Synopsis
The α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) subfamily of ionotropic glutamate receptors (iGluRs) is essential for fast excitatory neurotransmission in the central nervous system. The malfunction of AMPARs has been implicated in many neurological diseases including Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. The active channels of AMPARs and other iGluR subfamilies are tetramers formed exclusively by assembly of subunits within the same subfamily. It has been proposed that the assembly process is controlled mainly by the extracellular amino-terminal domain (ATD) of iGluR. In addition, ATD has also been implicated in synaptogenesis, iGluR trafficking, and trans-synaptic signaling, through unknown mechanisms. We report here a 2.5 Å resolution crystal structure of the ATD of GluA1. Comparative analyses of the structure of GluA1-ATD and other subunits sheds light on our understanding of how ATD drives subfamily-specific assembly of AMPARs. In addition, analysis of the crystal lattice of GluA1-ATD suggests a novel mechanism by which the ATD might participate in inter-tetramer AMPAR clustering, as well as in trans-synaptic protein-protein interactions.
Keywords: ion channel, glutamate receptor, AMPA receptor, amino-terminal domain, structural biology
Introduction
Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels that form transmembrane, cation-permeable channels. iGluRs exist as three distinct subfamilies according to their agonist specificity and amino acid sequence: α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs), kainate receptors, and N-methyl-D-aspartate receptors (NMDARs). Activation of iGluRs is critical for induction of some forms of long-term potentiation (LTP), a type of synaptic plasticity associated with learning and memory [1, 2]. iGluRs play crucial roles in normal neuronal function and in neurological disease, and thus are being actively pursued as therapeutic targets for the treatment of amyotrophic lateral sclerosis, neuropathic pain, major depression, Alzheimer’s disease, and Parkinson’s disease.
Despite having divergent functional properties, iGluR family members adopt a common modular architecture. A typical iGluR subunit contains four distinct domains: an extracellular amino-terminal domain (~ 400 residues, ATD), an extracellular ligand-binding domain (~ 300 residues, LBD), three transmembrane domains (TM1-3) plus a re-entrant pore loop (P), and an intracellular carboxy-terminal domain (Figure 1a). Active iGluR channels are tetramers formed exclusively by assembly of subunits within the same subfamily. This process is thought to be mediated in part by the ATD through a mechanism that is not fully understood.
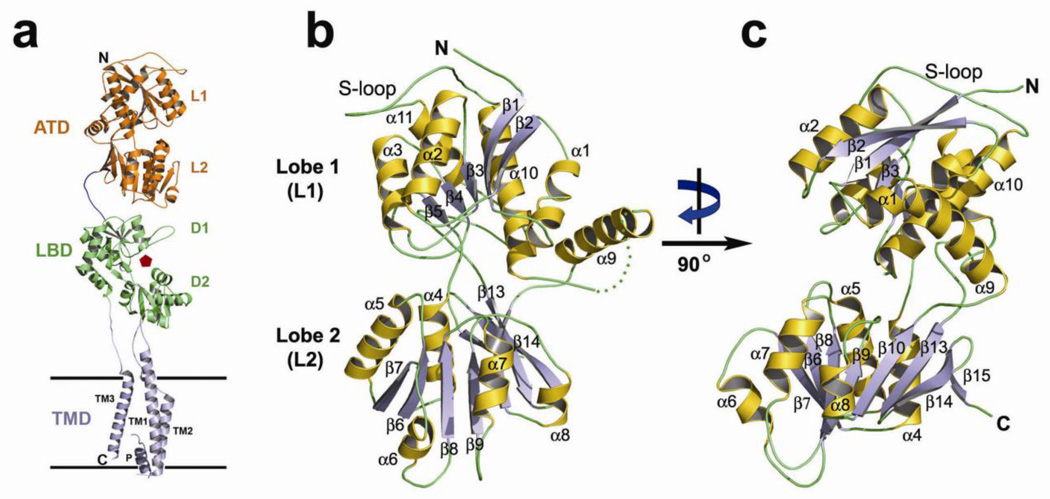
Figure 1. The structure of GluA1-ATD.
(a) iGluRs have a modular architecture that includes the amino-terminal domain (ATD), the ligand binding domain (LBD), the transmembrane ion channel domain (TMD) and the carboxy-terminal intracellular region. The ligand-binding pocket on LBD is defined as a red pentagon. Shown is one subunit based on crystal structure of the full length tetrameric GluA2 [6]. (b) Ribbon diagram of the structure of GluA1-ATD. The secondary structures are numbered in a sequential order and the α-helix, β-strand, and loop are colored in orange, light blue and green, respectively. The missing loop between helices α9 and α10 is shown as a dotted line. (c) View of the structure of GluA1-ATD from the side following a rotation of ~ 90° around a vertical axis.
Historically, structural biology has made significant contributions to our understanding of iGluR function. The first crystal structure of an iGluR, the LBD of GluA2, was determined in 1998 [3]. Since then, more than a hundred high-resolution crystal structures of iGluR LBD have been reported, including subunits from all three subfamilies of iGluR either in the apo form or in complex with a variety of agonists, antagonists, and modulators. Collectively, these structures have established the detailed molecular mechanisms underlying iGluR channel activation, inhibition, and desensitization [4, 5]. Nevertheless, the first crystal structure of a full length iGluR ion channel, the homomeric GluA2, was not accomplished until 2009 [6]. This extraordinary advance has yielded unprecedented information on the molecular architecture and symmetry of iGluR. Structural studies on full-length iGluRs are exceptionally difficult to perform, and to date have yielded only a single snapshot of a homomeric GluA2 bound with an antagonist. It is therefore not surprising that our understanding of iGluR structure and function continues to be derived from studies with isolated recombinant ATDs and LBDs.
