below average). Window and step sizes for the latter two rings were 10,000 and 200, respectively. Genes present in BBge and/or BPam and lost by BCpu are indicated. HP: hypothetical protein, ABCtr, ABC transporter, acps, putative acyl protein synthetase, mgag, mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase family protein, SAMd, radical SAM domain–containing protein, tscr, AsnC family transcriptional regulator.
The gene argH, coding for argininosuccinate lyase, has been lost as a single gene deletion. The neighboring genes gyrB and trmD are fully present with gene lengths similar to BBge and BPam, whereas of argH not even a remnant is present. ArgH converts L-argininosuccinate into L-arginine and fumarate. Furthermore, ArgH is one of the four enzymes constituting the urea cycle. The other three genes, rocF/speB, argF, and argG, as well as all other genes of the arginine biosynthesis pathway present in BBge and BPam, namely argABCDEF and carAB, have been retained. According to their amino acid sequences, all domains and active sites of these enzymes were present, thus, they seem to be fully functional. The isolated loss of argH results in an interruption of the urea cycle leaving argininosuccinate as a dead end metabolite. However, the BCpu genome codes for two other lyases that show moderate similarity to ArgH, PurB and FumC, although for none of the two enzymes it is reported that they could act with arginine succinate as natural substrate (BRENDA database). Structural analysis suggests substitution of ArgH activity by PurB-BCpu or FumC-BCpu therefore as unlikely.
Another functional gene missing was lysA, coding for diaminopimelate decarboxylase, which catalyses the last step in lysine biosynthesis. Like argH, lysA was lost as a single gene between the loci trnS and lon, but in this case, the gene region remained present as an intergenic region of 1,255 bp, with corresponding lengths in BBge and BPam of 1,267 and 1,244 bp, respectively (supplementary table 2, Supplementary Material online). However, in BCpu, the gene sequence has severely deteriorated, leaving only a pseudogenic remnant of former lysA. Despite significant gene erosion by more than 20 inline stops and a deletion of approximately 20 amino acids (aa), the remnant gives BlastP matches with an e value of approximately 10−60 with the orthologs of BBge and BPam covering more than 90% of the corresponding CDSs of 412 and 409 aa, respectively. The other genes of the lysine biosynthesis pathway present in BBge and BPam, dapABDEF, dapC/argD, and lysC, have been retained and appear to be functional. Exclusive loss of lysA and concomitant retention of all other lysine pathway enzymes is also reported for Wigglesworthia glossinidia, Candidatus Riesia pediculicola, and Baumannia cicadellinicola. Meso-diaminopimelate, the product of diaminopimelate epimerase (DapF), last but one enzyme in the pathway, is also involved in peptidoglycan synthesis as one of the two nonproteinogenic amino acids in the pentapeptide bridges. LysA-BBge and LysA-BPam contain a single domain PLPDE_III_DapDC[cd06828]. Arginine decarboxylase SpeA (EC 4.1.1.19) catalyses a homologous reaction, the decarboxylation of arginine to agmatine, and the enzyme is coded in the BCpu genome. SpeA-BCpu contains the domain PLPDE_III_ADC[cd06830]. Both domains belong to the Type III pyridoxal 5-phosphate (PLP)–dependent enzymes superfamily, cl00261 (homologous to Pfam clan CL0036 TIM_barrel) and share a common architecture. Despite this similarity between SpeA-BCpu and LysA-BBge/BPam, there is conflicting evidence that SpeA might also utilize diaminopimelate as a substrate and decarboxylate it to lysine. In Selenomonas ruminantium, it was shown that the enzyme acts on L-lysine at a rate of about 10% that corresponding to L-arginine (Km values 50 and 5.6 mM, respectively; Liao et al. 2008), whereas Giles and Graham (2007) reported that in the intracellular pathogen Chlamydophila pneumoniae arginine decarboxylase showed no activity on L-lysine. In Escherichia coli and Pseudomonas sp. L-lysine is reported as an inhibitor of arginine decarboxylase (Blethen et al. 1968; Boeker and Snell 1971; Rosenfeld and Roberts 1976).
For arginine and lysine, whose syntheses are both blocked in the last step of their biosynthetic pathways, there is no clear support that the action of PurB/FumC and SpeA, respectively, could substitute the lost enzyme activities of ArgH and LysA. Under this presumption, the loss of the terminal reactions also renders the Cryptocercus symbiont auxotrophic for these two amino acids, in addition to the seven whose pathways were completely deleted in BCpu as compared with BBge and BPam. Losses of argH and lysA differ regarding their genomic context suggesting that they have occurred at rather different time points in BCpu lineage evolution. Loss of argH can be addressed as a relatively old deletion because no remnants of the gene are present in the genome, whereas in the case of lysA an intergenic region of 1.3 kb containing a pseudogenic remnant indicates that loss of function and initiation of gene erosion occurred much more recently (Silva et al. 2001).
Beside the amino acid routes, gene losses affected other pathways: In the porphyrinogen and the folate biosynthesis pathways, four genes were lost in BCpu: cysH, sirA, hemC, and metF (supplementary table 2A, Supplementary Material online). Apart from these losses of complete or partial pathways, there are several other gene losses in BCpu as compared with BBge and BPam, usually as isolated deletions of one CDS gene without further affecting gene order. These “single gene” deletions account for 24 of a total of 31 deletion events.