In contrast to the well-characterized LBD, much less is known about the molecular properties of the ATD, even though this domain is crucial for the physiological function of iGluRs. For example, the spontaneous mouse mutation hotfoot is a recessive mutation characterized by cerebellar ataxia and jerky movement of the hind limbs, and is often caused by in-frame deletions of various regions of the ATD of GluRδ2 [7]. The functional activities of ATD fall into three general categories. (i) It is widely accepted that the iGluR-ATD guides receptor subfamily-specific assembly, ensuring that only subunits within the same subfamily assemble with one another [8–11]. Interestingly, the subfamily-specific assembly of tetrameric voltage-gated Shaker K+ channels is also determined by the amino-terminal T1 domain [12, 13]. (ii) The NMDAR-ATD modulates receptor function by providing binding sites for allosteric modulators that include protons, zinc ions, polyamines, and small organic molecules such as ifenprodil [1, 14–16]. However, no small molecules have been shown to bind the AMPAR-ATD or the ATD of kainate receptors. (iii) The ATD resides within the synaptic cleft, and thus may be involved in protein-protein interactions. By doing so, the ATD could regulate both dendritic spine morphogenesis and presynaptic stability [17, 18]. Some ATD binding partners have been identified, including neuronal pentraxins (Narp, NP1, and NPR) [19, 20] and N-cadherin [21] that bind to AMPAR-ATD, and an ephrin receptor that binds to NMDAR [22].
Our limited knowledge of the structure and function of the ATD is in part due to the challenge associated with recombinant protein production. Large-scale protein expression of the ATD using insect and mammalian cells has been successful only recently, and has resulted in a number of ATD structures, including the ATDs of GluA2, GluK2, GluK3, GluK5, and GluN2B [23–28]. Structural analyses have revealed several unique features of ATDs within each subfamily of iGluRs with respect to the protomer structure, subfamily-specific subunit assembly, and competence for ligand binding. These findings have clarified how the ATD prevents subunits from different subfamilies from assembling into a tetramer. However, the role played by the ATD in driving assembly of different subunits within a subfamily remains unclear. This is a critical process, as most native AMPAR channels are heteromers that co-assemble with the GluA2 subunit, which serves to decrease channel permeability to calcium ions. This problem has motivated us to perform structural studies on other subunits of AMPARs to complement the structure of the GluA2-ATD, which we reported in 2009 [23]. Here, we report the structure of the GluA1-ATD at 2.5 Å resolution. The crystal structures of GluA3 and GluN1-ATD were reported while this manuscript was in preparation [29, 30].
Experimental
Protein expression and purification
The rat GluA1-ATD (P4-A374) (amino acid numbering corresponds to the mature polypeptide after cleavage of endogenous signal peptide) was cloned into a modified pFastBac vector (Invitrogen). The human placental alkaline phosphatase signal peptide was added to the N-terminus of GluA1-ATD. A C-terminal Myc-tag and His8-tag following a PreScission protease cleavage site were introduced to facilitate purification and characterization. The protein was expressed and secreted from Spodoptera frugiperda (sf9) insect cells, and purified from media by Ni-NTA affinity chromatography (Qiagen). The C-terminal tags were removed by PreScission protease treatment after buffer exchange into 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA. The cleaved protein was then applied to an ion-exchange column (Mono S™ 5/50 GL, GE Healthcare) after a 5-fold dilution to pH 6.0, and subsequently eluted with a NaCl gradient. The peak fractions containing GluA1-ATD were exchanged into a buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 1 mM EDTA, and concentrated to ~ 10 mg/ml for crystallization.
For co-immunoprecipitation studies, GluA1-ATD was expressed and purified as described above, except that the Myc-tag and His-tag were not excised. Cloning, expression, and purification of GluA2-ATD were carried out as previously described [23]. As a negative control, a ~ 40 kDa bacterial protein with a C-terminal Myc-tag was expressed in Escherichia coli, and purified to homogeneity. All proteins were exchanged into the same buffer (25 mM Tris-HCl, pH 8.0, 200 mM NaCl, 0.2% NP-40) before assay.
Crystallization and diffraction data collection
Initial crystallization screens were carried out using a Phoenix crystallization robot (Art Robbins Instrument) and commercial high-throughput crystallization screen kits. After extensive manual optimization, the best GluA1-ATD crystals were grown by hanging-drop vapor diffusion at 18°C, in which the protein (10 mg/ml) was mixed in 1:1 ratio with a reservoir solution containing 100 mM sodium acetate, pH 5.0, 24% polyethylene glycol (PEG) 3350, and 200 mM MgCl2. Single crystals were obtained only by micro-seeding. The crystals were cryoprotected in the reservoir solution supplemented with 15% glycerol. The X-ray diffraction data were collected at −173°C at the microfocus beam line 12-2, Stanford Synchrotron Radiation Lightsource (SSRL), using a DECTRIS PILATUS 6M PIXEL detector. All data sets were processed and scaled by using iMOSFLM [31]. Data collection statistics are summarized in Table S1.
Structure determination and refinement
The structure of GluA1-ATD was determined by molecular replacement using Phaser [32]. One protomer of GluA2-ATD (PDB code 3H5V) was used as the search model to locate the four molecules of GluA1-ATD in one asymmetric unit. The preliminary structural model was subsequently refined with Phenix 1.5 [33] and re-built with COOT [34] in an iterative manner. Refinement progress was monitored with the free R value using a 5% randomly selected test set [35]. The GluA1-ATD structure was refined to 2.5 Å with Rwork/Rfree = 0.22/0.28. Structural refinement statistics are listed in Table S1. The coordinates and diffraction data for GluR1-ATD has been deposited in the Protein Data Bank (PDB code 3SAJ). Figures were prepared with PyMol (http://www.pymol.org).
Co-immunoprecipitation
Co-immunoprecipitation (co-IP) assays were carried out in a buffer containing 25 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 0.2% NP-40. Two antibodies were used for co-IP and Western immunoblotting (IB): a mouse anti-GluA2-ATD antibody (MAB397, Millipore) and a mouse anti-Myc antibody (9B11, Cell Signaling). The same amounts of proteins, antibodies, and Protein A resins were used in all experiments. The immunoprecipitation reactions were performed at 4°C overnight. After extensive washing, the immunocomplexes were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with anti-GluA2 or anti-Myc antibody. Westerns were visualized with an alkaline phosphatase-conjugated rabbit anti-mouse secondary antibody (Bio-Rad).
Results and Discussion
Purification, crystallization, and structure determination
Of all the modular domains of iGluR, the ATD has the largest sequence diversity across different families, showing only 0.2% sequence identity. The ATD also has the lowest sequence identity within each subfamily; for example, AMPARs have ~ 35% identity in the ATD, compared with 80% identity in the LBD and 87% in the transmembrane ion channel region [5]. Interestingly, a structure similarity search showed that the ATD adopts a structural fold similar to that of the bacterial leucine/isoleucine/valine-binding protein (LIVBP, a member of the periplasmic ligand binding protein family) [36], the LBD of metabotropic glutamate receptor (mGluR) [37], and the LBD of natriuretic peptide receptor (NPR) [38]. Nevertheless, the overall sequence identity between the ATD and these proteins is quite low (below 15%).
We expressed the GluA1-ATD (residues P4 to A374; numbering based on the mature GluA1) as a secreted form in sf9 insect cells. The secreted protein was directly purified from the media by Ni-NTA affinity chromatography followed by ion-exchange chromatography. The final yield was about 0.3 mg of purified protein per liter of cell culture.
Multiple crystal forms of GluA1-ATD were identified by robotic high-throughput screening followed by manual optimization. Most of the crystals diffracted weakly to ~ 6 Å resolution, consistent with our previous finding [23]. After extensive optimization of crystallization conditions, we focused on a condition using PEG 3350 as a precipitant, which yielded crystals that routinely diffracted to ~ 4 Å. Interestingly, the plate-like crystals grew from this condition only after micro-seeding. The original seed crystals were obtained from a drop set up by robot. This drop showed severe precipitation after setup; tiny crystals grew after a month at 18°C. Micro-seeding using these crystals yielded plate-like crystals that were further optimized. Single crystals grown under these conditions showed great variation in their thickness, and easily stacked on top of each other. This property significantly slowed the progress of structure determination. A large number of crystals have been screened and the best data set diffracted to about 2.5 Å (Table S1). The structure of GluA1-ATD was solved by molecular replacement (MR) using the structure of GluA2-ATD (PDB code 3H5V) as the search model. The structure was refined at 2.5 Å resolution (Rwork/Rfree= 0.22/0.28) and shows excellent crystallographic and stereochemical statistics (Table S1).
Overall architecture of GluA1-ATD
The crystal of GluA1-ATD shows four molecules in one asymmetric unit forming two pairs of dimers. Excellent electron densities were observed for 365 residues, but residues H260-W265 were not observed. GluA1-ATD has a two-domain flytrap-like structure (Figure 1b). The N-terminal lobe (L1) and the C-terminal lobe (L2) each have an α/β topology with a central β-sheets surrounded by α-helices, and are connected by three short inter-domain loops. All four crystallographically independent protomers have a similar conformation. Close inspection reveals that the overall structure of GluA1-ATD is similar to that of GluA2 and GluA3 [23–25, 29]. Pair-wise comparisons of Cα atoms yielded root mean square deviations (rmsd) of 0.8 Å (GluA1 vs. GluA2) and 0.9 Å (GluA1 vs. GluA3) in the core of the L1 domain. More diversity was observed in the L2 domain; ~ 1.3 Å between GluA1 and A2, and ~ 1.5 Å between GluA1 and A3.
Despite the conserved structural cores, there are five prominent differences between GluA1–3. First, a loop that connects helices α9 and α10 (H260–W265) has no visible electron density on GluA1-ATD, suggesting a highly flexible conformation in this area (Figure 1b). As a result, the helix α9 on GluA1-ATD swings away from the core of L1 by as far as 4.5 Å at the C-terminus of α9, compared to GluA1 and A2. Second, GluA1 has a shorter helix α6 (E165-F171) on L2 than does GluA2 (E169–E179). Furthermore, in comparison to GluA2, GluA1 has a shorter loop linking β7 and α6 (I159–E164 on GluA1 vs. V159-D168 on GluA2) and a longer linker between α6 and β8 (Q172-E179 on GluA1 vs. L180-R184 on GluA2). In contrast, GluA2 and GluA3 adopt very similar conformations in this region.
The third structural difference between GluA1-3 is in the specificity loop (S-loop) that links helices α10 and α11 and is attached to the core structure on L1 through a disulfide bond (Figure 1b). It is termed the specificity loop because it plays a key role in interactions at the ATD dimer interface, and because its sequence and length is more conserved within than between receptor subfamilies [23, 26]. This loop adopts a different conformation in GluA1 (I294-W313) than in GluA2 (I298-W317) or GluA3 (V301-W320), despite an almost identical amino acid sequence [23]. Unexpectedly, the S-loop in GluA1 participates in extensive inter-dimer interactions. The potential physiological relevance of this unique feature will be discussed further below. Fourth, a loop connecting helices α7 and β9 on L2 adopts a different conformation to the equivalent loop in GluA2. It is worth noting that in GluA2 this loop is mainly responsible for the dimer-of-dimers interaction in the ATD in the context of a tetrameric channel [6]. Such assembly in the ATD will not form if this loop adopts the conformation as observed in GluA1-ATD.