Besides the genes lost by BCpu that are present in both BBge and BPam, there are also eight protein genes, which have been differentially lost. These gene losses appear to have occurred independently in the different lineages. In the sulfate reduction pathway, the genes cysDIN, present in BBge but absent in BPam have been lost. In this pathway, the only functional gene present in BCpu is the one coding for sulphite reductase (NADPH) flavoprotein subunit alpha CysJ, a flavin reductase (EC 1.5.1.30), which is described to have additional activities such as electron relay (Siegel and Davis 1974) and has, therefore, been retained. DesA, a fatty acid desaturase, present in BPam as BPLAN_488 but absent in BBge is also absent in BCpu. DesA catalyses the insertion of double bonds in the delta position of fatty acids.
A particular case of metabolic reduction is the loss of one of two functions of the bifunctional enzyme ThrA, which has aspartate kinase (EC 2.7.2.4.) and homoserine dehydrogenase (EC 1.1.1.3.) activities in BBge and BPam. In BCpu, the corresponding protein BLBCPU_469 is only 530 aa long compared with 815 amino acids in the other two genomes, which is due to the loss of the C-terminal domains responsible for homoserine dehydrogenase activity. The monofunctionalization of the bifunctional ThrA is consistent with the loss of the threonine and methionine pathway genes. A homoserine dehydrogenase activity is no longer necessary for BCpu (see metabolic model in supplementary fig. 1, Supplementary Material online). The monofunctional aspartate kinase was annotated as LysC.
On the other hand, BCpu possesses three additional genes coding for functional proteins that are not present in BBge and BPam (supplementary table 2C, Supplementary Material online): a 179 amino acid long flavodoxin-like fold protein, which presumably acts as a NAD(P)H dehydrogenase on quinones (BLBCPU_583, ywrO), a 283 amino acid hypothetical membrane spanning protein (BLBCPU_006), and the gene for the protein component of RNase P, rnpA (BLBCPU_480), present as a functional ORF, whereas both the rnpA genes of BBge and BPam are pseudogenized. Finally, there are ten genes present in BCpu but only in one of the other two genomes (supplementary table 2B, Supplementary Material online). Most prominent are three genes for enzymes of the Krebs cycle that are fully present in BCpu and BBge, aconitate hydratase, acnA, isocitrate dehydrogenase (NADP+), icd, and citrate synthase, gltA, which are missing in BPam.
Pan-Genome of Blattabacterium sp.
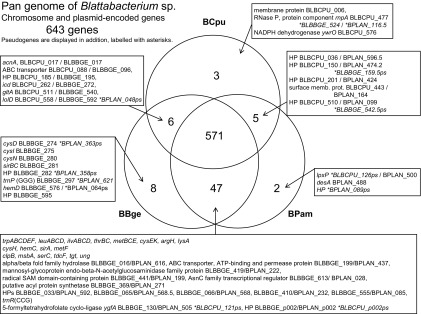
In total, 643 genes were found to be present, constituting the pan-genome of Blattabacterium sp. (fig. 2), whereas the core genome common to all three organisms consists of 571 genes (88.8% of all genes). The largest group of differentially present genes is those absent in BCpu but present in both BBge and BPam, which are 47 in total (supplementary table 2A, Supplementary Material online). Another ten genes present in either BBge or BPam are also absent in BCpu. The losses occurred as deletions of up to six consecutive genes.
FIG. 2.—
Pan and core genome of the three compared blattabacteria, BCpu, BBge, and BPam. The gene counts are based on the orthology table (supplementary table 1, Supplementary Material online). Pseudogenes are considered absent.
The core genome contains all genes belonging to the “informational” COG categories J, K, and L, which are present in BBge and BPam. With the exception of a putative transcription regulator (BLBBGE_613/BPLAN_028), they are all also present in BCpu.
Genetic Imprint of Amino Acid Pathway Losses
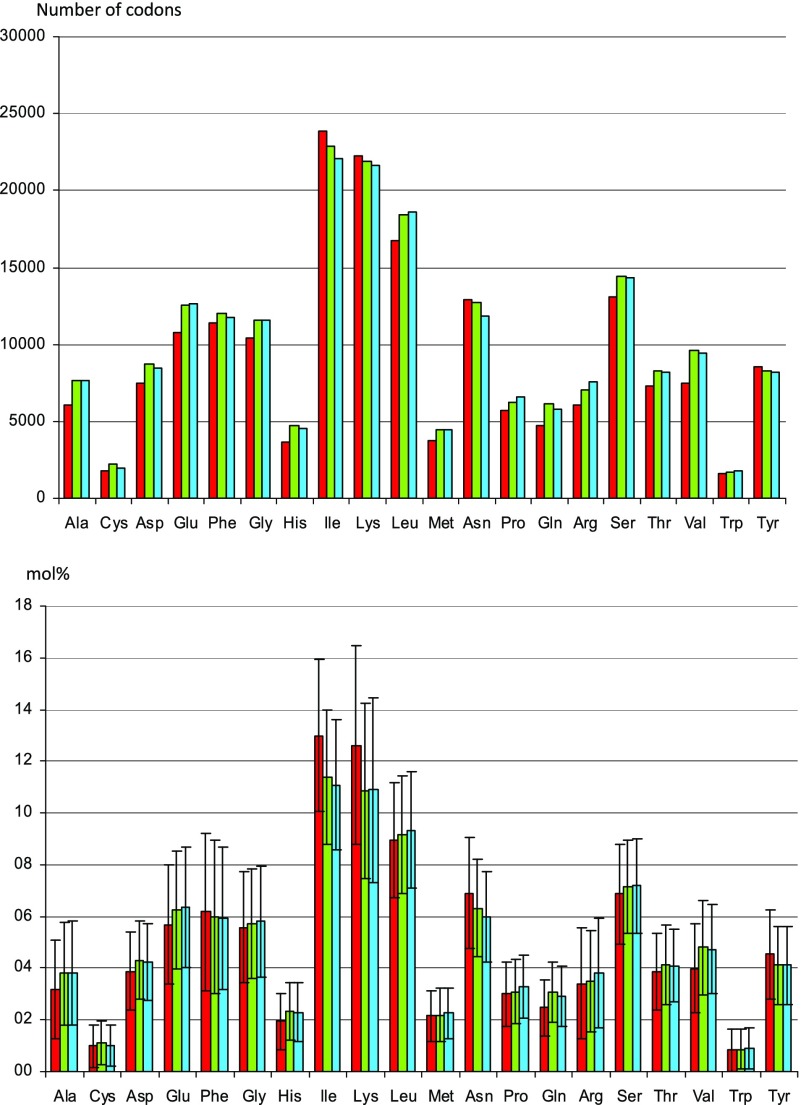
There are no significant differences in the utilization of the amino acids whose biosynthetic pathways have been lost in BCpu compared with BBge and BPam (fig. 3). Indeed, the two essential amino acids, isoleucine and lysine, are present in a higher percentage (12.8% and 12.0% vs. 11.3/11.1% and 10.8/10.9% in BBge and BPam, respectively).
FIG. 3.—
Distribution of amino acids in the deduced proteomes of BCpu, BBge, and BPam. All functional CDS (not considering pseudogenes) coded on the chromosome were analyzed. Columns in red BCpu, green BBge, and blue BPam. Upper graph shows total number of codons, lower graph average values ± standard deviation.
Nor is the loss of the amino acid pathways reflected in the codon usage: BCpu possesses only 32 tRNAs in comparison to 34 annotated tRNAs in BBge and 33 in BPam. The three strains possess one tRNA-Pro (anticodon UGG), which is most frequently observed for decoding Pro codons in bacterial genomes (Rocha 2004) and is able to decode the four proline codons. In addition, in BBge there is a second tRNA-Pro (anticodon GGG), able to decode CCC and CCU codons. This gene is absent in BCpu, whereas in BPam, it is annotated as a pseudogene, BPLAN_621, probably because it is not recognized by most programs detecting tRNA genes like tRNAscan-SE and Aragorn. The software TFAM1.3, specialized in tRNA classification, detects the gene as valid tRNA and classifies it as proline type. Also Rfam recognizes a tRNA gene with an e value of about 10−7, as compared with values between 10−10 and 10−14 for other tRNAs. These data indicate that the tRNA-Pro gene BPLAN_621 could in fact be functional in BPam. Interestingly, the anticodon sequence of BPLAN_621 has changed from GGG to AGG. This anticodon, however, also codes for proline and is putatively able to decode the four Pro codons. Pro-tRNAs with anticodon AGG are quite rare; however, the close relative of blattabacteria Candidatus Amoebophilus asiaticus 5a2, an obligate endosymbiont of Acanthamoeba, possesses a tRNA of this type. Distribution of relative synonymous codon usage (RSCU) values for the four Pro coding (Table 2) is slightly different in BCpu relative to BBge and BPam. The loss of tRNA-Pro (GGG) in BCpu may explain the slight decrease in the use of CCC and CCU codons. tRNA-Arg (CCG) is the other tRNA gene that has been lost in BCpu but is present in both BBge and BPam (BLBBGE_041 and BLAN_592). This reduces the decoding of CGN codons to a single tRNA-Arg (anticodon ACG). This tRNA is not only able to decode the four codons but again it is the most frequently observed fulfilling this role in bacterial genomes (Rocha 2004). There are no substantial differences in the distribution of RSCU values for the six Arg triplets among the protein-coding genes of the three symbionts. This is likely due to the binding of anticodons of alternative tRNAs charged with the same amino acid to synonymous codons via wobble base pair recognition. This could buffer the tRNA losses without leading to a changed codon usage. Alternatively, the unaffected codon distribution for Arg can be interpreted as an indication that this loss is relatively recent and is not reflected in codon usage alterations yet. On the other hand, the effect in codon adaptation for the Pro codons suggests an ancient loss of tRNA-Pro (GGG).
Table 2.