Finally, we observed extensive trans-domain interactions in the clamshell cleft on GluA1-ATD (Figure S1). Four pairs of hydrogen bonds and salt bridges stabilize a partially closed conformation involving residues R267 and Y270 on L1 and S188, D219, and Q236 on L2. In addition, F95 on L1 and R135 on L2 form a cation π interaction at the front edge of the clamshell (Figure S1). Among these interactions, only one pair of hydrogen bond formed between D219 and Y270 is conserved on GluA2; none of the interactions is observed on GluA3. Nevertheless, the ATD of GluA1-3 all adopt similar partially closed conformations, with only moderate domain twisting movement of about 3 to 6 degrees between L1 and L2, which suggests that the trans-domain interactions are not the major force stabilizing the closed clamshell conformation. In sharp contrast, the homologous structures, e.g. mGluR-LBD or LIVBP, could have a domain closure up to 50 degrees upon ligand binding in the cleft. A similar mechanism is proposed for NMDAR, in which binding of a modulator in the ATD cleft could trigger the close-open movement of the ATD and subsequently induce structural rearrangement at the ATD-ATD and ATD-LBD interfaces [39, 40]. However, direct evidence for such a mechanism has not yet been obtained. Interestingly, an unassigned electron density was observed in the ATD cleft of GluA2-ATD using the 1.75 Å resolution data, which could potentially be a ligand [29]. However, no similar density was observed in another high resolution GluA2-ATD structure (1.8 Å) or in any of the known structures of the kainate receptor ATD at resolutions up to 1.4 Å (GluK5) [24, 27].
Dimeric organization of the ATD is not rigid
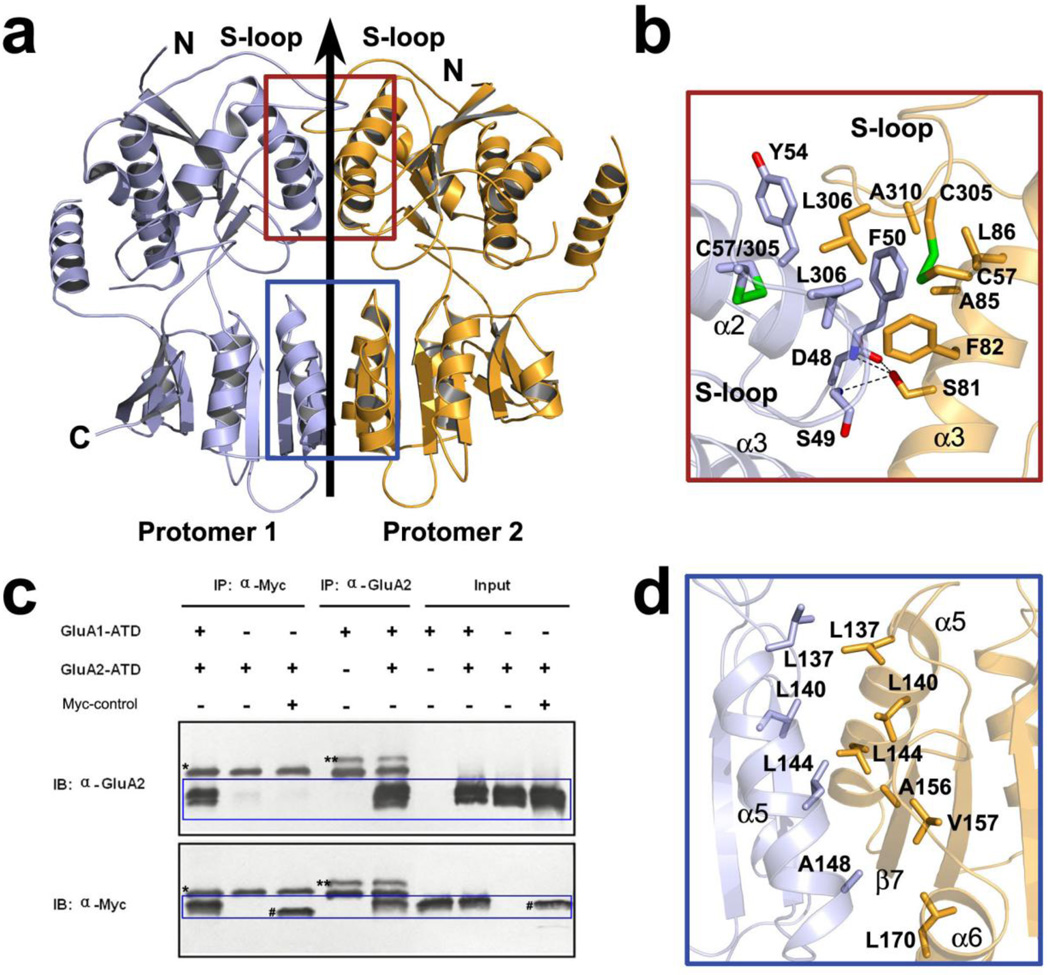
Both the GluA1 and GluA2 ATDs form tight dimers in solution [23, 24]. In the crystal, the GluA1-ATD forms two pairs of indistinguishable dimers, with each dimer possessing a non-crystallographic 2-fold symmetry. The L1-L1 dimer interface is formed primarily by helices α2 and α3 in combination with the S-loop lying on the top (Figure 2a). The aromatic ring of F50 of the α2 helix of one protomer inserts into a hydrophobic pocket formed by residues F82/A85/L86 on the α2 helix and L306/A310 of the S-loop on the other protomer. Three hydrogen bonds are observed on the L1-L1 interface, involving S81 on α3 and D48/S49/F50 on α2. Although the L1-L1 interface of GluA1-ATD is almost identical to that of GluA2, two key differences stand out. First, GluA2-N54 on α2 that forms a hydrogen bond with the main chain carbonyl group of L310 on the S-loop is replaced with Y54 on GluA1. The large side chain of Y54 not only abolishes this hydrogen bond but also pushes back the S-loop and thus negatively affects intra-dimer interaction. Second, GluA2-T78 forms water-mediated hydrogen bonding between L1 domains, but this polar interaction is abolished when T78 is replaced with M78 on GluA1. On the GluA1 dimer interface, the two M78 residues are in close contact with a distance of 6.6 Å between Cα atoms. The relatively large hydrophobic side chain of methionine instead of threonine at this position is not optimal for a tight L1-L1 interaction. Collectively, these observations suggest that Y54 and M78 on GluA1 contribute to the ~ 2-fold reduction in dimer association affinity of GluA1 compared to GluA2 [23].