Distribution of RSCU Values for Proline and Arginine Codons in BCpu, BBge, and BPam
| BCpu |
BBge |
BPam |
||||
| Count | RSCU | Count | RSCU | Count | RSCU | |
| Pro | ||||||
| CCU | 2848 | 2 | 3354 | 2.15 | 3395 | 2.07a |
| CCC | 295 | 0.21 | 469 | 0.3 | 507 | 0.31a |
| CCA | 2395 | 1.68 | 2124 | 1.36 | 2287 | 1.4 |
| CCG | 170 | 0.12 | 290 | 0.19 | 363 | 0.22 |
| Arg | ||||||
| CGU | 1399 | 1.39 | 1491 | 1.27 | 1632 | 1.29 |
| CGC | 35 | 0.03 | 103 | 0.09 | 154 | 0.12 |
| CGA | 640 | 0.64 | 585 | 0.5 | 633 | 0.5 |
| CGG | 140 | 0.14 | 102 | 0.09 | 174 | 0.14 |
| AGA | 3449 | 3.43 | 4370 | 3.72 | 4352 | 3.44 |
| AGG | 378 | 0.38 | 393 | 0.33 | 656 | 0.52 |
Note.—Presence of complementary acceptor tRNA anticodons is indicated in bold. Lost tRNA genes in BCpu are indicated in italics.
Annotated as pseudogene BPLAN_621ps in BPam, functional according to TFAM with anticodon AGG (see text).
Discussion
Comparative Genome Evolution in Blattabacteria
The symbiont of C. punctulatus contains a reduced blattabacterial genome characterized by the loss of 47 functional genes present in both BBge and BPam. Twenty-three of them are involved in amino acid biosynthesis, making the decline in amino acid production capacity the most distinctive difference compared with BBge and BPam genomes. BCpu has completely lost the pathways for seven amino acids, six essential ones, isoleucine, leucine, valine, threonine, methionine and tryptophan, and the nonessential cysteine. The pathways of two more amino acids, essential lysine and nonessential arginine, are inactivated by the loss of single genes, lysA and argH. All other genes involved in arginine and lysine biosynthesis and present in the genomes of the other two symbionts are retained in the BCpu genome and, according to domain and active center analysis, appear to be functional.
Compared with BBge and BPam, the differential absence of genes in BCpu must be a consequence of different adaptations in the respective cockroach lineages. Lo et al. (2003) concluded a radiation period during the early Cretaceous (approximately 135–150 Mya) resulted in the establishment of several independent lineages, among them Cryptocercidae, Blattellidae, and Blattidae. The latter family includes P. americana. Among the genes differentially present in the three strains are those for the porphyrinogen synthesis pathway, sirABC (=cysG) and hemCD, and for sulfate reduction, cysDIN, which are only present in BBge. According to the phylogenetic scenario of Lo et al. (2003), these genes would have been lost in the symbionts of the Cryptocercidae and Blattidae lineages independently, by convergent evolution. In reduced symbiont genomes, independent gene losses are frequently observed in parallel evolving lineages (Silva et al. 2001; Gómez-Valero et al. 2004). An alternative explanation is that Cryptocercidae and Blattidae evolved together for a time after the split from the Blattellidae ancestor, and sirABC and hemCD were lost during this time. However, because the gene losses are likely consequences of adaptive processes, the most parsimonious option is the convergent evolution scenario in congruence with the phylogenetic model showing cocladogenesis between symbionts and hosts (Lo et al. 2003).
The synteny of gene order is completely conserved among all three blattabacterial genomes, except a singular inversion of a 19-kb fragment in the origin region containing 16 (BCpu) or 17 genes (BBge and BPam). The rest of the chromosome shows identical gene order among all three blattabacteria studied. No indications were found of other recombination events, thus supporting the general model for genome evolution (Moran et al. 2008; Moya et al. 2008), involving gene inactivation and subsequent loss. The 47 genes lost on the chromosome correspond to 31 regions of loss spanning between one and seven consecutive genes present in BBge and/or BPam (supplementary table 2A, Supplementary Material online). Here, the 23 absent genes for the amino acid synthesis pathways correspond to just nine regions spanning between 1.2 and 6.5 kb in the BBge and BPam genomes.
BBge and BPam show biosynthetic capacities for 16 amino acids and cannot produce asparagine, glutamine, proline, or glycine (López-Sánchez et al. 2009). Auxotrophy for the nine additionally lost amino acids (fig. 4) implies that these amino acids must be provided by the host. The seven essential ones cannot be synthesized by the insect. If access to these amino acids by BCpu is negatively affected it would be reflected in the overall use of these amino acids in the proteome. However, the relatively unaltered overall distribution of amino acids in BCpu indicates that the symbiont also has unrestricted access to these nine unsynthesized amino acids. Therefore, the host C. punctulatus must assimilate the essential amino acids either from the diet or from provisioning by intestinal microorganisms.
FIG. 4.—
Biosynthetic capacities of BCpu for production of amino acids. Amino acids produced by all three blattabacteria (BBge, BPam, and BCpu) are shown in normal font. Amino acids that cannot be produced by any of the blattabacteria are underlined with dashes. Losses in BCpu are indicated in italics. Losses of complete pathways are double underlined and partial losses due to loss of terminal step enzymes (argH, lysA) are shown in bold.
Substitution of Amino Acid Synthesis Capacities
This wood roach lives in habitats in moderate climates and feeds primarily on rotten logs, that is, decaying wood (Nalepa and Bandi 1999; Nalepa et al. 2002; Kambhampati and Peterson 2007). Generally, wood decomposition (Leonowicz et al. 1999; Martínez et al. 2005) is launched by different types of filamentous fungi, basically, basidiomycetes and ascomycetes, which trigger lignocellulose breakdown. Once wood putrefaction has started, a complex secondary microbiota colonizes it, speeding up degradation. Cryptocercus punctulatus ingests the colonizing microbiota together with the wood (Bell et al. 2007). In contrast to the limited nutrient supply and recalcitrance of the wood fraction, the decomposing microorganisms are a rich and easily degradable nutrient source. This resource may be sufficient to provide the host and blattabacterial symbiont with all necessary amino acids and cofactors. Alternatively, surplus production of amino acids and cofactors by members of the hindgut microbiota could be also a source. Potentially, both sources could contribute to the amino acid provision of BCpu.
In wood-feeding Cryptocercidae and termites, two features evolved in parallel: first, the acquisition of a highly specific intestinal microbiota of protists and second, the development of sub- and eusocial behavior in Cryptocercus and termites, respectively. Lower termites and Cryptocercus harbor unique anaerobic protists that are required for the host to feed on cellulose (reviewed in Lo and Eggleton 2011; Ohkuma and Brune 2011). As a consequence of coevolution between the host and its hindgut microbiota, the highly specific protist community became additionally involved in the host's development and life cycle and therefore connected to the evolution of sociality (Nalepa et al. 2001). For example, an ancestral detritivore cockroach first developed intraspecific coprophagy, an early and basic adaptation of gregarious detritivores to feed on feces of their conspecifics and subsequently developed proctodeal trophallaxis, the feeding of nymphs on hindgut material that assures the transmission of the symbiotic protists. Both these nutritional alterations led to further behavioral changes to a subsocial insect.
Wood feeding by Cryptocercus is in contrast with the feeding habits of most other cockroaches. Many cockroaches, especially the so-called urban pests like B. germanica and P. americana, are considered omnivores and thus less specialized than Cryptocercus. Combined feeding on rotten material and the acquired hindgut community permitted the loss of the amino acid-biosynthetic pathways in the BCpu genome, following the general tendency of reductive genome evolution found in obligatory insect endosymbionts (Moran et al. 2008; Moya et al. 2008). The genera Salganea and Panesthia (Blaberidae: Panesthiinae) in Eastern Asia are also wood-feeding cockroaches (Nalepa 2011) and might contain blattabacteria with similar adaptations like Blattabacterium BCpu. Likewise, most termites are xylophageous (Thorne and Traniello 2003; Korb 2007; Bignell 2011; Lo and Eggleton 2011). Most higher termites feed on drywood. However, some species use also dampwood as a nest and food source, like Termopsidae and Stolotermitidae. Rosengaus et al. (2003) compared drywood and dampwood termite species belonging to the families Termopsidae and Kalotermitidae, respectively, and found that microbial loads of ingested material, measured as colony-forming units cultured from nest material and cuticular washes, are positively correlated with ambient moisture. This supports the hypothesis that microbiota colonizing the rotten wood contribute to the uptake of essential amino acids by C. punctulatus.
Genome reduction of Blattabacterium BCpu appears to be a coherent process in this coevolutionary scenario involving C. punctulatus and both symbiotic partners, the Blattabacterium endosymbiont and the gut microbiota. An ancestral omnivorous cockroach possessed a Blattabacterium symbiont able to produce the essential amino acids and to provide them to the host. During the Cryptocercidae lineage evolution, feeding on rotten wood might have conveyed an additional input of readily digestible nutrients and simultaneously led to the evolution of a symbiotic hindgut microbiota. Bacterial and archaeal symbionts of the hindgut flagellates, as well as microorganisms freely living in the hindgut lumen, are likely to possess amino acid synthesis potential like that described for termite symbionts (Hongoh 2011). Thus, the essential amino acids supply from the diet or the hindgut symbionts may have become constant and copious enough to substitute the original amino acid provision from the fat body symbiont. Amino acid synthesis capacities of Blattabacterium BCpu became therefore redundant and were lost. In termites, Hongoh et al. (2008a, 2008b) report on the presence of phylotypes of bacterial protist symbionts with ample synthesis capacities for amino acids that are even able to fix N2. Hongoh (2011) describes the primary role of these endosymbionts as the production of amino acids and cofactors. During the course of evolution toward the termite lineage, this substitution process likely resulted in a complete replacement of all functions of the fat body symbiont by the hindgut microbiota. In termites, the role of blattabacteria in cockroach uric acid metabolism is adopted by uricolytic bacteria in the gut (reviewed in Brune and Ohkuma 2011). Cockroach endosymbionts (Blattabacterium sp.) may help in the N recycling from the fat body-stored urates (López-Sánchez et al. 2009). In this scenario, the C. punctulatus symbiont BCpu has an intermediary role compared with “high capacity” blattabacteria, like BBge and BPam, and the entire loss of the Blattabacterium symbiont in extant termites.