Figure 2. GluA1-ATD forms a homodimer that is stabilized by extensive intra-dimer interactions.
(a) Ribbon diagram of the structure of a GluA1-ATD homodimer viewed perpendicular to the molecular two-fold axis. The two protomers are colored light blue and orange, respectively. The dimer interface can be divided into two sections that are outlined in red and blue boxes, respectively. (b, d) Close view of the dimer interface located in L1 (red frame) and L2 (blue frame). Key residues that participate in intra-dimer interactions are shown as sticks. Dashed lines indicate hydrogen bonds. (c) ATD mediates heterodimerization between GluA1 and GluA2. Purified recombinant GluA1-ATD with a C-terminal Myc-tag and GluA2-ATD were used for immunoprecipitation (IP). A specific anti-GluA2-ATD antibody (MAB397, Millipore) and an anti-Myc antibody (9B11, Cell Signaling) were used for IP and Western immunoblotting (IB). A non-related Myc-tagged protein (~40 kDa) was used as a negative control (labeled as “#” in lanes 3 and 9). Please note that the ATD bands were smeared due to glycosylation. Signals outside the blue boxes are non-specific due to antibody heavy chains (*) or an unknown protein that came from the anti-GluA2 antibody (**). “Input” represents the starting materials for IP.
Interactions at the L2 dimer interface are primarily hydrophobic, and are mediated by residues L137/L140/L144/A148 on helix α5, all of which protrude into the L2-L2 interface, together with A156/V157 on β7 (Figure 2d). This pattern is almost identical to that of the GluA2 structure with one subtle difference being that GluA2-I157 is replaced with GluA1-V157. The highly conserved intra-dimer interactions between GluA1 and GluA2 suggest that the predominant hetero-assembly of GluA1 and GluA2 into the same tetrameric channel in vivo is unlikely to be determined solely at the ATD level. Other domains, e.g. LBD and TMD, are likely to be required to fulfill the complex assembly [8].
Surprisingly, the L2-L2 interface on GluA3-ATD is quite different to that of GluA1 or GluA2. The key interacting residues on GluA1, L137 and V157, are replaced with F143 and R163 respectively, on GluA3. In this case, the closely packed L2-L2 interface that is conserved on GluA1 and GluA2 cannot be maintained because the hydrophobic patch is disrupted by insertion of the large side chain of F143 and by a positively charged R163. As a result, the GluA3 L2-L2 interface breaks up, and the dimer is maintained primarily by the conserved L1-L1 interface (Figure S2c). This could explain the fact that GluA3-ATD has the lowest affinity for homo-dimerization among all AMPARs (Kd is more than 600-fold lower than that of GluA2-ATD) [29].
To better understand the flexibility of dimer organization in the AMPAR family, we superimposed the dimeric ATDs of GluA1–3 based on Cα atoms of one protomer (Mol-A) and examined the conformational changes of the other protomer (Mol-B) (Figure 3). The protomer structures of GluA1–3 are all very similar (Mol-A, Figure 3a–b). The Mol-B of GluA1 and A2 in a dimer are almost identical, as could be predicted from their highly conserved L1-L1 and L2-L2 interfaces. Significant dimer rearrangement is observed on GluA3-ATD where the Mol-B of GluA3 is rotated clockwise along an axis perpendicular to the dimer interface (Figure 3c–d). Rotation and translation up to ~8° and ~7.4 Å are observed on GluA3-ATD, using as references helices α3 and α5 that mediate key interactions on L1 and L2, respectively.
Figure 3. Dimeric organization of the ATD is not rigid.
The structures of the ATD are shown as cartoons with α-helices shown as cylinders. The ATD of GluA1, A2, and A3 are colored in orange, gray, and green, respectively. (a, b) One protomer (Mol-A) of each of GluA1-3 is superimposed based on Cα atoms. The two views differ by a rotation of ~ 90° around a vertical axis. (c, d) The dimeric ATDs of GluA1–3 are superimposed based on Cα atoms on one protomer (Mol-A) while the other protomer (Mol-B) is left free to move. For clarity, only Mol-A of GluA1 is shown in (c) and it is omitted in (d). Helices α3 and α5 are selected as references for comparison since they mediate key interactions on L1 and L2, respectively.