A situation was recently described for the leaf-cutter ant Atta, which is comparable to the massive loss of amino acid synthesis capacities in a symbiotic scenario (Suen et al. 2011). This insect group, native in tropic New World forests, feeds on fungus cultivated on freshly cut leaves. Obligate ant–fungus mutualism permitted genetic modifications, among them the loss of arginine biosynthesis genes. In comparison to other Hymenoptera, Atta is missing ArgH and ArgG (Argininosuccinate synthase, EC 6.3.4.5), the two last steps of arginine synthesis. Authors have also suggested that the nutrient-rich diet of the fungus permitted these specific gene losses. Interestingly, another member of Formicidae, the omnivorous carpenter ant Camponotus floridanus, lives in a mutual symbiosis with the endosymbiont Blochmannia floridanus, a Gammaproteobacterium, in which arginine (a supposed nitrogen reservoir for the insect) plays a key role because its biosynthesis is shared between bacterium and host (Gil et al. 2003). Blochmannia floridanus disposes of the gene repertoire to transform glutamine into citrulline, argABCDEF, whereas the host C. floridanus performs the last two steps of Arg biosynthesis, the conversion of citrulline to arginine, catalyzed by ArgG and ArgH.
Concluding Remarks
The lifestyle of Cryptocercus as temperate forest dweller is characterized by a diet that includes probable access to essential amino acids and the presence of a hindgut microbiota as potential amino acid producers. These sources might have permitted an evolutionary substitution process and thus a highly adaptive genome reduction of its fat body symbiont. Cryptocercus is an attractive model organism due to its phylogenetically intermediate position between other Blattaria and Isoptera. The particular lifestyle and 2-fold symbiosis with fat body flavobacteria and hindgut flagellates make this family a complex ecological model for further interdisciplinary studies.
Supplementary Material
Supplementary tables 1 and 2 and figure 1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Ministerio de Ciencia e Innovación, Spain (BFU2009-12895-C02-01/BMC to A.L.) and the Generalitat Valenciana, Spain (Prometeo/2009/092 to A.M.). A.N. was supported by a Marie Curie Reintegration grant from the European Commission (FP7-PEOPLE-2007-2-2-ERG-206104). The authors are grateful to Christina Nalepa, University of North Carolina, for provisioning C. punctulatus specimen and for helpful discussions. The expert support by the sequencing service of the University of Valencia, Servei Central de Suport a la Investigació Experimental (SCSIE, Dr Amparo Martínez) is highly acknowledged.
References
- Aksoy S, Rio RV. Interactions among multiple genomes: tsetse, its symbionts and trypanosomes. Insect Biochem Mol Biol. 2005;35:691–698. doi: 10.1016/j.ibmb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Bandi C, et al. Flavobacteria as intracellular symbionts in cockroaches. Proc R Soc Lond B Biol Sci. 1994;257:43–48. doi: 10.1098/rspb.1994.0092. [DOI] [PubMed] [Google Scholar]
- Bandi C, et al. The establishment of intracellular symbiosis in an ancestor of cockroaches and termites. Proc Biol Sci. 1995;259:293–299. doi: 10.1098/rspb.1995.0043. [DOI] [PubMed] [Google Scholar]
- Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Belda E, Moya A, Bentley S, Silva FJ. Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics. 2010;11:449. doi: 10.1186/1471-2164-11-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell WJ, Roth LM, Nalepa CA. Cockroaches: ecology, behaviour, and natural history. Baltimore (MD): Johns Hopkins University Press; 2007. [Google Scholar]
- Besemer J, Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell DE. Morphology, physiology, biochemistry and functional design of the termite gut: an evolutionary wonderland. In: Bignell DE, Roisin Y, Lo N, editors. Biology of termites: a modern synthesis. Dordrecht (the Netherlands): Springer; 2011. pp. 375–412. [Google Scholar]
- Blethen SL, Boeker EA, Snell EE. Arginine decarboxylase from Escherichia coli. I. Purification and specificity for substrates and coenzyme. J Biol Chem. 1968;243:1671–1677. [PubMed] [Google Scholar]
- Boeker EA, Snell EE. Arginine decarboxylase (Escherichia coli B) Methods Enzymol. 1971;17B:657–662. [Google Scholar]
- Brinza L, et al. Systemic analysis of the symbiotic function of Buchnera aphidicola, the primary endosymbiont of the pea aphid Acyrthosiphon pisum. C R Biol. 2009;332:1034–1049. doi: 10.1016/j.crvi.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Brune A, Ohkuma M. Role of the termite gut microbiota in symbiotic digestion. In: Bignell DE, Roisin Y, Lo N, editors. Biology of termites: a modern synthesis. Dordrecht (the Netherlands): Springer; 2011. pp. 439–475. [Google Scholar]
- Chevreux B, Wetter T, Suhai S. In: Proceedings of the German Conference on Bioinformatics (GCB), 1999 Oct 4–6; Hannover, Germany. Braunschweig (Germany): GBF, Dep. of Bioinformatics. 1999. Genome sequence assembly using trace signals and additional sequence information; pp. 45–56.. Available from: http://www.bioinfo.de/isb/gcb99/talks/chevreux/ [Google Scholar]