When we compared all six available structures of the non-NMDARs, GluA1, A2, A3, K2, K3, and K5, we observed a correlation between the stability of the ATD dimer interface and the receptor’s ability to form a functional homomeric ion channel. Receptors that have large and stable dimer interfaces on both L1 and L2 domains, such as GluA1, A2, K2, and K3, all form functional channels. In contrast, GluA3 and GluK5 both have twisted and significantly weakened dimer interfaces, and interestingly, GluA3 has a high propensity to form heteromeric assemblies with other AMPARs, while GluK5 requires obligate co-assembly with GluK1–3 to form functional channels [27, 29]. This suggests that formation of a stable ATD dimer through homo- or hetero-dimerization is a key determinant for the assembly of a functional tetrameric channel.
We have previously shown that the ATD of GluA1 and GluA2 form homodimers in solution, with Kd of 270 nM and 152 nM, respectively [23]. Here, we observed that GluA1 and GluA2 ATD could directly interact with each other (Figure 2c). A robust interaction was observed between the ATD of GluA1 and GluA2 by co-immunoprecipitation (co-IP) when relatively high concentrations of recombinant proteins were used (~ 4 µM for each). It is likely that GluA1 and GluA2-ATD form heterodimers, because no tetrameric species was observed when a mixture of GluA1 and GluA2 ATDs was analyzed by analytic ultra-centrifugation at a similar concentration (data not shown). The interaction was much weaker when co-IP was performed at relatively low protein concentration, suggesting only a weak interaction between GluA1 and GluA2 at the ATD region. These findings are consistent with a model that heteromerization of AMPAR is mediated cooperatively by interactions at multiple regions, including the ATD, the LBD and the transmembrane channel [8]. Interestingly, it has been reported recently that GluA1 and GluA2 ATDs form heterodimers with a very high affinity (Kd ~ 0.4 nM) [25], and the same group estimated that GluA2-ATD forms homodimers with a similar Kd (~ 1.8 nM). It raises a question as to how these two high-affinity binding events, the homodimerization of GluA2 and the heterodimerization between GluA1 and GluA2, are fine-tuned.
L2 domain has a high degree of flexibility
To investigate further the structural flexibility of the ATD, we analyzed the B-factor distribution in each of the three members of AMPAR with known structures. The B-factors in a protein crystal structure reflect the fluctuation of each atom about its average position, and are important indicators of the flexibility and dynamics of a protein. Since measured B-factors in different structures are affected by differences in crystal handling, data collection, and structure refinement, we examined only the distribution of B-factors within each structure.
The B-factor analysis shows that, across GluA1–3, the core of the L1 domain is the most stable while the L2 domain has greater flexibility (Figure S2). This is consistent with the dynamic analysis of GluA3-ATD showing that its L2 domain is very mobile [29]. As expected, the peripheral regions of GluA1-ATD that show large structural differences with GluA2 and A3, including α6, α9, and the S-loop, all show high B-factors. Interestingly, GluA1-ATD seems to have higher intrinsic flexibility on the L2 domain compared to GluA2, despite their highly conserved intra-dimer interactions. While the precise explanation for this difference awaits further study, the high degree of flexibility of the L2 domain of GluA1 may explain its preference to form heteromeric channels with GluA2 rather than homomeric channels. The flexible structure of GluA1-ATD could also explain the difficulty in crystallizing the proteins that we have encountered.
The combination of a stable L1 domain and a mobile L2 domain on the ATD is reminiscent of the ligand binding property of the natriuretic peptide receptor (NPR), which has an LBD closely related to the ATD. NPR-LBD shares a similar dimmer assembly to the ATD except that the L2 domains are separated in a manner similar to that observed on GluA3-ATD (Figure S2c) [38]. The natriuretic peptide ligand binds in the center of the L2-L2 dimer interface on NPR-LBD, and subsequently pulls the L2 lobe in the dimer into closer proximity. Since the dimer of NPR is fixed by the stable L1-L1 interface, the movement of the L2 domain induces a 13.5° more open clamshell for each protomer [38].
Interestingly, the binding site for spermine, an allosteric modulator for NMDAR, is predicted to locate near the L2-L2 dimer interface of the ATD [14, 41, 42]. In the context of the full length tetrameric GluA2, the L2 of ATD is in direct contact with the D1 domain of LBD, and the stability of the LBD D1-D1 interface is a key determinant of the kinetics of channel activation and desensitization [4, 43]. It is intriguing to propose that modulator binding on the L2-L2 interface of AMPAR might change the mobility of the L2 domain, which in turn would affect the D1-D1 interface of the LBD and allosterically modulate channel activity. Thus, the L2-L2 dimer interface of AMPAR could provide a good target for development of therapeutically active allosteric modulators.
Crystal lattice suggests a novel mode of ATD-mediated AMPAR association
Remarkably, the isolated ATDs of three different iGluR subunits (GluA2, K2 and K3) all crystallize from various crystal forms in a similar dimer-of-dimers assembly, replicating the structure found in the full-length GluA2 [6, 23, 24, 26, 27]. This is another example showing that physiological protein-protein interacting interfaces are frequently used for crystal packing. As shown in Figure 4d, only the two “proximal” protomers within a tetramer (Mol-A and A’) bind to each other via the L2 domain. The two “distal” protomers (Mol-B and B’) stay further away. The tetramerization of the isolated ATD is consistent with the suggestion that the ATD plays a key role in tetramer assembly of iGluR in vivo [8–11].
Figure 4. Molecular packing in GluA1-ATD crystal reveals a novel head-to-head arrangement of the dimeric ATD.
(a) Two pairs of ATD dimer are shown as cartoons with α-helices shown as cylinders. The dimers are composed of molecule-A (Mol-A, green) and B (Mol-B, gray) and molecule-A’ (Mol-A’, light blue) and B’ (Mol-B’, orange), respectively. The S-loop is colored in red and labeled as a, b, a’, and b’. The blue parallelogram, using helices α9 and α10 as references, outlines a molecular surface that locates on the top of the dimmer-of-dimers assembly. Two different views of this head-to-head dimer-of-dimers arrangement of the ATD are shown following a rotation of ~ 90° around a horizontal axis (a, c). A close view on the inter-dimer interface, highlighted in the red box, is shown in (b). Key interacting residues are shown as sticks, and dashed lines represent hydrogen bonding. (d) The crystal structure of the full length GluA2 is shown on the left where the tail-to-tail assembly on the ATD is mediated by Mol-A and A’. In the plausible head-to-head ATD arrangement, all four protomers are involved in binding that could lead to inter-tetramer association of AMPAR on the synaptic membrane. The S-loops are depicted by red arrows.
Interestingly, another bona fide protein-protein interacting interface on iGluR, the LBD dimer interface, was originally identified as a crystal-packing “artifact”. For example, AMPAR-LBD forms a functionally authentic dimer in crystal lattice but is predominantly monomeric in solution [6, 43–45]. The LBDs of GluN1 and GluN2A do not oligomerize in solution, but crystallize as a physiologically relevant hetero-dimer [46].
Motivated by these observations, we inspected the crystal lattice of GluA1-ATD and found a novel inter-dimer interface on the ATD (Figure 4). This interface shows several significant differences from the known dimer-of-dimers ATD interface. First, the known interface is mediated by the L2 domain whereas the new interface is mediated by the L1 domain. For simplicity, we refer to the known interface as the tail-to-tail interface and the novel packing as the head-to-head interface. Second, both ATD protomers in a dimmer are involved in the head-to-head interface while only one of the two protomers makes contact in the tail-to-tail interface. Third, a large solvent-accessible surface of ~1,900 Å2 is buried in the head-to-head packing on GluA1-ATD, which is even larger than the intra-dimer interface (~1,500 Å2). It is generally accepted that a buried interface larger than 700 Å2 is likely to have physiological relevance [47]. In contrast, the tail-to-tail inter-dimer interface in the GluA2 crystal structure is only ~330 Å2. Finally, the head-to-head interface is mainly formed by the S-loop (Figure 4b). D304 and N308 on the S-loop form five pairs of hydrogen bonds with N6, P39, and I41 on its binding partner, and P309 and V311 on the S-loop form hydrophobic interactions with L28, P39, and I41 (Figure 4b). Since the composition of the S-loop is subfamily-specific, such head-to-head assembly will likely lead to association of iGluRs within each subfamily.
Structural modeling suggests that the novel head-to-head packing of the ATD is unlikely to exist within a tetramer, due to constraints from the downstream LBD [6]. However, the head-to-head interface could mediate the inter-tetramer interaction upon a slight conformational change on the ATD through a flexible linker (~17 residues) connecting the ATD and LBD (Figure 4d). Indeed, a similar Y-shape conformation of a tetrameric AMPAR was observed by single particle electron microscopy, where the two ATD dimers swing away from each other while the remaining tetramer structure remains the same [48, 49].
The ATD-mediated inter-tetramer aggregation of AMPAR raises the intriguing possibility that ATD might play a role in clustering of the highly concentrated AMPAR at the synaptic membrane. This would be consistent with the observation that neuronal pentraxins (NARP, NP1, and NPR) interact with the ATD of AMPAR and promote clustering of the receptors [19, 20, 50]. Furthermore, the head-to-head assembly of the ATD generates a large molecular surface on top of the ATD in the shape of a parallelogram (diagonals 107 Å × 56 Å), which stands ~130 Å away from the post-synaptic membrane, making it an ideal surface to interact with presynaptic proteins (Figures 4 & S3). In keeping with this hypothesis, it has been shown that AMPARs play a crucial structural role in regulating the stability of presynaptic inputs, which is mediated by the ATD and is independent of receptor-mediated channel activity [18].
Conclusion
This study reports the first crystal structure of the ATD of GluA1 subunit of iGluRs. Detailed structural analyses comparing GluA1–3 suggest that homo-dimerization of ATD is mediated mainly by interactions between the L1 domain, which are highly conserved within the AMPAR family. The L2 domain has greater relative mobility and is likely responsible for the different affinities for homodimerization among the AMPAR ATDs. It is notable that the intra-dimer interface is highly conserved between GluA1 and GluA2 and we observe weak hetero-dimerization between them. Thus, the isolated ATD likely is insufficient to drive the preferred hetero-assembly of AMPAR as observed in vivo. This conclusion is consistent with a model in which the LBD and the transmembrane channel also play important roles in heteromerization of AMPARs [8].
Our results suggest a plausible head-to-head arrangement of the dimeric ATD that is mostly mediated by the specificity loop. This unique inter-tetramer association of AMPAR provides novel suggestion as to how highly concentrated AMPARs form clusters on the synaptic membrane, and how the ATD might be involved in trans-synaptic signal transduction. Several synaptically localized molecules function across the synaptic cleft to reciprocally coordinate differentiation on both sides of the synapse, including neurexin-neuroligin, SynCAM, cadherin, EphrinB-EphB, and Liprin-α/LAR. The ATD crystal structure reported here will allow investigations of the role played by ATD in trans-synaptic signaling and in regulating presynaptic stability, and will add a new dimension to our understanding of iGluR function.
ACKNOWLEDGEMENT
Portions of this research were performed at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
FUNDING
This work was supported by NIH awards 1R21AG033813 and 1R01GM090023, by the Alfred P. Sloan Research Fellowship, and by the start-up research fund from the Sanford-Burnham Medical Research Institute.