- Clark JW, Hossain S, Burnside CA, Kambhampati S. Coevolution between a cockroach and its bacterial endosymbiont: a biogeographical perspective. Proc Biol Sci. 2001;268:393–398. doi: 10.1098/rspb.2000.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland L, Hall SR, Sanders EP, Collier J. The wood feeding roach Cryptocercus, its protozoa, and the symbiosis between protozoa and roach. Mem Am Acad Arts Sci. 1934;17:185–342. [Google Scholar]
- Cochran DG. Nitrogen excretion in cockroaches. Annu Rev Entomol. 1985;30:29–49. [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, et al. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc Natl Acad Sci U S A. 2003;100:9388–9393. doi: 10.1073/pnas.1533499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles TN, Graham DE. Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae. J Bacteriol. 2007;189:7376–7383. doi: 10.1128/JB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Valero L, Latorre A, Silva FJ. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol Biol Evol. 2004;21:2172–2181. doi: 10.1093/molbev/msh232. [DOI] [PubMed] [Google Scholar]
- Harrison CP, Douglas AE, Dixon AFG. A rapid method to isolate symbiotic bacteria from aphids. J Invertebr Pathol. 1989;53:427–428. [Google Scholar]
- Hongoh Y. Toward the functional analysis of uncultivable, symbiotic microorganisms in the termite gut. Cell Mol Life Sci. 2011;68:1311–1325. doi: 10.1007/s00018-011-0648-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongoh Y, et al. Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc Natl Acad Sci U S A. 2008a;105:5555–5560. doi: 10.1073/pnas.0801389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongoh Y, et al. Genome of an endosymbiont coupling N2 fixation to cellulolysis within protist cells in termite gut. Science. 2008b;322:1108–1109. doi: 10.1126/science.1165578. [DOI] [PubMed] [Google Scholar]
- Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inward D, Beccaloni G, Eggleton P. Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol Lett. 2007;3:331–335. doi: 10.1098/rsbl.2007.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhampati S, Peterson AT. Ecological niche conservation and differentiation in the wood-feeding cockroach, Cryptocercus, in the United States. Biol J Linn Soc. 2007;90:457–466. [Google Scholar]
- Klass KD, Nalepa C, Lo N. Wood-feeding cockroaches as models for termite evolution (Insecta: Dictyoptera): Cryptocercus vs. Parasphaeria boleiriana. Mol Phylogenet Evol. 2008;46:809–817. doi: 10.1016/j.ympev.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Korb J. Termites. Curr Biol. 2007;17:R995–R999. doi: 10.1016/j.cub.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre A, Gil R, Silva FJ, Moya A. Chromosomal stasis versus plasmid plasticity in aphid endosymbiont Buchnera aphidicola. Heredity. 2005;95:339–347. doi: 10.1038/sj.hdy.6800716. [DOI] [PubMed] [Google Scholar]
- Leonowicz A, et al. Biodegradation of lignin by white rot fungi. Fungal Genet Biol. 1999;27:175–185. doi: 10.1006/fgbi.1999.1150. [DOI] [PubMed] [Google Scholar]
- Liao S, et al. Occurrence of agmatine pathway for putrescine synthesis in Selenomonas ruminantium. Biosci Biotechnol Biochem. 2008;72:445–455. doi: 10.1271/bbb.70550. [DOI] [PubMed] [Google Scholar]
- Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol Biol Evol. 2003;20:907–913. doi: 10.1093/molbev/msg097. [DOI] [PubMed] [Google Scholar]
- Lo N, Beninati T, Stone F, Walker J, Sacchi L. Cockroaches that lack Blattabacterium endosymbionts: the phylogenetically divergent genus Nocticola. Biol Lett. 2007;3:327–330. doi: 10.1098/rsbl.2006.0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, Eggleton P. Termite phylogenetics and co-cladogenesis with symbionts. In: Bignell DE, Roisin Y, Lo N, editors. Biology of termites: a modern synthesis. Dordrecht (the Netherlands): Springer; 2011. pp. 27–50. [Google Scholar]
- Lo N, et al. Save Isoptera: a comment on Inward et al. Biol Lett. 2007;3:562–563. doi: 10.1098/rsbl.2007.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez MJ, et al. Blattabacteria, the endosymbionts of cockroaches, have small genome sizes and high genome copy numbers. Environ Microbiol. 2008;10:3417–3422. doi: 10.1111/j.1462-2920.2008.01776.x. [DOI] [PubMed] [Google Scholar]
- López-Sánchez MJ, et al. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 2009;5:e1000721. doi: 10.1371/journal.pgen.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin A, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa K, Park YC, Lo N. Phylogeny of endosymbiont bacteria harbored by the woodroach Cryptocercus spp. (Cryptocercidae: Blattaria): molecular clock evidence for a late Cretaceous–early Tertiary split of Asian and American lineages. Mol Phylogenet Evol. 2005;36:728–733. doi: 10.1016/j.ympev.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Martínez AT, et al. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol. 2005;8:195–204. [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci U S A. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci U S A. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moya A, Peretó J, Gil R, Latorre A. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet. 2008;9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa CA. Colony composition, protozoan transfer and some life history characteristics of the woodroach Cryptocercus punctulatus Scudder. Behav Ecol Sociobiol. 1984;14:273–279. [Google Scholar]
- Nalepa CA. Altricial development in wood-feeding cockroaches: the key antecedent of termite euscociality. In: Bignell DE, Roisin Y, Lo N, editors. Biology of termites: a modern synthesis. Dordrecht (the Netherlands): Springer; 2011. pp. 69–95. [Google Scholar]
- Nalepa CA, Bandi C. Phylogenetic status, distribution, and biogeography of Cryptocercus (Dictyoptera: Cryptocercidae) Ann Entomol Soc Am. 1999;92:292–330. [Google Scholar]
- Nalepa CA, Bignell DE, Bandi C. Detritivory, coprophagy, and the evolution of digestive mutualisms in Dictyoptera. Insect Soc. 2001;48:194–201. [Google Scholar]
- Nalepa CA, Luykx P, Klass KD, Deitz LL. Distribution of karyotypes of the Cryptocercus punctulatus species complex (Dictyoptera: Cryptocercidae) in the Southern Appalachians: relation to habitat and history. Ann Entomol Soc Am. 2002;95:276–287. [Google Scholar]
- O'Donnell M. Insect excretory mechanisms. In: Simpson SJ, editor. Advances in insect physiology. Vol. 35. New York: Academic Press; 2008. pp. 1–122. [Google Scholar]
- Ohkuma M, Brune A. Diversity, structure, and evolution of the termite gut microbial community. In: Bignell DE, Roisin Y, Lo N, editors. Biology of termites: a modern synthesis. Dordrecht (the Netherlands): Springer; 2011. pp. 413–438. [Google Scholar]
- Pérez-Brocal V, Gil R, Moya A, Latorre A. New insights on the evolutionary history of aphids and their primary endosymbiont Buchnera aphidicola. Int J Evol Biol. 2011;2011:250154. doi: 10.4061/2011/250154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP. Codon usage bias from tRNA's point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004;14:2279–2286. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld HJ, Roberts J. Arginine decarboxylase from a Pseudomonas species. J Bacteriol. 1976;125:601–607. doi: 10.1128/jb.125.2.601-607.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengaus RB, Moustakas JE, Calleri DV, Traniello JF. Nesting ecology and cuticular microbial loads in dampwood (Zootermopsis angusticollis) and drywood termites (Incisitermes minor, I. schwarzi, Cryptotermes cavifrons) J Insect Sci. 2003;3:31. doi: 10.1093/jis/3.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sabree ZL, Degnan PH, Moran NA. Chromosome stability and gene loss in cockroach endosymbionts. Appl Environ Microbiol. 2010;76:4076–4079. doi: 10.1128/AEM.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabree ZL, Kambhampati S, Moran NA. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci U S A. 2009;106:19521–19526. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Esser S, et al. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. J Bacteriol. 2010;192:1045–1057. doi: 10.1128/JB.01379-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu S, Wilson AC. Genomic revelations of a mutualism: the pea aphid and its obligate bacterial symbiont. Cell Mol Life Sci. 2011;68:1297–1309. doi: 10.1007/s00018-011-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel LM, Davis PS. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of enterobacteria. IV. The Escherichia coli hemoflavoprotein: subunit structure and dissociation into hemoprotein and flavoprotein components. J Biol Chem. 1974;249:1587–1598. [PubMed] [Google Scholar]
- Silva FJ, Belda E, Talens SE. Differential annotation of tRNA genes with anticodon CAT in bacterial genomes. Nucleic Acids Res. 2006;34:6015–6022. doi: 10.1093/nar/gkl739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva FJ, Latorre A, Moya A. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- Suen G, et al. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 2011;7:e1002007. doi: 10.1371/journal.pgen.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tåquist H, Cui Y, Ardell DH. TFAM 1.0: an online tRNA function classifier. Nucleic Acids Res. 2007;35:W350–W353. doi: 10.1093/nar/gkm393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne BL, Traniello JF. Comparative social biology of basal taxa of ants and termites. Annu Rev Entomol. 2003;48:283–306. doi: 10.1146/annurev.ento.48.091801.112611. [DOI] [PubMed] [Google Scholar]
- Van Domselaar GH, et al. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33:W455–W459. doi: 10.1093/nar/gki593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Wheeler DE. Remaining flexible in old alliances: functional plasticity in constrained mutualisms. DNA Cell Biol. 2009;28:371–382. doi: 10.1089/dna.2009.0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Wernegreen JJ. Unprecedented loss of ammonia assimilation capability in a urease-encoding bacterial mutualist. BMC Genomics. 2010;11:687. doi: 10.1186/1471-2164-11-687. [DOI] [PMC free article] [PubMed] [Google Scholar]