Abbreviations used
- ATD
amino-terminal domain
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- iGluR
ionotropic glutamate receptor
- LBD
ligand-binding domain
- NMDA
N-methyl-D-aspartate
Footnotes
AUTHOR CONTRIBUTION
Jie ZHOU cloned GluA1-ATD and optimized expression conditions. Guorui YAO performed the protein expression, purification, characterization, and crystallization. Irimpan MATHEWS collected the diffraction data. Yinong ZONG and Shenyan GU performed structure determination and analysis. Rongsheng JIN supervised the project. All authors are involved in manuscript preparation.
REFERENCES
- 1.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Malenka RC, Nicoll RA. Long-term potentiation--a decade of progress? Science. 1999;285:1870–1874. doi: 10.1126/science.285.5435.1870. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 4.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 5.Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Matsuda S, Drews V, Torashima T, Meisler MH, Yuzaki M. A hot spot for hotfoot mutations in the gene encoding the delta2 glutamate receptor. Eur. J. Neurosci. 2003;17:1581–1590. doi: 10.1046/j.1460-9568.2003.02595.x. [DOI] [PubMed] [Google Scholar]
- 8.Ayalon G, Stern-Bach Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron. 2001;31:103–113. doi: 10.1016/s0896-6273(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 9.Ayalon G, Segev E, Elgavish S, Stern-Bach Y. Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J. Biol. Chem. 2005;280:15053–15060. doi: 10.1074/jbc.M408413200. [DOI] [PubMed] [Google Scholar]
- 10.Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O'Hara PJ, Heinemann SF. Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 11.Leuschner WD, Hoch W. Subtype-specific assembly of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their n-terminal domains. J. Biol. Chem. 1999;274:16907–16916. doi: 10.1074/jbc.274.24.16907. [DOI] [PubMed] [Google Scholar]
- 12.Kreusch A, Pfaffinger PJ, Stevens CF, Choe S. Crystal structure of the tetramerization domain of the Shaker potassium channel. Nature. 1998;392:945–948. doi: 10.1038/31978. [DOI] [PubMed] [Google Scholar]
- 13.Zerangue N, Jan YN, Jan LY. An artificial tetramerization domain restores efficient assembly of functional Shaker channels lacking T1. Proc. Natl. Acad. Sci. U S A. 2000;97:3591–3595. doi: 10.1073/pnas.060016797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masuko T, Kashiwagi K, Kuno T, Nguyen ND, Pahk AJ, Fukuchi J, Igarashi K, Williams K. A regulatory domain (R1–R2) in the amino terminus of the N-methyl-D-aspartate receptor: effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol. Pharmacol. 1999;55:957–969. doi: 10.1124/mol.55.6.957. [DOI] [PubMed] [Google Scholar]
- 15.Zheng F, Erreger K, Low CM, Banke T, Lee CJ, Conn PJ, Traynelis SF. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat. Neurosci. 2001;4:894–901. doi: 10.1038/nn0901-894. [DOI] [PubMed] [Google Scholar]
- 16.Perin-Dureau F, Rachline J, Neyton J, Paoletti P. Mapping the binding site of the neuroprotectant ifenprodil on NMDA receptors. J. Neurosci. 2002;22:5955–5965. doi: 10.1523/JNEUROSCI.22-14-05955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- 18.Ripley B, Otto S, Tiglio K, Williams ME, Ghosh A. Regulation of synaptic stability by AMPA receptor reverse signaling. Proc. Natl. Acad. Sci. U S A. 2011;108:367–372. doi: 10.1073/pnas.1015163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 20.Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, Sheng M, Passafaro M. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 22.Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 23.Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. Embo. J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J. Mol. Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- 25.Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM, Greger IH. Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. Embo. J. 2011;30:959–971. doi: 10.1038/emboj.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat. Struct. Mol. Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar J, Mayer ML. Crystal structures of the glutamate receptor ion channel GluK3 and GluK5 amino-terminal domains. J. Mol. Biol. 2010;404:680–696. doi: 10.1016/j.jmb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. Embo. J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sukumaran M, Rossmann M, Shrivastava I, Dutta A, Bahar I, Greger IH. Dynamics and allosteric potential of the AMPA receptor N-terminal domain. Embo. J. 2011;30:972–982. doi: 10.1038/emboj.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J. Neurosci. 2011;31:3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 36.Trakhanov S, Vyas NK, Luecke H, Kristensen DM, Ma J, Quiocho FA. Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity. Biochemistry. 2005;44:6597–6608. doi: 10.1021/bi047302o. [DOI] [PubMed] [Google Scholar]
- 37.Jingami H, Nakanishi S, Morikawa K. Structure of the metabotropic glutamate receptor. Curr. Opin. Neurobiol. 2003;13:271–278. doi: 10.1016/s0959-4388(03)00067-9. [DOI] [PubMed] [Google Scholar]
- 38.He X, Chow D, Martick MM, Garcia KC. Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone. Science. 2001;293:1657–1662. doi: 10.1126/science.1062246. [DOI] [PubMed] [Google Scholar]
- 39.Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traynelis SF, Hartley M, Heinemann SF. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- 42.Huggins DJ, Grant GH. The function of the amino terminal domain in NMDA receptor modulation. J. Mol. Graph. Model. 2005;23:381–388. doi: 10.1016/j.jmgm.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 44.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 45.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat. Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 46.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 47.Janin J. Specific versus non-specific contacts in protein crystals. Nat. Struct. Biol. 1997;4:973–974. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 49.Nakagawa T. The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors. Mol. Neurobiol. 2010;42:161–184. doi: 10.1007/s12035-010-8149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song I, Huganir RL. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]